VISUAL, an ectopic induction system for phloem differentiation in Arabidopsis, allows the molecular mechanism underlying this crucial process to be analyzed, opening up new avenues of research.
Abstract
Cell differentiation is a complex process involving multiple steps, from initial cell fate specification to final differentiation. Procambial/cambial cells, which act as vascular stem cells, differentiate into both xylem and phloem cells during vascular development. Recent studies have identified regulatory cascades for xylem differentiation. However, the molecular mechanism underlying phloem differentiation is largely unexplored due to technical challenges. Here, we established an ectopic induction system for phloem differentiation named Vascular Cell Induction Culture System Using Arabidopsis Leaves (VISUAL). Our results verified similarities between VISUAL-induced Arabidopsis thaliana phloem cells and in vivo sieve elements. We performed network analysis using transcriptome data with VISUAL to dissect the processes underlying phloem differentiation, eventually identifying a factor involved in the regulation of the master transcription factor gene APL. Thus, our culture system opens up new avenues not only for genetic studies of phloem differentiation, but also for future investigations of multidirectional differentiation from vascular stem cells.
INTRODUCTION
Cell differentiation is a crucial process performed by multicellular organisms to generate a variety of functional cells. In general, master regulators are considered to determine cell differentiation during various developmental processes (reviewed in De Rybel et al., 2016; reviewed in Simmons and Bergmann, 2016; Takada et al., 2013). In plants, the xylem and phloem function as two different conductive tissues to transport water, nutrients, and signaling molecules. It is widely recognized that xylem and phloem cells are commonly derived from procambial/cambial cells (reviewed in Miyashima et al., 2013). The master regulators VASCULAR-RELATED NAC-DOMAINs (VNDs) induce xylem cell differentiation (Kubo et al., 2005; Zhou et al., 2014; Endo et al., 2015), and ALTERED PHLOEM DEVELOPMENT (APL) plays a crucial role in phloem cell differentiation (Bonke et al., 2003). Recent studies have identified diverse downstream cascades of VNDs that regulate secondary cell wall thickening and programmed cell death during xylem differentiation (Ohashi-Ito et al., 2010; Taylor-Teeples et al., 2015; Yamaguchi et al., 2011). In addition, recent work identified downstream components of APL including NAC DOMAIN-CONTAINING PROTEIN45 (NAC045), NAC086, and NAC45/86-DEPENDENT EXONUCLEASE-DOMAIN PROTEINs (NENs), which regulate enucleation during phloem differentiation (Furuta et al., 2014). Despite our advanced understanding of downstream factors during xylem and phloem development, upstream components that spatiotemporally regulate these master genes remain to be identified.
Vascular tissues are deeply embedded inside the plant body, which makes it difficult to analyze their sequential developmental process. Culture systems have helped advance our understanding of the regulatory mechanisms for these processes, especially xylem differentiation (Fukuda and Komamine, 1980; Kubo et al., 2005; Oda and Fukuda, 2012; Derbyshire et al., 2015). To date, many key factors that regulate xylem differentiation from procambial/cambial cells have been identified (Motose et al., 2004; Ito et al., 2006; reviewed in Růžička et al., 2015). By contrast, only a limited number of regulators have been identified for phloem cell differentiation. Phloem differentiation involves unique subcellular events, such as enucleation, callose deposition, and P-protein accumulation (reviewed in Heo et al., 2014). Nevertheless, it is very difficult to distinguish phloem cells from other cell types without the use of phloem-specific markers, due to the absence of marked morphological changes. Therefore, a culture system that can be used to induce phloem differentiation with phloem-specific markers is needed, especially using model plants such as Arabidopsis thaliana and rice (Oryza sativa).
We recently established a tissue culture system for ectopic induction of xylem cells using Arabidopsis leaves, which enabled us to perform genetic studies of xylem cell differentiation using mutants and marker lines (Kondo et al., 2015). Here, with the use of phloem markers, we found that both phloem sieve element (SE)-like cells and xylem tracheary elements (TEs) are formed in this tissue culture system. We named this culture system Vascular Cell Induction Culture System Using Arabidopsis Leaves (VISUAL). In this study, we used VISUAL to thoroughly analyze phloem differentiation processes. Detailed observations and transcriptome analysis revealed that phloem differentiation in VISUAL mimics the process of in vivo phloem SE differentiation. Genetic analysis confirmed that APL plays a central role in SE differentiation in VISUAL. VISUAL transcriptome data with high temporal resolution enabled us to construct a coexpression network for SE-related genes, which led to the identification of a transcription factor that can regulate early phloem SE development.
RESULTS
Appearance of Phloem Markers in VISUAL
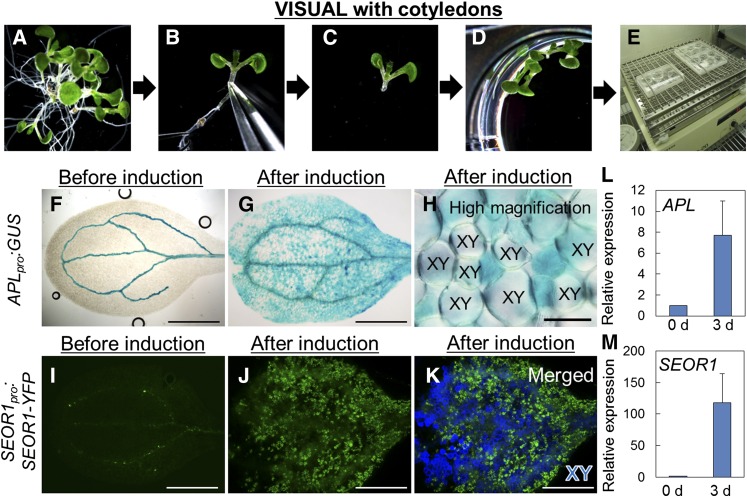
Glycogen synthase kinase 3 proteins (GSK3s) play central roles in vascular meristem maintenance by suppressing xylem cell differentiation (Kondo et al., 2014; reviewed in Kondo and Fukuda, 2015). Inhibition of GSK3 activity with the kinase inhibitor bikinin induces ectopic xylem cell differentiation, which allowed us to establish a culture system for vascular cell differentiation using Arabidopsis cotyledons and leaf disks (Figures 1A to 1E; Supplemental Figures 1A to 1E) (Kondo et al., 2015, 2014). We utilized this culture system to examine ectopic induction of phloem cell differentiation using APLpro:GUS (Bonke et al., 2003) and SEOR1pro:SEOR1-YFP (Froelich et al., 2011), which are markers of phloem precursor cells (fused to the β-glucuronidase gene) and differentiating SEs (SIEVE ELEMENT OCCLUSION-RELATED1 fused to the yellow fluorescent protein gene), respectively. These marker signals strongly appeared within 3 to 4 d in the culture system using cotyledons (Figures 1F to 1K) and leaf disks (Supplemental Figures 1F and 1G). RT-qPCR analysis confirmed the increased levels of APL and SEOR1 expression after culture (Figures 1L and 1M). Reexamination of our previous microarray data on leaf disk culture indicated that APL and SEOR1 transcript levels increase rapidly between 24 and 48 h of culture (Supplemental Figures 1H and 1I) (Kondo et al., 2015). Phloem marker signals (YFP fluorescence or GUS) and xylem marker signals (autofluorescence or thickened secondary cell walls) were detected simultaneously in leaves but were differentially observed at the cellular level (Figures 1H and 1K; Supplemental Figure 1G). These results strongly suggest that both xylem and phloem cell differentiation can be induced in this culture system, named VISUAL.
Figure 1.
Phloem Marker Expression in Cotyledons Revealed by VISUAL.
(A) to (E) Workflow of VISUAL using cotyledons.
(A) Arabidopsis seedlings were grown in MS liquid medium for 6 d under continuous light.
(B) Seedling at an appropriate growth stage for VISUAL.
(C) An explant whose bottom half was removed by forceps.
(D) Transfer of explants to liquid induction medium in a 12-well plate.
(E) Explant cultivation with shaking under continuous light for 4 d.
(F) and (G) Expression of APLpro:GUS before (F) and after (G) induction.
(H) High-magnification image of (G). XY, xylem tracheary elements.
(I) and (J) Expression of SEOR1pro:SEOR1-YFP before (I) and after (J) induction.
(K) A merged image of (J) and autofluorescent image with CFP excitation. Blue signal indicates autofluorescence from xylem cells.
(L) and (M) Expression levels of phloem marker genes in VISUAL. Relative expression levels of APL (L) and SEOR1 (M) were calculated by comparing samples before and after induction. Error bars indicate sd (n ≥ 4; biological replicates).
Bars = 1 mm in (F), (G), (I), (J), and (K) and 100 μm in (H). See also Supplemental Figure 1.
Cell Division Is Required for Phloem Cell Differentiation in VISUAL
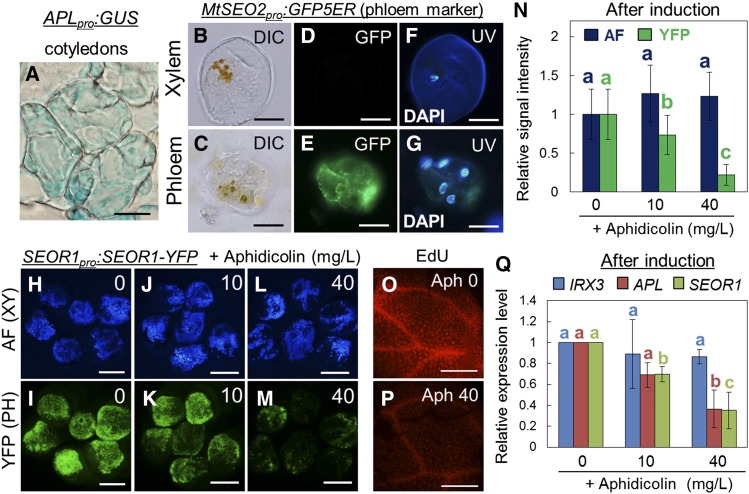
To visualize the histological features of phloem-like cells, cross sections of cultured cotyledons harboring a phloem marker were produced. In these sections, each APLpro:GUS-positive cell appeared to be separated into small compartments (Figure 2A). Dual-color imaging with a GFP phloem marker (MtSEO2pro:GFP5ER, i.e., GFP tagged to the endoplasmic reticulum under the control of the Medicago truncatula SEO2 promoter) (Froelich et al., 2011) and 4′,6-diamidino-2-phenylindole (DAPI) staining clearly indicated that a GFP-positive cell lump possessed multiple nuclei, whereas one nucleus was observed in a differentiating xylem cell lump with thick secondary cell walls (Figures 2B to 2G). These results suggest that phloem cells induced in VISUAL undergo multiple rounds of cell division. To reveal the relationship between cell division and phloem cell differentiation, the effects of the specific DNA synthesis inhibitor, aphidicolin, were examined in VISUAL. We previously reported that VISUAL involves two distinct differentiation processes, i.e., differentiation from mesophyll cells to procambial cells and from procambial cells to xylem or phloem cells (Kondo et al., 2015) (Supplemental Figure 2A). To examine the effect with a focus on the latter process, we added aphidicolin to the culture medium after inducing procambial cells (Kondo et al., 2015) (Supplemental Figure 2B) and then examined SEOR1-YFP signals and autofluorescence as indicators of phloem and xylem cells, respectively. Aphidicolin treatment significantly suppressed SEOR1-YFP signals, but not CFP autofluorescence (Figures 2H to 2N). Indeed, an EdU (5-ethynyl-2'-deoxyuridine) assay clearly confirmed that DNA replication is blocked by aphidicolin application in VISUAL cotyledons (Figures 2O and 2P). Consistent with our observation, aphidicolin downregulated APL and SEOR1 expression, whereas the expression levels of the xylem-specific marker gene IRREGULAR XYLEM3 (IRX3) were not significantly reduced (Figure 2Q). These results strongly suggest that cell division is required prior to phloem cell differentiation in VISUAL. We also investigated the effects of auxin and cytokinin treatment on phloem differentiation. However, both had much less of an effect on the ratio of xylem to phloem cell differentiation than aphidicolin (Supplemental Figure 3).
Figure 2.
Cell Division Is a Key Process Associated with Induced Phloem Cells.
(A) Cross-section image of APLpro:GUS cotyledons cultured for 4 d.
(B) to (G) Fluorescence images from DAPI staining and phloem marker (MtSEO2pro:GFP5ER) expression in an isolated immature xylem lump ([B], [D], and [F]) and an isolated phloem lump ([C], [E], and [G]). Differential interference contrast (DIC) images ([B] and [C]), GFP fluorescence images ([D] and [E]), and DAPI staining images ([F] and [G]) were obtained 3 d after induction in VISUAL.
(H) to (M) Effects of aphidicolin on autofluorescence (AF) of xylem secondary cell wall and YFP signal of SEOR1pro:SEOR1-YFP. Aphidicolin (0, 10, and 40 mg/L) was added to the culture medium at 24 h after the start of preculture (see also Supplemental Figure 2).
(N) Quantification of fluorescence signal intensities for (H) to (M). Relative signal intensity was calculated by comparison to a sample cultured without aphidicolin. Error bars indicate sd (n = 12; number of analyzed cotyledons). Significant differences (P < 0.05) are indicated by different letters (Tukey’s test).
(O) and (P) Effects of aphidicolin on cell division in VISUAL. DNA replication in VISUAL was detected by labeling with the modified thymidine analog EdU after no (O) or 40 mg/L aphidicolin treatment (P). EdU accumulation is indicated by red fluorescent signals.
(Q) Effects of aphidicolin on expression levels of xylem- (IRX3) and phloem-related (APL and SEOR1) genes. Relative expression levels were calculated by comparison to a sample cultured without aphidicolin. Error bars indicate sd (n = 3; biological replicates). Significant differences (P < 0.05) are indicated by different letters (Tukey’s test).
Bars = 50 μm in (A), 20 μm in (B) to (G), 2 mm in (H) to (M), and 500 μm in (O) and (P).
Electron Microscopy Observation of Phloem Cells Induced in VISUAL
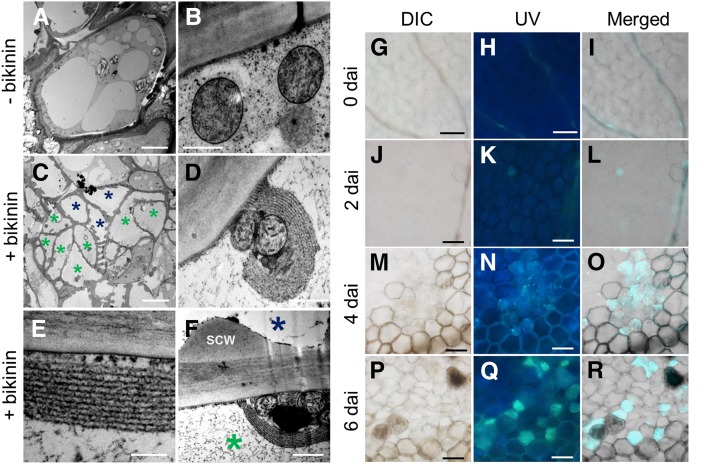
Next, we observed the subcellular structures of phloem cells induced in VISUAL using a high-pressure freezing method and transmission electron microscopy (TEM). As our previous observation indicated that cell division is the characteristic feature of induced phloem cells (Figure 2), we focused here on divided cells. Careful observation revealed that divided small cells were formed only in bikinin-treated cotyledons (Figures 3A and 3C). These divided cells have unique subcellular structures surrounding mitochondria (Figure 3D) similar to those observed in in vivo SEs (reviewed in Sjolund, 1997; Froelich et al., 2011). These structures were not observed in cotyledons cultured without bikinin (Figure 3B). Highly magnified images suggested that these clamp-like structures are similar to sieve element reticulum (Sjolund and Shih, 1983) or bundles of P-protein filaments (Froelich et al., 2011) (Figure 3E). TEM also indicated that some of the SE-like cells developed thick primary cell walls (Figures 3A to 3F). On the other hand, phloem marker-positive cells often exhibited distorted DAPI-stained structures or lacked DAPI staining signals at 4 d after induction (Supplemental Figure 4), suggesting that an enucleation-like event occurs in VISUAL SE-like cell differentiation (reviewed in Sjolund, 1997; Furuta et al., 2014). Although aniline blue staining indicated that callose accumulated in dot-like patterns after 4 d after induction, sieve plate-like structures were not detected as far as we observed (Figures 3G to 3R; Supplemental Figure 5). These results suggest that VISUAL-induced phloem differentiation shares most of the cellular events associated with in vivo SE differentiation. Interestingly, we observed a cell cluster produced by multiple divisions, in which small cells had differentiated into not only SE-like cells (Figure 3C, green asterisks) but also TEs (Figures 3C and 3F, blue asterisks), which can be distinguished by filamentous structures and secondary cell walls, respectively (Supplemental Figure 6A). This type of cluster with both SEs and TEs accounted for less than 20% of the clusters (Supplemental Figures 6B to 6F).
Figure 3.
Subcellular Structures of Induced Phloem Cells in VISUAL.
(A) to (F) Ultrastructure of cells in cotyledons cultured for 4 d without ([A] and [B]) and with ([C] to [F]) bikinin. Structures of cells originating from mesophyll cells were observed under TEM.
(A) Cell in a cotyledon cultured without bikinin.
(B) Subcellular structures of the cell in (A).
(C) A cell lump undergoing multiple cell divisions in a cotyledon cultured with bikinin. In the cell lump, most cells had clamp-like structures (green asterisks), and other cells developed secondary cell walls (blue asterisks).
(D) Subcellular structures of the cell in (C). Clamp-like structures surround mitochondria.
(E) High-magnification image of a layered clamp-like structure.
(F) High-magnification image showing a SE-like (green asterisk) and TE-like (blue asterisk) cell. SCW indicates developing secondary cell wall.
(G) to (R) Aniline blue staining of cells in cotyledons cultured for 0 ([G] to [I]), 2 ([J] to [L]), 4 ([M] to [O]), and 6 d ([P] to [R]) with bikinin. DIC images ([G], [J], [M], and [P]), UV-irradiated images ([H], [K], [N], and [Q]), and their merged images ([I], [L], [O], and [R]) were obtained.
Bars = 10 μm in (A) and (C), 500 nm in (B), (D), and (F), 200 nm in (E), and 100 μm (G) to (Q).
Gene Expression Profiles of SE-Like Cells Induced in VISUAL
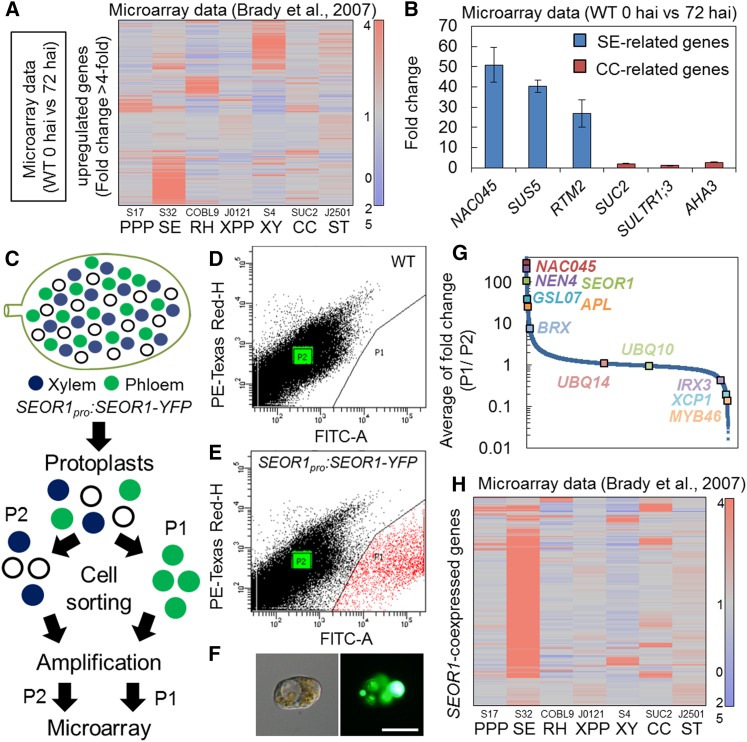
Next, we investigated the gene expression profiles of phloem cells induced in VISUAL. First, we compared microarray data before and after induction in VISUAL and determined that many upregulated genes (>4-fold) are preferentially expressed in SEs or xylem cells according to root cell type-specific microarray data (Brady et al., 2007) (Figure 4A). This result supports the proposal that xylem and SE-like cells are concomitantly differentiated in VISUAL. In contrast to SE-related genes (Chisholm et al., 2001; Barratt et al., 2011; Furuta et al., 2014), the expression levels of companion cell (CC)-related genes (DeWitt and Sussman, 1995; Stadler and Sauer, 1996; Yoshimoto et al., 2003) were not upregulated in VISUAL (Figures 4A and 4B), suggesting that induced phloem cells primarily consist of SEs but not CCs.
Figure 4.
Cell Sorting of Induced SE-like Cells Using SEOR1pro:SEOR1-YFP.
(A) Expression profiles of genes that were upregulated in VISUAL microarray data (>4-fold compared with cotyledons cultured for 0 and 72 h). Expression was visualized with a heat map image according to their expression levels in root cell type-specific microarray data (Brady et al., 2007).
(B) Changes in expression levels of SE- (NAC045, SUS5, and RTM2) and CC-related (SUC2, SULTR1;3, and AHA3) genes. Fold changes were calculated by comparing samples at 0 and 72 h after induction (hai). Error bars indicate sd (n = 3; biological replicates).
(C) Schematic illustration of the procedure used for cell-sorting analysis.
(D) and (E) Plots of fluorescent signal intensities for a total of 100,000 protoplasts from wild-type (D) and SEOR1pro:SEOR1-YFP cotyledons (E). The x axis indicates the signal intensity for FITC-A (excitation, 488 nm; detection, 515 to 545 nm). The y axis indicates the signal intensity for PE-Texas Red-H (excitation, 561 nm; detection, 600 to 620 nm). P1 (red dots) and P2 (green dots) indicate YFP-positive and YFP-negative cells, respectively.
(F) DIC image (left) and YFP image (right) of a protoplast gated in P1 after cell sorting. Bar = 50 μm.
(G) Overview of microarray data from sorted cells. The average fold change (P1/P2) for ∼28,500 genes was calculated from three independent experiments and arranged in descending order.
(H) Expression profiles of SEOR1-coexpressed genes, which were highly enriched in positive cells (P1/P2 >16-fold), visualized with a heat map image according to their expression levels in root cell type-specific microarray data (Brady et al., 2007). Color-coded heat maps for (A) and (H) were generated by the Subio Platform according to the color scale shown in the right panel.
Our transcriptome data using whole cotyledons contain mixed gene expression profiles derived from multiple types of vascular cells. Therefore, we performed cell-sorting analyses using the FACSAria III cell sorter (BD Biosciences) to specifically collect induced phloem SEs expressing the differentiating SE marker SEOR1pro:SEOR1-YFP (Figure 4C). Protoplasts isolated from cultured cotyledons of Arabidopsis wild-type and SEOR1pro:SEOR1-YFP seedlings were plotted according to levels of green YFP signal intensity (indicated on the x axis) and red autofluorescent signal (indicated on the y axis) to define the group of YFP-positive cells (Figures 4D and 4E). A cell population displaying high YFP signal fluorescence (marked by red dots in Figure 4E) in protoplasts extracted from SEOR1pro:SEOR1-YFP cotyledons was considered to contain fluorescence-positive cells (P1 cells) (Figures 4D and 4E). Indeed, bright YFP signal fluorescence was observed only in the sorted P1-positive cells (Figure 4F). To determine the contribution of original phloem SEs in cotyledon veins to the counts of P1-positive cells, we compared the frequencies of P1-positive cells in uninduced (− bikinin) and induced (+ bikinin) samples. The number of P1 cells in uninduced samples was ∼300 times smaller than that in induced samples (Supplemental Figure 7), indicating that more than 99% of P1 cells were derived from ectopically induced phloem SE-like cells in VISUAL. Next, mRNA extracted from P1 cells and P2 (fluorescence-negative) cells was subjected to microarray analysis. Microarray data revealed that phloem-specific genes such as NAC045, NEN4 (Furuta et al., 2014), SEOR1 (Froelich et al., 2011), GLUCAN SYNTHASE-LIKE7 (GSL07) (Barratt et al., 2011), APL (Bonke et al., 2003), and BREVIS RADIX (BRX) (Depuydt et al., 2013) were highly enriched in P1 cells (Figure 4G). By contrast, the expression of xylem-specific genes such as IRX3, XYLEM CYSTEINE PEPTIDASE1 (XCP1), and MYB46 (Taylor et al., 1999; Zhao et al., 2000; Zhong et al., 2008) was much lower in P1 than in P2 cells (Figure 4G). The expression of regulatory genes such as UBIQUITIN10 (UBQ10) and UBQ14 (Kubo et al., 2005; Kondo et al., 2015) did not differ between P1 and P2 cells (Figure 4G). To characterize the gene expression profiles of P1 cells, we extracted genes that were enriched more than 16-fold in P1 cells (SEOR1-coexpressed genes; listed in Supplemental Data Set 1). These SEOR1-coexpressed genes were expressed preferentially in root SEs (Figure 4H).
APL Is a Central Regulator of SE Differentiation in VISUAL
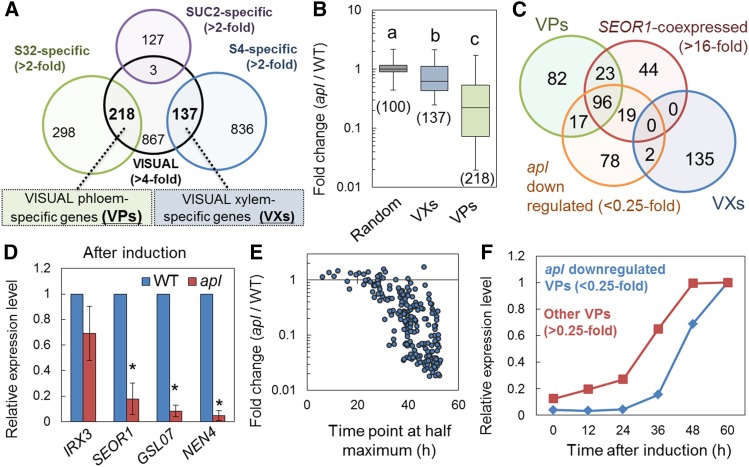
APL is a well-known transcription factor governing phloem development (Bonke et al., 2003). A previous study reported that the Arabidopsis apl mutant displays abnormal root growth and a seedling lethal phenotype, which prevents analysis of the primary effect of apl mutation on phloem development (Bonke et al., 2003). However, VISUAL using young apl cotyledons can overcome such disadvantage and allows us to examine the impact of apl mutation on the phloem SE differentiation process. For this purpose, we first performed microarray analysis of wild-type and apl cotyledons in VISUAL. A total of 213 genes were identified as strongly (<0.25-fold) downregulated genes in the apl mutant (genes are listed in Supplemental Data Set 1), when compared with the wild type. To determine the extent to which apl affects the expression of vascular-specific genes, we extracted 218 VISUAL phloem-specific genes (VPs) and 137 VISUAL xylem-specific genes (VXs) from the upregulated genes in VISUAL of the wild type (>4-fold) (Figures 4A and 5A) with reference to root cell type-specific expression data (Brady et al., 2007) (genes are listed in Supplemental Data Set 1). CC-specific genes that are preferentially coexpressed with SUC2 in roots (Brady et al., 2007) did not overlap with upregulated genes in VISUAL (Figure 5A), supporting the idea that CCs are rarely induced in VISUAL. In the apl mutant, VXs were slightly downregulated compared with 100 randomly selected genes (Figure 5B). By contrast, fold changes (apl/wild type) in expression of VPs were much lower than those of VXs and random genes (Figure 5B). Furthermore, apl down-regulated genes (< 0.25-fold) included many VPs but only two VXs (Figure 5C). Indeed, SEOR1-coexpressed genes highly overlapped with apl downregulated genes (Figure 5C), suggesting that APL preferentially upregulates many phloem-specific genes in VISUAL. We performed RT-qPCR analysis, confirming that the expression of phloem-specific genes such as SEOR1, GSL07, and NEN4 is strongly suppressed in VISUAL with apl cotyledons, whereas the expression of the xylem-specific gene IRX3 does not significantly differ in VISUAL between wild-type and apl cotyledons (Figure 5D).
Figure 5.
Transcriptome Analysis of the apl Mutant.
(A) Definition of VP and VX. Among the upregulated genes in VISUAL (>4-fold), phloem-specific genes (VPs), and xylem-specific genes (VXs) were extracted according to their S32 specificity (>2-fold) and S4 specificity (>2-fold) in root cell type-specific microarray data (Brady et al., 2007). Note that upregulated genes in VISUAL did not overlap with CC-specific genes extracted according to their SUC2 specificity (>2-fold).
(B) Box plot diagram showing fold changes for 100 randomly selected genes (random), VXs, and VPs in the wild type and apl at 72 h after induction. The average fold change was calculated from three independent experiments. Boxes indicate upper and lower quartiles, the central line within the boxes signifies median, and upper and lower bars indicate maximum and minimum fold change, respectively. Significant differences (P < 0.05) are indicated by different letters (Tukey’s test). The number of analyzed genes is shown below the box.
(C) Venn diagram for VPs, apl downregulated genes (<0.25-fold), SEOR1-coexpressed genes (>16-fold), and VXs.
(D) Expression of xylem- (IRX3) and phloem-related (SEOR1, GSL07, and NEN4) genes in the wild type and apl. Relative expression levels were calculated by comparison with the wild type. Error bars indicate sd (n = 3; biological replicates). Significant differences were examined by Student’s t test (*P < 0.05).
(E) and (F) Relationship between expression patterns in VISUAL and apl downregulation scores for VPs.
(E) Plot of fold changes (apl/wild type) and time point at half maximum for all VPs (see also Supplemental Figure 8).
(F) Comparison of expression patterns between VPs downregulated in apl (<0.25-fold) and other VPs (>0.25-fold). Relative expression levels were calculated from the average expression and then normalized with respect to the maximum expression, which was set to 1.
However, not all VPs were downregulated in the apl mutant (Figures 5B and 5C). To identify which VPs were downregulated in apl, we investigated the relationship between VP expression patterns and apl downregulation scores (apl/wild type) for all VPs. Here, we evaluated the temporal VP expression in VISUAL by calculating the time point when their expression first reaches the half maximum based on time-course transcriptome data (Supplemental Figure 8). The data clearly indicated that VPs with a later time point at half maximum tended to be more severely downregulated in the apl than those with earlier time points (Figure 5E; Supplemental Figure 8). Consistent with this result, VPs downregulated in apl (<0.25-fold) displayed later expression patterns than other VPs (>0.25-fold) (Figure 5F). These combined results suggest that APL regulates the expression of genes acting during the later process of phloem SE-like cell differentiation in VISUAL.
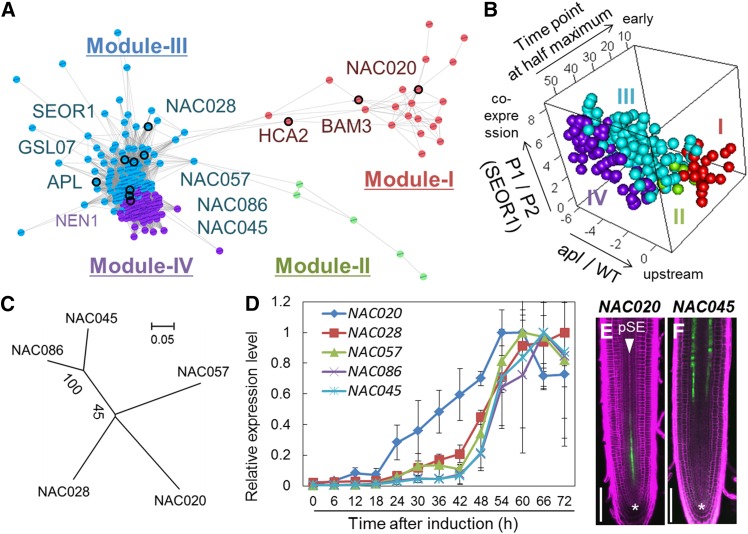
Network Analysis of SE-Specific Genes in VISUAL
We obtained three different sets of VISUAL transcriptome data, including time-course, SEOR1 cell sorting, and apl mutant data, all of which are useful for predicting the timing of gene expression (Supplemental Figure 9). Using these data, we constructed a coexpression network for VPs to dissect the process of phloem SE differentiation (Supplemental Figure 9). The VP network contained four distinct modules (I to IV) named according to the order of gene expression timing (Figures 6A and 6B; Supplemental Data Set 2). Modules III and IV contained differentiation-related genes such as APL, SEOR1, and GSL07 (Figure 6A; Supplemental Data Set 2). On the other hand, module II consisted of only five uncharacterized genes. By contrast, module I contained well-known regulators BARELY ANY MERISTEM3 (BAM3) and HIGH CAMBIAL ACTIVITY2 (HCA2) (Figure 6A; Supplemental Figure 10 and Supplemental Data Set 2). Genes in module I displayed earlier expression patterns, lower SEOR1 coexpression levels, and higher apl/wild type scores than those in any other modules (Figure 6B). These results suggest that module I genes function during early phloem differentiation and may act upstream of APL. Among module I genes, we focused on NAC020, which shares high sequence similarity with phloem-specific NAC members (NAC028, NAC057, NAC086, and NAC045) (Figure 6C). In contrast to NAC020, other phloem-specific NAC genes were categorized into module III (Figure 6A; Supplemental Data Set 2). Quantitative time-course expression analysis indicated that NAC020 displays the earliest expression pattern among five phloem-specific NAC genes in VISUAL (Figure 6D). Consistent with this result, NAC020 was expressed in protophloem cell files at the root apical meristem (Figure 6E), whereas NAC045 was expressed in phloem SEs of the root elongation and differentiation zones (Furuta et al., 2014) (Figure 6F). In addition, other module III NAC genes, NAC028 and NAC057, were expressed slightly later than NAC020 during root phloem development (Supplemental Figure 11). NAC020 expression was initiated a few cells above the quiescent center and then disappeared gradually before initiating SE differentiation (Figure 6E). These results indicate that NAC020 is expressed during the early stage of SE development in roots and in VISUAL.
Figure 6.
Classification of VPs Based on Coexpression Network Analysis.
(A) Coexpression network for VPs constructed from three different VISUAL transcriptome data sets using the WGCNA package (Supplemental Figure 6). Four distinct modules are highlighted with different colors. A node represents a gene. An edge indicates high correlation between nodes (TOM > 0.05).
(B) 3D plots for VPs with three different axes indicating SEOR1 coexpression scores (P1/P2 in log2 scale), apl downregulation scores (apl/wild type in log2 scale), and time point at half maximum.
(C) Phylogenetic tree of phloem-specific NAC family members constructed with the neighbor-joining method. The scale bar indicates the number of amino acid changes per site. Numbers shown next to the branches indicate the percentage of bootstrap values (1000 replicates).
(D) Time-course expression profiles of phloem-specific NACs in VISUAL. Relative expression levels were calculated by comparison with the maximum expression levels for each individual gene. Error bars indicate sd (n = 3; biological replicates).
(E) and (F) GFP expression patterns of NAC020pro:2xsGFP (E) and NAC045pro:GFP-GUS (F) lines with PI staining. Asterisks indicate quiescent center. Bars = 100 μm.
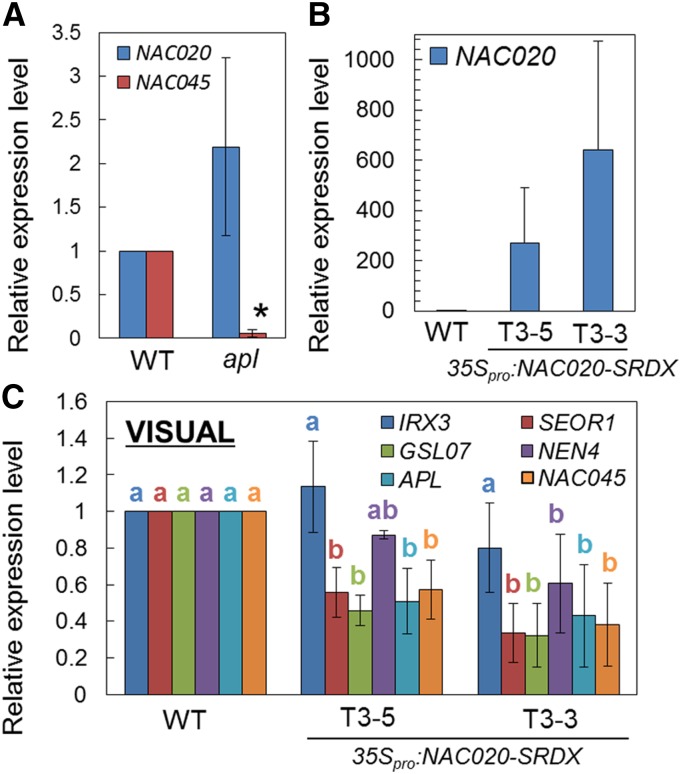
NAC020 Is an Early Regulator of SE Differentiation
In VISUAL, NAC045 expression was much lower in the apl mutant than in the wild type, whereas NAC020 expression was not affected by apl mutation (Figure 7A), suggesting that NAC020 acts upstream of APL. Because no loss-of-function mutant for NAC020 was available, to reveal the role of NAC020, we produced transgenic plants harboring NAC020 fused with a chimera repression domain (35Spro:NAC020-SRDX). We selected two lines, the weak line T3-5 and the strong line T3-3, and further examined their phenotypes in VISUAL (Figure 7B). APL and its downstream genes were downregulated in both lines, whereas the expression levels of the xylem-specific gene IRX3 did not differ between wild-type and SRDX lines (Figure 7C). These results suggest that NAC020 is involved in SE-like cell differentiation in VISUAL as an early phloem regulator. In vivo, however, 35Spro:NAC020-SRDX did not exhibit abnormal phloem development (Supplemental Figures 12A and 12B).
Figure 7.
Functional Analysis of NAC020 in VISUAL.
(A) Expression of NAC020 and NAC045 at 72 h after induction in VISUAL with the wild type and apl. Relative expression levels were calculated by comparison with the wild type. Error bars indicate sd (n = 3; biological replicates). Significant differences were examined by Student’s t test (*P < 0.05).
(B) Expression levels of NAC020 in 7-d-old seedlings of the wild type and two different 35Spro:NAC020-SRDX lines.
(C) Expression levels of xylem- (IRX3) and phloem-related genes (SEOR1, GSL07, NEN4, APL, and NAC045) in 35Spro:NAC020-SRDX lines at 72 h after induction. Relative expression levels were calculated by comparison with the wild type. Error bars indicate sd (n = 3; biological replicates). Significant differences (P < 0.05) are indicated by different letters (Tukey’s test).
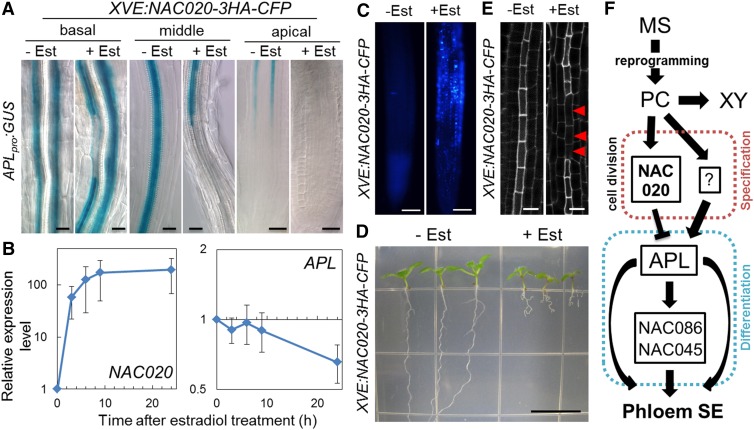
Next, we produced plants harboring both an estradiol-inducible NAC020-CFP construct and the APLpro:GUS reporter construct. Unexpectedly, NAC020 overexpression caused partial discontinuity of APLpro:GUS expression in the basal part of the roots and abolished APLpro:GUS expression in the meristematic and differentiation zones, where metaxylem vessels had already differentiated (Figure 8A). RT-qPCR analysis revealed that NAC020 expression was highly induced within 3 h after estradiol treatment (Figure 8B), which is consistent with the accumulation of CFP signal upon estradiol application (Figure 8C). Subsequently, APL expression decreased between 9 and 24 h after treatment (Figure 8B). NAC020 overexpression also caused severe root growth defects (Figures 8C and 8D) and discontinuous SE differentiation in roots (Figure 8E), which are similar to the phenotypes of the apl mutant. These results suggest that overexpression of NAC020 leads to the inhibition of phloem SE differentiation, probably through a decrease in APL expression (Figure 8F). Although apl produces ectopic xylem cells at the phloem position (Bonke et al., 2003), NAC020 overexpression did not induce such a phenotype, but it reduced cambium activity in hypocotyls (Supplemental Figures 12C to 12E).
Figure 8.
Functional Analysis of NAC020 in Phloem Development.
(A) GUS expression in the basal, middle, and apical regions of APLpro:GUS/XVE:NAC020-3HA-CFP roots treated without (−Est) or with (+Est) 10 μM estradiol for 7 d.
(B) Expression levels of NAC020 and APL in estradiol-treated seedlings harboring XVE:NAC020-3HA-CFP. Relative expression levels were calculated by comparing samples before and after estradiol treatment. Error bars indicate sd (n = 3; biological replicates).
(C) CFP fluorescent signals of XVE:NAC020-3HA-CFP seedlings treated without or with 10 μM estradiol for 24 h.
(D) Root growth of XVE:NAC020-3HA-CFP seedlings treated without or with estradiol for 7 d.
(E) SE differentiation in roots of XVE:NAC020-3HA-CFP treated without or with 10 μM estradiol. Red arrowheads indicate cells in which SE differentiation was suppressed.
(F) Schematic illustration of the sequential differentiation processes in VISUAL with a focus on phloem SE differentiation. SE differentiation process was divided into specification (red dotted line) and differentiation (blue dotted line) phases by coexpression network and genetic analysis.
Bars = 20 μm in (A), 100 μm in (C), 1 cm in (D), and 10 μm in (E).
DISCUSSION
VISUAL Can Induce Ectopic Xylem and Phloem SE-Like Cell Differentiation
Bikinin inhibits the GSK3 activity of plant kinases of the SKI (SK11, SK12, and SK13) and SKII (BIN2, BIL1, and BIL2) families (De Rybel et al., 2009). We previously reported that SKI and SKII family members redundantly suppress xylem differentiation by inhibiting BRI1-EMS-SUPPRESSOR1 (BES1) activity downstream of tracheary element differentiation inhibitory factor (TDIF)–TDIF RECEPTOR signaling (Kondo et al., 2014). Therefore, bikinin induces ectopic xylem differentiation by inhibiting GSK3 activity (Kondo et al., 2015). In this study, we achieved bikinin-mediated ectopic induction of xylem and phloem SE differentiation. A recent study reported that OCTOPUS (OPS) negatively regulates BIN2 activity to promote protophloem differentiation by promoting BES1 and BRASSINAZOLE RESISTANT1 activity (Anne et al., 2015). This finding is consistent with our result that bikinin can induce not only xylem but also phloem SE-like cell differentiation.
Here, we established the VISUAL system for analyzing vascular development. In VISUAL, mesophyll cells change their fate into procambial cells (Kondo et al., 2015), which in turn differentiate into TEs or SE-like cells. Although many convenient ectopic xylem induction systems have been established, there are few culture systems available for detailed study and analysis of phloem differentiation. Calli culture sometimes induces ectopic formation of SE-like cells in several plant species (Wetmore and Rier, 1963; Aloni, 1980; reviewed in Sjolund, 1997). Regeneration of phloem tissues after bark girdling has also been used to study phloem differentiation (Pang et al., 2008). VISUAL has several advantages beyond these conventional methods. One of the biggest advantages of VISUAL is that Arabidopsis resources such as mutants and marker lines can be utilized for analyses. Our genetic analysis of the apl mutant revealed the importance of APL for phloem SE-like cell differentiation in VISUAL. The observations and cell-sorting experiment with phloem marker lines indicate that differentiation in VISUAL and in vivo SE differentiation are similar. The combined use of mutants and marker lines with VISUAL is a powerful tool for molecular genetic analyses of phloem SE differentiation as well as xylem TE differentiation.
Cellular Events during SE Differentiation in VISUAL
SE-specific genes including NEN4 and GSL07, which are implicated in enucleation (Furuta et al., 2014) and callose deposition (Barratt et al., 2011; Xie et al., 2011), respectively, were significantly upregulated in VISUAL. Indeed, VISUAL caused enucleation and callose deposition in SE-like cells (Figures 3G to 3R; Supplemental Figures 3 to 5), indicating that changes in gene expression in VISUAL correspond with cellular events underlying SE differentiation. In VISUAL, inhibition of cell cycle progression reduced APL expression, suggesting that cell division is necessary for SE-like cell fate specification. Indeed, module I genes included a cell cycle-related gene, CDC2C, which may be involved in the process of cell division during SE-like cell differentiation (Supplemental Data Set 2). During phloem development, asymmetric cell division plays a crucial role in SE and CC production from phloem precursor cells (reviewed in Sjolund, 1997). However, CC differentiation-related genes were not upregulated in VISUAL, suggesting that cell division in VISUAL may not induce SE-CC separation. Previous studies have shown that protophloem precursor cells in roots divide twice to form the procambium, protophloem, and metaphloem cell files (Rodriguez-Villalon et al., 2014). Further genetic studies of module I genes such as CDC2C may provide insights into the role of cell division in phloem development.
Coexpression Network Highlights Regulatory Components Acting Upstream of APL
Recent reports combining cell-sorting and mutant studies have revealed several candidates for downstream components of APL, such as NAC086, NAC045, and NENs (Furuta et al., 2014). However, the regulatory components acting upstream of APL have remained unclear. VISUAL has an advantage in that differentiation from uncommitted cells to mature phloem SEs can be analyzed with clear time-course resolution due to the high level of cell synchrony. A gene expression network constructed with microarray data from VISUAL displayed early phloem genes included BAM3, which is expressed in SE cell files of the root apical meristem (Rodriguez-Villalon et al., 2014). BAM3 is a receptor for CLAVATA3/EMBRYO SURROUNDING REGION-RELATED45 (CLE45), and CLE45-BAM3 signaling suppresses APL expression, resulting in the inhibition of protophloem differentiation (Depuydt et al., 2013). OPS and COTYLEDON VASCULAR PATTERN2 (CVP2) are known as early regulators of protophloem differentiation (Truernit et al., 2012; Rodriguez-Villalon et al., 2014). These genes displayed similar expression profiles to those of module I genes (Supplemental Figure 10), which validates the categorization of early phloem genes. Among the early phloem SE genes, we identified the NAC transcription factor NAC020. Both overproduction of NAC020 and NAC020-SRDX caused the reduction of APL expression and led to the partial inhibition of SE differentiation. To further investigate the functions of NAC020, genetic analysis with loss-of-function mutants of NAC020 and its related genes is required.
In this study, we classified VPs into the categories specification- and differentiation-related genes. However, the molecular mechanisms underlying the regulation of phloem SE differentiation have not been completely elucidated. NAC086 and NAC045 were reported to function in enucleation but not in callose deposition (Furuta et al., 2014), suggesting that other transcription factors downstream of APL regulate SE differentiation (Figure 8F). Furthermore, NAC020 and BAM3-CLE45 signaling play negative roles in phloem cell specification, suggesting the existence of other as yet unidentified factors that positively regulate APL expression and SE differentiation (Figure 8F). Detailed analysis of early and late phloem-related genes with VISUAL will generate a full view of the regulatory network for SE differentiation. Another important issue to be elucidated is the switching mechanisms by which xylem or phloem differentiation is determined. It will be interesting to analyze early xylem development as well as early phloem development in VISUAL in order to further understand vascular stem cell fates.
METHODS
Plant Materials
SEOR1pro:SEOR1-YFP and MtSEO2pro:GFP5ER were provided by Michale Knoblauch (Froelich et al., 2011). The Arabidopsis thaliana mutant apl and NAC045pro:GUS-GFP were obtained from Ykä Helariutta and Kaori Furuta-Miyashima (Bonke et al., 2003; Furuta et al., 2014). To generate NACpro:2xsGFP, the ∼2-kb promoter region of NAC020, NAC028, and NAC057 was cloned and then introduced into the pGWB1 vector (Nakagawa et al., 2007). DNA fragments of 2xsGFP were introduced into the NAC020pro:pGWB1, NAC028pro:pGWB1, and NAC057pro:pGWB1 vectors by LR reaction (Invitrogen). The NAC020 coding sequence and the 3HA and CFP fragment were cloned into the pMDC7 vector (Curtis and Grossniklaus, 2003) to generate XVE:NAC020-3HA-CFP. The floral dip method (Clough and Bent, 1998) was used to generate transgenic plants in Arabidopsis. All Arabidopsis mutants in this study are in the Col-0 background.
VISUAL
See Supplemental Methods.
Observation of Ectopic Vascular Cells
GUS staining was performed according to a previously published method (Kondo et al., 2014). YFP fluorescence and autofluorescence were observed under a fluorescent stereomicroscope (Leica) and BX51 fluorescent microscope (Olympus). Fluorescence intensity was calculated from the obtained images using ImageJ (http://imagej.nih.gov/ij/). DAPI staining in VISUAL was performed after isolation of single cells from cultured leaves. Images of fluorescence signals resulting from UV excitation were obtained using the BX51 microscope (Olympus). For aniline blue staining, cultured cotyledons were fixed in a mixture of acetic acid:ethanol (1:3, v/v). Cotyledons were incubated in 2 M NaOH for 1 h and then stained overnight in 0.005% aniline blue solution (in 50 mM phosphate buffer, pH 6.8). Fluorescence signals resulting from UV excitation were obtained using the BX51 (Olympus).
RT-qPCR
Total RNA was extracted from cultured cotyledons (6 to 10 cotyledons) using an RNeasy Plant Mini Kit (Qiagen). After reverse transcription, relative gene expression was calculated by quantitative PCR with TaqMan probes using a light cycler (Roche Diagnostics). UBQ14 was used as an internal control for the assay. For the statistical analysis, quantitative PCR was repeated with at least three independent biological sets.
Aphidicolin Treatment and EdU Assay
First, cotyledons were precultured for 24 h with auxin, cytokinin, and bikinin. After preincubation, the cotyledons were treated with aphidicolin at various concentrations and were further cultured for 72 h. The EdU assay was performed with a Click-iT EdU Alexa Fluor 594 imaging kit (Life Technologies). Cotyledons cultured for 24 h were treated with aphidicolin, and 3 h later, with 5 µM EdU. The cotyledons were further cultured for 24 h before EdU detection. EdU detection was performed according to the manufacturer’s instructions, except that tissue fixation and permeabilization were performed as previously described (Paciorek et al., 2006).
TEM Analysis
For TEM analysis, cotyledon leaf disks were subjected to high-pressure freezing and freeze-substitution fixation to preserve the fine subcellular ultrastructure. Wild-type cotyledons cultured with or without bikinin for 4 d were used. Small (1 mm in diameter) leaf disks were cut out with a hole punch, quickly transferred to the specimen holder, and rapidly frozen in a high-pressure freezer (HPM010; Bal-Tec). Frozen samples were transferred to frozen 4% osmium tetroxide in anhydrous acetone at the temperature of liquid nitrogen. Samples were maintained at −80°C for 8 d, −20°C for 2.5 h, 4°C for 1.5 h, and finally room temperature for 30 min. The samples were washed several times with anhydrous acetone, infiltrated with increasing concentrations of Spurr’s resin (Nisshin EM) in anhydrous acetone at room temperature, and embedded in Spurr’s resin. Ultrathin sections (50 to 90 nm) were cut using an ultramicrotome (Ultracut; Leica), and the sections were stained with uranium acetate and lead citrate and observed under TEM (JEM1010; JEOL) at 80 kV. Images were acquired with a CCD camera (Veleta; Olympus).
Cell-Sorting Analysis
At least 200 cotyledons were prepared for protoplast isolation. Cotyledons cultured for 3 d were cut in half and washed on a 70-μm nylon cell strainer (BD Falcon) with Solution A (0.6 M mannitol, 10 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 0.039% MES, and 0.1% BSA). Washed cotyledons were collected in a conical tube filled with 10 mL of Solution B (150 mg of cellulase Onozuka R-10 and 40 mg of macerozyme R-10 in 10 mL of Solution A) and incubated under a vacuum for 30 to 60 min. The tubes were further incubated at 27°C for 3 to 4 h with slow shaking (40 to 50 rpm). After incubation, the samples were filtered through a 70-μm nylon cell strainer (BD Falcon) to eliminate undigested leaves. Filtered samples were transferred to two centrifuge tubes and centrifuged at 300g for 5 min at 4°C. The supernatants were discarded, and the pellets were suspended in 5 mL of Solution A per tube. This washing process was repeated. Finally, the pellets were suspended in 1.5 mL of Solution A and placed on ice for cell-sorting analysis. The FACSAria III (BD) was prepared according to the manufacturer’s instructions. The samples were loaded after they were filtered through a 70-μm nylon cell strainer (BD Falcon). For transcriptome analysis, protoplasts gated in P1 and P2 were collected in tubes filled with RLT buffer (RNeasy Plant Micro Kit; Qiagen) until the total cell count reached 20,000.
Microarray Experiments
Total RNA was extracted from cultured cotyledons (6 to 10 cotyledons) using an RNeasy Plant Mini Kit (Qiagen). Microarray experiments for wild-type and apl cotyledons were performed using the Arabidopsis Gene 1.0 ST Array (Affymetrix) according to the standard Affymetrix protocol. For cell sorting of samples, RNA was extracted using an RNeasy Plant Micro Kit (Qiagen). Extracted RNA was amplified with the Amplification Ovation RNA Amplification System V2 (NuGEN) and purified using a MinElute Reaction Cleanup Kit (Qiagen). Purified cDNA was fragmented and labeled with the Biotin EnCore Biotin Module (NuGEN). Hybridization, scanning, and data normalization were performed according to the standard Affymetrix instructions. The resulting data were analyzed using the Subio Platform (Subio) to generate heat map images. To extract the gene sets (SEOR1-coexpressed genes, VPs, VXs, and apl downregulated genes), normalized expression values were compared (see Supplemental Data Set 1).
Construction of the Coexpression Network
For all VPs (218 genes), median normalized values in log2 scale were obtained from three sets of transcriptome data: time-course, wild type versus apl, and SEOR1 cell-sorting data. Based on these values, the phloem coexpression network was constructed using the weighted gene coexpression network analysis (WGCNA) package (Langfelder and Horvath, 2008). The adjacency matrix was calculated with soft thresholding power, which was chosen based on the criterion of scale-free topology (fit index = 0.9). To minimize the effects of noise and spurious associations, the adjacency matrix was transformed into a topological overlap matrix (TOM). A fast greedy modularity optimization algorithm was used to determine modules. The network was constructed using the TOM and visualized by the igraph package.
Molecular Phylogenetic Analysis
Coding sequence data of phloem-related NAC genes were obtained from TAIR (https://www.arabidopsis.org/). Multiple sequence alignment was conducted with ClustalX (Supplemental File 1). The phylogenetic tree was constructed with the obtained alignments using the neighbor-joining method (through MEGA7). Evolutionary distances were computed using the p-distance method.
Observation of Root Vasculature
To observe continuous SE formation, samples were subjected to modified pseudo-Schiff-propidium iodide (mPS-PI) staining (Truernit et al., 2008). Briefly, roots of 7-d-old seedlings were dipped in PI solution (20 μg/mL in water) for 10 s and washed to remove excess PI. They were immediately mounted with water to prevent drying and observed under a FV1200 confocal microscope (Olympus).
Accession Numbers
Sequence data from this study can be found in the Arabidopsis Genome Initiative data library under the following accession numbers: APL (AT1G79430), SEOR1 (AT3G01680), NAC045 (AT3G03200), SUS5 (AT5G37180), RTM2 (AT5G04890), SUC2 (AT1G22710), SULTR1;3 (AT1G22150), AHA3 (AT5G57350), NEN4 (AT4G39810), GSL07 (AT1G06490), BRX (AT1G31880), UBQ14 (AT4G02890), UBQ10 (AT4G05320), IRX3 (AT5G17420), XCP1 (AT4G35350), MYB46 (AT5G12870), VND6 (AT5G62380), NAC020 (AT1G54330), NAC028 (AT1G65910), NAC057 (AT3G17730), NAC086 (AT5G17260), NEN1 (AT5G07710), HCA2 (AT5G62940), BAM3 (AT4G20270), OPS (AT3G09070), and CVP2 (AT1G05470). Microarray data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Accession numbers are GSE80027 (for SEOR1 cell sorting) and GSE80026 (for wild type versus apl).
Supplemental Data
Supplemental Figure 1. Phloem Marker Expression in Leaf Disks Revealed by VISUAL.
Supplemental Figure 2. Experimental Procedure for Aphidicolin Assay.
Supplemental Figure 3. Effects of Auxin and Cytokinin on Xylem and Phloem Cell Differentiation in VISUAL.
Supplemental Figure 4. Enucleation during SE-Like Cell Differentiation in VISUAL.
Supplemental Figure 5. A Confocal image of Differentiated Cells after Aniline Blue Staining in VISUAL.
Supplemental Figure 6. TE and SE Clusters in VISUAL.
Supplemental Figure 7. Bikinin-Dependent Increase in the Number of YFP-Positive Cells.
Supplemental Figure 8. Definition of Time Point at Half Maximum.
Supplemental Figure 9. Strategies for Constructing Phloem SE Coexpression Networks.
Supplemental Figure 10. Comparison between Known Early Phloem Regulators and Module Genes.
Supplemental Figure 11. Expression Patterns of NAC020, NAC028, and NAC057 in Roots.
Supplemental Figure 12. Cross Sections of Hypocotyl Vasculature in NAC020ox and NAC020-SRDX.
Supplemental Data Set 1. Genes Identified by Microarray Analyses.
Supplemental Data Set 2. Lists of Phloem-Related Genes.
Supplemental File 1. Alignment Used for Phylogenetic Analysis.
Supplementary Material
Acknowledgments
We thank Yasuko Ozawa and Yukiko Sugisawa for technical support. We also thank Michale Knoblauch, Ykä Helariutta, and Kaori Furuta-Miyashima for providing plant materials. This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (15H01226 to Y.K., and 15H05958 and NC-CARP project to H.F.), from the Japan Society for the Promotion of Science (26891005 to Y.K. and 23227001 to H.F.), and from Naito Foundation to H.F. This work was also supported by Japan Advanced Plant Science Network.
AUTHOR CONTRIBUTIONS
Y.K. designed the experiments. Y.K., A.M.N., C.S., Y.I., M.S., K.Y., N.M., and M.O.-T. conducted the experiments. Y.K. and H.F. wrote the article.
Glossary
- SE
sieve element
- TE
tracheary element
- DAPI
4′,6-diamidino-2-phenylindole
- EdU
5-ethynyl-2'-deoxyuridine
- TEM
transmission electron microscopy
- CC
companion cell
- DIC
differential interference contrast
References
- Aloni R. (1980). Role of auxin and sucrose in the differentiation of sieve and tracheary elements in plant tissue cultures. Planta 150: 255–263. [DOI] [PubMed] [Google Scholar]
- Anne P., Azzopardi M., Gissot L., Beaubiat S., Hématy K., Palauqui J.C. (2015). OCTOPUS negatively regulates BIN2 to control phloem differentiation in Arabidopsis thaliana. Curr. Biol. 25: 2584–2590. [DOI] [PubMed] [Google Scholar]
- Barratt D.H., Kölling K., Graf A., Pike M., Calder G., Findlay K., Zeeman S.C., Smith A.M. (2011). Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis. Plant Physiol. 155: 328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonke M., Thitamadee S., Mähönen A.P., Hauser M.T., Helariutta Y. (2003). APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186. [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806. [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Parra M.A., Anderberg R.J., Carrington J.C. (2001). Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long-distance movement of tobacco etch virus. Plant Physiol. 127: 1667–1675. [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S., Rodriguez-Villalon A., Santuari L., Wyser-Rmili C., Ragni L., Hardtke C.S. (2013). Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc. Natl. Acad. Sci. USA 110: 7074–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire P., Ménard D., Green P., Saalbach G., Buschmann H., Lloyd C.W., Pesquet E. (2015). Proteomic analysis of microtubule interacting proteins over the course of xylem tracheary element formation in Arabidopsis. Plant Cell 27: 2709–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., et al. (2009). Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 16: 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., Mahonen A.P., Helariutta Y., Weijers D. (2016). Plant vascular development: from early specification to differentiation. Nat. Rev. Mol. Cell Biol. 17: 30–40. [DOI] [PubMed] [Google Scholar]
- DeWitt N.D., Sussman M.R. (1995). Immunocytological localization of an epitope-tagged plasma membrane proton pump (H(+)-ATPase) in phloem companion cells. Plant Cell 7: 2053–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo H., Yamaguchi M., Tamura T., Nakano Y., Nishikubo N., Yoneda A., Kato K., Kubo M., Kajita S., Katayama Y., Ohtani M., Demura T. (2015). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56: 242–254. [DOI] [PubMed] [Google Scholar]
- Froelich D.R., Mullendore D.L., Jensen K.H., Ross-Elliott T.J., Anstead J.A., Thompson G.A., Pélissier H.C., Knoblauch M. (2011). Phloem ultrastructure and pressure flow: Sieve-Element-Occlusion-Related agglomerations do not affect translocation. Plant Cell 23: 4428–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H., Komamine A. (1980). Establishment of an experimental system for the study of tracheary element differentiation from single cells isolated from the mesophyll of Zinnia elegans. Plant Physiol. 65: 57–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K.M., et al. (2014). Plant development. Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science 345: 933–937. [DOI] [PubMed] [Google Scholar]
- Heo J.O., Roszak P., Furuta K.M., Helariutta Y. (2014). Phloem development: current knowledge and future perspectives. Am. J. Bot. 101: 1393–1402. [DOI] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845. [DOI] [PubMed] [Google Scholar]
- Kondo Y., Fujita T., Sugiyama M., Fukuda H. (2015). A novel system for xylem cell differentiation in Arabidopsis thaliana. Mol. Plant 8: 612–621. [DOI] [PubMed] [Google Scholar]
- Kondo Y., Fukuda H. (2015). The TDIF signaling network. Curr. Opin. Plant Biol. 28: 106–110. [DOI] [PubMed] [Google Scholar]
- Kondo Y., Ito T., Nakagami H., Hirakawa Y., Saito M., Tamaki T., Shirasu K., Fukuda H. (2014). Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat. Commun. 5: 3504. [DOI] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19: 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S., Sebastian J., Lee J.Y., Helariutta Y. (2013). Stem cell function during plant vascular development. EMBO J. 32: 178–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motose H., Sugiyama M., Fukuda H. (2004). A proteoglycan mediates inductive interaction during plant vascular development. Nature 429: 873–878. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Oda Y., Fukuda H. (2012). Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337: 1333–1336. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Oda Y., Fukuda H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22: 3461–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Zhang J., Cao J., Yin S.Y., He X.Q., Cui K.M. (2008). Phloem transdifferentiation from immature xylem cells during bark regeneration after girdling in Eucommia ulmoides Oliv. J. Exp. Bot. 59: 1341–1351. [DOI] [PubMed] [Google Scholar]
- Paciorek T., Sauer M., Balla J., Wiśniewska J., Friml J. (2006). Immunocytochemical technique for protein localization in sections of plant tissues. Nat. Protoc. 1: 104–107. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Villalon A., Gujas B., Kang Y.H., Breda A.S., Cattaneo P., Depuydt S., Hardtke C.S. (2014). Molecular genetic framework for protophloem formation. Proc. Natl. Acad. Sci. USA 111: 11551–11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K., Ursache R., Hejátko J., Helariutta Y. (2015). Xylem development - from the cradle to the grave. New Phytol. 207: 519–535. [DOI] [PubMed] [Google Scholar]
- Simmons A.R., Bergmann D.C. (2016). Transcriptional control of cell fate in the stomatal lineage. Curr. Opin. Plant Biol. 29: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolund R.D. (1997). The phloem sieve element: A river runs through it. Plant Cell 9: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolund R.D., Shih C.Y. (1983). Freeze-fracture analysis of phloem structure in plant tissue cultures. I. The sieve element reticulum. J. Ultrastruct. Res. 82: 111–121. [DOI] [PubMed] [Google Scholar]
- Stadler R., Sauer N. (1996). The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot. Acta 109: 299–306. [Google Scholar]
- Takada S., Takada N., Yoshida A. (2013). ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development 140: 1919–1923. [DOI] [PubMed] [Google Scholar]
- Taylor-Teeples M., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517: 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N.G., Scheible W.R., Cutler S., Somerville C.R., Turner S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Belcram K., Barthélémy J., Palauqui J.C. (2012). OCTOPUS, a polarly localised membrane-associated protein, regulates phloem differentiation entry in Arabidopsis thaliana. Development 139: 1306–1315. [DOI] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J.C. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore R.H., Rier J.P. (1963). Experimental induction of vascular tissues in callus of angiosperms. Am. J. Bot. 50: 418–430. [Google Scholar]
- Xie B., Wang X., Zhu M., Zhang Z., Hong Z. (2011). CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. 65: 1–14. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Mitsuda N., Ohtani M., Ohme-Takagi M., Kato K., Demura T. (2011). VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 66: 579–590. [DOI] [PubMed] [Google Scholar]
- Yoshimoto N., Inoue E., Saito K., Yamaya T., Takahashi H. (2003). Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 131: 1511–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Johnson B.J., Kositsup B., Beers E.P. (2000). Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol. 123: 1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Zhou J., McCarthy R.L., Ye Z.H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zhong R., Ye Z.H. (2014). Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One 9: e105726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.