ABA-mediated desiccation tolerance in P. patens depends on a trimodular protein kinase, found only in early land plants and aquatic algae, proposed to have facilitated plant colonization of land.
Abstract
The anatomically simple plants that first colonized land must have acquired molecular and biochemical adaptations to drought stress. Abscisic acid (ABA) coordinates responses leading to desiccation tolerance in all land plants. We identified ABA nonresponsive mutants in the model bryophyte Physcomitrella patens and genotyped a segregating population to map and identify the ABA NON-RESPONSIVE (ANR) gene encoding a modular protein kinase comprising an N-terminal PAS domain, a central EDR domain, and a C-terminal MAPKKK-like domain. anr mutants fail to accumulate dehydration tolerance-associated gene products in response to drought, ABA, or osmotic stress and do not acquire ABA-dependent desiccation tolerance. The crystal structure of the PAS domain, determined to 1.7-Å resolution, shows a conserved PAS-fold that dimerizes through a weak dimerization interface. Targeted mutagenesis of a conserved tryptophan residue within the PAS domain generates plants with ABA nonresponsive growth and strongly attenuated ABA-responsive gene expression, whereas deleting this domain retains a fully ABA-responsive phenotype. ANR orthologs are found in early-diverging land plant lineages and aquatic algae but are absent from more recently diverged vascular plants. We propose that ANR genes represent an ancestral adaptation that enabled drought stress survival of the first terrestrial colonizers but were lost during land plant evolution.
INTRODUCTION
Plants successfully colonized terrestrial habitats ∼500 million years ago and changed the face of the planet (Kenrick and Crane, 1997; Kenrick et al., 2012; Wickett et al., 2014). The first plants to colonize land were derived from an aquatic algal ancestor (Delwiche and Cooper, 2015), and fossil evidence obtained of the earliest land plants indicates many features in common with extant bryophytes, lineages that diverged prior to the acquisition of vascular tissue that are often referred to as “basal” land plants. What is clear is that the earliest land plants lacked the anatomical adaptations to a terrestrial environment characteristic of today’s dominant vascular plant flora: extensive ramifying root systems scavenging water to be transported to aerial organs via a highly differentiated vascular system reinforced by lignin and waterproofed by cuticular wax and suberin. In order to survive frequent cycles of dehydration and rehydration, the first land plants must necessarily have deployed efficient molecular/biochemical strategies to withstand drought. Even today, the existence of vegetative desiccation tolerance is widespread throughout the bryophytes but was lost following the emergence of the vascular plants, although its occasional reacquisition suggests that mutations in relatively few genes are required for loss or gain of this key adaptive trait (Oliver et al., 2005).
In fact, the tracheophytes still display widespread desiccation tolerance, although this is typically restricted to the reproductive propagules: spores, pollen grains, and seeds all have the ability, and frequently the necessity, to survive extreme dehydration (Franchi, et al., 2011). Throughout the land plant lineage, water stress responses are coordinated by the plant growth regulator abscisic acid (ABA). ABA is synthesized in response to water deficit, and it elicits a range of responses to water stress, from the execution of stomatal closure to the induction of gene expression leading to the accumulation of protective compounds in desiccated tissues (typically disaccharide sugars and Late Embryogenesis Abundant [LEA] proteins required for the stabilization of cellular macromolecules and membranes in the dry state) (Koster and Leopold, 1988; Tunnacliffe and Wise, 2007; Buitink and Leprince, 2008; Shimizu et al., 2010; Clarke et al., 2015; Popova et al., 2015). The response to ABA is achieved through a highly conserved core signaling pathway, in which a family of receptor proteins (the PYRABACTIN RESISTANCE1 [PYR1]/PYR1-LIKE [PYL]/REGULATORY COMPONENTS OF ABA RECEPTORS [RCAR] proteins) bind ABA, which potentiates a molecular interaction with protein phosphatase 2C (PP2C) enzymes (e.g., those encoded by the ABSCISIC ACID INSENSITIVE1 [ABI1] and ABI2 genes). Sequestration of these phosphatases by the ABA-bound receptors allows the phosphorylation of SnRK2 protein kinases that in turn activate phosphorylation of transcription factors (e.g., the ABI5 bZip transcription factor) that trigger ABA-mediated transcription of the genes encoding dehydration tolerance-associated proteins (Park et al., 2009; Cutler et al., 2010).
Using Physcomitrella patens, we can investigate the evolution of gene function in extant land plants representative of the earliest diverging groups (∼500 million years ago). The availability of the P. patens genome sequence (Rensing et al., 2008) coupled with the ability to undertake direct mutagenesis through highly efficient homologous recombination-mediated gene targeting (Kamisugi et al., 2005) has enabled the functional dissection of the ABA signal transduction pathway in nonvascular land plants. The P. patens genome encodes all of the components of the core ABA signaling pathway identified in angiosperms, and the use of gene-targeted knockout mutants has identified the fundamental conservation of the elicitation of stress-mediated ABA synthesis, of PP2C and SnRK2 functions in growth responses, stomatal closure and gene expression, and the role of conserved transcription factors in eliciting ABA- and stress-mediated gene expression (Komatsu et al., 2009, 2013; Khandelwal et al., 2010; Chater et al., 2011; Takezawa et al., 2015).
Gene targeting is a powerful tool for comparative genomic studies, through the use of reverse genetics, but relies on a priori assumptions derived from sequence homologies and thus is unsuitable for the identification of novel components that may be taxon-specific in nature. To determine whether there were any additional features of the ABA signal transduction pathway unique to basal plants, we undertook a genetic analysis of the ABA response in P. patens. This identified a previously unsuspected regulator of ABA- and drought stress responses: a trimodular MAP3 kinase that was recently demonstrated to also play a role in ethylene signaling (Yasumura et al., 2015), a key component of flooding tolerance, thus representing a potential point of integration for the transduction of water-deficit and water-surfeit stress responses.
RESULTS
Characterization of an ABA Nonresponsive Mutant
When P. patens protoplasts or protonemal explants are regenerated in the presence of ABA, the cells exhibit a characteristic change in growth habit characterized by cellular differentiation producing brachycytes (brood cells)—small, round, thick-walled, nonvacuolate cells in which vacuolation and cell expansion is suppressed (Figure 1)—interspersed with tmema cells (empty, thin-walled cells that enable dispersal of dehydrated brood cells that can act as vegetative spores) (Decker et al., 2006). The consequence of this cellular change is a highly reduced growth rate in which regenerating plants have a dwarf, twisted, protonemal morphology.
Figure 1.
ABA-Mediated Cellular Differentiation.
Wild-type chloronemal cells have a typically elongated appearance, containing a single large vacuole enclosed by the tonoplast, here marked by the expression of the vacuolar membrane marker At-Vam3-GFP. Bars = 20 μm.
(A) Bright-field image.
(B) GFP fluorescence. In the presence of ABA (10−5 M), smaller rounded brood cells are formed. The plastids are densely packed and the vacuole unexpanded.
(C) Bright-field image.
(D) GFP fluorescence.
We used UV-C irradiation to generate mutants in the ABA response. By regenerating UV-irradiated protoplasts from the Gransden (Gr) strain in the presence of ABA, we identified mutants that retained the rapid growth habit of regenerating protonemata, comparable to that normally observed in the absence of ABA (Figure 2). This clearly contrasted with the wild-type response in which brood cells were formed (Figures 2A and 2B). These mutants also exhibited drought hypersensitivity, with the aerial gametophore tissue undergoing premature senescence unless continually irrigated (Figures 2C and 2D). A number of independent mutants, designated aba non-responsive (anr1-7), were cross-fertilized by the genetically divergent ‘Villersexel K3’ (Vx) strain to generate segregating populations (Kamisugi et al., 2008). Hybrid diploid sporogonia recovered from the mutant (maternal, Gr) plants were identified by spore germination and growth testing of the sporelings on ABA-supplemented medium; hybrid sporogonia yielded haploid spores that segregated 1:1 for the anr and wild-type phenotypes. The spores obtained from hybrid sporogonia represent a segregating population containing recombinant chromosomes—the equivalent of recombinant inbred lines in diploid species.
Figure 2.
Phenotype of anr Mutants.
(A) Identification of an anr mutant regenerant following UV mutagenesis (arrowed).
(B) The region enclosed in the rectangle in (A).
(C) Wild-type plant growing on compost.
(D) Premature senescence of aerial gametophore leaves in an anr mutant.
Ninety-six segregants (48 mutant and 48 wild type) from the anr4 × Vx cross were genotyped at 4309 single nucleotide polymorphism (SNP) markers, using an Illumina GoldenGate genotyping platform, to identify a genetic interval containing the mutant locus. Plotting the SNP genotype ratio of Gr alleles for the 48 anr4 mutants across each chromosome in the P. patens V3.0 sequence assembly revealed a single peak on chromosome 12 (Figure 3). The inverse relationship was observed between the wild-type phenotype and the Vx alleles (Figure 3A). The minimum genetic interval contained three mapped SNPs, encompassing a region of 88,366 bp containing eight gene models (Figure 3B). Candidate genes were sequenced from sets of overlapping PCR fragments, and a C>T point mutation at chromosome 12 sequence coordinate 2,814,230 generating a premature termination codon (CAG>TAG: Q712Ter) was identified in anr mutant segregants in the V3.0 locus Phpat.012G009800 (also known as Pp1s462_10V6, Phypa_30352, and Pp3c12_3550 in the V1.6, 1.2, and 3.1 assemblies, respectively) (Zimmer et al., 2013; http://phytozome.jgi.doe.gov; www.cosmoss.org).
Figure 3.
Identification of ANR.
(A) SNP genotype ratios for wild-type and anr segregants along chromosome 12.
(B) Gene models located within the genetic interval identified in (A).
(C) Targeting strategy to generate anrKO and anrPTC4 mutants. Untranslated region sequences are indicated in blue and protein-coding sequences in red. The position of the anr4 premature termination codon is shown in exon 7.
We designated this locus ANR. Wild-type ANR encodes a MAP3 kinase-like protein containing two additional conserved domains: a central EDR domain, found in the Arabidopsis thaliana Raf3-like MAP3 kinases CTR1 and EDR1, which regulate ethylene and pathogen responses, respectively (Kieber et al.,1993; Frye et al., 2001); and an N-terminal PAS domain, a small and ancient sensor domain best described in bacterial two-component systems but also found in a range of eukaryotic response factors (Henry and Crosson, 2011). The anr4 mutation was predicted to introduce a premature termination codon (Q712Ter) located between the EDR and kinase domains (Figure 3C). Amplification and sequencing of this region in 10 additional segregants (five anr and five wild type) showed the termination codon occurring exclusively in the mutant segregants, strongly suggesting it as the causative mutation arising from the UV mutagenesis performed in the Gransden background. Functional confirmation was obtained by targeted mutagenesis of the wild-type locus. First, a null mutant (anrKO) was obtained by targeted replacement of all but the first and last two exons of the gene (a deletion corresponding to 813 amino acid residues) with a neomycin phosphotransferase selection cassette (Figure 3C; Supplemental Figure 1). The anrKO mutant demonstrated a clear anr phenotype by growing uninhibitedly in the presence of ABA (Figure 4). Additionally, we generated a targeted point mutation in which the wild-type exon 7 CAG was converted to TAG, thus generating an anr4 Q712Ter point mutant line otherwise isogenic with the Gr background (anr4PTC; Supplemental Figure 2). This similarly displayed an anr growth phenotype (Figure 4A), confirming the identification of the point mutation as sufficient to cause ABA nonresponsiveness.
Figure 4.
anr Mutants Grow Freely and Are Desiccation-Sensitive.
(A) Growth rates of the wild type, anrKO, and anr4PTC mutants in the presence (upper panel) or absence (lower panel) of 10−5 M ABA.
(B) and (C) Dehydration sensitivity of mutants. Chloronemal homogenate cultures grown on cellophane overlays were divided into four quadrants and pretreated with ABA at 10−6 M (A−6), 10−5 M (A−5), and 10−4 M (A−4) ABA (B) or were untreated (C) for 24 h prior to complete dehydration. Rehydrated tissue was allowed to recover on medium lacking ABA. Wild-type tissue treated with ABA survives dehydration, whereas untreated tissue does not. Neither anrKO nor anr4PTC mutant lines exhibit ABA-mediated desiccation tolerance.
anr Mutants Do Not Acquire ABA-Induced Desiccation Tolerance
The anr mutants do not undergo ABA-dependent cellular differentiation but continue to show normal growth kinetics. The anr mutant lines additionally display dehydration hypersensitivity. In addition to the drought hypersensitivity of mutant gametophores (Figure 2), the mutant protonemal tissue is also dehydration intolerant. Pretreatment with ABA for 24 h is sufficient to enable wild-type protonemal tissue to survive complete desiccation and recover normal growth upon rehydration (Oldenhof et al., 2006; Khandelwal et al., 2010). By contrast, both the anrKO and the anr4PTC mutants remain drought-intolerant, notwithstanding pretreatment with ABA (Figure 4B).
ANR Is Required to Coordinate Widespread Molecular Responses to Stress
ABA-mediated desiccation tolerance is associated with transcriptional induction of conserved plant stress-response genes through the action of a well-characterized set of transcription factors (designated ABI3, ABI4, and ABI5 in Arabidopsis), and this pathway also appears to be activated in P. patens (Komatsu et al., 2009; Khandelwal et al., 2010, Sakata et al., 2010). RNA-seq transcriptome-wide profiling of the hormone and stress response of chloronemal tissue demonstrates that ANR is required to elicit a molecular response to ABA, and osmotic and drought stresses required for the acquisition of ABA-dependent desiccation tolerance. Wild-type chloronemata rapidly accumulated transcripts encoding an overlapping spectrum of protective gene products when treated with ABA, exposed to moderate osmotic stress (10% mannitol), or dehydrated to 70% water loss. We identified 911 genes significantly upregulated across the three treatments in wild-type plants, among which those encoding LEA proteins, membrane channel proteins, and transporters, and photosynthesis-related stress proteins comprised a significant fraction (Figure 5; Supplemental Data Set 1 and Supplemental Figures 3 and 4). There was considerable overlap between the genes induced by ABA and stress treatments (Supplemental Figures 5 to 9), with 575 (63%) of these being upregulated in response to either two or (in the case of the most highly responsive genes) three treatments. There were 903 unique genes downregulated, with those encoding components of secondary metabolism (notably enzymes in the phenylpropanoid pathway) being a prominent class (Supplemental Data Set 2 and Supplemental Figure 10). By contrast, the anrKO mutant showed a massively reduced molecular response to ABA and stress and failed to accumulate the characteristic spectrum of products required for dehydration tolerance (Figure 5; Supplemental Data Set 3). Of the 119 genes upregulated in the mutant, only 16 were in common with those upregulated in the wild type. A larger number of genes (341) was downregulated in the anr mutant (Supplemental Data Set 4), of which over half (205) were in common with the genes downregulated in the wild type (Supplemental Figure 11). The majority of these common genes (194) comprised genes downregulated in response to mannitol-induced osmotic stress, of which only five showed a reduction in transcript abundance following ABA treatment, suggesting that whereas the induction of stress-induced gene expression is strongly correlated with the action of ABA, the early responses to osmotic stress may involve a stress-associated turnover of transcripts that is largely ABA independent.
Figure 5.
anrKO Mutants Do Not Accumulate LEA Transcripts in Response to ABA or Stress.
(A) Numbers of genes up- and downregulated in response to ABA, osmotic stress (10% mannitol), and dehydration in wild-type chloronemal tissue.
(B) Total numbers of differentially regulated genes in wild-type and mutant plants. Pale-gray bars indicate downregulated genes, and dark-gray bars indicate upregulated genes
(C) Change in abundance (log2) of transcripts corresponding to 46 LEA genes (listed in Supplemental Table 2) following treatment of chloronemal tissue with 10−5 M ABA in wild-type (blue bars) and anrKO (red bars) plants.
The ABA Response Is Self-Reinforcing
Among the genes strongly upregulated by ABA, osmotic and drought stresses are known components of the ABA signal transduction pathway. These include the ABI1/ABI2 protein phosphatases, the SnRK2 kinase PpOST1-1, genes encoding transcription factors implicated in ABA- and drought-responsive gene expression (including three ABI3 paralogs, two genes encoding drought response element binding proteins [DREBs], and homologs of the bZip class ABI5 transcription factors), and a histidine kinase homologous with the Arabidopsis osmosensory protein AtHK1 (Table 1; Supplemental Table 1). None of these genes are upregulated in the anrKO mutant, suggesting that the ANR gene is important to implement a powerful positive feedback loop to rapidly reinforce the ABA/drought response.
Table 1. ABA Biosynthetic and Signaling Genes Are Upregulated by ABA and Stress Treatments.
| Gene Product | ABA Fold | Dehydration Fold | Mannitol Fold | KO Fold | V3.0 Gene Model | |||
|---|---|---|---|---|---|---|---|---|
| WT | KO | WT | KO | WT | KO | KO | ||
| Core components | ||||||||
| PYL4-2 | 2.4 | – | 1.8 | – | 1.6 | −1.7 | – | Phpat.013G023700 |
| PYL5 | 7.2 | 1.8 | 4.0 | 1.6 | 4.6 | 1.5 | – | Phpat.009G076800 |
| ABI1 | 10.8 | – | 10.9 | – | 9.2 | – | – | Phpat.007G024200 |
| ABI2 | 16.7 | – | 8.8 | – | 8.6 | −2.1 | – | Phpat.011G067300 |
| OST1-1 | 27.6 | 2.6 | 21.7 | – | 29.7 | – | – | Phpat.005G078000 |
| OST1-2 | 7.1 | – | 5.8 | – | 2.7 | −1.6 | 1.5 | Phpat.006G041000 |
| OST1-4 | 4.8 | – | 2.5 | −1.8 | 2.0 | −1.5 | – | Phpat.005G065600 |
| ABI3A | 4.1 | – | 1.8 | – | 4.3 | – | 1.8 | Phpat.002G014700 |
| ABI3B | 4.3 | 1.5 | 4.3 | – | 7.3 | 1.7 | – | Phpat.017G059900 |
| ABI3C | 4.0 | 1.6 | 2.6 | – | 1.7 | −1.9 | – | Phpat.004G026900 |
| ABI4-like | 2.8 | 1.5 | 2.1 | – | 2.5 | – | 1.6 | Phpat.013G044400 |
| ABI5-like 1 | 10.0 | – | 6.1 | −1.7 | 2.5 | −2.3 | – | No good model |
| ABI5-like 2 | 18.8 | – | 16.9 | −1.6 | 5.6 | −2.0 | −1.7 | No good model |
| ABA biosynthetic enzymes | ||||||||
| NCED | 22.3 | – | 14.9 | −3.1 | 13.2 | – | −2.7 | Phpat.025G017600 |
| NCED | 24.5 | −3.1 | 16.1 | – | 14.8 | – | – | Phpat.016G070400 |
| AAO | 2.8 | – | 1.9 | – | – | −1.8 | 2.6 | Phpat.020G059900 |
| CYP707A | −5.3 | −3.5 | −38.4 | −13.3 | −93.3 | −12.1 | −2.1 | Phpat.015G014800 |
| Putative ABA-dependent signaling components | ||||||||
| HK1-like | 27.4 | −1.7 | 33.6 | −1.6 | 38.6 | −1.6 | 1.7 | Phpat.010G014900 |
| PP2C; C subfamily | 95.6 | 1.8 | 79.4 | 2.0 | 22.8 | – | −3.3 | Phpat.005G075000 |
| PP2C; C subfamily | 34.2 | 2.1 | 18.5 | 1.6 | 25.6 | 2.7 | −2.8 | Phpat.017G037200 |
| GRF (14-3-3) | 129.2 | 2.6 | 70.4 | 1.7 | 25.2 | – | −6.1 | Phpat.003G031200 |
| CBF/NF-Y | 183.5 | – | 89.0 | – | 37.8 | – | – | Phpat.009G024400 |
| DREB; subfamily A-2 | 141.8 | – | 120.2 | – | 32.8 | – | – | Phpat.010G046900 |
| DREB; subfamily A-4 | 16.5 | – | 26.6 | – | 17.9 | – | – | Phpat.008G027400 |
ANR-Like Genes Are a Feature of All Basal Plant Lineages
A feature of the ANR protein is its trimodular structure, containing a PAS, EDR, and kinase domain (an architecture we describe as PEK). A search of the available genomic and transcriptomic databases for similar genes failed to identify any gene products among angiosperms, gymnosperms, or ferns with a comparable structure, although proteins comprising an EDR domain and kinase domain (EK), including the CTR1 and EDR1 regulators of ethylene and pathogen responses, respectively, are widespread, as are proteins containing a PAS domain and kinase domain (PK). By contrast, PEK proteins were identified in the genome sequences of the lycophyte Selaginella moellendorfii, the moss Ceratodon purpureus, and the liverwort Marchantia polymorpha, and searching the 1000 Plant Transcriptomes database (www.onekp.com) identifies additional PEK proteins within the hornworts and the green algae (Supplemental Table 3).
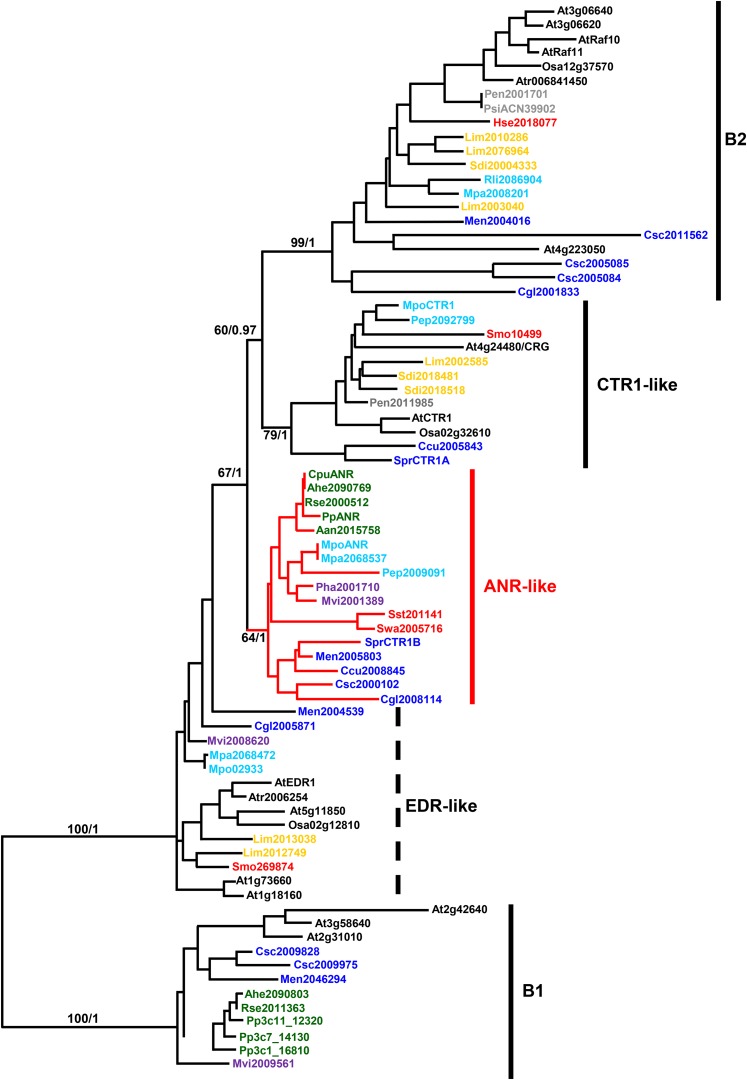
Phylogenetic analysis using the individual domains clarifies the relationships among these proteins (Figure 6). The ANR kinase domain is most closely related to the B-group (Raf-like) MAP3K family of protein kinases (Ichimura et al., 2002; Champion, et al., 2004). This is subdivided into subfamilies in which several members have been implicated in hormone and stress signaling in angiosperms. In addition to the EDR and CTR1 EK kinases, these include two PK kinase paralogs (Raf10 and Raf11) that positively regulate ABA responses in seed development and growth in Arabidopsis (Lee et al., 2015). A third PK kinase (encoded by At4g23050) is ABA activated and confers salt tolerance in Arabidopsis (Shitamichi et al., 2013). The B-group MAP3K family is subdivided into the B1, B2, B3, and B4 subfamilies, of which the B4 subfamily is the most divergent and is not included in our analysis. The previously defined B1 and B3 subfamilies contain EK kinases, whereas the B2 subfamily contains PK kinases (Ichimura et al., 2002). Multispecies phylogenetic analysis based on the conserved kinase domains suggests that the classification of the B3 subfamily should be revised, as we find the CTR1-like and EDR1-like EK kinases to be part of distinct subfamilies (Figure 6; Supplemental Data Set 5). The PEK kinases form a distinct and newly described subfamily of Raf-like MAP3 kinases that include members from the charophytes, hornworts, liverworts, mosses, and Selaginella spp. Crucially, inclusion of sequences from charophytic algae, in addition to those from all of the major lineages of land plants, demonstrates that these subfamilies were already established in the green algal ancestors of terrestrial plants and that Raf-like PK kinases and CTR1- and EDR1-like EK kinases occur in all taxonomic groups analyzed, except the mosses. Significantly, the PEK kinases are absent in later-diverging taxa. The recovery of the EK B1 subfamily, which contains all EK kinases from P. patens, as sister to the EDR1-, CTR1-, ANR-like, and B2 subfamilies, suggests that EK kinases were ancestral to the subfamilies analyzed. The inference of evolutionary events suggests that acquisition of a PAS domain by an EK kinase generated the PEK kinase subfamily that subsequently spawned the CTR1-like (EK) and B2 (PK) kinase subfamilies through the subsequent losses of the PAS and EDR domains, respectively. Phylogenetic analysis based on the EDR domain sequences supports the separation of the CTR1-like and ANR-like kinases, with the two groups appearing as distinct sister clades (Supplemental Figure 12 and Supplemental Data Set 6). As noted above, while other bryophytes retain members representing each kinase subfamily, none of the moss species for which sufficient sequence resources are available contain either CTR1-like EK kinases or any B2 PK kinases (Supplemental Figure 13 and Supplemental Data Set 7). Thus, the dual functionality of the ANR kinase in mediating both ABA and ethylene responses described by Yasumura et al. (2015) may be a unique feature of the moss lineage. More generally, the presence of PEK ANR-like genes in all of the early green plant lineages suggests that ANR-mediated regulation of drought responses is an ancestral adaptation, essential for the survival of dehydration by early land plants, but subsequently lost in the euphyllophytes.
Figure 6.
Phylogenetic Analysis of ANR Homologs.
Protein tree of the kinase domains from Raf-like B-group MAP3 kinases that show the highest homology to ANR. The amino acid sequences from species across all major plant lineages were analyzed using both a Bayesian (MrBayes) and Maximum Likelihood (RAxML) approaches. Both methods produced nearly identical topologies, and both bootstrap (BS) and posterior probability (PP) values are shown for key clade branches (BS/PP). Note the generally lower values for BS than PP. The actual tree shown is that produced by RAxML. Entries are color-coded so that light blue = algae, dark blue = liverworts, green = mosses, purple = hornworts, red = lycophytes, orange = ferns, gray = gymnosperms, and black = angiosperms. Species are indicated by three-letter codes where the first letter is the first letter of the genus followed by first two letters of the species name. Exceptions are At = Arabidopsis thaliana and Pp = Physcomitrella patens. All species are listed in Supplemental Table 3. The Raf-like MAP3K subfamilies are indicated by vertical lines. The branches in red indicate the ANR-like kinases. The B1 subfamily of EK kinases is found as sister to the other kinases included.
The PAS Domain of ANR: An Enigmatic Conserved Feature
Since a defining feature of the ANR protein and its homologs is the presence of the N-terminal PAS domain, we sought to further characterize its function. PAS domains are widespread in all branches of the tree of life and are associated with the regulation of responses to external stimuli. They have been identified variously as small ligand binding domains, mediators of protein-protein interactions, and as protein dimerization domains and have become attractive regulatory modules in synthetic biology (see Möglich and Moffat, 2010). Characterized by a highly conserved three-dimensional structure (the PAS-fold), they are nevertheless highly diverse in amino acid sequence; consequently, attempts to infer functional properties through sequence conservation have been largely unsuccessful. To define the role of the ANR PAS domain in more detail, we therefore undertook its structural determination by x-ray crystallographic analysis.
Crystals were obtained that diffracted to 1.7-Å resolution, enabling the crystal structure to be determined. The structure was a homodimer (chain A and chain B) in the C2 space group with a cell volume of 505,920 Å3 (Supplemental Table 4). A single disulphide bond is formed between residues C4 and C98 covalently binding the first and last β-strand of each chain. The domains in each homodimer are in a 180° rotation relative to each other such that the Fα helices are on the same side of the cell and a primary dimerization interface is formed between antiparallel Gβ strands (Figures 7A and 7B). This dimerization orientation, as well as packing effects, introduces a number of asymmetries between the two chains in each homodimer (Supplemental Figure 14). This can be clearly visualized by the B-factor scores of the residues whereby greater values (heat-mapped and visualized as thicker regions on the B-factor putty image; Figure 7B) represent regions of greater flexibility. The Fα of Chain B has higher B-factor scores than that of Chain A, averaging 40.8 Å2 compared with 22.7 Å2 across positions 43 to 59. The Hβ-Iβ loop region shows similar differences with Chain B, averaging 53.4 Å2 compared with 26.3 Å2 across positions 86 to 91 (Figure 7B). The Fα helix is known to be variable and flexible in many PAS domains often regulating the internal cavity size and, therefore, specificity of potential ligands and cofactors.
Figure 7.
The ANR PAS Domain Forms a Homodimer in the Crystal Structure.
(A) Solving of the ANR PAS domain structure finds a homodimer in each asymmetric unit (C2 space group). The two chains (A = green; B = blue) are found in a 90° rotation from each other in this plane of view. Each monomer shows the typical PAS-fold structure with the typically large Fα helix opposing the β-sheet. Chain B has an extended C terminus likely due to the stabilizing effects of interactions with Chain A. The primary dimerization interface is shown between the antiparallel Gb strands, with the interacting residues and their hydrogen bonds (black dashed lines) including one water-mediated bond (red sphere). The secondary structures are labeled for Chain A.
(B) Visualization of the B-factor scores for the structure reveals two regions on Chain B with markedly higher flexibility (thicker red/orange regions) than the relative positions on Chain A and to the rest of the structure. The Fα of Chain B has an average B-factor score of 40.8 Å2 compared with 22.7 Å2 on Chain A for positions 43 to 59. The Hβ-Iβ loop region has an average B-factor score of 53.4 Å2 compared with 26.3 Å2 on Chain A for positions 86 to 91.
The dimerization interaction appears to be primarily mediated by multiple hydrogen bonds along the antiparallel Gβ strands (Figures 7A and 7B). Analysis with the PISA program reveals that this dimerization interface is made up of six hydrogen bonds between four residues from each domain (Supplemental Figure 15). This interface is also augmented by water-mediated hydrogen bonds between G59 on chain A and Q65 on chain B. The PISA analysis also revealed the dimerization between Chains A and B to be moderately weak with a total buried surface area of 751.9 Å2 and a complexation significance score of only 0.031, suggesting the dimerization is primarily due to crystal packing. This supported the finding that the ANR PAS protein appears to be a mix of mostly monomers and small amounts of dimers in solution as assessed by gel filtration (Supplemental Figure 16). However, the crystal structure homodimers are found in a novel PAS dimer orientation that shows no similarity to known PAS homodimer structures (Supplemental Table 5). Structural comparisons of the monomers show the ANR PAS domain to be highly similar to all of a diverse set of PAS structures as shown by RMSD scores between monomers, making these comparisons unrevealing for functional predictions. Intriguingly, this structure does represent the only non-light-sensing plant PAS domain whose role and mode of action remain a mystery.
To probe the role of the PAS domain in mediating ABA- and stress-responsiveness, we used targeted mutagenesis to create novel variants (Supplemental Figures 17 and 18). A highly conserved tryptophan at position 16 (W16) in the PAS domain (W105 in the full-length ANR sequence) mediates hydrogen bond bridges that enable a tertiary loop region to form (Supplemental Figure 19). Mutation of this residue to alanine (a W16A mutant) was predicted to collapse the domain, and W16A mutant lines were ABA nonresponsive in growth tests (Figures 8A and 8B), indicating the requirement for a correctly folded PAS domain for ABA-mediated brood cell formation. Molecular responses to ABA were quantitatively attenuated in the W16A mutant, with a subset of signature ABA- and stress-responsive genes showing only an ∼10-fold increase in transcript levels in response to ABA (Figure 9). By contrast, a gene-targeted deletion mutant, lacking the entire PAS domain, displayed a normal response to ABA and stress, at both the phenotypic (Figure 8C) and molecular levels (Figure 9).
Figure 8.
Growth Responses of WT and W16A Mutant Lines.
(A) Growth of wild-type and W16A mutant lines (M30 and M45) 20 d following inoculation on unsupplemented BCDAT growth medium and on medium supplemented with 10 μM ABA, as indicated.
(B) Growth index of the wild type over the 20-d incubation period.
(C) Growth index of mutant lines over the 20-d incubation period.
(D) Growth of wild-type and PAS-deletion (C11 and D17) lines after 21 d on 10 μM ABA.
(E) Growth index of lines on control and ABA-supplemented medium over the 21-d incubation period. Growth index values are means ± sd (n = 8 to 9)
Figure 9.
Transcript Abundance in WT and PAS Domain Mutants.
Quantitative real-time PCR was used to determine the consequences of the W16A mutation in the PAS domain for ABA- and stress-mediated gene expression. Transcript abundance is expressed as the number of molecules per 10 ng total RNA. The genes tested were (from top) Phpat.010G025800 (TspO), Phpat.004G114600 (AWPM), Phpat.005G044800 (Dhn), Phpat.008G073700 (Up3), and Phpat.012G077600 (LEA3). Left-hand panels show transcript abundance in wild-type P. patens; right-hand panels show transcript abundance in two independently derived W16A mutant lines, M30 and M45. Treatments were with 10 μM ABA and 10% (w/v) mannitol, for 1 h, and dehydration to 50% fresh weight loss (∼5 h in an atmosphere of 80% relative humidity). Values are means ± sd for three biological replicates with two technical replicates each.
DISCUSSION
P. patens has been established as model for comparative genomics through its capacity for reverse genetic functional analysis of genes homologous with those previously identified in flowering plants (principally Arabidopsis). We now demonstrate the capacity for inferring ancestral gene functions by implementing a genetics approach in P. patens to identify a novel regulator of ABA responses, not predictable from studies in flowering plants.
The regulator encoded by ANR, a multidomain protein kinase, appears to have evolved in the aquatic algal ancestors of the land plants. In P. patens, it plays a central role in coordinating the molecular processes required for the acquisition of dehydration tolerance, as well as affecting the growth and developmental responses (brood cell differentiation) required to enable the survival of vegetative tissues that can be dispersed as vegetative spores. As such, we propose that this gene represents an early solution to the problems faced by the first land plants in colonizing terrestrial habitats, which was subsequently lost in the euphyllophytes.
A recent report, published while this article was in review, showed that the ANR kinase acts upstream of the established core ABA response pathway, directly phosphorylating the SnRK2 kinases that activate ABA-mediated gene expression, growth responses, and the acquisition of desiccation tolerance (Saruhashi et al., 2015). ABA acts through binding the PYR/PYL/RCAR ABA receptor proteins, which then sequester the protein phosphatases (PP2Cs) encoded by the ABI1 and ABI2 genes, thereby preventing their dephosphorylation of the SnRK2 kinases that activate downstream ABA responses (Park et al., 2009; Cutler et al., 2010). The core of this signal transduction pathway is ancient, with PP2C and SnRK2 families being present in unicellular chlorophytes, filamentous green algae, and all land plants (Ju et al., 2015). However, the ABA receptor gene family is not found in the aquatic ancestors of the land plants. While ABA synthesis occurs in a wide range of basal taxa, including cyanobacteria, fungi, and algae, ABA signaling has not been convincingly detected (Hirsch et al., 1989; Kobayashi et al., 1997, Hartung, 2010) in any of these taxa.
Nevertheless, biological desiccation tolerance occurs widely in nature, depending on the accumulation of cellular components with a protective function in maintaining macromolecular integrity in the face of cellular water loss. Many of the components of this response, such as LEA proteins, are of ancient evolutionary origin, being found in prokaryotes and invertebrates, as well as in both embryophytes and aquatic algae (Tunnacliffe and Wise, 2007; Tunnacliffe et al., 2010): The drought stress response in the semiterrestrial charophyte alga Klebsormidium crenulatum involves a transcriptional response similar in most respects to that seen in all other land plants that leads to the acquisition of a desiccation-tolerant state (Holzinger et al., 2014). It is thus likely that ANR-like genes, conserved in the earliest land plant lineages and their immediate ancestors, coordinated an ancient drought stress response that subsequently came under the control of the ABA response pathway when the PYR/PYL/RCAR gene products evolved their regulatory interaction with ABA in the land plant lineages. Komatsu et al. (2013) showed that in PP2C-deficient P. patens mutants, the ABA-mediated phosphorylation of SnRK2 kinases was only slightly greater than that observed in wild-type plants, suggesting these phosphatases might act downstream of SnRK2 activity. However, an alternative possibility is that the ANR kinase acts as an overdrive in the basal land plants, phosphorylating the SnRK2 effectors to an extent that the PP2Cs are relatively less important in ABA-mediated responses. The evolutionary loss of ANR orthologs in vascular plants would further bring drought stress responses more stringently under the control of ABA perception, with the hormone progressively regulating a suite of responses including restriction of water loss (e.g., stomatal closure) and osmoregulation to ameliorate the acute effects of drought. The diminished importance of vegetative desiccation tolerance is strongly correlated with increased anatomical complexity; plants with highly branched root systems and well-developed vascularization are able to efficiently retrieve water within the soil and transport it throughout the aerial plant body, enabling growth and photosynthetic production to be maintained under conditions of moderate drought (Alpert, 2005). Furthermore, vascularization and the attendant reinforcement of cell walls allow an increase in the size of the plant body. Thus, while in anatomically simple plants survival through vegetative desiccation tolerance (poikilohydry) is a selective advantage, as complexity and body size increase, it becomes a competitive disadvantage, since highly vascularized plants will outgrow their poikilohydric neighbors.
The distinguishing feature of ANR is its trimodular PAS-EDR-kinase (PEK) domain structure, and this modularity may be a key feature of its biological function. Interestingly, this tripartite PEK class of regulator has been lost in the more recently diverged groups of vascular plants, although dimodular EK kinases are common between all clades in the land plant lineage, while PK and EDR1- and CTR1-like EK Raf-like MAP3 kinases appear to be absent in mosses.
In P. patens, which lacks an EK CTR1-like kinase, ANR has a dual function, as both an ABA response regulator and an ethylene response regulator. The central EDR domain interacts with the ETR ethylene receptor (Yasumura et al., 2015). However, this characteristic appears to be unique to mosses, since while all other plant groups contain both EDR1-like and CTR1-like EK kinases, the mosses appear not to have retained an EK kinase specialized for ethylene response regulation (Figure 6; Supplemental Data Set 5), although such a kinase is clearly structurally and functionally conserved throughout 500 million years of evolution, in both algae (Ju et al., 2015) and angiosperms (Kieber et al., 1993). Significantly, however, whereas in Spirogyra pratensis a specific interaction was shown between its EK CTR1 homolog (Sp-CTR1a) and the algal ETR ethylene receptor, no such interaction was observed between Sp-ETR and the algal PEK kinase (Sp-CTR1b) that is an ANR ortholog (Ju et al., 2015).
The PAS domain, conserved in ANR orthologs in the basal lineages of the land plants and their charophytic ancestors, appears to be an important feature of this class of protein kinases, but its role remains enigmatic. PAS domains function as sensor domains in a wide range of environmental sensor kinases and have been found to sense light signals via a flavin ligand (Christie et al., 1999; Krauss et al., 2009), oxygen via a heme ligand (Gong et al., 1998; Delgado-Nixon et al., 2000), to directly bind small molecule effectors, including ions, as in divalent cation recognition by the bacterial transmembrane receptor PhoQ (García Véscovi et al., 1996; Véscovi et al., 1997; Cho et al., 2006) and to function in metabolite sensing (Golby et al., 1999) and redox state regulation, as in the NifL-mediated regulation of nitrogen fixation (Little et al., 2006).
PAS-mediated dimerization is also frequently a feature of PAS-dependent signal responses, including in the eponymous PAS-containing transcription factors PER, ARNT, and SIM (Huang et al., 1993; Card et al., 2005), in the Bacillus subtilis sporulation-inducing kinase KinA (Lee et al., 2008) and in the NifL kinase (Key et al., 2007). ANR dimerization mediated by the moderately weak interactions was identified in the PAS domain crystal structure, although not to a great extent in solution. However, it is plausible that the weak dimer interface identified by this domain in crystals could contribute to a more stable, cumulative dimer interface in the context of the full-length protein, a possibility that awaits resolution by more extensive structural analysis. The strong attenuation of the ABA and stress responses following the targeted mutagenesis of a structurally key residue suggest that the integrity of this domain is important for ANR activity, but since a PAS-deleted anr mutant retained a completely wild-type phenotype, it is likely that the structural collapse of the PAS domain in the W16A mutant engendered a partial denaturation or degradation of the mutant kinase. At present, the status of this domain remains enigmatic and will require further mutagenic analysis to fully elucidate its function.
Evolutionary change requires the acquisition of new gene functions and the evolution of such functions is typified by gene duplication and subfunctionalization (Ohno, 1970; Zhang, 2003; Flagel and Wendel, 2009), often accelerated through the acquisition and loss of conserved functional modules. The stress-related Raf-like B group MAP3K families in plants now provide an example of how this can occur, with the gain and loss of PAS and EDR domains apparent throughout their evolutionary history. Additionally, the emergence of the B2 PK proteins may have created redundancy in ABA signaling among the MAP3Ks, illustrated by the role recently demonstrated for Raf10/11 as positive ABA regulators in Arabidopsis (Lee et al., 2015). In P. patens, the loss of canonical CTR1 and the absence of Raf10/11 orthologs may have necessitated the retention of the role of ANR in ethylene signaling, although the status of these MAP3Ks in other basal plant species, such as liverworts (in which all B group subfamilies are found), needs resolution to understand fully this relationship in basal plants.
METHODS
Plant Material
Physcomitrella patens plants were propagated on BCDAT agar medium as previously described (Knight et al., 2002). Protoplasts were generated for mutagenesis and transformation using standard procedures (Schaefer, et al., 1991; Kamisugi et al., 2005). Strains used to generate segregating populations for genetic mapping were the ‘Gransden2004’ and ‘Villersexel K3’ laboratory strains (Kamisugi et al., 2008).
Mutagenesis
Protoplasts of the Gransden2004 strain were embedded in PRM-T agar medium (BCDAT containing 6% [w/v] mannitol, 10 mM CaCl2, and 0.4% agar) on cellophane overlaying the same medium containing 0.55% (w/v) agar in Petri dishes. Approximately 50,000 protoplasts were suspended in 3 mL PRM-T for each 9-cm Petri dish. These were exposed to 25,000 μJ⋅cm−2 UV radiation (280 nm) in a UV Stratalinker 2400 and incubated in darkness for 24 h. This dose resulted in an ∼20% survival rate. The plates were then incubated at 25°C under continuous illumination. After 2 d to permit cell wall regeneration, the cellophane discs bearing the embedded regenerants were transferred to plates containing standard BCDAT-agar medium (1 mM CaCl2, no mannitol) supplemented with 10−5 M ABA for 13 d, by which time anr mutants were distinguishable and could be routinely subcultured.
Establishment of Segregating Populations
Isolated mutants were crossed with the Villersexel K3 (Vx) wild type by inoculating explants adjacent to each other on BCD agar medium (lacking ammonium tartrate) as described previously (Kamisugi et al., 2008). The Gransden strain exhibits low levels of male fertility; consequently, the appearance of developing sporophytes on this strain was generally indicative of cross-fertilization by the Vx parent. Mature spore capsules on the Gransden parent were harvested and surface-sterilized before releasing the spores by crushing in sterile water. Spores were germinated and the progeny were replica-picked onto medium with and without 10−5 M ABA.
Genetic Mapping
DNA was extracted from individual segregants using a CTAB protocol (Knight et al., 2002) and 96 segregants (48 anr: 48 wild type) were genotyped on a custom SNP marker array (GoldenGate; Illumina) comprising 4309 loci at the Joint Genome Institute Oak Ridge National Laboratory. Data were analyzed by a custom Python script following chromosome scale mapping to identify Gransden-specific SNPs cosegregating with the anr phenotype and the reciprocal Vx-specific SNPs cosegregating with the wild-type phenotype. These SNPs were then located on the V3.0 P. patens genome assembly to define the physical limits of the genetic interval. SNP-specific primers from within this region were used to confirm the genotyping. The location of SNPs on chromosome 12, the SNP IDs, and SNP sequences are listed in Supplemental Data Set 8. Sequencing of candidate genes was performed by amplification of fragments from a single anr segregant using KOD polymerase (Takara) to amplify overlapping segments, which were cloned in pBluescriptKS− and sequenced (Source Bioscience) using both universal and custom primers. Identification of a CAG>TAG nonsense mutation in the V3.0 locus Phpat.012G009800 (Pp1s462_10V6, Phypa_30352, and Pp3c12_3550 in the V1.6, 1.2, and 3.1 assemblies) was confirmed by amplification and direct sequencing of a 350-bp PCR product containing this site in multiple anr and wild-type segregants.
Gene Targeting
The candidate anr4 locus was functionally confirmed by (1) targeted deletion of the protein-coding sequence and (2) targeted point mutagenesis of the mutated base to generate a CAG>TAG mutant in an isogenic Gransden genetic background. For targeted deletion, a strain anrKO was created by transforming wild-type Gransden2004 protoplasts with a DNA fragment containing a central CaMV35S-nptII-CaMVter selection cassette flanked by regions of homology corresponding to the 5′- and 3′-ends of the V3.0 Phpat.012G009800 locus. The targeting sequences comprised 1116 bp (V3.0 chromosome 12 sequence coordinates 2,809,033 to 2,810,198, comprising the first two exons of the gene) at the 5′-end and 878 bp (coordinates 2,817,434 to 2,818,311 comprising the last two exons of the gene). The transgene was amplified from a pBluescript KS−-based clone and used to transform protoplasts (Schaefer et al., 1991). Transformants containing a single copy replacement of the central 13 exons, no wild-type Pp1s462_10V6.1 sequence, and no ectopic insertions were identified by standard methodology using gene- and transgene-specific PCR and DNA gel blot analysis (Supplemental Figure 1; Kamisugi et al., 2005).
Targeted point mutagenesis was undertaken by marker-free transformation of wild-type protoplasts to generate two mutant alleles. First, a mutation corresponding to the anr4 premature termination codon was generated by transformation with a 1246-bp PCR amplicon derived from the anr4 mutant (sequence coordinates 2,813,530 to 2,814,775) containing the C>T mutation at position 2,814,230 approximately centrally located. Correctly targeted transformants were identified by the ABA-nonresponsive phenotype of regenerating protoplasts, followed by PCR and DNA gel blot analyses (Supplemental Figure 2).
Second, a mutation of a key residue in the PAS domain was generated by transformation of protoplasts with a 1484-bp fragment corresponding to sequence coordinates 2,808,882 to 2,810,365, in which the terminal dinucleotide (TG) of exon 1 was altered to CG. This results in the introduction of a W-to-A substitution at a key position in the PAS domain and destroys an AluI restriction site in the ANR locus. Regenerants exhibiting an ABA-nonresponsive phenotype were then analyzed by PCR amplification of the targeted region using primers external to the transforming DNA, digestion with AluI to identify mutated individuals, and DNA gel blot analysis to identify clean gene-targeted mutants. For both classes of single-copy targeted point mutants, the entire gene was resequenced to verify that the only mutation present was that introduced by gene targeting (Supplemental Figure 17).
Targeted deletion of the entire PAS domain was accomplished by transformation with a 1779-bp fragment comprising a fusion of the genomic sequences between coordinates 2,808,616 to 2,809,364 and 2,810,128 to 2,811,157. Protoplasts were cotransformed with this fragment and a circular pMBL5 plasmid encoding kanamycin resistance. Regenerating plants resistant to G418 were screened by PCR amplification using primers external to the transforming sequences to identify transformants in which the PAS domain sequence had been deleted. Selection was then relaxed, so that unintegrated selectable plasmid sequences would be lost. Correctly targeted ΔPAS mutants (a deletion of 119 amino acids) were confirmed by DNA sequence analysis (to ensure a correct in-frame deletion had been generated and DNA gel blotting; Supplemental Figure 18).
Growth Testing
Growth tests of wild-type and mutant plants were performed by measuring the increase in size of explants regenerating on BCDAT agar medium in the presence and absence of 10−5 M ABA. The growth index was calculated by image analysis of digital photographs using ImageJ as previously described (Kamisugi et al., 2012), with average plant area normalized either to the area of the Petri dish (Figure 4) or to a 5-cm line (Figure 8).
Desiccation Testing
The ability of wild-type and anr mutant P. patens protonemal tissue to acquire desiccation tolerance was tested as described previously (Khandelwal et al., 2010). Cellophane overlays bearing 6-d regenerated homogenate cultures were transferred onto filter paper discs soaked with BCDAT medium supplemented with ABA at concentrations of 10−6, 10−5, and 10−4 M, respectively, and incubated for 24 h. Control treatments lacked the addition of ABA. The cellophane overlays were then transferred to empty Petri dishes and allowed to dry in a laminar flow tissue culture cabinet until constant mass was achieved (>12 h). The tissue was rehydrated by returning to BCDAT agar medium and incubated for a further 7 d.
Gene Expression Analysis
Global transcriptomic analysis was undertaken by Illumina RNA-seq. Tissue treatments and RNA isolation was as previously described for microarray analysis of the P. patens stress response (Cuming et al., 2007). Tissue treatments included control (BCDAT medium), ABA-treated (BCDAT supplemented with 10−5 M ABA, 1 h), osmotically stressed (BCDAT supplemented with 10% [w/v] mannitol, 1 h), and dehydrated (incubated in an atmosphere at 80% relative humidity until a fresh weight loss of 70% was achieved, ∼12 h).
Three replicate samples for each treatment were then combined for the construction and analysis of RNA-seq libraries (0.5 μg RNA per treatment; Illumina HiSeq single-end 50-base reads; GATC Biotech). At least 30 million sequence reads were obtained for each sample. Raw reads of individual libraries were first viewed with fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to identify any biases and issues to guide preprocessing with trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic). Trimmed reads were aligned to the P. patens V3.0 genome and transcriptome assembly (http://phytozome.jgi.doe.gov/pz/portal.html#!bulk?org=Org_Ppatens) using TopHat2. Tophat_out files in .bam format were processed to obtain count values for each gene locus first using samtools (http://samtools.sourceforge.net/). The eight processed, aligned, and formatted libraries then had count data extracted using bedtools (http://bedtools.readthedocs.org/en/latest/). Tabular output files were trimmed for use in the Trinity Differential Expression package (http://trinityrnaseq.github.io/), which wraps the EdgeR Bioconductor package. Data were analyzed as described by the Trinity package guidelines using EdgeR and by normalizing count data by TMM scaling. Significance was set at 2-fold change and false discovery rate P value < 0.001. Cluster analysis was performed by manual cluster definition as described in the Trinity package. Library statistics are provided in Supplemental Table 7.
The expression of signature genes found to be significantly upregulated by ABA in the transcriptomic analysis was monitored by real-time PCR in mutant lines in which an amino acid substitution had been generated in the ANR PAS domain. Transcript abundance was determined using a dilution series of cDNA fragments amplified for each test gene and normalized relative to a reference gene encoding a Clathrin Coat Assembly Protein AP50 (CAP50; Phpat.027G008500). Primers used in this work are listed in Supplemental Table 6.
Phylogenetic Analysis
Sequences for analysis were retrieved from a number of databases, listed in Supplemental Table 3, using both BLASTp and tBLASTn searches to recover amino acid sequences. The use of Onekp involved tBLASTn searches, the retrieval of DNA scaffold sequences, and de novo protein translation prediction using the ExPASy tool (http://web.expasy.org/translate/). Ceratodon purpureus sequences were identified in the C. purpureus transcriptome data (Szövényi et al., 2015) and Marchantia polymorpha sequences assembled from genomic sequence data deposited in the NCBI Trace archive (accession number PRJNA251267) or obtained from previously published sources (Yasumura et al., 2012). Sequences were aligned using the Clustal omega tool (http://www.ebi.ac.uk/Tools/msa/clustalo/) and converted to nexus format for use with MrBayes. Alignments were 249, 339, and 138 amino acids long for the kinase, EDR, and PAS domain analyses, respectively. Analyses with MrBayes were set to sample across fixed amino acid rate matrices (with the Jones model selected as best for the kinase and EDR analyses and the WAG model for the PAS domain analysis), and rate variation was set to the gamma distribution. The mcmc run was set to stop automatically when the average sd of split frequencies dropped below 0.01 showing convergence between runs. Complete runs were summarized by sump and sumt commands with default settings. RAxML was run using the combined rapid bootstrap (using autoFC, which selected 300 replicates for the kinase [Figure 6] and EDR domain [Supplemental Figure 12] trees and 600 replicates for the PAS domain [Supplemental Figure 13] tree) and ML tree search method to produce the best tree with bootstrap values, using the LG matrix for all analyses. The trees were viewed and formatted using Tree Graph 2 (http://treegraph.bioinfweb.info/).
Structural Analysis of the PAS Domain
A 339-bp fragment encoding the PAS domain was amplified from a full-length ANR cDNA using the primers PAS_F and PAS_R (Supplemental Table 6). This was ligated into an Ecl136I site in a modified pET-28a plasmid generating a fusion with an N-terminal SUMO-His6 sequence. This was used to transform Escherichia coli BL21 cells for recombinant protein expression. Protein was isolated from IPTG-induced 2-liter cultures following lysis of the cell pellet resuspended in 20 mM Tris-Cl, pH 8, and 0.5 M NaCl. Recombinant protein was recovered by affinity purification on a 5 mL Ni2+-sepharose HisTrap HP column (GE Healthcare) and eluted with imidazole. The initial fusion protein was cleaved by a SUMO protease and dialyzed against lysis buffer for further purification on the 5 mL Ni2+-sepharose HisTrap HP column. Purified PAS protein was concentrated at 4°C in 15-mL Centriprep centrifugal filters (EDM Millipore; 10-kD molecular mass cutoff) at 2770g until the desired concentration or volume was reached and further purified by size exclusion chromatography on a Superdex 75 (26/60) column (GE Healthcare) (20 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM DTT, and 5% glycerol).
Crystals of the PAS domain were grown at 18°C by the sitting-drop vapor diffusion method. Drops consisted of 1 μL of protein (at 10 mg/mL) and 1 μL mother liquor containing 0.1 M Tris, pH 8.5 (HCl), 2 M Li2SO4, and 2% PEG 400. Crystals typically grew to 25 × 50 × 50 µm3 and were transferred to a cryoprotectant solution containing the mother liquor and 20% (v/v) glycerol (final concentration) before being mounted in loops and flash-cooled directly into liquid nitrogen.
Data were recorded to a resolution of 1.7 Å from a single crystal at 100K on the macromolecular crystallography beamline station i24 at Diamond Light Source. The diffraction images were processed using XIA2 (Winter, 2010), and the processing and crystallographic statistics are summarized in Supplemental Table 4. The single crystal belong to space group C2, with unit cell parameters of a = 85.2 Å, b = 64.9 Å, c = 51.9 Å, and b = 121.4°. There are two PAS domains per asymmetric unit.
The crystal structure was determined by molecular replacement using the program PHASER (McCoy et al., 2007) with the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea PDB entry 3LYX as the search models. Iterative manual model building and restrained refinement were performed using COOT (Emsley et al., 2010) and REFMAC5 (Murshudov et al., 2011). The polypeptide chains were checked against both 2Fo-Fc and Fo-Fc electron density maps during model building in COOT. Water molecules were added in COOT for peaks over 2.0ơ in the Fo-Fc map, and the structure validation was performed with MOLPROBITY (Chen et al., 2010). The final structure of the PAS domain was refined to Rfactor = 16.1% and Rfree = 19.9%. The refinement statistics are summarized in Supplemental Table 6. The atomic coordinates and structure factors have been deposited into the Protein Data Bank (www.pdb.org) with the accession code 5IU1.
Accession Numbers
Transcriptomic data obtained in this study have been deposited in the Gene Expression Omnibus database under accession number GSE72583 and thence to the NCBI Sequence Read Archive (accession number SRP063055; BioProject PRJNA294412). The structure of the PpANR PAS domain has been deposited in the Protein Structure Database under accession number 5IU1.
Supplemental Data
Supplemental Figure 1. Deletion of ANR.
Supplemental Figure 2. Targeted point mutagenesis (anr4PTC).
Supplemental Figure 3. Expression changes of 12 genes encoding ELIPs.
Supplemental Figure 4. Expression changes of 34 genes encoding membrane transporters.
Supplemental Figure 5. Correlation of gene expression between treatments.
Supplemental Figure 6. Clusters of genes strongly upregulated by ABA and stress.
Supplemental Figure 7. Clusters of genes specifically upregulated by mannitol.
Supplemental Figure 8. Clusters of genes strongly downregulated by ABA and stress.
Supplemental Figure 9. Clusters of genes downregulated by mannitol.
Supplemental Figure 10. Expression changes of 39 genes encoding enzymes of phenylpropanoid metabolism.
Supplemental Figure 11. Numbers of genes up- and downregulated in PpanrKO.
Supplemental Figure 12. Phylogeny based on the conserved EDR domain.
Supplemental Figure 13. Phylogeny based on the conserved PAS domain.
Supplemental Figure 14. Asymmetries in the PAS dimer.
Supplemental Figure 15. PAS dimerization interface.
Supplemental Figure 16. PAS monomers and dimers in solution.
Supplemental Figure 17. Targeted point mutagenesis of PAS domain.
Supplemental Figure 18. Targeted deletion of PAS domain.
Supplemental Figure 19. A key structural role for a conserved tryptophan residue.
Supplemental Table 1. Expression data for ABA biosynthetic genes.
Supplemental Table 2. LEA genes represented in Figure 5C.
Supplemental Table 3. Sequences used in phylogenetic analysis.
Supplemental Table 4. Parameters of PAS domain crystal structure.
Supplemental Table 5. PAS homodimer orientation comparisons.
Supplemental Table 6. Primer sequences used.
Supplemental Table 7. RNA-seq library statistics.
Supplemental Data Set 1. Genes upregulated in the wild type.
Supplemental Data Set 2. Genes downregulated in the wild type.
Supplemental Data Set 3. Genes upregulated in anrKO.
Supplemental Data Set 4. Genes downregulated in anrKO.
Supplemental Data Set 5. Sequence alignments of kinase domains used in Figure 6
Supplemental Data Set 6. Sequence alignments of EDR domains used in Supplemental Figure 12.
Supplemental Data Set 7. Sequence alignments of PAS domains used in Supplemental Figure 13.
Supplemental Data Set 8. SNP loci and sequences mapped in chromosome 12.
Supplementary Material
Acknowledgments
S.R.S. was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) doctoral training grant to the University of Leeds Centre for Plant Sciences. A.C.C. and Y.K. gratefully acknowledge the receipt of BBSRC Research Grant BB/F0017971/1 to develop a P. patens forward genetics platform. The work conducted by the U.S. Department of Energy Joint Genome Institute (J.W.J., J.S., J.G., W.M., and G.A.T.) was supported by the Office of Science of the U.S. Department of Energy under Contract DE-AC02-05CH11231. S.A.R., D.L., and R.R. acknowledge funding from the Deutsche Forschungsgemeinschaft in the form of DFG Grant RE 837/10-2, and R.R. was supported by the Excellence Initiative of the German Federal and State Governments. The 1000 Plants (1KP) initiative, led by G.K.-S.W., is funded by the Alberta Ministry of Innovation and Advanced Education, Alberta Innovates Technology Futures (AITF), Innovates Centres of Research Excellence (iCORE), Musea Ventures, BGI-Shenzhen, and China National Genebank (CNGB). We thank Shengqiang Shu (Joint Genome Institute) for providing the cross-referencing between the V3.0, V3.1, and V3.3 P. patens locus IDs.
AUTHOR CONTRIBUTIONS
A.C.C. conceived the research and with Y.K. generated and screened the mutagenized populations, established segregating populations, isolated DNA for genotyping, and undertook RT-PCR analyses of gene expression in wild-type and mutant lines. J.S. led the whole-genome sequencing and assembly, J.W.J. the chromosome assembly and analysis, and J.G. the end sequencing of BAC clones generated by S.A.R., D.L., and R.R. W.M. and G.A.T. undertook the Illumina SNP genotyping as part of the P. patens Flagship V3 genome assembly program, which was led by S.A.R., J.S., and R.R. Resources and informatics tools for the 1KP initiative were contributed by M.M., C.J.R., F.-W.L., and A.L., led by G.K.-S.W. S.R.S. mapped the ANR gene and generated and analyzed the targeted knockout and point mutant lines phenotypically and by Illumina RNA-seq. S.R.S. undertook the phylogenetic analyses and the crystallographic analysis of the PAS domain, under the direction of C.H.T. and T.A.E. S.R.S. and A.C.C. wrote the manuscript.
Glossary
- ABA
abscisic acid
- SNP
single nucleotide polymorphism
Footnotes
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Alpert P. (2005). The limits and frontiers of desiccation-tolerant life. Integr. Comp. Biol. 45: 685–695. [DOI] [PubMed] [Google Scholar]
- Buitink J., Leprince O. (2008). Intracellular glasses and seed survival in the dry state. C. R. Biol. 331: 788–795. [DOI] [PubMed] [Google Scholar]
- Card P.B., Erbel P.J., Gardner K.H. (2005). Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization. J. Mol. Biol. 353: 664–677. [DOI] [PubMed] [Google Scholar]
- Champion A., Picaud A., Henry Y. (2004). Reassessing the MAP3K and MAP4K relationships. Trends Plant Sci. 9: 123–129. [DOI] [PubMed] [Google Scholar]
- Chater C., Kamisugi Y., Movahedi M., Fleming A., Cuming A.C., Gray J.E., Beerling D.J. (2011). Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr. Biol. 21: 1025–1029. [DOI] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B. III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho U.S., Bader M.W., Amaya M.F., Daley M.E., Klevit R.E., Miller S.I., Xu W. (2006). Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J. Mol. Biol. 356: 1193–1206. [DOI] [PubMed] [Google Scholar]
- Christie J.M., Salomon M., Nozue K., Wada M., Briggs W.R. (1999). LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96: 8779–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M.W., Boddington K.F., Warnica J.M., Atkinson J., McKenna S., Madge J., Barker C.H., Graether S.P. (2015). Structural and functional insights into the cryoprotectioin of membranes by the intrinsically disordered dehydrins. J. Biol. Chem. 290: 26900–26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Cuming A.C., Cho S.H., Kamisugi Y., Graham H., Quatrano R.S. (2007). Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens. New Phytol. 176: 275–287. [DOI] [PubMed] [Google Scholar]
- Decker E.L., Frank W., Sarnighausen E., Reski R. (2006). Moss systems biology en route: phytohormones in Physcomitrella development. Plant Biol. (Stuttg.) 8: 397–405. [DOI] [PubMed] [Google Scholar]
- Delgado-Nixon V.M., Gonzalez G., Gilles-Gonzalez M.A. (2000). Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 39: 2685–2691. [DOI] [PubMed] [Google Scholar]
- Delwiche C.F., Cooper E.D. (2015). The evolutionary origin of a terrestrial flora. Curr. Biol. 25: R899–R910. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L.E., Wendel J.F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183: 557–564. [DOI] [PubMed] [Google Scholar]
- Franchi G.G., Piotto B., Nepi M., Baskin C.C., Baskin J.M., Pacini E. (2011). Pollen and seed desiccation tolerance in relation to degree of developmental arrest, dispersal, and survival. J. Exp. Bot. 62: 5267–5281. [DOI] [PubMed] [Google Scholar]
- Frye C.A., Tang D., Innes R.W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98: 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Véscovi E., Soncini F.C., Groisman E.A. (1996). Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84: 165–174. [DOI] [PubMed] [Google Scholar]
- Golby P., Davies S., Kelly D.J., Guest J.R., Andrews S.C. (1999). Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J. Bacteriol. 181: 1238–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W., Hao B., Mansy S.S., Gonzalez G., Gilles-Gonzalez M.A., Chan M.K. (1998). Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. USA 95: 15177–15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung W. (2010). The evolution of abscisic acid (ABA) and ABA function in lower plants, fungi and lichen. Funct. Plant Biol. 37: 806–812. [Google Scholar]
- Henry J.T., Crosson S. (2011). Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 65: 261–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch R., Hartung W., Gimmler H. (1989). Abscisic acid content of algae under stress. Bot. Acta 102: 326–334. [Google Scholar]
- Holzinger A., Kaplan F., Blaas K., Zechmann B., Komsic-Buchmann K., Becker B. (2014). Transcriptomics of desiccation tolerance in the streptophyte green alga Klebsormidium reveal a land plant-like defense reaction. PLoS One 9: e110630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.J., Edery I., Rosbash M. (1993). PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature 364: 259–262. [DOI] [PubMed] [Google Scholar]
- Ichimura K., et al. MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7: 301–308. [DOI] [PubMed] [Google Scholar]
- Ju C., Van de Poel B., Cooper E.D., Thierer J.H., Gibbons T.R., Delwiche C.F. Chang C. (2015). Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants. 1: 14004. [DOI] [PubMed] [Google Scholar]
- Kamisugi Y., Cuming A.C., Cove D.J. (2005). Parameters determining the efficiency of homologous recombination mediated gene targeting in the moss Physcomitrella patens. Nucleic Acids Res. 33: e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi Y., Schaefer D.G., Kozak J., Charlot F., Vrielynck N., Holá M., Angelis K.J., Cuming A.C., Nogué F. (2012). MRE11 and RAD50, but not NBS1, are essential for gene targeting in the moss Physcomitrella patens. Nucleic Acids Res. 40: 3496–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi Y., von Stackelberg M., Lang D., Care M., Reski R., Rensing S.A., Cuming A.C. (2008). A sequence-anchored genetic linkage map for the moss, Physcomitrella patens. Plant J. 56: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P., Crane P.R. (1997). The origin and early evolution of plants on land. Nature 389: 33–39. [Google Scholar]
- Kenrick P., Wellman C.H., Schneider H., Edgecombe G.D. (2012). A timeline for terrestrialization: consequences for the carbon cycle in the Palaeozoic. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 519–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key J., Hefti M., Purcell E.B., Moffat K. (2007). Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: signaling, dimerization, and mechanism. Biochemistry 46: 3614–3623. [DOI] [PubMed] [Google Scholar]
- Khandelwal A., Cho S.H., Marella H., Sakata Y., Perroud P.-F., Pan A., Quatrano R.S. (2010). Role of ABA and ABI3 in desiccation tolerance. Science 327: 546. [DOI] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441. [DOI] [PubMed] [Google Scholar]
- Knight C.D., Cove D.J., Cuming A.C., Quatrano R.S. (2002). Moss gene technology. In Molecular Plant Biology, Vol. 2, P.M. Gilmartin and C. Bowler, eds (Oxford, UK: Oxford University Press), pp. 285–299. [Google Scholar]
- Kobayashi M., Hirai N., Kurimura Y., Ohigashi H., Tsuji Y. (1997). Abscisic acid-dependent algal morphogenesis in the unicellular green alga Haematococcus pluvialis. Plant Growth Regul. 22: 79–85. [Google Scholar]
- Komatsu K., Nishikawa Y., Ohtsuka T., Taji T., Quatrano R.S., Tanaka S., Sakata Y. (2009). Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens. Plant Mol. Biol. 70: 327–340. [DOI] [PubMed] [Google Scholar]
- Komatsu K., Suzuki N., Kuwamura M., Nishikawa Y., Nakatani M., Ohtawa H., Takezawa D., Seki M., Tanaka M., Taji T., Hayashi T., Sakata Y. (2013). Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat. Commun. 4: 2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster K.L., Leopold A.C. (1988). Sugars and desiccation tolerance in seeds. Plant Physiol. 88: 829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss U., Minh B.Q., Losi A., Gärtner W., Eggert T., von Haeseler A., Jaeger K.E. (2009). Distribution and phylogeny of light-oxygen-voltage-blue-light-signaling proteins in the three kingdoms of life. J. Bacteriol. 191: 7234–7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Tomchick D.R., Brautigam C.A., Machius M., Kort R., Hellingwerf K.J., Gardner K.H. (2008). Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry 47: 4051–4064. [DOI] [PubMed] [Google Scholar]
- Lee S.-J., Lee M.H., Kim J.-I., Kim S.Y. (2015). Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response. Plant Cell Physiol. 56: 84–97. [DOI] [PubMed] [Google Scholar]
- Little R., Martinez-Argudo I., Dixon R. (2006). Role of the central region of NifL in conformational switches that regulate nitrogen fixation. Biochem. Soc. Trans. 34: 162–164. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. (2007). Phaser crystallographic software. J. Appl. Cryst. 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A., Moffat K. (2010). Engineered photoreceptors as novel optogenetic tools. Photochem. Photobiol. Sci. 9: 1286–1300. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1970). Enormous diversity in genome sizes of fish as a reflection of nature’s extensive experiments with gene duplication. Trans. Am. Fish. Soc. 99: 120–130. [Google Scholar]
- Oldenhof H., Wolkers W.F., Bowman J.L., Tablin F., Crowe J.H. (2006). Freezing and desiccation tolerance in the moss Physcomitrella patens: an in situ Fourier transform infrared spectroscopic study. Biochim. Biophys. Acta 1760: 1226–1234. [DOI] [PubMed] [Google Scholar]
- Oliver M.J., Velten J., Mishler B.D. (2005). Desiccation tolerance in bryophytes: a reflection of the primitive strategy for plant survival in dehydrating habitats? Integr. Comp. Biol. 45: 788–799. [DOI] [PubMed] [Google Scholar]
- Park S.-Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova A.V., Rausch S., Hundertmark M., Gibon Y., Hincha D.K. (2015). The intrinsically disordered protein LEA7 from Arabidopsis thaliana protects the isolated enzyme lactate dehydrogenase and enzymes in a soluble leaf proteome during freezing and drying. Biochim. Biophys. Acta 1854: (10 Pt A) 1517–1525. [DOI] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69. [DOI] [PubMed] [Google Scholar]
- Sakata Y., Nakamura I., Taji T., Tanaka S., Quatrano R.S. (2010). Regulation of the ABA-responsive Em promoter by ABI3 in the moss Physcomitrella patens: role of the ABA response element and the RY element. Plant Signal. Behav. 5: 1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saruhashi M., et al. (2015). Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signalling upstream of SNF1-related protein kinase 2. Proc. Natl. Acad. Sci. USA 112: E6388–E6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer D., Zrÿd J.-P., Knight C.D., Cove D.J. (1991). Stable transformation of the moss Physcomitrella patens. Mol. Gen. Genet. 226: 418–424. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Kanamori Y., Furuki T., Kikawada T., Okuda T., Takahashi T., Mihara H., Sakurai M. (2010). Desiccation-induced structuralization and glass formation of group 3 late embryogenesis abundant protein model peptides. Biochemistry 49: 1093–1104. [DOI] [PubMed] [Google Scholar]
- Shitamichi N., Matsuoka D., Sasayama D., Furuya T., Nanmori T. (2013). Over-expression of MAP3Kδ4, an ABA-inducible Raf-like MAP3K that confers salt tolerance in Arabidopsis. Plant Biotechnol. 30: 111–118. [Google Scholar]
- Szövényi P., Perroud P.-F., Symeonidi A., Stevenson S., Quatrano R.S., Rensing S.A., Cuming A.C., McDaniel S.F. (2015). De novo assembly and comparative analysis of the Ceratodon purpureus transcriptome. Mol. Ecol. Resour. 15: 203–215. [DOI] [PubMed] [Google Scholar]
- Takezawa D., Watanabe N., Ghosh T.K., Saruhashi M., Suzuki A., Ishiyama K., Somemiya S., Kobayashi M., Sakata Y. (2015). Epoxycarotenoid-mediated synthesis of abscisic acid in Physcomitrella patens implicating conserved mechanisms for acclimation to hyperosmosis in embryophytes. New Phytol. 206: 209–219. [DOI] [PubMed] [Google Scholar]
- Tunnacliffe A., Hincha D.K., Leprince O., Macherel D. (2010). LEA proteins: versatility of form and function. In Dormancy and Resistance in Harsh Environments, Lubzens E., Cerda A., Clark M.S., eds (Berlin: Springer; ), pp. 91–108. [Google Scholar]
- Tunnacliffe A., Wise M.J. (2007). The continuing conundrum of the LEA proteins. Naturwissenschaften 94: 791–812. [DOI] [PubMed] [Google Scholar]
- Véscovi E.G., Ayala Y.M., Di Cera E., Groisman E.A. (1997). Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+. J. Biol. Chem. 272: 1440–1443. [DOI] [PubMed] [Google Scholar]
- Wickett N.J., et al. (2014). Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 111: E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G. (2010). XIA2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43: 186–190. [Google Scholar]
- Yasumura Y., Pierik R., Fricker M.D., Voesenek L.A., Harberd N.P. (2012). Studies of Physcomitrella patens reveal that ethylene-mediated submergence responses arose relatively early in land-plant evolution. Plant J. 72: 947–959. [DOI] [PubMed] [Google Scholar]
- Yasumura Y., Pierik R., Kelly S., Sakuta M., Voesenek L.A., Harberd N.P. (2015). An ancestral role for Constitutive Triple Response 1 (CTR1) proteins in both ethylene and abscisic acid signaling. Plant Physiol. 169: 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer A.D., Lang D., Buchta K., Rombauts S., Nishiyama T., Hasebe M., Van de Peer Y., Rensing S.A., Reski R. (2013). Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genomics 14: 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18: 292–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.