Scanning electron microscopy and three-dimensional immunolocalization reveal that leaves of C4 monocots have more plasmodesmata per pitfield area and greater pitfield area coverage than C3 monocots.
Abstract
C4 photosynthesis is characterized by a CO2-concentrating mechanism between mesophyll (M) and bundle sheath (BS) cells of leaves. This generates high metabolic fluxes between these cells, through interconnecting plasmodesmata (PD). Quantification of these symplastic fluxes for modeling studies requires accurate quantification of PD, which has proven difficult using transmission electron microscopy. Our new quantitative technique combines scanning electron microscopy and 3D immunolocalization in intact leaf tissues to compare PD density on cell interfaces in leaves of C3 (rice [Oryza sativa] and wheat [Triticum aestivum]) and C4 (maize [Zea mays] and Setaria viridis) monocot species. Scanning electron microscopy quantification of PD density revealed that C4 species had approximately twice the number of PD per pitfield area compared with their C3 counterparts. 3D immunolocalization of callose at pitfields using confocal microscopy showed that pitfield area per M-BS interface area was 5 times greater in C4 species. Thus, the two C4 species had up to nine times more PD per M-BS interface area (S. viridis, 9.3 PD µm−2; maize, 7.5 PD µm−2; rice 1.0 PD µm−2; wheat, 2.6 PD µm−2). Using these anatomical data and measured photosynthetic rates in these C4 species, we have now calculated symplastic C4 acid flux per PD across the M-BS interface. These quantitative data are essential for modeling studies and gene discovery strategies needed to introduce aspects of C4 photosynthesis to C3 crops.
INTRODUCTION
The superior photosynthetic performance of C4 crop plants is largely due to the biochemical and anatomical specialization that results in concentration of CO2 at the active site of Rubisco, reducing photorespiration and permitting Rubisco to operate close to its catalytic optimum. Separation of the biochemical CO2 pump in the mesophyll (M; where atmospheric CO2 is initially fixed into C4 acids by PEP carboxylase) from the site of decarboxylation and refixation by Rubisco in the bundle sheath (BS) means that C4 acids must move into the BS and C3 products return to the M at rates greater than the net rate of photosynthesis (Hatch and Osmond, 1976; von Caemmerer and Furbank, 2003). Due to the high degree of secondary thickening and often suberization of the BS walls, metabolite movement is limited to the symplasm and abundant plasmodesmata (PD) at this cell interface have been demonstrated (Hatch, 1987).
For accurate modeling of C4 photosynthetic flux, it is essential to quantify the number of PD between the M and BS, which facilitate the bidirectional movement of assimilates. However, due to their minute size (30 to 50 nm), individual PD can only be seen under the electron microscope (Ding et al., 1992; Robards, 1976). Clusters of PD, called pitfields, are at the limit of detectability for light microscopy (Carr, 1976; Robards, 1976). Transmission electron microscopy (TEM) has been routinely used to study details of PD structure (Robards, 1976; Evert et al., 1977; Ding et al., 1992; Overall and Blackman, 1996), but extracting quantitative data requires careful serial sectioning and reconstruction. There have been very few reports quantifying PD density at the M-BS cell interface in C4 (or C3) plants (Olesen, 1975). This has severely limited our ability to model C4 photosynthetic flux and strategies for determining the genetics and evolution of the anatomical specialization for C4 metabolite flux.
In some cases, estimates of PD frequency via TEM in other plants have employed proportionality constant, originally derived by Gunning (1978). This value is 1/(t + 1.5R), where t is the section thickness and R is the average radius of PD. However, the use of the proportionality constant for quantification is limited to randomly distributed, nonclustered PD such as those found in cell plates of Azolla roots, from which the constant was derived (Gunning, 1978). However, in most monocot leaves, PD are clustered in pitfields (Evert et al., 1977; Faulkner et al., 2008; Robinson-Beers and Evert, 1991a, 1991b), making the use of this proportionality constant for PD quantification invalid (Gunning, 1978).
Another common approach to quantify PD density is the plasmodesmogram, in which the total number of PD detected along a given length of cell wall interface is expressed as a PD frequency (Botha and Evert, 1988; Botha, 1992; Botha and van Bel, 1992). The main drawback of this technique is that TEM sections provide only a thin (200 nm at most) 2D slice of a complex 3D cell wall interface, so the number of PD detected is dependent on the angle at which the pitfield was cut. Furthermore, the 2D TEM images capture only a fraction of the total PD within a pitfield and do not allow for the mostly noncircular shape of the pitfields.
In this study, we report a new method that combines scanning electron microscopy and 3D immunolocalization by confocal microscopy to better quantify PD connections in the leaf. Scanning electron microscopy has been used in PD-related studies but not as routinely as TEM due to its inability to capture PD ultrastructural details. With recent advances in high-resolution scanning electron microscopy, capturing the 3D morphology of PD in cell walls of algae, ferns, and vascular plants is now possible (Brecknock et al., 2011; Barton and Overall, 2015). The advantage of scanning electron microscopy to elucidate PD and pitfield distribution on cell surfaces (Botha and Evert, 1988; Faulkner et al., 2008; Sage and Sage, 2009) is that the whole pitfield and all the individual PD within it can be seen in a single image. PD frequency per pitfield area can then be obtained simply by measuring the pitfield area and counting individual PD. In fact, one of the earliest studies that made use of scanning electron microscopy in tandem with TEM looked into the PD frequency at the M-BS cell interface in C4 species (Olesen, 1975), but these measurements covered only a small portion of the leaf. Scanning electron microscopy alone is impractical for PD quantification because only a small portion of the entire cell interface can be captured at one time.
Confocal microscopy can be used to visualize pitfields using dyes or fluorescent probes known to label proteins or molecules colocalizing with PD (Faulkner et al., 2008). A common method for detecting PD is to use an antibody against callose, a β-1,3-glucan, which is known to be deposited around the PD neck (Turner et al., 1994). However, in intact tissues, the absorption and scattering of light by cell walls and cell contents limits detection of PD to the outer layers such as the leaf epidermis and trichomes (Faulkner et al., 2008). Recent success in 3D confocal imaging of intact plant tissues using a clearing technique (PEA-CLARITY; Palmer et al., 2015) enables the quantification of pitfield distribution and abundance within whole, cleared tissue over large areas of cell interfaces by callose immunofluorescence. By combining details of PD frequency per pitfield by scanning electron microscopy imaging, with the density of pitfields per cell interface derived from 3D confocal imaging, we can more accurately calculate the PD density per cell interface.
Using this improved quantitative method, we report here that C4 leaves have up to 9-fold higher PD density at the interface between M and BS cells than C3 leaves. This is due to both an increase in number of PD per pitfield area and an increase in pitfield density at this interface. Quantification of PD at this interface will enable both modeling of C4 metabolic flux and the design of experiments to determine the genetic regulation and evolution of the symplastic transport mechanisms of C4 plants.
RESULTS
Leaf Anatomy of Two C3 and Two C4 Crop Species
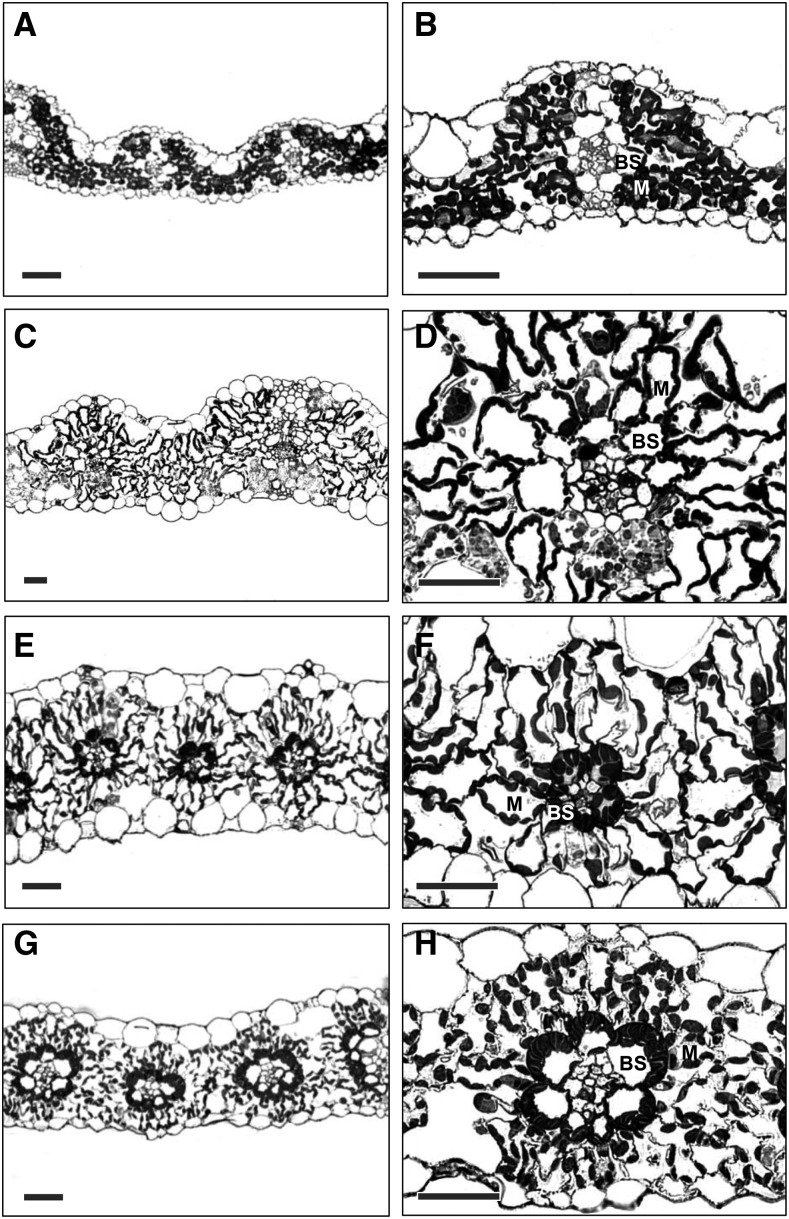
Transverse sections of leaves of the two C3 species (rice [Oryza sativa] and wheat [Triticum aestivum]) showed that, as is typical for C3 plants, the chloroplasts were abundant in M cells with very few or no chloroplasts visible in BS cells (Figures 1A to 1D). In the C4 photosynthetic leaves, such as Setaria viridis and maize (Zea mays), both M cells and BS cells have abundant chloroplasts (Figures 1E to 1H). In both C3 and C4 species studied, chloroplasts in the M cells were distributed along the cell periphery. Moreover, in the C4 species studied, the BS cell chloroplasts were positioned centrifugally toward the M cells, characteristic of the NADP-malic enzyme decarboxylation type found in grasses (Hatch, 1987). In the transverse sections shown, the shape of BS cells in both the C3 and C4 species was cylindrical with a smooth surface. M cell shape, on the other hand, varied from lobed in rice (Figure 1B) to elongated in S. viridis (Figure 1F).
Figure 1.
Light Micrographs of Transverse Sections of Leaves of the Two C3 Species and Two C4 Species Examined.
(A) and (B) Rice, C3.
(C) and (D) Wheat, C3.
(E) and (F) S. viridis, C4.
(G) and (H) Maize, C4.
Bars = 50 µm.
Plasmodesmata of C3 and C4 Species Viewed under Transmission Electron Microscope
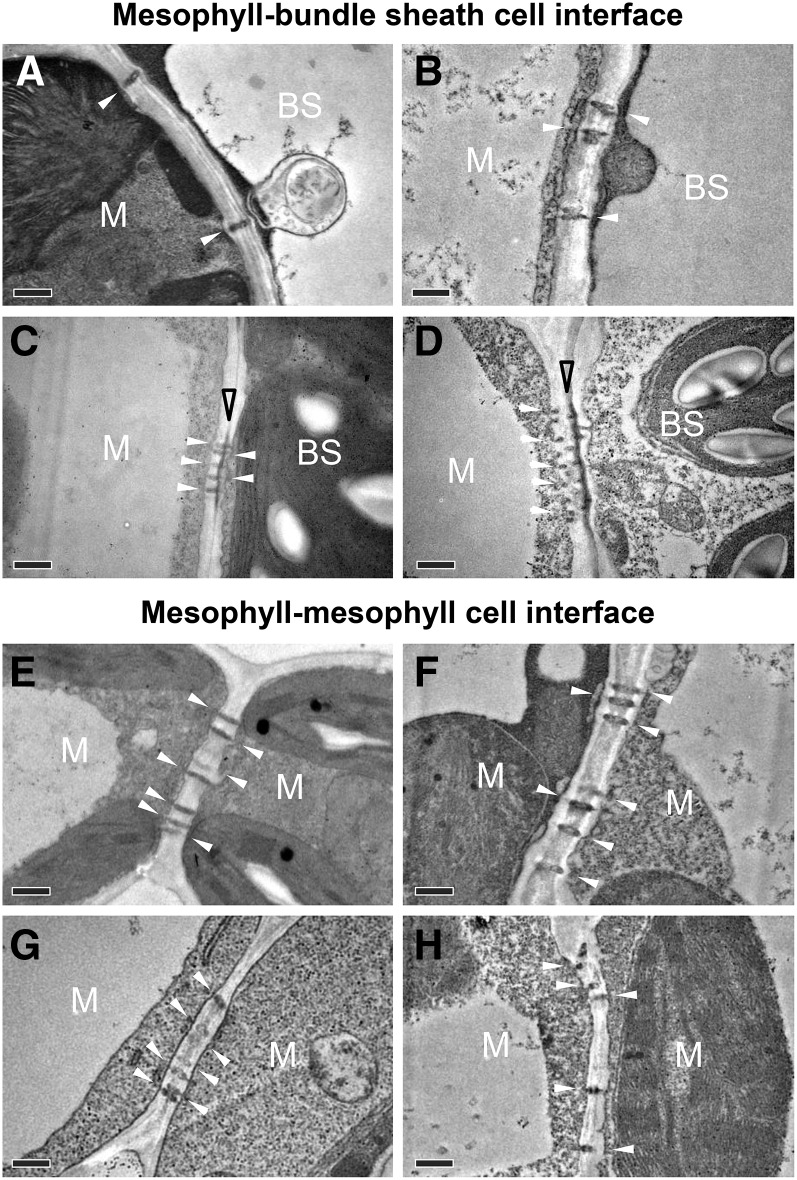
PD at cell interfaces were readily identified in TEM ultrathin sections (Figure 2), but only a small portion of the total PD within a pitfield could be captured this way. A thicker suberin lamella in the area where PD lie between the M cell and BS cell was observed only in the C4 species (S. viridis and maize; Figures 2C and 2D) but not in the C3 species (rice and wheat; Figures 2A and 2B). No suberized layer was found between M cells in either C3 or C4 species (Figures 2E to 2H). The areas of individual PD were similar in the two C4 species, S. viridis (0.007 ± 0.0002 µm2) and maize (0.007 ± 0.0002 µm2) while in C3 species, a larger PD area was observed in wheat (0.008 ± 0.0002 µm2) than rice (0.006 ± 0.0001 µm2).
Figure 2.
Transmission Electron Micrographs of Plasmodesmata at Cell Interfaces in Leaves of C3 and C4 Species.
(A) and (E) Rice, C3.
(B) and (F) Wheat, C3.
(C) and (G) S. viridis, C4.
(D) and (H) Maize, C4.
White and open black arrowheads indicate plasmodesmata and suberin lamella, respectively. Bars = 0.5 µm.
Quantification of Pitfield Area and Plasmodesmata Frequency per Pitfield Area
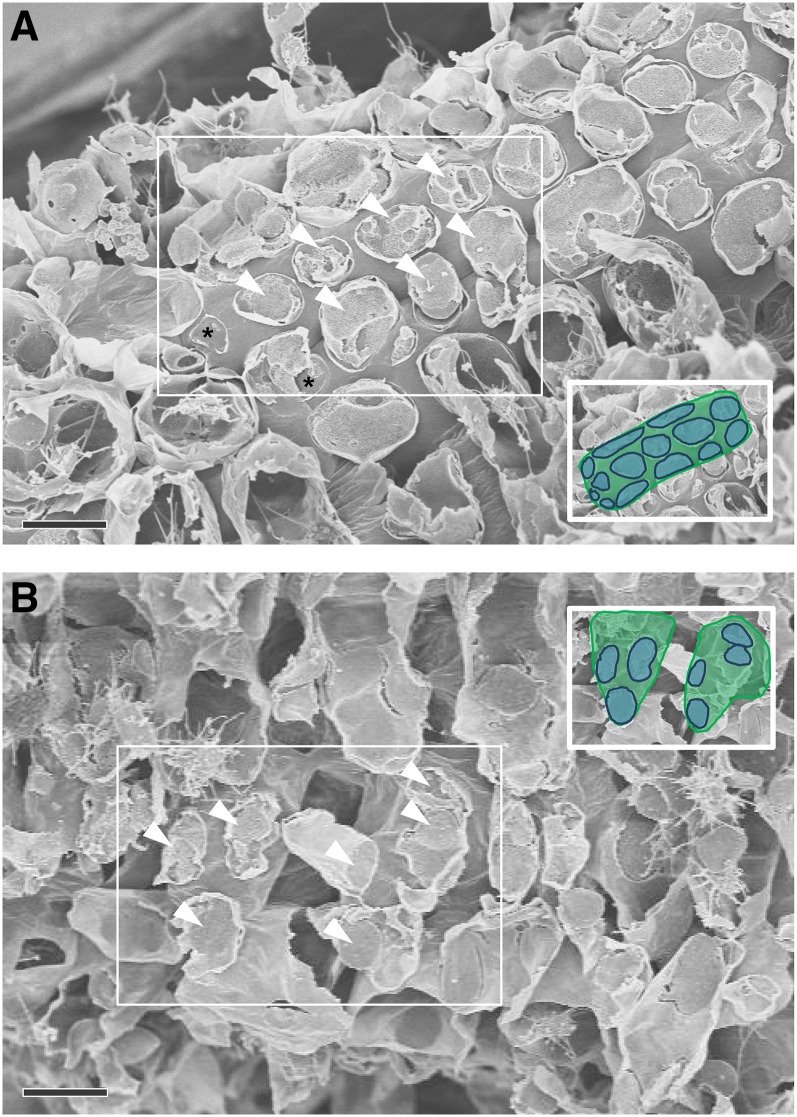
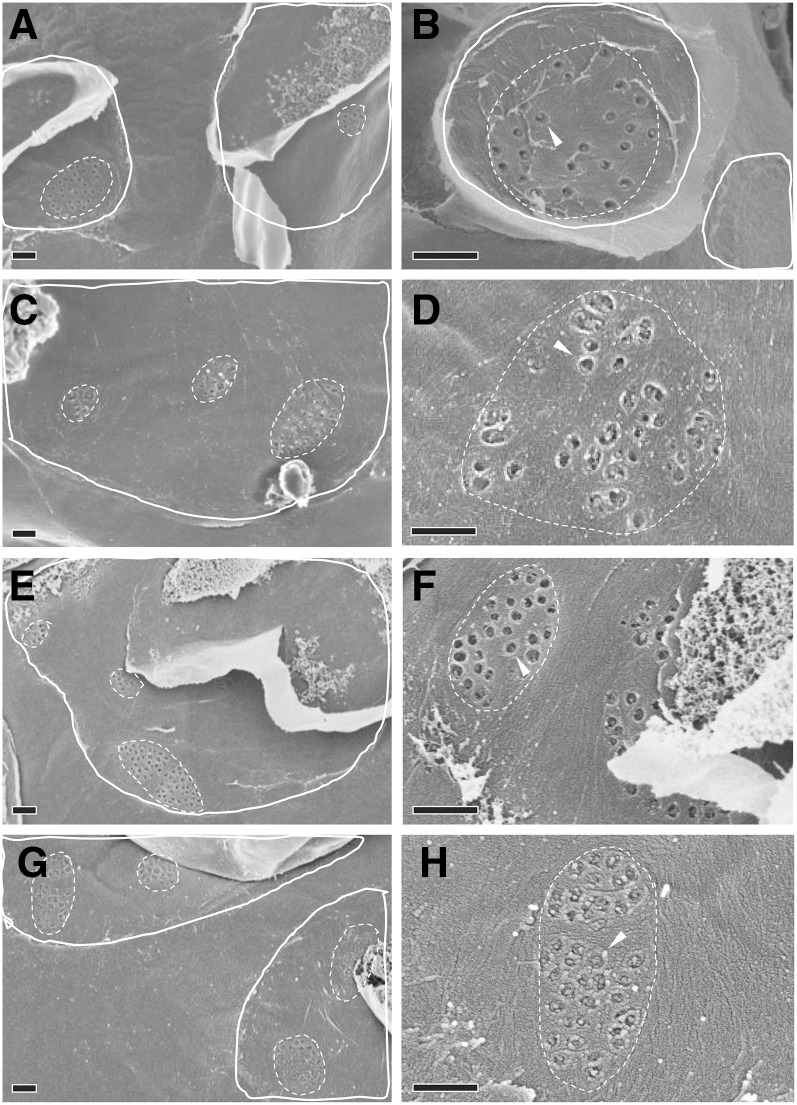
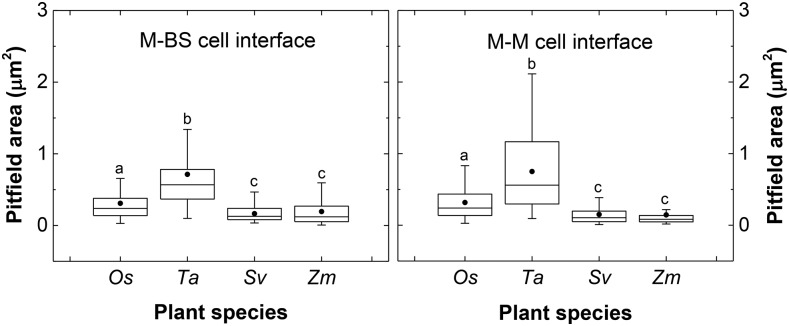
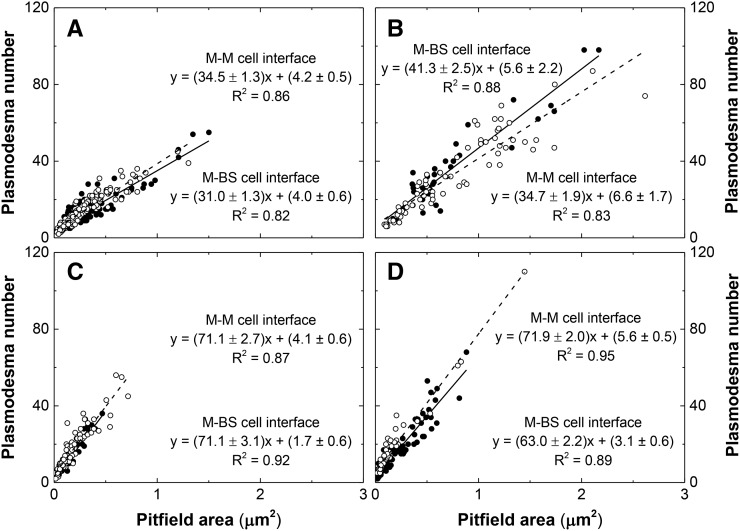
Regions of contact between M cells and BS cells that could be visualized with scanning electron microscopy (Figure 3) were exposed by tearing critical point dried leaf tissue parallel to the veins. On closer inspection, clusters of pitfields were seen (Figures 4A, 4C, 4E, and 4G), and within each pitfield, all PD could be counted (Figures 4B, 4D, 4F, and 4H). PD in C3 species were generally spaced farther apart than PD in C4 species (Figure 4). C3 species also had larger pitfield areas on both the M-BS and M-M cell interfaces compared with the C4 species examined (Figure 5). Because there was considerable variation in pitfield area, we plotted the number of PD as a function of pitfield area and found strong linear correlations for all four species (Figure 6). Plotting values obtained from M-BS cell interface with values from M-M cell interface for the same species resulted in statistically different regression lines in rice, wheat, and maize but not in S. viridis (Figure 6). Calculated PD per µm2 pitfield based on the linear regressions obtained revealed that the C4 species, S. viridis and maize, had almost double the PD per µm2 pitfield in the M-BS cell interface compared with C3 species, rice and wheat (Figure 6, Table 1). Similarly, a higher PD per µm2 pitfield in the M-M cell interface was found in C4 species, S. viridis and maize, compared with the C3 species, rice and wheat (Figure 6, Table 1).
Figure 3.
Field Emission Scanning Electron Micrograph of Cell Interfaces in S. viridis Leaf.
(A) M-BS cell interface. The torn patches (arrowheads) are mesophyll cell remnants on the surface of underlying cylindrical bundle sheath cells.
(B) M-M cell interface. The torn patches (arrowheads) are mesophyll cell remnants on the sides of highly lobed mesophyll cells.
In each case, the torn patches (arrowheads) on the cell surfaces are remnants of attachment sites with neighboring mesophyll cells. Most patches are covered in mesophyll cell debris, and only sites lacking cell debris (asterisk) were used for plasmodesmata quantification. Inset is the boxed area with underlying bundle sheath (A) or mesophyll (B) cell area in green and attachment sites in blue. Bars = 10 µm.
Figure 4.
Field Emission Scanning Electron Micrographs of Plasmodesmata at the M-BS Cell Interface of C3 and C4 Species.
(A) and (B) Rice, C3.
(C) and (D) Wheat, C3.
(E) and (F) S. viridis, C4.
(G) and (H) Maize, C4.
(A), (C), (E), and (G) Example enlargements from M-BS attachment sites (solid white outlines) free of overlying mesophyll cell debris, containing pitfields (dotted white outlines). The underlying bundle sheath cell surface outside the attachment sites would normally face an intercellular space within the leaf. The clear areas within the attachment sites indicate M-BS cell wall connections that lack plasmodesmata.
(B), (D), (F), and (H) Individual pitfields (dotted outlines) within M-BS attachment sites, showing pitfield plasmodesmata (arrowheads). Bars = 0.5 µm.
Figure 5.
Pitfield Area at Cell Interfaces of Leaves of C3 and C4 Species Derived from Field Emission Scanning Electron Micrograph Measurements.
The box and whisker represent the 25 to 75 percentile and minimum-maximum distributions of the data, respectively. Closed circles give the overall means. Statistical differences according to MATLAB two-sample t test at P value < 0.05 are indicated by letters. Values indicated by the same letter are not statistically different. Os, Oryza sativa; Ta, Triticum aestivum; Sv, Setaria viridis; Zm, Zea mays.
Figure 6.
Plasmodesmata Frequency per Pitfield Area at Cell Interfaces in Leaves of C3 and C4 Species.
(A) Rice, C3.
(B) Wheat, C3.
(C) S. viridis, C4.
(D) Maize, C4.
Closed circles correspond to the values obtained from the M-BS cell interface. Open circles correspond to the values obtained from the M-M cell interface. Solid line and dashed line correspond to the regression lines generated using the plotted values from M-BS cell interface and M-M cell interface, respectively. Analysis using SPSS Statistics software with P value < 0.05 revealed M-BS cell interface and M-M cell interface regression lines to be statistically different in (A), (B), and (D) but not in (C). The plasmodesmata frequency per pitfield area was calculated using the linear equation, y = mx + b, where y is the plasmodesmata frequency, m is the slope, x is the pitfield area, and b is the intercept. R2 is coefficient of determination, where a value of 1 indicates that the regression line perfectly fits the data while a value of 0 indicates that the line does not fit the data at all.
Table 1. Density of Plasmodesmata on Cell Interfaces of C3 and C4 Species.
| Species | Mean Cell Interface Area (µm2) | Mean Pitfield Area per Cell Interface (µm2) | Mean Pitfield Area per Cell Interface Area (%) | PD per µm2 Pitfield | PD per µm2 Cell Interface | PD Area per Cell Interface Area (%) |
|---|---|---|---|---|---|---|
| M-BS cell interface | ||||||
| Rice, C3 | 609 ± 25a | 17 ± 1a | 2.8 ± 0.1a | 35.0 ± 0.4a | 1.0 ± 0.1a | 0.6 ± 0.02a |
| Wheat, C3 | 2704 ± 150b | 149 ± 28b | 5.5 ± 0.3b | 46.9 ± 1.0b | 2.6 ± 0.1b | 2.0 ± 0.03b |
| S. viridis, C4 | 486 ± 14c | 61 ± 5c | 12.7 ± 0.3c | 72.8 ± 0.9c | 9.3 ± 0.2c | 6.2 ± 0.07c |
| Maize, C4 | 1203 ± 32d | 137 ± 11b | 11.4 ± 0.3d | 66.1 ± 0.7d | 7.5 ± 0.2d | 5.4 ± 0.06d |
| M-M cell interface | ||||||
| Rice, C3 | 249 ± 9e | 10 ± 2d | 4.1 ± 0.1e | 38.7 ± 0.4e | 1.6 ± 0.1e | 1.0 ± 0.02e |
| Wheat, C3 | 2246 ± 183b | 83 ± 16c | 3.7 ± 0.2e | 41.3 ± 0.7f | 1.5 ± 0.1e | 1.2 ± 0.02f |
| S. viridis, C4 | 375 ± 7f | 32 ± 2e | 8.5 ± 0.1f | 75.2 ± 0.8c | 6.4 ± 0.1f | 4.2 ± 0.05g |
| Maize, C4 | 810 ± 17g | 117 ± 6b | 14.4 ± 0.2g | 77.5 ± 0.6g | 11.2 ± 0.2g | 8.0 ± 0.07h |
Numbers in the “PD per μm2 Pitfield” column are the sum of slopes and intercepts shown in Figure 6. Statistical differences according to MATLAB two-sample t test or SPSS Statistics software at P value < 0.05 are indicated by letters. Values indicated by the same letter within a column are not statistically different.
Using 3D Immunolocalization to Quantify Pitfield Area per Cell Interface Area
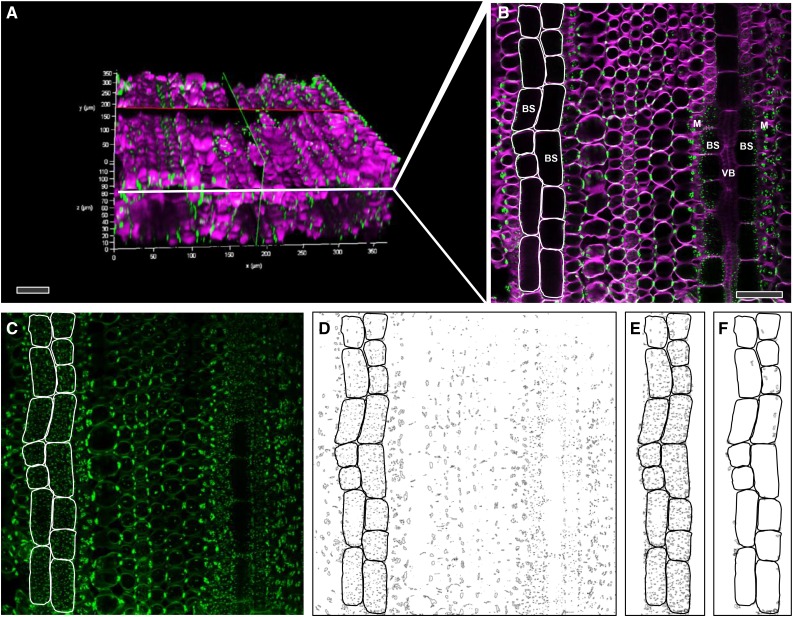
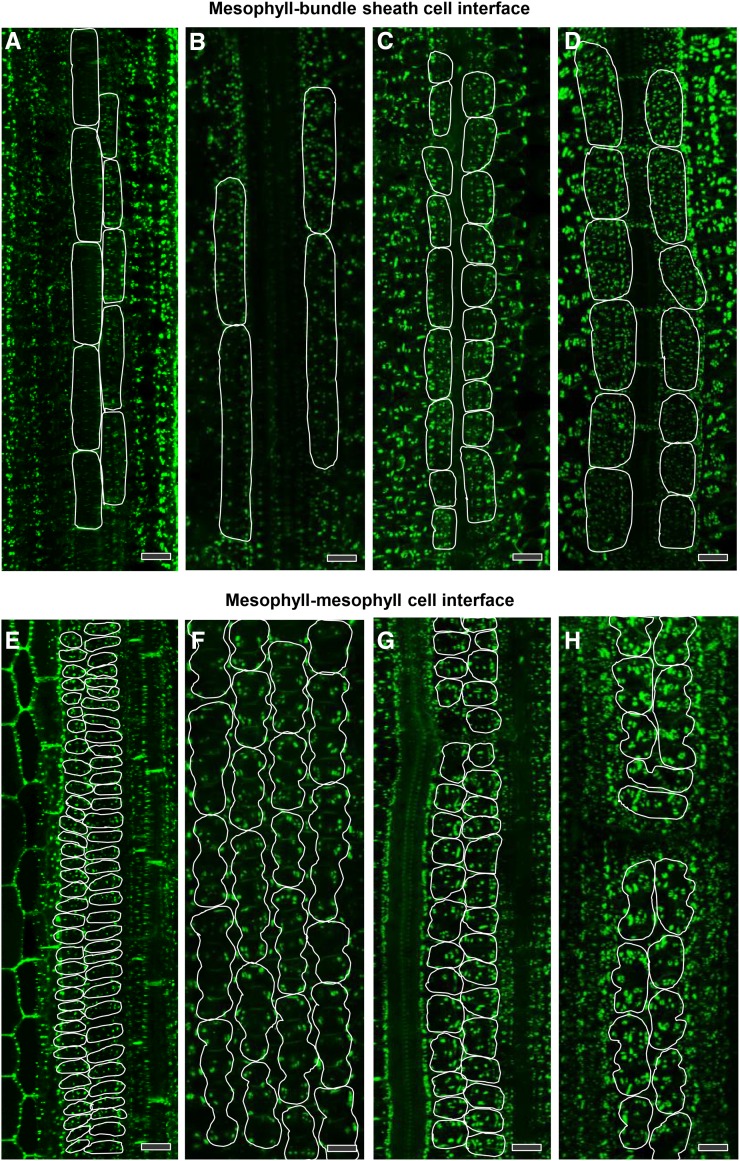
3D immunolocalization of pitfields allowed quantification of total pitfield area on cell interfaces over large surface areas in leaves (Figure 7). The area corresponding to cell/cell interfaces was derived from the 3D image by selecting a subset of images from the complete z-stack that included all focal planes of the cell file of interest. Maximum intensity projections generated a single in-focus image that captured all the pitfield signals within that cell interface. In cases where there was an overlap of pitfield signals from different cell interfaces, it was possible to eliminate extraneous signals by filtering based on the size and shape of typical pitfields. The pitfields on the M-BS cell interface (parallel to the surface) were smaller than those on the M-M cell interface (perpendicular to the surface) mainly because of the pitfield orientation (Figure 7). The pitfields on the M-BS cell interface were also more evenly distributed than those on the M-M cell interface, which appeared to be clustered (Figure 8). Quantification using ImageJ software revealed that the two C4 species had more pitfields per cell interface area than the two C3 species (Figure 8, Table 1). On the M-BS cell interface, S. viridis and maize had 12.7% ± 0.3% and 11.4% ± 0.3% pitfield area coverage, respectively, while rice had 2.8% ± 0.1% and wheat had 5.5% ± 0.3% (Table 1). Similarly, on the M-M cell interface, the two C4 species had higher pitfield area coverage than the two C3 species. S. viridis was recorded to have 8.5% ± 0.1% pitfield area per cell interface area while maize had 14.4% ± 0.2%. Only 4.8% ± 0.1% and 3.7% ± 0.2% were found for rice and wheat, respectively (Table 1).
Figure 7.
Determination of Total Pitfield Area per M-BS Cell Interface of Maize Leaf Using 3D Immunolocalization Confocal Micrographs.
(A) 3D reconstructed image (from 154 single focal planes) of maize leaf hybridized with primary antibody to β-1,3-glucan and secondary antibody tagged with Alexa Fluor 488 (green) and stained with calcofluor white (magenta) to show cell walls.
(B) A single optical section derived from (A) showing pitfields (green) between cells. The BS cells for which pitfields were quantified in this example are outlined in white.
(C) Maximum intensity projection image of 19 consecutive single focal planes (a subset of [A]) comprising that region of BS cell surface in contact with the neighboring M cells, outlined in white. Note the pitfield signal size difference between M-BS cell interface, with pitfields parallel to the image plane, and M-M cell interface, with pitfields perpendicular to the image plane.
(D) Corresponding binary image of (C) after processing (Supplemental Figure 2).
(E) Selected area of interest from (D) used for pitfield quantification.
(F) Area from (E) corresponding to M-M cell interface pitfields detected at the edges of certain focal planes. This area is subtracted from the total pitfield area obtained in (E) to obtain the final area of M-BS pitfields.
VB, vascular bundle. Bars = 50 µm.
Figure 8.
Pitfield Distribution at Cell Interfaces in Leaves of C3 and C4 Species after Immunofluorescence Detection of β-1,3-Glucan.
(A) and (E) Rice, C3
(B) and (F) Wheat, C3.
(C) and (G) S. viridis, C4.
(D) and (H) Maize, C4.
Pitfields are in green (Alexa Fluor 488 fluorescence). In (A) to (D), bundle sheath cell surface areas in direct contact with mesophyll cells are outlined in white. In (E) to (H), mesophyll cell surface areas in direct contact with other mesophyll cells are outlined in white. Bars = 20 µm.
Calculation of Plasmodesmata Density on Cell Interface Areas
By combining the values for PD per µm2 pitfield with pitfield area per cell interface area, we could calculate PD density per cell interface area (Table 1). Between M and BS cells, the C4 species had up to 9 times more PD per cell interface area (S. viridis had 9.3 ± 0.2 PD µm−2; maize had 7.5 ± 0.2 PD µm−2) than the C3 species (rice had 1.0 ± 0.1 PD µm−2; wheat had 2.6 ± 0.1 PD µm−2) (Table 1). Similarly, the PD density per cell interface area difference between M cells was larger in the C4 species than in the C3 species (Table 1). The C3 species, rice and wheat, had 1.6 ± 0.1 PD µm−2 and 1.5 ± 0.1 PD µm−2, respectively, whereas of the two C4 species, S. viridis had 6.4 ± 0.1 PD µm−2 and maize had the highest at 11.2 ± 0.2 PD µm−2.
Plasmodesmatal Flux Calculation
Measuring CO2 assimilation rates in leaves of the C3 and C4 monocot species examined allowed estimates of PD flux between M cells and BS cells to be calculated. In C4 species, the flux rate of C4 acids into the bundle sheath has to equal or slightly exceed the CO2 assimilation rate. In C3 species, the estimates give the fluxes per PD needed if a C4 photosynthetic pathway were installed without changes in PD frequency (Table 2). The CO2 assimilation rate per leaf area was found to be lowest in rice (27.1 ± 0.96 µmol CO2 m−2 s−1), but this value is not statistically different from that of S. viridis (29.5 ± 1.50 µmol CO2 m−2 s−1). Maize had the highest CO2 assimilation rate per leaf area at 38.6 ± 1.14 µmol CO2 m−2 s−1, but this is not statistically different from wheat at 35.0 ± 1.48 µmol CO2 m−2 s−1 (Table 2). However, C4 species were found to have twice as much BS surface area per unit leaf area (Sb) compared with C3 species (Table 2). Consequently, CO2 assimilation rate (a surrogate for C4 and C3 metabolite flux) per BS surface area was greater in C3 species (rice, 24.7 ± 0.87 µmol CO2 m−2 s−1; wheat, 29.8 ± 1.26 µmol CO2 m−2 s−1) than in C4 species (S. viridis, 18.1 ± 0.92 µmol CO2 m−2 s−1; maize, 19.5 ± 0.58 µmol CO2 m−2 s−1) (Table 2). If a C4 photosynthetic pathway were to be installed in a C3 species, which has up to 9 times lower PD density (Table 1), based on the CO2 assimilation rate per PD (expressed as ×10−18 mol CO2 s−1) in the C3 species (rice, 24.7 ± 0.87; wheat, 11.5 ± 0.49) plasmodesmatal flux per PD would need to be up to 12 times greater than in the C4 species (S. viridis, 1.9 ± 0.10; maize, 2.6 ± 0.08). Assuming that sucrose passes from M to BS cells symplastically in C3 leaves, the sucrose flux across the M-BS interface of the C3 species per PD would be similar to that calculated for C4 metabolite flux through PD in the C4 species (Table 2).
Table 2. Estimates of Plasmodesmal Flux between Mesophyll and Bundle Sheath Cells in Leaves of C3 and C4 Species.
| Species | CO2 Assimilation Rate per Leaf Area (µmol CO2 m−2 s−1) | Mean Sb (m2 m−2) | CO2 Assimilation Rate per Bundle Sheath Surface Area (µmol CO2 m−2 s−1) | CO2 Assimilation Rate per PD (×10−18 mol CO2 s−1) | Sucrose Flux per PD (×10−18 mol Sucrose s−1) |
|---|---|---|---|---|---|
| Rice, C3 | 27.1 ± 0.96a | 1.1 ± 0.06a | 24.7 ± 0.87a | 24.7 ± 0.87a | 2.1 ± 0.07a |
| Wheat, C3 | 35.0 ± 1.48b | 1.2 ± 0.17a | 29.8 ± 1.26b | 11.5 ± 0.49b | 1.0 ± 0.04b |
| S. viridis, C4 | 29.5 ± 1.50a | 1.6 ± 0.04b | 18.1 ± 0.92c | 1.9 ± 0.10c | NA |
| Maize, C4 | 38.6 ± 1.14b | 2.0 ± 0.08c | 19.5 ± 0.58c | 2.6 ± 0.08d | NA |
For the “CO2 Assimilation Rate per PD” column, assuming that in C4 species the minimum flux of C4 acids through the PD needs to be equal to or greater than the CO2 assimilation rate. For C3 species, the numbers indicate the fluxes required if a C4 photosynthetic pathway were installed without anatomical changes. For the “Sucrose Flux per PD” column, assuming sucrose must pass from mesophyll to bundle sheath cells for phloem loading in C3 monocot leaves at a rate in equilibrium with CO2 assimilation rates. Statistical differences according to MATLAB two-sample t test at P value < 0.05 are indicated by letters. Values indicated by the same letter within a column are not statistically different. NA, not applicable.
DISCUSSION
We developed a new method to quantify the opportunities for cell-to-cell transport of key photosynthates between M and BS in both C3 and C4 species. The combination of scanning electron microscopy and 3D immunolocalization confocal microscopy allowed us to quantify both the PD distribution and pitfield distribution on these cell interfaces, and to calculate PD density.
Plasmodesmata Frequency in Pitfields and Pitfield Distribution on Cell Interfaces Both Contribute to Higher Plasmodesmata Density in C4 Species
Scanning electron microscopy measurements revealed that PD were at higher frequency in pitfields in the C4 species, S. viridis and maize, compared with the C3 species, rice and wheat (Figure 4). Conversely, the two C3 species had larger pitfields than C4 species (Figure 5). Interestingly, this pattern of PD frequency was not specific to the M-BS interface but was also seen in the M-M cell interfaces, suggesting that this may be a more general phenomenon throughout the leaf (Figure 6). This may be of particular significance in regard to the evolution of the C4 photosynthetic mechanism if the genetic basis of the high symplasmic connectivity of the M-BS is also reflected in other cells in the leaf.
3D immunolocalization provided quantitative information on how the pitfields are distributed on cell interfaces. This method served as the key technique allowing us to quantify PD density over much larger cell surface areas than reported previously (Olesen, 1975). It also provided a new and potentially improved method to measure BS and M cell size, an important consideration in quantifying pitfield distribution on a cell interface area basis and important parameters for modeling C4 photosynthesis (von Caemmerer, 2000; von Caemmerer and Furbank, 2003; Wang et al., 2014).
Images collected showed an even distribution of pitfields on M-BS cell interfaces, while pitfields found on M-M cell interfaces appeared clustered (Figure 8). BS cells are close to cylindrical in shape and arranged in cell files, enclosing the vascular tissue. This allowed visualization of pitfields on the outer surface of BS cells, parallel to the field of view. By contrast, M cells are irregularly shaped and in most cases arranged less regularly with air spaces in between. With one M cell having connections with other M cells in different directions, pitfields captured in the images acquired here are usually at a particular angle relative to the field of view, which would also contribute to the clustered appearance.
Measurements using 3D immunolocalization images revealed that not only did C4 species have higher PD frequency within pitfields but they also had more abundant pitfields on cell interfaces (Figure 8, Table 1). These two factors combined resulted in C4 species having higher PD density per cell interface area compared with C3 species, consistent with the findings of Botha (1992). More importantly, the established PD density values for M-BS and M-M cell interfaces are species specific and therefore can be used for transport-related studies as well as in modeling.
Comparison with Traditional Methods for Quantification of Plasmodesmata Frequency
Prior to the development of the method described here, TEM was routinely used to provide semiquantitative data on PD distribution and density. Calculations using the Gunning constant (Gunning, 1978) and the plasmodesmogram method (Botha and Evert, 1988; Botha, 1992; Botha and van Bel, 1992) are common in the literature. However, it appears that this method has limited use in calculations of PD density in vascular plants, given the requirement for a random distribution of PD (Gunning, 1978). In most vascular plants, PD are clustered in pitfields, as found in our study, which makes estimates of PD density at the M-BS interface calculated using this method unreliable (Sowiński et al., 2003, 2007). Plasmodesmograms provide semiquantitative data on symplastic connectivity between cells, based on PD that can be detected using TEM and yield values in relative units (Botha and Evert, 1988; Botha, 1992; Botha and van Bel, 1992). However, these data are difficult to compare with the quantitative data derived here from combined scanning electron microscopy and 3D immunolocalization methods. Our new data will now allow quantitative modeling of metabolite transport in a range of C3 and C4 species to improve our understanding of C4 evolution and efficiencies of the C4 pathway.
Role of Plasmodesmata in Metabolite Exchange at the Mesophyll-Bundle Sheath Cell Interface
Even in early modeling and analysis of C4 photosynthesis, it was suggested that rapid transport of C4 acids between M and BS cells and return of C3 products of decarboxylation was essential for the evolution of C4 plants (Osmond, 1971; Hatch and Osmond, 1976). The cell walls of the BS abutting the M are thickened and often heavily suberized, and it has been argued that this barrier serves to minimize CO2 leakage from the site of decarboxylation (Hatch, 1987; von Caemmerer and Furbank, 2003). This thickening and suberization has long been regarded as an insurmountable obstacle to simple diffusion or apoplastic transport of C4 acids and the pivotal role of PDs in this pathway was proposed soon after the discovery of the C4 pathway (Osmond, 1971).
Structures within the PD, where they penetrate the suberin lamella, are also viewed as possible regulators of PD metabolite transport in C4 species (Robinson-Beers and Evert, 1991a), although evidence is circumstantial. The species examined here, S. viridis and maize, belong to the C4 NADP-malic enzyme subtype. Examining C4 species belonging to other subtypes, such as NAD-malic enzyme (which do not possess suberized M-BS walls) and phosphoenolpyruvate carboxykinase types, presents an opportunity to determine if suberin lamellae are involved in regulation of metabolite transport via PD in the BS cell wall and have a role in determining C4 acid flux.
Assuming that BS cell walls are a barrier to metabolite diffusion between M and BS, the calculation of fluxes of C4 acids and C3 metabolites across the M-BS interface depends on accurate estimates of the cross sectional area of PD available for diffusion at this interface (Osmond, 1971; Hatch and Osmond, 1976; Stitt and Heldt, 1985; Wang et al., 2014). The absence of reliable estimates of this parameter has substantively hindered modeling studies. In early work, assumptions of the proportion of M-BS interface area populated by PD varied from 1 to 10% (Osmond, 1971); subsequently, these values and those of Hatch and Osmond (1976), based on median literature values for C3 plant cell PD data, have been routinely used. In recent work using reaction diffusion modeling, Wang et al. (2014) used values between 0.3% and 3% for the percentage of M-BS cell wall interface area occupied by PD. The values obtained here for PD area as a proportion of M-BS cell interface area equate to between 5.4% ± 0.06 and 6.2% ± 0.07% of the cell/cell interface (Table 1) and are at the higher end of values used in models to date.
Increasing Plasmodesmata Density Might Be Necessary for a Functional C4 Rice
This quantitative comparison of M-BS and M-M cell interface PD density between C3 and C4 species is of particular relevance to the creation of functional C4 rice. In a C3 system like rice, the M-BS interface is most likely a conduit for delivery of sucrose to the phloem for export from the leaf (Aoki et al., 2012). Fluxes of sucrose (a 12-carbon sugar) will be an order of magnitude less than the rate of photosynthesis if sucrose is continuously exported from the M tissues in the light. In C4 plants, the CO2-concentrating mechanism, and consequently the flux of C4 and C3 metabolites to and from the BS cells, must equal or exceed the rate of net photosynthesis. We used measurements of CO2 assimilation rates to calculate the flux of C4 and C3 metabolites across the M-BS interface for C4 species, the sucrose flux across the M-BS interface in the C3 species, as well as the hypothetical flux that would need to cross the M-BS interface in C3 species if a C4 photosynthetic pathway were installed without anatomical changes (Table 2). The values in Table 2 will actually underestimate the malate/aspartate and pyruvate/alanine fluxes required to support these net rates of photosynthesis by ∼20%. This is an average figure for “overcycling” of the C4 pump predicted to compensate for leakage of CO2 from the bundle sheath compartment from gas exchange, carbon isotope discrimination, and modeling studies (von Caemmerer and Furbank, 2003). If the properties of PD are similar between C3 and C4 plants (physical cross sectional areas of individual C3 and C4 PDs measured here were similar; 0.006 to 0.008 μm2), one would predict that the PD density at this interface would need to increase by up to 12-fold for effective exploitation of a C4 mechanism in rice. This study revealed that the C4 species, S. viridis and maize, have PD densities per M-BS cell interface area around 9-fold higher than the C3 species, rice and wheat. The scanning electron microscopy images showed that these C3 species commonly had regions of M-BS cell interface lacking PD entirely, suggesting that there is sufficient wall contact area available to allow the introduction of additional PD to support C4 fluxes.
In the context of C4 evolution and engineering, it is interesting to note that PD density is equally elevated in both M-BS and M-M interfaces of C4 leaves compared with C3 leaves. This suggests that in the evolution of C4 plants, a general increase in foliar symplastic connections may have occurred, not specific to the C4 mechanism, and a phylogenetic analysis of this hypothesis is currently underway. While the genes determining PD density are not currently known, the method reported here provides a more rapid, quantitative tool to probe the developmental biology of PD formation.
METHODS
Plant Material and Growth Conditions
Rice (Oryza sativa cultivar Kitaake), wheat (Triticum aestivum cultivar Yecora 70), Setaria viridis cultivar A10, and maize (Zea mays cultivar B73) were grown in a growth cabinet (High Resolution Plant Phenomics Centre, CSIRO Black Mountain) equipped with white fluorescent tubes (Philips TL5 HO) and maintained at 28°C day/22°C night temperatures, 60% relative humidity, and 16 h light/8 h dark with peak at 1000 µmol quanta m−2 s−1 light intensity and ambient CO2 concentration. The youngest fully expanded leaves from seedlings, 9 d after germination, were used.
Light Microscopy and Transmission Electron Microscopy
The middle portion of each leaf was cut into 1-mm2 strips and fixed in 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.3, overnight at 4°C. The tissue was postfixed with 1% osmium tetroxide for 2 h at room temperature. After dehydration in a graded ethanol series, the tissue was infiltrated with LR White resin and polymerized overnight at 60°C (Gunning et al., 1978). For light microscopy, semithin sections (0.5- to 1.0-µm thick) were cut with glass knives using a Reichert Jung ultramicrotome and stained with hot aqueous 0.05% Toluidine blue, pH 5.0. Imaging was done under 10× and 40× objectives using Nikon Eclipse 50i upright microscope (Nikon Instruments). For TEM, ultrathin sections (70- to 90-nm thick) were cut with a diamond knife using a Leica EM UC7 Ultramicrotome (Leica Microsystems) and examined using a Hitachi HA7100 transmission electron microscope (Hitachi High Technologies America) at 75 to 100 kV.
Scanning Electron Microscopy
Whole leaves were cut into 10-mm strips and fixed in 4% paraformaldehyde and 0.2% glutaraldehyde in 25 mM sodium phosphate buffer, pH 7.2, overnight at 4°C. The tissue was dehydrated in a graded ethanol series and critical point dried (Balzers CPD 030). Dried tissue was ripped open parallel to the veins using forceps and mounted onto copper holders using nail polish. Scanning electron microscopy preparations were sputter coated with platinum for 4 min using an Emitech EMS550X sputter and examined using a Zeiss Ultra Plus field emission scanning electron microscope at 3 kV.
3D Immunolocalization Confocal Microscopy
The 3D immunolocalization was performed using a modification of the PEA-CLARITY protocol of Palmer et al. (2015). Leaf tissue was cut directly into hydrogel monomer solution (4% acrylamide, 0.05% bis, 0.25% VA-044 initiator, 1× PBS, and 4% paraformaldehyde) and vacuum-infiltrated until the tissue sank. The gel solution was polymerized by incubating in a 37°C water bath overnight. Polymerized tissue was carefully removed from the gel using lint-free paper and transferred to clearing solution (200 mM boric acid and 4% SDS, pH 8.5). Tissue was incubated in the clearing solution with gentle shaking at room temperature for at least 6 to 8 weeks. Cleared tissue was digested in an enzyme cocktail containing 1 μL mL−1 β-xylanase M6 (Megazyme), 1 μL mL−1 α-L-arabinofuranosidase (Megazyme), 1 μL mL−1 pectate lyase (Megazyme), 0.5 μL mL−1 of 2 mg mL−1 cellulase (Sigma-Aldrich), 1 μL mL−1 α-amylase (Sigma-Aldrich), and 1 μL mL−1 pullulanase (Sigma-Aldrich) for at least 8 d at 37°C with gentle shaking. For rice leaf, 5× enzyme cocktail was used. Incubation in clearing solution followed by enzyme digestion was repeated three to four times until rice leaf is fully clarified. Tissues were then immunolabeled by incubating the clarified and digested tissue first in 1:500 primary antibody [β(1-3)-glucan antibody (catalog no. 400-2; Biosupplies) in 1× Tris-buffered saline with Tween 20 (TBST)] at 4°C for 5 to 7 days with 3 × 5 min vacuum infiltration each day, rinsed 5 × 30 min in 1× TBST, pH 7.4, then incubated in 1:500 secondary antibody (Alexa Fluor 488; catalog no. ab150117, lot no. GR181100-1 [Abcam] in TBST) at 4°C for 5 to 7 d with 3 × 5 min vacuum infiltration each day. Labeled tissue was poststained with 0.05% aqueous calcofluor white (Sigma-Aldrich) for 30 min at room temperature, washed twice with distilled water, and mounted onto glass slides with 80% glycerol. Tissues were then examined with a Leica SP8 multiphoton confocal microscope (Leica Microsystems) using long-distance dipping lens objectives (HCX APO L U-V-I 40×/0.80 water). Excitation at 488 nm allowed visualization of pitfields (Alexa Fluor 488) with emission at 522 to 550 nm. Simultaneously, fluorescence from calcofluor white-stained cell walls was detected at 434 to 445 nm following excitation at 405 nm. Corresponding background controls were obtained by hybridizing the tissue with 1:500 secondary antibody with and without calcofluor white poststaining (Supplemental Figure 1).
Plasmodesmata Area Measurement
TEM images of transverse sections of at least 40 PD from each type of cell interface were used to quantify the PD area enclosed by the plasma membrane. Measurement was performed with ImageJ software (National Institutes of Health) and a Wacom Cintiq graphics tablet (Wacom Technology).
Plasmodesmata Frequency in Pitfields
PD were counted and pitfield area was measured in scanning electron microscopy images using ImageJ software and a Wacom Cintiq graphics tablet. Data from at least 40 pitfields located either in the M-BS cell interface or M-M cell interface were used to generate regressions of PD numbers versus pitfield area. PD frequency per µm2 pitfield area was calculated using the linear equation, y = mx + b, where m is the slope, b is the intercept, and y is the PD frequency when pitfield area, x = 1 µm2. This frequency value is a reflection of how the PD are distributed within the pitfield. Ultimately, a higher value means that the PD are positioned in close proximity to each other while a lower value means PD are farther apart.
Pitfield Area Quantification on Cell Interfaces
A subset of 10 to 30 consecutive single focal planes was derived from the 3D confocal image z-stack to reveal BS cell surfaces in contact with M cells and M cell surfaces in contact with adjacent M cells (Leica Application Suite X software; Supplemental Figure 2). The maximum intensity projection images generated from the cropped stack were processed in ImageJ software (Supplemental Figure 2). The cell interface area in focus was selected and the total pitfield area (pfa) was quantified. The number of cell interfaces (ci) covered by the focused area were counted and individual cell interface area (cia) was measured. Pitfield area per cell interface area was obtained using this equation: pfa/(ci × cia). Pitfields parallel to the cell interface were seen as individual pits while pitfields perpendicular to the cell interface were seen as clusters and were therefore larger. In cases where pitfield signals coming from different cell interfaces were captured, size discrimination was applied.
Plasmodesmata Density on Cell Interfaces
The PD density per cell interface area was obtained from the product of PD frequency per µm2 pitfield area (scanning electron microscopy) and pitfield area per cell interface area (3D immunolocalization). By multiplying this value by the PD area measured using TEM images, the percentage of PD area per cell interface area was calculated.
Gas Exchange Measurement
Gas exchange was measured on the youngest fully expanded leaf of 9 d after germination seedlings using a LI-6400 equipped with a blue-red LED light source (LI-COR). Leaves were initially equilibrated for 30 min in a standard environment of 380 µmol mol−1 CO2, 25°C leaf temperature, flow rate of 500 µmol s−1, and an irradiance of either 1500 (for C3 species) or 2000 (for C4 species) µmol quanta m−2 s−1. Photosynthetic rates measured under these conditions were used for flux calculations.
CO2 Assimilation Rate and Sucrose Flux Calculations per PD
The BS surface areas per unit leaf area (Sb) for each species were calculated using the equation described by Pengelly et al. (2010). The CO2 assimilation rate (a surrogate for C4 acid fluxes) per BS surface area was obtained by dividing the photosynthetic rate derived from gas exchange measurements by Sb. Then, the CO2 assimilation rate per PD was calculated by dividing CO2 assimilation per BS surface area by the PD density per M-BS cell interface area. For the C3 species, rice and wheat, sucrose flux per PD was obtained by dividing CO2 assimilation rate per PD by 12.
Statistical Analysis
The statistical differences between most quantitative measurements were assessed using a MATLAB two-sample t test (The MathWorks) at 5% significance level (P value < 0.05). Statistical differences at P value < 0.05 between the M-BS cell interface and M-M cell interface regression lines in Figure 6 were evaluated using SPSS Statistics software.
Supplemental Data
Supplemental Figure 1. Confocal Micrograph Background Controls for 3D Immunolocalization.
Supplemental Figure 2. Image Processing Workflow for Pitfield Area Measurement Using 3D Immunolocalization Confocal Micrographs.
Acknowledgments
We thank Brian Gunning for helpful discussion. We thank the ANU Centre for Advanced Microscopy, Australian Microscopy and Microanalysis Research Facility (AMMRF) and CSIRO Microscopy Centre for providing support and technical assistance. F.R.D. is supported by scholarship awards from Lee Foundation (IRRI) and Centre of Excellence for Translational Photosynthesis (ANU).
AUTHOR CONTRIBUTIONS
S.v.C. and R.T.F. conceived the project. F.R.D. conducted all the experiments, imaging, quantification, and data analysis in consultation with S.v.C., R.T.F., R.G.W, and W.P.Q. All authors contributed to writing the manuscript.
Glossary
- M
mesophyll
- BS
bundle sheath
- PD
plasmodesmata
- TEM
transmission electron microscopy
Footnotes
Articles can be viewed without a subscription.
References
- Aoki N., Hirose T., Furbank R. (2012). Sucrose transport in higher plants: from source to sink. In Photosynthesis,Vol. 34, J.J. Eaton-Rye, B.C. Tripathy, T.D. Sharkey, eds (Dordrecht, The Netherlands: Springer), pp. 703–729. [Google Scholar]
- Barton D., Overall R. (2015). Imaging plasmodesmata with high-resolution scanning electron microscopy. In Plasmodesmata, Vol. 1217, M. Heinlein, ed (New York: Springer), pp. 55–65. [DOI] [PubMed] [Google Scholar]
- Botha C.E.J. (1992). Plasmodesmatal distribution, structure and frequency in relation to assimilation in C3 and C4 grasses in southern Africa. Planta 187: 348–358. [DOI] [PubMed] [Google Scholar]
- Botha C.E.J., Evert R.F. (1988). Plasmodesmatal distribution and frequency in vascular bundles and contiguous tissues of the leaf of Themeda triandra. Planta 173: 433–441. [DOI] [PubMed] [Google Scholar]
- Botha C.E.J., van Bel A.J.E. (1992). Quantification of symplastic continuity as visualised by plasmodesmograms: diagnostic value for phloem-loading pathways. Planta 187: 359–366. [DOI] [PubMed] [Google Scholar]
- Brecknock S., Dibbayawan T.P., Vesk M., Vesk P.A., Faulkner C., Barton D.A., Overall R.L. (2011). High resolution scanning electron microscopy of plasmodesmata. Planta 234: 749–758. [DOI] [PubMed] [Google Scholar]
- Carr D.J. (1976). Plasmodesmata in growth and development. In Intercellular Communication in Plants: Studies on Plasmodesmata, B.E.S. Gunning and A.W. Robards, eds (Berlin, Heidelberg: Springer), pp. 243–289. [Google Scholar]
- Ding B., Turgeon R., Parthasarathy M.V. (1992). Substructure of freeze-substituted plasmodesmata. Protoplasma 169: 28–41. [Google Scholar]
- Evert R.F., Eschrich W., Heyser W. (1977). Distribution and structure of the plasmodesmata in mesophyll and bundle-sheath cells of Zea mays L. Planta 136: 77–89. [DOI] [PubMed] [Google Scholar]
- Faulkner C., Akman O.E., Bell K., Jeffree C., Oparka K. (2008). Peeking into pit fields: a multiple twinning model of secondary plasmodesmata formation in tobacco. Plant Cell 20: 1504–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning B.E.S. (1978). Age-related and origin-related control of the numbers of plasmodesmata in cell walls of developing Azolla roots. Planta 143: 181–190. [DOI] [PubMed] [Google Scholar]
- Gunning B.E.S., Hughes J.E., Hardham A.R. (1978). Formative and proliferative cell divisions, cell differentiation, and developmental changes in the meristem of Azolla roots. Planta 143: 121–144. [DOI] [PubMed] [Google Scholar]
- Hatch M.D. (1987). C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 895: 81–106. [Google Scholar]
- Hatch M.D., Osmond C.B. (1976). Compartmentation and transport in C4 photosynthesis. In Transport in Plants, Vol. 3, Stocking C.R., Heber U., eds (Berlin, Heidelberg: Springer; ), pp. 144–184. [Google Scholar]
- Olesen P. (1975). Plasmodesmata between mesophyll and bundle sheath cells in relation to the exchange of C4-acids. Planta 123: 199–202. [DOI] [PubMed] [Google Scholar]
- Osmond C.B. (1971). Metabolite transport in C4 photosynthesis. Aust. J. Biol. Sci. 24: 159–163. [DOI] [PubMed] [Google Scholar]
- Overall R.L., Blackman L.M. (1996). A model of the macromolecular structure of plasmodesmata. Trends Plant Sci. 1: 307–311. [Google Scholar]
- Palmer W.M., Martin A.P., Flynn J.R., Reed S.L., White R.G., Furbank R.T., Grof C.P.L. (2015). PEA-CLARITY: 3D molecular imaging of whole plant organs. Sci. Rep. 5: 13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly J.J.L., Sirault X.R.R., Tazoe Y., Evans J.R., Furbank R.T., von Caemmerer S. (2010). Growth of the C4 dicot Flaveria bidentis: photosynthetic acclimation to low light through shifts in leaf anatomy and biochemistry. J. Exp. Bot. 61: 4109–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robards A.W. (1976). Plasmodesmata in higher plants. In Intercellular Communication in Plants: Studies on Plasmodesmata, Gunning B.E.S., Robards A.W., eds (Berlin, Heidelberg: Springer; ), pp. 15–57. [Google Scholar]
- Robinson-Beers K., Evert R.F. (1991a). Fine structure of plasmodesmata in mature leaves of sugarcane. Planta 184: 307–318. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K., Evert R.F. (1991b). Ultrastructure of and plasmodesmatal frequency in mature leaves of sugarcane. Planta 184: 291–306. [DOI] [PubMed] [Google Scholar]
- Sage T.L., Sage R.F. (2009). The functional anatomy of rice leaves: implications for refixation of photorespiratory CO2 and efforts to engineer C4 photosynthesis into rice. Plant Cell Physiol. 50: 756–772. [DOI] [PubMed] [Google Scholar]
- Sowiński P., Bilska A., Barańska K., Fronk J., Kobus P. (2007). Plasmodesmata density in vascular bundles in leaves of C4 grasses grown at different light conditions in respect to photosynthesis and photosynthate export efficiency. Environ. Exp. Bot. 61: 74–84. [Google Scholar]
- Sowiński P., Rudzińska-Langwald A., Kobus P. (2003). Changes in plasmodesmata frequency in vascular bundles of maize seedling leaf induced by growth at sub-optimal temperatures in relation to photosynthesis and assimilate export. Environ. Exp. Bot. 50: 183–196. [Google Scholar]
- Stitt M., Heldt H.W. (1985). Generation and maintenance of concentration gradients between the mesophyll and bundle sheath in maize leaves. Biochim. Biophys. Acta 808: 400–414. [Google Scholar]
- Turner A., Wells B., Roberts K. (1994). Plasmodesmata of maize root tips: structure and composition. J. Cell Sci. 107: 3351–3361. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S. (2000). Biochemical Models of Leaf Photosynthesis. (Collingwood, Australia: CSIRO Publishing; ). [Google Scholar]
- von Caemmerer S., Furbank R.T. (2003). The C4 pathway: an efficient CO2 pump. Photosynth. Res. 77: 191–207. [DOI] [PubMed] [Google Scholar]
- Wang Y., Long S.P., Zhu X.-G. (2014). Elements required for an efficient NADP-malic enzyme type C4 photosynthesis. Plant Physiol. 164: 2231–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]