A newly identified effector of RAB5 GTPases helps these molecular switch-like proteins regulate endosomal trafficking events during early Arabidopsis development.
Abstract
RAB5 GTPases act as molecular switches that regulate various endosomal functions in animal cells, including homotypic fusion of early endosomes, endosomal motility, endosomal signaling, and subcompartmentalization of the endosomal membrane. RAB5 proteins fulfill these diverse functions through interactions with downstream effector molecules. Two canonical RAB5 members, ARA7 and RAB HOMOLOG1 (RHA1), are encoded in the Arabidopsis thaliana genome. ARA7 and RHA1 play crucial roles in endocytic and vacuolar trafficking pathways. Plant RAB5 GTPases function via interactions with effector molecules, whose identities and functions are currently unclear. In this study, we searched for canonical RAB5 effector molecules of Arabidopsis and identified a candidate, which we called ENDOSOMAL RAB EFFECTOR WITH PX-DOMAIN (EREX). The intimate genetic interaction between EREX and RAB5 members, the results from subcellular colocalization experiments, and the direct interaction observed in an in vitro pull-down assay strongly suggest that EREX is a genuine effector of canonical RAB5s in Arabidopsis. We further found that close homologs of EREX play partially redundant functions with EREX in the transport of seed storage proteins. Our results indicate that canonical plant RAB5s acquired distinct effector molecules from those of non-plant systems to fulfill their functions.
INTRODUCTION
Endosomal trafficking plays pivotal roles in various cellular functions in animals and plants, which include the maintenance of cell polarity, selective degradation and recycling of membrane proteins, and nutrient utilization (Jolliffe et al., 2005; Miao et al., 2008; Grant and Donaldson, 2009; Friml, 2010; Reyes et al., 2011; Contento and Bassham, 2012). RAB GTPases are small GTPases that act as molecular switches by cycling between GTP-bound active and GDP-bound inactive states, serving as key regulators of membrane trafficking, including endosomal trafficking (Saito and Ueda, 2009). When RAB GTPases are activated by the replacement of GDP with GTP, which is mediated by guanine nucleotide exchange factors (GEFs), they interact with specific sets of interacting partners collectively called RAB effectors. Through their interaction with effector molecules, RAB GTPases evoke a wide spectrum of downstream events, including the tethering of transport vesicles to target organelles, the formation of subdomains on organelle membranes, organelle movement, alteration of lipid composition in organelle membranes, and organelle maturation (Stenmark, 2009; Mizuno-Yamasaki et al., 2012). Once GTP is hydrolyzed by the action of GTPase activating proteins, the GDP-bound RAB proteins are detached from the membranes by forming complexes with GDP dissociation inhibitors and are retained in the cytosol until the next round of the GTPase cycle (Seabra and Wasmeier, 2004; Goody et al., 2005).
RAB5 is one of the best-characterized groups of RAB proteins in animal systems (Somsel Rodman and Wandinger-Ness, 2000; Benmerah, 2004; Galvez et al., 2012). Animal RAB5s interact with more than 20 proteins when in the GTP-bound state (Christoforidis and Zerial, 2000), including class I and class III phosphatidylinositol-3 kinases and phosphoinositide phosphatases (Christoforidis et al., 1999; Shin et al., 2005). In addition to these enzymes, various proteins with phosphoinositide binding moieties such as Early Endosome Antigen1 (EEA1), Rabenosyn-5, Rabankyrin-5, and APPL1 and 2, which shuttle between the endosomal membrane and nucleus, have also been identified as RAB5 effectors (Simonsen et al., 1998; Nielsen et al., 2000; Miaczynska et al., 2004; Schnatwinkel et al., 2004). These lines of evidence strongly suggest that there is a tight correlation between RAB5 function and phosphoinositides in animal cells.
RAB GTPases also play key roles in membrane trafficking pathways in plant cells (Lycett, 2008; Nielsen et al., 2008; Pedrazzini et al., 2013). The Arabidopsis thaliana genome encodes 57 RAB GTPases, which are classified into eight subgroups according to their similarity to animal Rab GTPases (Woollard and Moore, 2008). Among these subgroups, the RAB5 group (also called RABF) consists of three members, which are further classified into two subtypes: plant-unique ARA6 (also known as RABF1) and the canonical RAB5 group (ARA7 and RAB HOMOLOG1 [RHA1], also known as RABF2b and RABF2a, respectively) (Ueda et al., 2001, 2004; Ebine and Ueda, 2009). Despite the differences in their primary structures, these RAB5 members are activated by the same GEF, VACUOLAR PROTEIN SORTING 9a (VPS9a), whose loss of function results in embryonic lethality (Goh et al., 2007; Uejima et al., 2010, 2013). The double mutant of canonical RAB5, ara7 rha1, also exhibits gametophytic lethality (Dhonukshe, 2009). These lines of evidence indicate that RAB5 GTPases fulfill an essential function in Arabidopsis, although the molecular basis underlying this function remains mostly unknown.
Analyzing plant RAB5 effectors is an effective approach for unraveling the molecular mechanisms of RAB5-mediated membrane trafficking. However, clear homologs of animal RAB5 effectors such as Rabaptin-5 (Stenmark et al., 1995) and EEA1 (Simonsen et al., 1998) are not found in plants, suggesting that the operation of RAB5-dependent membrane trafficking in plants is distinct from that in non-plant systems. The unique mechanism of the RAB5-mediated trafficking pathway is also highlighted by the divergent functions of the two RAB5 subtypes in plants. Canonical RAB5s and ARA6 are localized to distinct but overlapping populations of multivesicular endosomes (Ueda et al., 2004; Haas et al., 2007; Ebine et al., 2011), suggesting diversification of their functions. ARA6 acts in the endosome-to-plasma membrane trafficking pathway (Ebine et al., 2011) and may also be involved in a vacuolar trafficking pathway (Bottanelli et al., 2011). ARA7 and RHA1, in contrast, redundantly regulate endocytic and vacuolar trafficking pathways (Sohn et al., 2003; Kotzer et al., 2004; Dhonukshe et al., 2006; Ebine et al., 2011, 2014; Beck et al., 2012). The unique organization of endosomal trafficking in plants is also demonstrated by the observation that the trans-Golgi network (TGN) acts as an early endosome in plants (Dettmer et al., 2006; Lam et al., 2007; Viotti et al., 2010), from which multivesicular endosomes are proposed to be directly generated by maturation (Niemes et al., 2010; Scheuring et al., 2011). The RAB11 group (also known as RABA in Arabidopsis), which is localized to TGN-related secretory compartments and is involved in cell plate formation and tip growth (Preuss et al., 2004; Chow et al., 2008; Szumlanski and Nielsen, 2009; Kang et al., 2011; Feraru et al., 2012; Asaoka et al., 2013; Berson et al., 2014; Kirchhelle et al., 2016), is also implicated in endosomal/vacuolar trafficking (Bottanelli et al., 2011; Choi et al., 2013). These lines of evidence indicate that endosomal trafficking in plant cells operates in a different manner from that in non-plant organisms.
Recently, a complex consisting of SAND/MONENSIN SENSITIVITY1 and CALCIUM CAFFEINE ZINC SENSITIVITY1 (CCZ1) was demonstrated to act as an effector of canonical plant RAB5s (Cui et al., 2014; Ebine et al., 2014; Singh et al., 2014). The SAND-CCZ1 complex interacts with GTP-bound RAB5s and serves as a GEF for RAB7, which leads to RAB5-to-RAB7 conversion on the endosome. This finding clearly demonstrates that plants harbor an endosomal/vacuolar trafficking pathway in which RAB5 and RAB7 act sequentially, as proposed previously (Bottanelli et al., 2012). In addition to this pathway, plants are also equipped with at least two distinct trafficking pathways for vacuoles, which are distinctly dependent on RAB5 and RAB7 (Bottanelli et al., 2011; Ebine et al., 2014). This complexity in the vacuolar trafficking system also indicates that plants have evolved elaborate endosomal trafficking pathways in a unique manner.
To begin characterizing the roles of canonical plant RAB5s in the endosomal trafficking pathways, we performed yeast two-hybrid screening for interacting partners of canonical RAB5s from Arabidopsis. We isolated an interacting partner of RAB5s that harbors a phosphoinositide binding domain. Our results indicate that this protein acts as a RAB5 effector and mediates vacuolar trafficking of seed storage proteins by interacting with canonical RAB5.
RESULTS
Screening for Proteins That Interact with Canonical RAB5
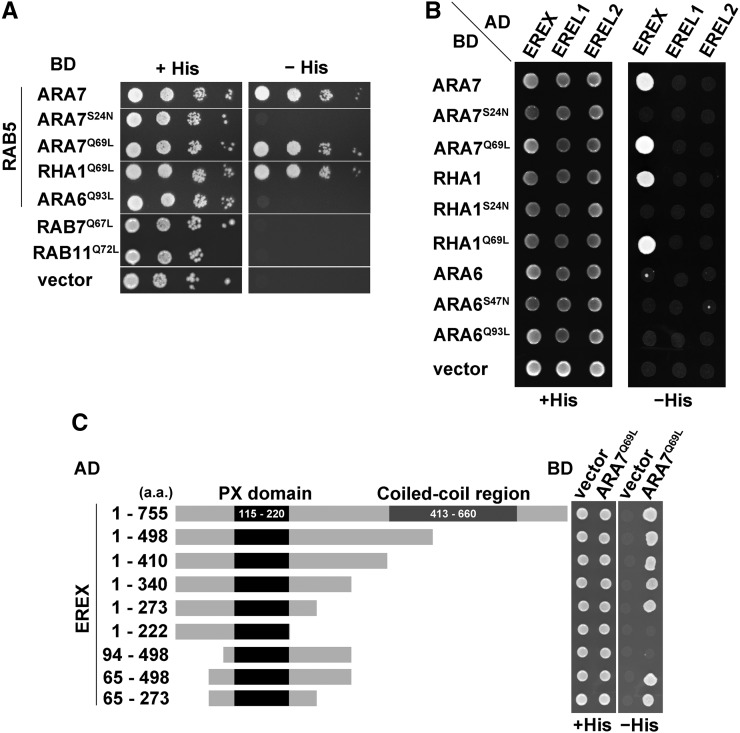
To identify candidates for effectors of canonical RAB5s in Arabidopsis, we performed yeast two-hybrid screening using constitutively active ARA7 (ARA7Q69L) as bait. In this study, C-terminal cysteine residues of RAB GTPases were deleted to ensure their nuclear localization in yeast cells. We screened a cDNA library prepared from 4-d-old Arabidopsis root tissues and identified a positive clone containing a partial sequence of cDNA for At3g15920, which encodes a partial protein lacking the N-terminal 27 amino acids of the predicted full-length protein (Figure 1A). This candidate protein exhibited a positive interaction with the other Arabidopsis canonical RAB5, RHA1, but did not interact with constitutively active mutants of RAB7/RABG3b, RAB11/RABA1e, or ARA6/RABF1 (Figure 1A). Thus, the identified candidate protein specifically interacted with canonical RAB5. At3g15920 encodes a protein of unknown function, which contains a phox homology (PX) domain known as a phosphoinositide binding module. We named this protein ENDOSOMAL RAB EFFECTOR WITH PX-DOMAIN (EREX) and investigated the possibility that this protein acts as an effector of canonical RAB5. There are 11 genes encoding PX domain-containing proteins in the Arabidopsis genome, which are classified into four subgroups (van Leeuwen et al., 2004). Among these genes, At4g32160 and At2g25350 encode structurally similar proteins to EREX, which we named EREX-LIKE1 (EREL1) and EREL2, respectively. We isolated open reading frame sequences for full-length EREX and EREL proteins and used the yeast two-hybrid method to determine whether they interacted with RAB5. Full-length EREX also interacted with wild-type and constitutively active ARA7 and RHA1. In contrast, an interaction between EREL proteins and RAB GTPases was not detected by this method (Figure 1B).
Figure 1.
Interaction of EREX and EREL Members with RAB GTPases Detected by the Yeast Two-Hybrid Method.
(A) to (C) RAB proteins were expressed as fusion proteins with a DNA binding domain (BD). EREX and EREL members were expressed as fusions with a transcriptional activation domain (AD). Interactions between the two cotransformed proteins were tested using the HIS3 reporter gene in the AH109 yeast strain. Cells were spotted on synthetic medium lacking tryptophan and leucine (+His) or lacking tryptophan, leucine, and histidine (-His) and incubated at 30°C for 5 d.
(A) The originally isolated plasmid containing a partial sequence of cDNA for EREX was retransformed with the plasmids containing endosomal RABs with the GTP-freeze (Q-to-L mutations) or GDP-freeze (S-to-N mutation) mutations. The result of a 10-fold serial dilution is indicated.
(B) EREX, EREL1, and EREL2 were fused to the AD and cotransformed with BD-ARA7, BD-RHA1, and BD-ARA6 containing the indicated mutations.
(C) Deletion mutants of EREX were cotransformed with constitutively active ARA7.
Interaction of EREX with Canonical RAB5
To determine the region of EREX responsible for the interaction with RAB5, we constructed a deletion series of EREX and examined the interaction with constitutively active ARA7. We identified the polypeptide containing amino acids 65 to 273 of EREX (EREX65-273) as the minimum region required for the interaction with ARA7 (Figure 1C). Thus, the PX domain and its flanking regions are responsible for interaction with RAB5, and the coiled-coil region is dispensable for this interaction.
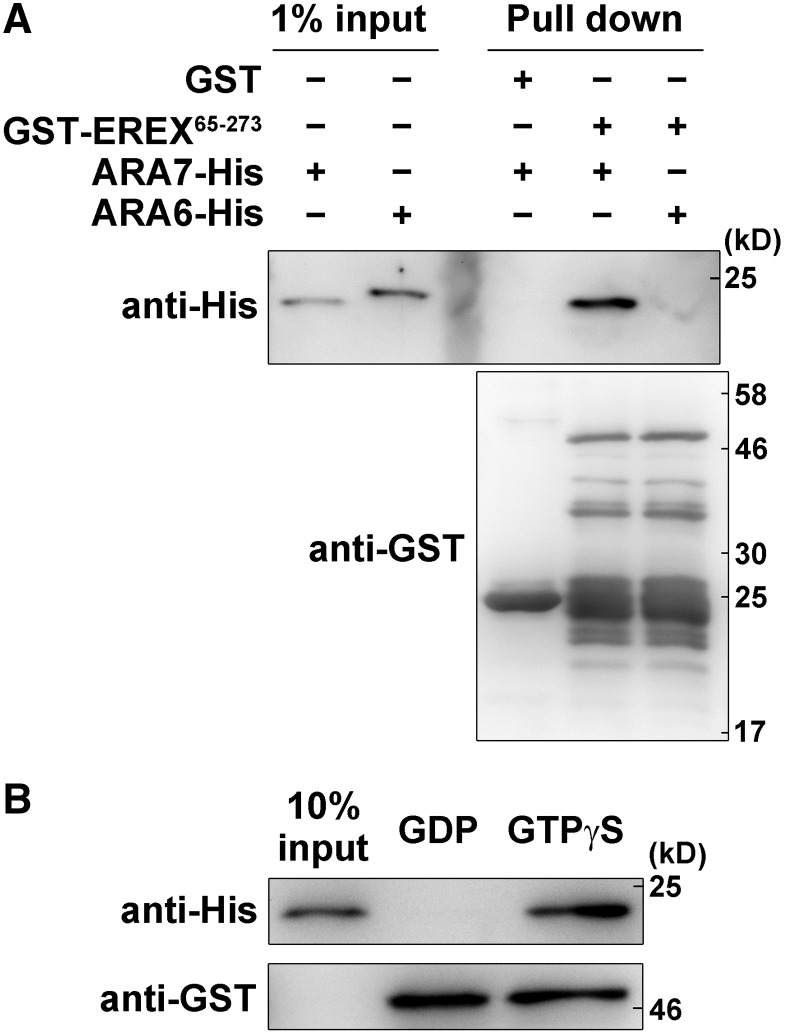
We then attempted to detect direct interaction between EREX and ARA7 with an in vitro pull-down assay using bacterially expressed purified proteins. At first, we used the full-length EREX protein tagged with GST for the assay, but we could not detect its binding with ARA7 because the full-length EREX protein was easily degraded during the experimental procedure. We then examined the interaction with GST-tagged EREX65-273. We incubated GST-EREX65-273 with His6-tagged ARA7 or ARA6 in the presence of GTPγS and examined whether these RAB proteins were pulled down using glutathione sepharose. As suggested from the results of the yeast two-hybrid assay, GST-EREX65-273 pulled down ARA7-His6 but not ARA6-His6 (Figure 2A). We confirmed that GST alone did not pull down ARA7-His6. When GTPγS in the binding buffer was replaced with GDP, the amount of ARA7-His6 pulled down by GST-EREX65-273 was drastically reduced (Figure 2B). These results further indicate that canonical RAB5s interact with EREX in the GTP-bound active state.
Figure 2.
Interactions between EREX65-273 and ARA7 Detected in the in Vitro Pull-Down Assay.
(A) The interaction between GST-tagged EREX65-273 and His6-tagged ARA7 or ARA6 was tested in the presence of 10 μM GTPγS. Proteins were precipitated using Glutathione-Sepharose 4B resin and analyzed by immunoblotting using anti-His6 antibody and anti-GST antibodies. EREX65-273 specifically and directly interacted with ARA7.
(B) GST-EREX65-273 and ARA7-His6 were incubated with Glutathione-Sepharose 4B resin in the presence of 25 μM GTPγS or GDP. GST-EREX65-273 interacted with GTPγS-bound ARA7.
EREX Depends on RAB5 Activity to Localize to Endosomes
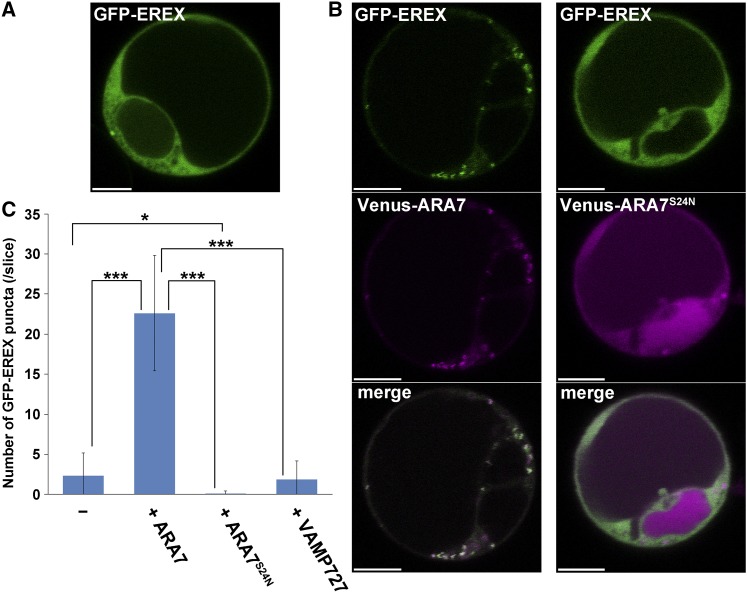
We then analyzed the subcellular localization of N-terminally GFP-tagged EREX in the protoplasts of suspension-cultured Arabidopsis cells. When GFP-EREX was expressed alone under the control of the CaMV35S promoter, fluorescence from GFP-EREX was predominantly observed in the cytosol and rarely localized to punctate structures (Figure 3A). When EREX was coexpressed with ARA7, EREX was targeted to the punctate compartments and colocalized with ARA7 (Figure 3B). In contrast, a similar effect on GFP-EREX was not observed for RABG3b, a member of the RAB7 group (Supplemental Figure 1). We then examined whether the nucleotide states of ARA7 affected the localization of GFP-EREX. When coexpressed with a GDP-fixed mutant of ARA7 (ARA7S24N) tagged with Venus, EREX dispersed into the cytosol (Figure 3B). Conversely, when coexpressed with GTP-fixed ARA7Q69L, which was localized to the punctate compartments and the vacuolar membrane, GFP-EREX was also targeted to the same puncta and the vacuolar membrane (Supplemental Figure 1). GTP-fixed ARA6Q93L, which was also localized to the punctate compartments and the vacuolar membrane, had no effect on GFP-EREX (Supplemental Figure 1). Thus, the endosomal localization of EREX depends on the activating state of ARA7, which suggests that EREX and canonical RAB5s actually act together in plant cells. This result was further supported by quantitative analysis. When GFP-EREX was coexpressed with wild-type ARA7, 22.6 ± 7.2 punctate structures were observed in a confocal section of a protoplast, whereas 2.33 ± 2.9 puncta of GFP-EREX were observed when GFP-EREX was expressed alone (Figure 3C). Only 0.111 ± 0.32 GFP-EREX-positive dots were observed when GFP-EREX was coexpressed with ARA7S24N. Coexpression of VAMP727, a soluble N-ethylmaleimide-sensitive factor attachment protein receptor that resides on the endosome with canonical RAB5s (Ueda et al., 2004; Ebine et al., 2008), did not affect the subcellular localization of GFP-EREX (1.86 ± 2.3 puncta were observed in a confocal section of a protoplast). These results indicate that EREX is specifically recruited to endosomes by canonical RAB5s in a nucleotide state-dependent manner. Intriguingly, the subcellular localizations of EREL1 and EREL2 were also affected by coexpressed ARA7 in a nucleotide state-dependent manner (Supplemental Figure 1), although the interaction between ARA7 and EREL proteins was not detected by the yeast two-hybrid method.
Figure 3.
ARA7-Dependent Recruitment of EREX to the Endosome.
(A) and (B) XFP-tagged proteins were expressed transiently in the protoplasts of suspension-cultured Arabidopsis cells. Bars = 5 μm.
(A) When expressed alone, GEP-EREX was mainly observed in the cytosol.
(B) When coexpressed, wild-type ARA7 and EREX colocalized to the same endosomes (left panels), whereas coexpression of EREX with dominant-negative ARA7 (ARA7S24N) resulted in the dispersal of EREX into the cytosol (right panels).
(C) Numbers of GFP-EREX-positive puncta in protoplasts expressing GFP-EREX and wild-type ARA7 or ARA7S24N. The results are presented as the means ± sd (n = 12 confocal sections of protoplasts for GFP-EREX alone, n = 13 for coexpression with Venus-ARA7, n = 18 for coexpression with Venus-ARA7S24N, and n = 7 for coexpression with Venus-VAMP727). *P < 0.1 and ***P < 0.001; Student’s t test.
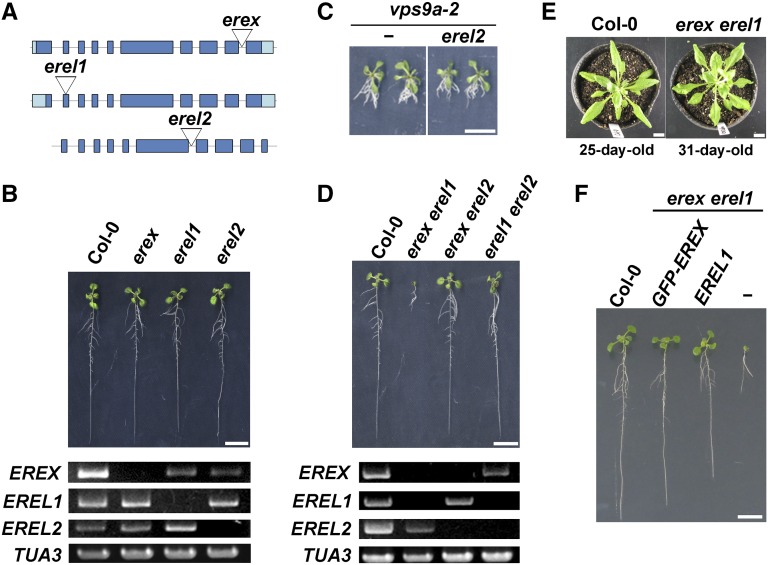
Characterization of erex and erel Mutants
To understand the functions of EREX, EREL1, and EREL2 in planta, we characterized T-DNA-tagged mutants of these genes (Figure 4A). We confirmed that the full-length transcript was not detected in each mutant plant (Figure 4B). Although the absence of EREX and EREL transcripts suggested that these mutants are null alleles (Figure 4B), abnormalities in plant morphology and fertility were not detected in these mutants. To determine the functional interactions between RAB5 and EREX and EREL, we examined the genetic interaction between erex or erel and vps9a-2, which is a weak mutant allele of the RAB5 GEF responsible for activating all RAB5 members in Arabidopsis (Goh et al., 2007; Sunada et al., 2016). We crossed vps9a-2 and erex or erel mutants to generate double mutants and found that vps9a-2 erex and vps9a-2 erel1 double mutant plants could not be established, most likely because these double mutants were defective in gametogenesis and/or embryogenesis (Supplemental Tables 1 and 2). To further understand the effects of these double mutations, we conducted cross-pollination analyses using the vps9a-2 erex double mutant. When we crossed female mutant plants with homozygous erex and hemizygous vps9a-2 mutations (vps9a-2+/−erex−/−) and male wild-type plants (Col-0), the double mutation was transferred to 34% of the progeny, suggesting that the vps9a-2 erex mutation had slightly reduced viability in the female gametophyte. We then crossed male vps9a-2+/−erex−/− and female Col-0 plants. The vps9a-2 erex mutation was transferred to only 7% of the progeny (Supplemental Table 3). Thus, the double mutation affected the function or formation of the male gametophyte more severely than it did the female gametophyte. However, the production of a double homozygous mutant is still theoretically possible at low frequency because the male and female gametophytes were not completely sterile, although we failed to establish a vps9a-2 erex double mutant. We examined cleared seedpods harvested from the vps9a-2+/−erex−/− mutants and found that embryos at retarded developmental stages were present at a low frequency. This result suggests that the double homozygous mutation of vps9a-2 erex results in embryonic lethality, which is consistent with the embryonic lethal phenotype of the vps9a-1 mutant, a null allele of vps9a (Supplemental Figure 2A; Goh et al., 2007).
Figure 4.
Characterization of erex and erel Mutants.
(A) Schematic structures of the EREX genes and the positions of T-DNA insertions.
(B) Phenotypes of erex single mutants. Ten-day-old wild-type and mutant plants (upper panel) and expression level of EREX genes in plants (lower panels) with the indicated genotypes are presented.
(C) The erel2 mutation did not enhance the vps9a-2 mutation. Twelve-day-old plants are presented.
(D) Phenotypes of double mutants of erex and erel members. Ten-day-old wild-type and mutant plants (upper panel) and expression levels of EREX genes in the plants (lower panels) are presented. The growth of the erex erel1 mutant was severely affected.
(E) The growth phenotype in the erex erel1 mutant was specific to the juvenile stage. Twenty-five-day-old wild-type and 31-d-old erex erel1 mutant are presented.
(F) Ten-day-old wild-type and erex erel1 double mutant seedlings expressing GFP-EREX or EREL1. The rightmost plant is the erex erel1 double mutant. Bars = 1 cm.
Similarly defective embryos were also observed in cleared seedpods from the vps9a-2+/−erel1−/− mutant (Supplemental Figure 2A). The double homozygous vps9a-2 erel1 mutation should result in embryonic lethality without having a deleterious effect on gametophyte viability based on the progeny segregation ratio derived from the vps9a-2+/−erel1−/− mutant plants (Supplemental Table 2). The vps9a-2 erel2 double mutant was indistinguishable from the vps9a-2 mutant (Figure 4C).
The synergistic genetic interaction between vps9a-2 and erex or erel1 strongly suggests that EREX and EREL1 act in the same genetic (and trafficking) pathway as VPS9a, which further supports the hypothesis that EREX acts as an effector of RAB5.
Because erex and erel1 exerted similar effects on the vps9a-2 mutation, we next examined the genetic interaction between erex and erel1. Consistent with the notion that EREX and EREL1 act in the same trafficking event, the erex erel1 double mutant exhibited severe growth retardation at a juvenile stage (Figure 4D; Supplemental Figure 2B). Interestingly, this growth defect was not persistent, and the erex erel1 double mutant eventually grew to a size similar to that of wild-type plants (Figure 4E). These phenotypes of the double mutant were restored by introducing either the genomic fragment of EREX containing the GFP cDNA insert or the genomic fragment of EREL1 into the mutant background (Figure 4F), indicating that the observed phenotypes could be attributed to defective EREX and EREL1. The erex erel2 and erel1 erel2 double mutants were indistinguishable from wild-type plants (Figure 4D). These results indicate that EREX and EREL1 play redundant functions, which are especially important in embryogenesis and/or early developmental processes after germination. Intriguingly, we could not recover the erex erel1 erel2 triple mutant, most likely because of the partial deficiencies in the male gametophyte with the triple mutation and embryonic lethality of the triple homozygous mutant plant (Supplemental Tables 4 and 5 and Supplemental Figure 2A). Thus, EREL2 might partly share a common function with EREX and EREL1, although its contribution cannot be substantial given the weak genetic interaction between vps9a-2 and erel2.
If EREX acts as an effector of canonical RAB5 members and the growth defect observed in the erex erel1 mutant is caused by defective membrane trafficking involving RAB5, it is possible that overexpression of constitutively active RAB5s could suppress the erex erel1 phenotypes. To test this possibility, we overexpressed constitutively active ARA6 or ARA7 tagged with GFP under the control of the 35S promoter in the erex erel1 mutant. As expected, the growth defects at the early developmental stages were partially, but significantly, suppressed by constitutively active ARA7 but not by constitutively active ARA6 (Supplemental Figure 2C). This result further supports the hypothesis that EREX acts as an effector of canonical RAB5s in RAB5-mediated trafficking events.
Expression and Subcellular Localization of EREX in Plants
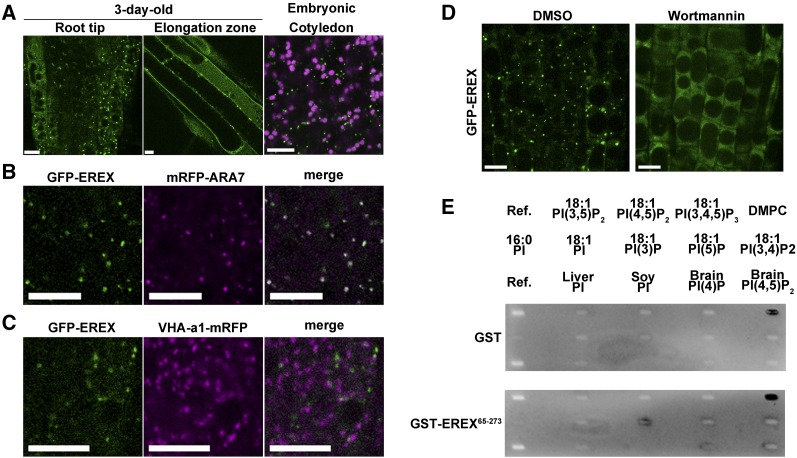
Because EREX performs an important function in early developmental processes as described above, we expected that EREX would be expressed at this stage. To examine the expression pattern of EREX, we constructed transgenic Arabidopsis plants expressing a translational fusion of the first exon of EREX and the cDNA for GUS under the control of the 1.6-kb EREX promoter. GUS staining indicated that EREX was abundantly expressed in young seedlings, and its expression was also observed in young leaves, root tips, and developing embryos (Supplemental Figure 3). A similar expression pattern was also observed in transgenic Arabidopsis plants expressing GFP-tagged EREX under the control of its own regulatory elements, including the promoter, introns, and terminator (Figure 5A). GFP-EREX was localized to punctate compartments and to the cytosol in the cells of root tips and embryos. This construct restored the growth retardation phenotype of the erex erel1 double mutant (Figure 4F), indicating that the GFP-tagged EREX protein retained its original function. To analyze the identity of the EREX-positive compartments, we crossed this transgenic plant with plants expressing fluorescently tagged organelle markers and found that GFP-EREX was almost completely colocalized with mRFP-ARA7 (Figure 5B). GFP-EREX was not colocalized with a TGN marker, VHA-a1-mRFP, whereas GFP-EREX and VHA-a1-mRFP were occasionally located in close proximity to each other (Figure 5C). These results indicate that EREX localized to multivesicular endosomes bearing ARA7 in planta in a manner that was consistent with the results from transient expression in protoplasts.
Figure 5.
Localization of EREX in Planta.
(A) GFP-EREX (green) was localized to punctate compartments in epidermis cells in the root tip (left panel) and root elongation zone (middle panel) in 3-d-old erex erel1 seedlings expressing GFP-EREX. Similar localization patterns were also observed in embryonic cotyledons (right panel). Autofluorescence from plastids is shown in magenta.
(B) GFP-EREX was colocalized with mRFP-ARA7 in 3-d-old root epidermis cells.
(C) GFP-EREX and VHA-a1-mRFP exhibited distinct localization in the root epidermal cells of 3-d-old plants.
(D) The 3-d-old GFP-EREX plants were treated with DMSO or 33 μM wortmannin for 100 min. GFP-EREX signals were dispersed in the cytosol after wortmannin treatment. Bars in (A) to (D) = 10 μm.
(E) EREX protein lipid binding assay. The truncated version of EREX (EREX65-273) comprising the PX domain and ARA7 binding activity was used for this assay. Purified GST or GST-tagged EREX65-273 was incubated with the lipid-spotted membrane, and bound protein was detected with anti-GST antibody.
EREX harbors a PX domain, which is known to bind phosphatidylinositol 3′-monophosphate [PtdIns(3)P] with a few exceptions (van Leeuwen et al., 2004). To examine the requirement of phosphoinositide binding for the endosomal localization of EREX, we treated transgenic plants expressing GFP-EREX under the control of its own promoters with wortmannin, an inhibitor of phosphoinositide 3- and 4-kinases in plants (Matsuoka et al., 1995). As shown in Figure 5D, GFP-EREX was dispersed into the cytosol upon wortmannin treatment. This result suggests that EREX binds PtdIns(3)P, as reported for PX domain-containing proteins in other organisms, which was further supported by a lipid binding assay. We performed a protein-lipid blot assay using membrane-immobilized phosphoinositides and a truncated version of EREX (GST-EREX65-273) and detected specific binding of purified GST-EREX65-273 to PtdIns(3)P (Figure 5E). These results indicate that PtdIns(3)P binding via the PX domain is required for the endosomal localization of EREX.
RAB5 Acts Upstream of EREX
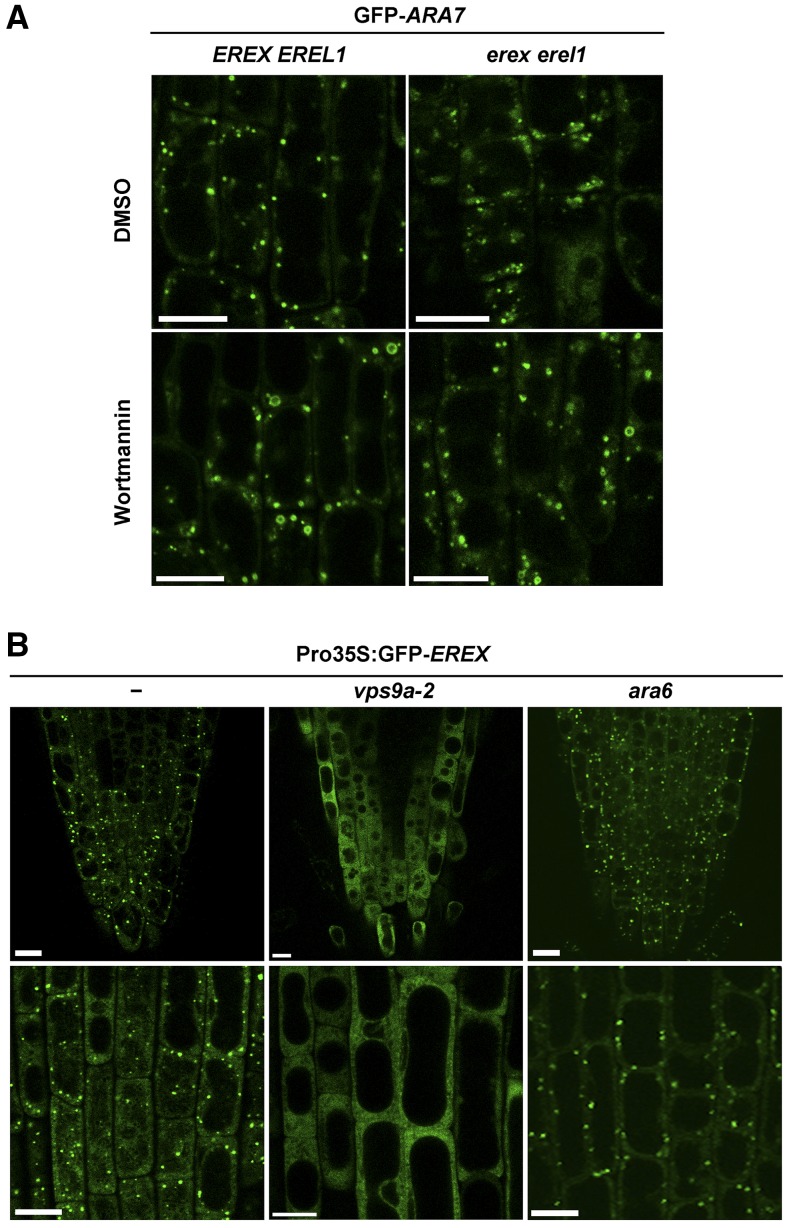
After wortmannin treatment, although EREX was dispersed into the cytosol, ARA7 was retained on endosomes, which had dilated morphology (Figure 6A), as previously reported (Jaillais et al., 2008; Ebine et al., 2011). This result suggests that the recruitment of EREX onto endosomes is not required for the endosomal localization of RAB5. We further confirmed this by examining the localization of GFP-ARA7 in the erex erel1 double mutant; endosomal localization of GFP-ARA7 was observed in erex erel1 in both the presence and absence of wortmannin (Figure 6A). Conversely, we then examined the requirement of RAB5 activity for the endosomal localization of EREX by expressing GFP-EREX under the control of the 35S promoter in the vps9a-2 mutant, which is partially defective in activating RAB5 members. Intriguingly, GFP-EREX was dispersed into the cytosol without localizing to endosomes (Figure 6B). We confirmed that the expression levels of GFP-EREX were comparable in the transgenic plants used in this experiment (Supplemental Figure 4A). We also examined GFP-EREX localization in the loss-of-function mutant of ARA6, which is also activated by VPS9a. We did not observe alterations in the subcellular localization of GFP-EREX in the ara6 mutant (Figure 6B), which indicates that the endosomal localization of GFP-EREX requires the activation of canonical RAB5 but not plant-specific ARA6. We also noticed that the expression of EREX in vps9a-2 mutant plants slightly but significantly stunted the growth of the plant, while overexpression of EREX did not alter the growth of wild-type plants (Supplemental Figures 4B and 4C). This result further suggests a functional interaction between EREX and canonical RAB5.
Figure 6.
EREX Functions Downstream of Canonical RAB5.
(A) Four-day-old erex erel1 and EREX EREL1 plants were treated with DMSO or 33 μM wortmannin for 100 min. The ara7 rha1 mutations and the rha1 mutation were also introduced into EREX EREL1 and erex erel1, respectively, to avoid ARA7 overexpression.
(B) Subcellular localization of GFP-EREX in root cells of 9-d-old wild-type, vps9a-2, or ara6 plants. Lower panels are magnified images of the other root epidermal cells in the same plant as those shown in the upper panels. Bars = 10 μm.
EREX and EREL1 Are Required for the Vacuolar Transport of Storage Proteins
If EREX actually acts as an effector of RAB5, we expect that EREX would be involved in endocytic and/or biosynthetic trafficking to vacuoles. We tested this by monitoring the endocytic transport of the dye FM4-64, a marker of endocytosis, and PIN2-GFP, an auxin transporter fused to GFP, in root epidermal cells of 3-d-old wild-type and erex erel1 plants, which exhibited no discernable difference (Supplemental Figure 5).
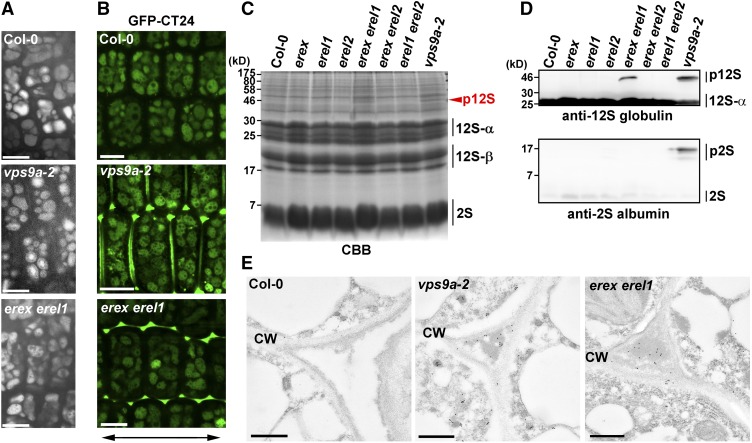
Given the high expression levels of EREX in developing embryos and young seedlings and the mutant phenotype observed at early developmental stages, it is highly likely that EREX plays crucial roles in vacuolar biogenesis and/or transport during embryogenesis and germination stages. We thus investigated the trafficking of storage proteins to protein storage vacuoles (PSVs) in embryos. The morphology of PSVs in mature erex erel1 seeds, which was visualized by their autofluorescence, was not markedly affected when compared with the PSVs of wild-type and vps9a-2 seeds (Figure 7A). We then monitored trafficking of an artificial cargo, GFP-CT24, which consists of a signal peptide, GFP, and 24 C-terminal amino acids of the α’-subunit of β-conglycinin (Nishizawa et al., 2003) to PSVs. In wild-type embryos, GFP-CT24 was observed in PSVs (Figure 7B). Conversely, GFP-CT24 was mis-secreted to extracellular spaces in the erex erel1 and vps9a-2 mutants (Figure 7B), indicating that the erex erel1 mutations impair the trafficking pathway to PSVs. We then investigated the native cargos for PSVs, 12S globulin and 2S albumin, which are major storage proteins in Arabidopsis seeds whose precursors are processed by vacuolar processing enzymes into mature forms in PSVs (Shimada et al., 2003a). We conducted SDS-PAGE of seed proteins prepared from mature seeds collected from wild-type, erex, and/or erel mutant and vps9a-2 mutant plants, which was followed by Coomassie blue staining or immunoblotting with anti-12S globulin or anti-2S albumin antibody. Wild-type seeds accumulated only mature forms of 12S globulin (α- and β-subunits) and 2S albumin (Figures 7C and 7D). In contrast, vps9a-2 mutant seeds accumulated precursors of both 12S globulin and 2S albumin (Figures 7C and 7D). Intriguingly, erex erel1 double mutant seeds accumulated precursors of 12S globulin, whereas substantial accumulation of 2S albumin precursors was not detected (Figures 7C and 7D). The mis-secretion of 12S globulin to the extracellular space in vps9a-2 and erex erel1 mutant seeds was also confirmed by immunoelectron microscopy with the anti-12S globulin antibody (Figure 7E). Precursor accumulation was not detected in seeds of erex and erel single mutants, or erex erel2 and erel1 erel2 double mutants. These results indicate that EREX and EREL1 are indeed involved in the transport of 12S globulin to PSVs, which is also known to involve RAB5.
Figure 7.
EREX Is Required for Transport to the PSV.
(A) The morphology of PSVs in wild-type (Col-0), vps9a-2, and erex erel1 embryos was visualized by autofluorescence. Embryonic hypocotyl cells were observed. Bars = 10 μm.
(B) GFP-CT24 localization was observed in wild-type, vps9a-2, and erex erel1 embryonic hypocotyl cells. The double-headed arrow indicates the apical-basal axis. Bars = 10 μm.
(C) Profiles of Coomassie blue-stained proteins were prepared from 40 grains of dry seeds with the indicated mutations. The arrowhead indicates precursors of 12S globulin.
(D) Immunoblot analysis of proteins from dry seeds from the indicated genotypes was performed using anti-12S globulin (upper panel) and anti-2S albumin (lower panel) antibodies. p12S, precursors of 12S globulin; p2S, precursors of 2S albumin.
(E) Immunoelectron micrographs of green Col-0, vps9a-2, and erex erel1 embryos prior to desiccation with the anti-12S globulin antibody. CW, cell wall. Bars = 500 nm.
DISCUSSION
Redundant Functions of EREX and EREL1 and 2
Two structurally related proteins, EREX and EREL1, exhibited distinct interaction patterns with canonical RAB5 in the yeast two-hybrid assay, in which EREX interacted with ARA7 and RHA1, whereas EREL1 was not shown to interact with these RAB5 members. However, the genetic interaction between EREX and EREL1 strongly suggested that EREX and EREL1 act in the same trafficking and developmental events. The erex erel1 double mutant exhibited deleterious phenotypes in the vacuolar transport of storage proteins and early developmental processes, whereas the single mutant of each gene was indistinguishable from the wild-type plant. Furthermore, erex and vps9a-2, as well as erel1 and vps9a-2, exhibited synthetic lethality (i.e., lethality only in combination). These lines of evidence strongly suggest that EREL1 coordinates with EREX to play a pivotal role in RAB5-mediated endosomal trafficking events. Another EREX-like protein, EREL2, also appears to possess at least a partially redundant function with EREX and EREL1. Although the interaction between EREL2 and canonical RAB5 was not detected in the yeast two-hybrid assay, and the loss of function of EREL2 did not enhance the phenotypes of the erex, erel1, and vps9a-2 mutations, we failed to recover the erex erel1 erel2 triple mutant. The synergistic enhancement of the growth defect of erex erel1 by erel2 suggested that EREL2 harbors a redundant function with EREX and EREL1, although its significance in endosomal/vacuolar trafficking remains unclear. The weak effect of the erel2 mutation compared with erel1 may reflect the slightly lower expression level of EREL2 than that of EREL1, which could be determined by examining public databases such as Arabidopsis eFP Browser (Winter et al., 2007).
Structural Features of EREX/EREL Members
The PX domain is a type of phosphoinositide binding module that preferentially binds PtdIns(3)P (Ellson et al., 2002). Eleven PX domain-containing proteins are encoded in the Arabidopsis genome and are classified into four subgroups based on the similarity of their PX domains (van Leeuwen et al., 2004). EREX and EREL1 and 2 constitute one of these subgroups. PtdIns(3)P is enriched in the multivesicular endosomal membrane in plants, to which RAB5 is also localized (Gillooly et al., 2000; Vermeer et al., 2006; Simon et al., 2014). In many cases, the affinity of the PX domain for PtdIns(3)P is not strong enough to direct proteins to the membrane, and cooperation with other interactors is needed for PX-containing proteins to bind with membranes (Lemmon, 2008). Consistently, in this study, the coexpression of ARA7 dramatically enhanced the endosomal localization of EREX. Meanwhile, wortmannin treatment resulted in the dissociation of EREX from the endosomal membrane. These results indicate that EREX must interact with both canonical RAB5 and the phosphoinositide to be efficiently recruited to the endosomal membrane. Although phosphoinositide metabolism on the endosomes has not been directly linked to RAB5 in plants, our results suggest that canonical RAB5 functions in close association with phosphoinositides, as is the case for other plant RAB GTPases and animal RAB5 (Grosshans et al., 2006; Preuss et al., 2006; Camacho et al., 2009; Antignani et al., 2015).
The coiled-coil region, which is known as a protein-protein interaction motif, was predicted to be present in the C termini of EREX and EREL members by the COILS program (Lupas et al., 1991). Because this motif is not required for the interaction between EREX and canonical RAB5, EREX may interact with some other molecules via the coiled-coil domain. Identifying and characterizing such molecules represents a promising approach for revealing the molecular mechanism of EREX-mediated trafficking to the PSV.
How Does EREX Mediate Vacuolar Trafficking with RAB5?
In this study, we focused on trafficking to PSVs because EREX was expressed abundantly in embryos and young seedlings. The phenotype of the erex erel1 double mutant, which consists of severe growth retardation only during germination and the seedling stage, also suggests that EREX and EREL1 predominantly function during embryogenesis and/or early development. This mutant phenotype might be accounted for by the presence of inappropriately deposited storage proteins in embryos, although it is also possible that this phenotype could be attributable to impairments in other endosomal/vacuolar functions specifically required in young seedlings. It should be noted that RAB5 members are constitutively expressed throughout development in Arabidopsis. Moreover, the vps9a-2 mutant exhibits pleiotropic abnormalities that are not restricted to the juvenile stage. These lines of evidence suggest that distinct effector sets are recruited to the vacuolar trafficking pathways during different developmental stages. EREX and EREL members might be tailored for the RAB5-mediated trafficking pathway to PSVs, and distinct effector sets are employed in trafficking pathways to lytic vacuoles. Ectopically expressed GFP-EREX had a negative effect on the growth of vps9a-2. This effect might be exerted by titrating canonical RAB5 by EREX, which could result in hindered interactions between RAB5 and effectors mediating lytic vacuolar trafficking.
GFP-CT24 and 12S globulin were mis-secreted in erex erel1, which helps confirm that EREX and EREL1 play critical roles in vacuolar trafficking in embryos in a manner consistent with the expected role of EREX as a RAB5 effector. Many mutants with similar phenotypes have been reported thus far, including mutants of vacuolar sorting receptors, vacuolar processing enzymes, and machinery components of membrane trafficking (Shimada et al., 2003a, 2003b, 2006; Tamura et al., 2007; Yamazaki et al., 2008; Ebine et al., 2008, 2014; Takahashi et al., 2010; Contento and Bassham, 2012; Shirakawa et al., 2014; Gao et al., 2015; Reguera et al., 2015; Teh et al., 2015). In these mutants, the accumulation of unprocessed precursors is also observed for 2S albumin, a major storage protein in Arabidopsis seeds. vps9a-2, a mutant defective in the activation of RAB5, was also shown to accumulate precursors of both 12S globulin and 2S albumin. However, we did not detect accumulation of precursors of 2S albumin in the erex erel1 mutant. This phenotypic difference suggests that EREX and EREL1 are required in a specific trafficking pathway among multiple RAB5-regulated vacuolar trafficking pathways. In the experiment presented in Figure 7B, GFP-CT24 accumulation in the horizontal sides of the cells was generally observed in vps9a-2 seeds; however, this accumulation pattern was not frequently observed in erex erel1 mutant seeds. This difference could also reflect distinct effects of the erex erel1 mutation on multiple vacuolar trafficking pathways in developing seeds, although it is also possible that these different patterns are due to the higher severity of the vps9a-2 mutation in the transport of GFP-CT24 compared with the erex erel1 mutation. Vacuolar storage proteins have been proposed to be transported through multiple trafficking pathways to PSVs (Jolliffe et al., 2005; Vitale and Hinz, 2005). Moreover, multiple trafficking pathways involving RAB5 have been proposed to operate in vacuolar trafficking in Arabidopsis (Ebine et al., 2014). One of these trafficking pathways, which is responsible for the transport of a subpopulation of 12S globulin, might specifically involve EREX and EREL1. In some mutants defective in transport between the ER and Golgi, 12S globulin and 2S albumin are reported to accumulate in different regions of the Maigo body, an abnormal compartment derived from the ER (Li et al., 2006; Takahashi et al., 2010). Although it is not clear whether the precursors of these storage proteins are segregated in the ER in wild-type plants, some populations of 12S globulin might be transported from the ER to the vacuole via a distinct pathway from 2S albumin in an EREX- and EREL1-dependent manner. Alternatively, canonical RAB5 may regulate sorting for PSVs on the TGN, which is similar to reports in rice (Oryza sativa) endosperm (Ren et al., 2014; Wen et al., 2015). A subpopulation of canonical RAB5 has been localized to the TGN, which is likely responsible for maturation from the TGN to the multivesicular endosomes (Stierhof and El Kasmi, 2010; Scheuring et al., 2011; Singh et al., 2014). EREX might coordinate cargo sorting and RAB5 activity during this event, which is consistent with the findings that the mutations in AP-4 and sorting nexins that are involved in vacuolar protein transport at the TGN and RAB5-positive endosomes, respectively, exert similar effects on transport of 12S globulin and 2S albumin to the erex erel1 mutation (Pourcher et al., 2010; Fuji et al., 2016).
As described above, we demonstrated that EREX, the plant-specific PX domain-containing protein, regulates specific aspects of vacuolar transport as an effector of canonical RAB5. This may also be true for EREL members. EREX mediates vacuolar transport of distinctive proteins to PSVs, which is indispensable for the proper development of Arabidopsis at the early developmental stages. Importantly, our findings indicate that a specific set of RAB5 effectors is responsible for a developmental stage-specific or cargo-specific endosomal/vacuolar trafficking pathway. This result suggests that plant RAB5 exerts its wide range of functions throughout the plant’s life cycle via interactions with various effector proteins specialized for trafficking pathways and/or vacuolar functions. Further experiments aimed at isolating other effector proteins and functional analyses of these molecules are needed to unravel the complete function of RAB5 GTPases in plants.
METHODS
Yeast Two-Hybrid Assay
cDNA fragments for wild-type and mutant versions of RAB proteins without isoprenylation sites were subcloned into the EcoRI or SalI site of the pBD-GAL4 Cam vector (Stratagene). Interacting partners of constitutively active ARA7 were screened using the yeast two-hybrid method from the cDNA library prepared from the roots of 4-d-old Arabidopsis thaliana plants and subcloned between EcoRI and XhoI sites of the pAD-GAL4-2.1 plasmid (provided by T. Demura, NAIST). Yeast AH109 cells (Clontech; trp1 leu2 his3 LYS2::GAL1UAS-GAL1TATA-HIS3) expressing constitutively active ARA7 (ARA7Q69L) fused to the Gal4 binding domain were transformed with the library, and positive clones were selected. After confirming reproducibility at least three times, false positives were eliminated by cotransformation with the empty vector for each candidate. Full-length cDNA fragments of EREX and EREL members were amplified by RT-PCR using total RNA prepared from Arabidopsis roots as a template, which were subcloned into the BamHI site of pAD-GAL4-2.1 vector (Stratagene) and used for subsequent experiments.
Purification of Bacterially Expressed Proteins
The GST-fused EREX65-273 protein and GST were expressed in the Escherichia coli Rosetta (DE3) strain (Merck) using the pGEX4T-1 vector (GE Healthcare). Cells expressing these proteins were suspended in lysis buffer (1× PBS, pH 7.4, 1% Triton X-100, 1 mg/mL Lysozyme [Wako], 0.5 units/mL DNase I [Invitrogen], and a protease inhibitor cocktail [GE Healthcare]), sonicated, and centrifuged at 10,000g for 40 min. Supernatants were loaded onto Glutathione-Sepharose 4B columns (GE Healthcare) and washed with three column volumes of washing buffer (1× PBS, pH 7.4, and 1% Triton X-100), and the proteins were then eluted with elution buffer (20 mM reduced glutathione, 1× PBS, pH 7.4, and 1% Triton X-100). Purification of His6-tagged ARA6 and ARA7 was conducted as described previously (Ebine et al., 2014).
In Vitro Pull-Down Assay
Purified GST-EREX65-273 and RAB5-His6 were coincubated for 2 h at 4°C with Glutathione-Sepharose 4B resin (GE Healthcare) in binding buffer (1× PBS, pH 7.4, and 0.03% Tween-20 containing either 10 μM GTPγS [Figure 2A] or 25 μM GTPγS or GDP [Figure 2B]). Protein complexes bound to resins were washed three times with the same buffer, boiled in sample buffer with bromophenol blue and mercaptoethanol for 5 min at 95°C, and subjected to immunoblot analysis.
Antibodies
The anti-GST polyclonal antibody (Z-5; Santa Cruz Biotechnology), anti-His6 monoclonal antibody (04905318001; Roche), and anti-GFP monoclonal antibody (GF200; Nacalai Tesque) were commercially available. Anti-2S albumin and anti-12S globulin antibodies were supplied by I. Hara-Nishimura (Kyoto University).
Plant Materials and Plasmids
erex (SALK_008160), erel1 (SALK_114362), and erel2 (SALK_139378) mutants were obtained from the ABRC (Alonso et al., 2003) and backcrossed at least three times with wild-type Arabidopsis. Arabidopsis ecotype Columbia (Col-0) was used as the wild type in this study. Transgenic plants expressing GFP-CT24, VHA-a1-mRFP, and PIN2-GFP were kindly provided by N. Maruyama (Kyoto University), K. Schumacher (Ruprecht Karls University), and J. Friml (IST Austria), respectively. Transgenic plants expressing GFP-ARA7 or mRFP-ARA7 and the vps9a-2 mutant were retrieved from the lab stock (Goh et al., 2007; Ebine et al., 2014). For erex complementation, the cDNA for GFP was inserted after the start codon of the genomic fragment of EREX, which contained 1581 bp upstream and 701 bp downstream sequences of the coding region. This sequence was subcloned into the binary vector pGWB1 provided by T. Nakagawa (Shimane University) (Nakagawa et al., 2007), and the resulting plasmid was used to transform the erex erel1 double mutant. For EREL1, the genomic fragment containing 1401 bp upstream and 879 bp downstream sequences of the coding region of EREL1 was subcloned into pGWB1 and used for transformation. Primer sequences used for amplification of genomic fragments are listed in Supplemental Table 6. The translational fusion between GFP and EREX was generated by fluorescently tagging full-length proteins (Tian et al., 2004). For the GUS-fusion expression analysis, the genomic fragment containing the promoter region used in the complementation experiment and the first exon of EREX were subcloned in-frame into pGWB3 (from T. Nakagawa; Nakagawa et al., 2007) and used to transform wild-type Arabidopsis. We cloned EREX cDNA into pGWB6 (from T. Nakagawa; Nakagawa et al., 2007) containing the 35S promoter for overexpression of EREX. Transformation of Arabidopsis plants was performed via floral dipping using Agrobacterium tumefaciens (strain GV3101::pMP90; Clough and Bent, 1998). Arabidopsis seeds were surface sterilized and sown onto Murashige-Skoog plates supplemented with Gamborg’s vitamins (Sigma-Aldrich), 30 mM sucrose, and 0.7% gellan gum (Wako). After a 3-d incubation period at 4°C to break seed dormancy, Arabidopsis plants were grown at 23°C under continuous white light with a fluorescent lamp (FL40SW; Toshiba) and transferred to soil after 2 to 3 weeks.
GUS Staining
Developed embryos, 3-d-old seedlings, and 15-d-old seedlings were soaked in staining solution (100 mM sodium phosphate buffer, pH 7.2, 10 mM EDTA, 0.1% Triton X-100, and 0.5 mg/mL X-glucuronide) and decompressed for 15 min, followed by incubation at 37°C for 15 h. Samples were incubated in 70% ethanol at 37°C for 2 h, transferred to fresh 70% ethanol and incubated for 1 to 2 weeks at 4°C, and then mounted in a clearing solution (8 g of chloral hydrate in 1 mL of glycerol and 2 mL of water) on glass microscope slides.
RT-PCR
Total RNA was extracted using the RNAqueous-4PCR kit (Ambion) and treated with DNase I (Invitrogen) to reduce genomic DNA contamination. cDNA was synthesized by SuperScript III reverse transcriptase (Invitrogen) with an oligo(dT) 18 primer. The primer sets used for PCR are listed in Supplemental Table 6. The PCR conditions were as follows: 95°C (2 min); 35 cycles of 95°C (30 s), 55°C (30 s), 72°C (90 s), and 72°C (10 min).
Light Microscopy
Transient expression in protoplasts was conducted as described previously (Ueda et al., 2001). For single-color imaging in root cells, GFP was excited using a diode-pumped solid-state laser (Cobolt Blues) at 473 nm and observed under a microscope (BX51; Olympus) equipped with a confocal scanner unit (CSU10; Yokogawa Electric) and a cooled CCD camera (ORCA-AG; Hamamatsu Photonics). Multicolor observation was performed using an LSM510, LSM710, or LSM780 confocal microscope (Carl Zeiss). The morphology of PSVs was observed as described previously (Ebine et al., 2008). Imaging of GUS-stained plants was conducted using a stereomicroscope (MZ16FA; Leica) and a light microscope (BX60; Olympus) equipped with a CCD camera (VB-7010 [Keyence] and DP73 [Olympus], respectively). Samples were treated with 33 μM wortmannin (Sigma-Aldrich) for phosphoinositide kinase inhibition experiments and 16 μM FM4-64 (Sigma-Aldrich) for tracing endocytosis. mRFP-ARA7, VHA-a1-mRFP, GFP-ARA7, and PIN2-GFP were observed in the root epidermal cells of 3- to 5-d-old plants. For observation of embryos, seedpods were cleared according to previously published methods (Aida et al., 1997). vps9a-2 erex, vps9a-2 erel1, and erex erel1 erel2 embryos were observed with either a microscope (BX52; Olympus) equipped with a confocal scanner (CSU10; Yokogawa Electric) and camera (ORCA-ER; Hamamatsu Photonics) or a microscope (DM4500B; Leica) equipped with a camera alone (DFC300FX; Leica).
Lipid Binding Assay
Thirteen lipid species spotted onto a membrane (Inositol Snoopers; Avanti) were incubated for 1 h at room temperature in wash buffer (20 mM Tris-HCl, pH 7.5, and 0.8% NaCl) containing 3% fatty acid-free BSA (Nacalai). Purified GST or GST-EREX65-273 was then added at a concentration of 1 μg/mL and incubated for 1 h at room temperature in wash buffer supplemented with 1% fatty acid-free BSA. The membrane was washed three times for 10 min each in wash buffer, and bound GST and GST-EREX65-273 were detected with anti-GST antibody (diluted 1000-fold).
Detection of Seed Storage Proteins
SDS-PAGE and immunoblot analyses were performed as described previously (Shimada et al., 2003a, 2003b). Blots were probed with anti-12S globulin α-subunit (anti-12S, diluted 1000-fold) and anti-2S albumin (anti-2S, diluted 1000-fold) antibodies. Signals were detected using the ECL detection system (GE Healthcare). Immunoelectron microscopy was performed as described previously (Ebine et al., 2014).
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are At3g15920 (EREX), At4g32160 (EREL1), At2g25350 (EREL2), At4g19640 (ARA7/RABF2b), At5g45130 (RHA1/RABF2a), At3g54840 (ARA6/RABF1), At3g19770 (VPS9a), At1g22740 (RAB75/RABG3b), At4g18430 (RABA1e), At5g57090 (PIN2), At3g54300 (VAMP727), and At2g28520 (VHA-a1).
Supplemental Data
Supplemental Figure 1. Colocalization of EREX and EREL with RAB5/RABF and RAB7/RABG3b.
Supplemental Figure 2. Characterization of the erex and erel mutants.
Supplemental Figure 3. Promoter-reporter assay of EREX.
Supplemental Figure 4. Overexpression of GFP-EREX in vps9a-2.
Supplemental Figure 5. The erex erel1 mutations did not affect the endocytic pathway.
Supplemental Table 1. Segregation analysis of the vps9a-2+/−erex−/− mutant.
Supplemental Table 2. Segregation analysis of the vps9a-2+/−erel1−/− mutant.
Supplemental Table 3. Cross-pollination analysis of the vps9a-2+/−erex−/− mutant.
Supplemental Table 4. Segregation analysis of the erex+/− erel1−/− erel2−/− mutant.
Supplemental Table 5. Cross-pollination analysis of the erex+/−erel1−/−erel2−/− mutant.
Supplemental Table 6. Primer sequences used in this study.
Acknowledgments
We thank the Salk Institute for providing the Arabidopsis T-DNA insertion mutants. We also thank T. Demura, T. Nakagawa, N. Maruyama, K. Schumacher, and I. Hara-Nishimura for sharing materials, H. Tsukaya for support in microscopy observation, E. Furuyama for backcrossing, cross-pollination, and generating multiple mutants of erex and erel members, and lab members for critical reading of the manuscript. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (A.N. and T.U.) and by a Grant-in-Aid for JSPS Fellows (T.I. and H.T.S.). This research was also supported by JST, PRESTO (T.U.).
AUTHOR CONTRIBUTIONS
H.T.S. performed almost all experiments presented in this article and wrote the manuscript. T.I. prepared His6-tagged RAB5s, which was essential for the pull down-assay. T.U. designed the research and wrote the manuscript, and A.N. supervised the study.
Glossary
- GEF
guanine nucleotide exchange factor
- TGN
trans-Golgi network
- PSV
protein storage vacuole
References
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Antignani V., Klocko A.L., Bak G., Chandrasekaran S.D., Dunivin T., Nielsen E. (2015). Recruitment of PLANT U-BOX13 and the PI4Kβ1/β2 phosphatidylinositol-4 kinases by the small GTPase RabA4B plays important roles during salicylic acid-mediated plant defense signaling in Arabidopsis. Plant Cell 27: 243–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka R., Uemura T., Ito J., Fujimoto M., Ito E., Ueda T., Nakano A. (2013). Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. Plant J. 73: 240–249. [DOI] [PubMed] [Google Scholar]
- Beck M., Zhou J., Faulkner C., MacLean D., Robatzek S. (2012). Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24: 4205–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A. (2004). Endocytosis: signaling from endocytic membranes to the nucleus. Curr. Biol. 14: R314–R316. [DOI] [PubMed] [Google Scholar]
- Berson T., von Wangenheim D., Takáč T., Šamajová O., Rosero A., Ovečka M., Komis G., Stelzer E.H., Šamaj J. (2014). Trans-Golgi network localized small GTPase RabA1d is involved in cell plate formation and oscillatory root hair growth. BMC Plant Biol. 14: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottanelli F., Gershlick D.C., Denecke J. (2012). Evidence for sequential action of Rab5 and Rab7 GTPases in prevacuolar organelle partitioning. Traffic 13: 338–354. [DOI] [PubMed] [Google Scholar]
- Bottanelli F., Foresti O., Hanton S., Denecke J. (2011). Vacuolar transport in tobacco leaf epidermis cells involves a single route for soluble cargo and multiple routes for membrane cargo. Plant Cell 23: 3007–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho L., Smertenko A.P., Pérez-Gómez J., Hussey P.J., Moore I. (2009). Arabidopsis Rab-E GTPases exhibit a novel interaction with a plasma-membrane phosphatidylinositol-4-phosphate 5-kinase. J. Cell Sci. 122: 4383–4392. [DOI] [PubMed] [Google Scholar]
- Choi S.W., Tamaki T., Ebine K., Uemura T., Ueda T., Nakano A. (2013). RABA members act in distinct steps of subcellular trafficking of the FLAGELLIN SENSING2 receptor. Plant Cell 25: 1174–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C.M., Neto H., Foucart C., Moore I. (2008). Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20: 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S., Zerial M. (2000). Purification and identification of novel Rab effectors using affinity chromatography. Methods 20: 403–410. [DOI] [PubMed] [Google Scholar]
- Christoforidis S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S.C., Waterfield M.D., Backer J.M., Zerial M. (1999). Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1: 249–252. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Contento A.L., Bassham D.C. (2012). Structure and function of endosomes in plant cells. J. Cell Sci. 125: 3511–3518. [DOI] [PubMed] [Google Scholar]
- Cui Y., Zhao Q., Gao C., Ding Y., Zeng Y., Ueda T., Nakano A., Jiang L. (2014). Activation of the Rab7 GTPase by the MON1–CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell 26: 2080–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Baluška F., Schlicht M., Hlavacka A., Šamaj J., Friml J., Gadella T.W.J. Jr. (2006). Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev. Cell 10: 137–150. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P. (2009). Cell polarity in plants: Linking PIN polarity generation mechanisms to morphogenic auxin gradients. Commun. Integr. Biol. 2: 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., Ueda T. (2009). Unique mechanism of plant endocytic/vacuolar transport pathways. J. Plant Res. 122: 21–30. [DOI] [PubMed] [Google Scholar]
- Ebine K., Inoue T., Ito J., Ito E., Uemura T., Goh T., Abe H., Sato K., Nakano A., Ueda T. (2014). Plant vacuolar trafficking occurs through distinctly regulated pathways. Curr. Biol. 24: 1375–1382. [DOI] [PubMed] [Google Scholar]
- Ebine K., Okatani Y., Uemura T., Goh T., Shoda K., Niihama M., Morita M.T., Spitzer C., Otegui M.S., Nakano A., Ueda T. (2008). A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., et al. (2011). A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13: 853–859. [DOI] [PubMed] [Google Scholar]
- Ellson C.D., Andrews S., Stephens L.R., Hawkins P.T. (2002). The PX domain: a new phosphoinositide-binding module. J. Cell Sci. 115: 1099–1105. [DOI] [PubMed] [Google Scholar]
- Feraru E., Feraru M.I., Asaoka R., Paciorek T., De Rycke R., Tanaka H., Nakano A., Friml J. (2012). BEX5/RabA1b regulates trans-Golgi network-to-plasma membrane protein trafficking in Arabidopsis. Plant Cell 24: 3074–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J. (2010). Subcellular trafficking of PIN auxin efflux carriers in auxin transport. Eur. J. Cell Biol. 89: 231–235. [DOI] [PubMed] [Google Scholar]
- Fuji K., Shirakawa M., Shimono Y., Kunieda T., Fukao Y., Koumoto Y., Takahashi H., Hara-Nishimura I., Shimada T. (2016). The Adaptor Complex AP-4 Regulates Vacuolar Protein Sorting at the trans-Golgi Network by Interacting with VACUOLAR SORTING RECEPTOR1. Plant Physiol. 170: 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez T., Gilleron J., Zerial M., O’Sullivan G.A. (2012). SnapShot: Mammalian Rab proteins in endocytic trafficking. Cell 151: 234–234.e2. [DOI] [PubMed] [Google Scholar]
- Gao C., Zhuang X., Cui Y., Fu X., He Y., Zhao Q., Zeng Y., Shen J., Luo M., Jiang L. (2015). Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. of Sci. USA 112: 1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., Stenmark H. (2000). Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19: 4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T., Uchida W., Arakawa S., Ito E., Dainobu T., Ebine K., Takeuchi M., Sato K., Ueda T., Nakano A. (2007). VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 19: 3504–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody R.S., Rak A., Alexandrov K. (2005). The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell. Mol. Life Sci. 62: 1657–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B.D., Donaldson J.G. (2009). Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10: 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans B.L., Ortiz D., Novick P. (2006). Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA 103: 11821–11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas T.J., Sliwinski M.K., Martínez D.E., Preuss M., Ebine K., Ueda T., Nielsen E., Odorizzi G., Otegui M.S. (2007). The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19: 1295–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miège C., Gaude T. (2008). Evidence for a sorting endosome in Arabidopsis root cells. Plant J. 53: 237–247. [DOI] [PubMed] [Google Scholar]
- Jolliffe N.A., Craddock C.P., Frigerio L. (2005). Pathways for protein transport to seed storage vacuoles. Biochem. Soc. Trans. 33: 1016–1018. [DOI] [PubMed] [Google Scholar]
- Kang B.H., Nielsen E., Preuss M.L., Mastronarde D., Staehelin L.A. (2011). Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329. [DOI] [PubMed] [Google Scholar]
- Kirchhelle C., Chow C.M., Foucart C., Neto H., Stierhof Y.D., Kalde M., Walton C., Fricker M., Smith R.S., Jérusalem A., Irani N., Moore I. (2016). The specification of geometric edges by a plant Rab GTPase is an essential cell-patterning principle during organogenesis in Arabidopsis. Dev. Cell 36: 386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzer A.M., Brandizzi F., Neumann U., Paris N., Moore I., Hawes C. (2004). AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J. Cell Sci. 117: 6377–6389. [DOI] [PubMed] [Google Scholar]
- Lam S.K., Siu C.L., Hillmer S., Jang S., An G., Robinson D.G., Jiang L. (2007). Rice SCAMP1 defines clathrin-coated, trans-Golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M.A. (2008). Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9: 99–111. [DOI] [PubMed] [Google Scholar]
- Li L., Shimada T., Takahashi H., Ueda H., Fukao Y., Kondo M., Nishimura M., Hara-Nishimura I. (2006). MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana. Plant Cell 18: 3535–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. (1991). Predicting coiled coils from protein sequences. Science 252: 1162–1164. [DOI] [PubMed] [Google Scholar]
- Lycett G. (2008). The role of Rab GTPases in cell wall metabolism. J. Exp. Bot. 59: 4061–4074. [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Bassham D.C., Raikhel N.V., Nakamura K. (1995). Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J. Cell Biol. 130: 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaczynska M., Christoforidis S., Giner A., Shevchenko A., Uttenweiler-Joseph S., Habermann B., Wilm M., Parton R.G., Zerial M. (2004). APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 116: 445–456. [DOI] [PubMed] [Google Scholar]
- Miao Y., Li K.Y., Li H.Y., Yao X., Jiang L. (2008). The vacuolar transport of aleurain-GFP and 2S albumin-GFP fusions is mediated by the same pre-vacuolar compartments in tobacco BY-2 and Arabidopsis suspension cultured cells. Plant J. 56: 824–839. [DOI] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E., Rivera-Molina F., Novick P. (2012). GTPase networks in membrane traffic. Annu. Rev. Biochem. 81: 637–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nielsen E., Cheung A.Y., Ueda T. (2008). The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol. 147: 1516–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Christoforidis S., Uttenweiler-Joseph S., Miaczynska M., Dewitte F., Wilm M., Hoflack B., Zerial M. (2000). Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 151: 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemes S., Langhans M., Viotti C., Scheuring D., San Wan Yan M., Jiang L., Hillmer S., Robinson D.G., Pimpl P. (2010). Retromer recycles vacuolar sorting receptors from the trans-Golgi network. Plant J. 61: 107–121. [DOI] [PubMed] [Google Scholar]
- Nishizawa K., Maruyama N., Satoh R., Fuchikami Y., Higasa T., Utsumi S. (2003). A C-terminal sequence of soybean beta-conglycinin alpha’ subunit acts as a vacuolar sorting determinant in seed cells. Plant J. 34: 647–659. [DOI] [PubMed] [Google Scholar]
- Pedrazzini E., Komarova N.Y., Rentsch D., Vitale A. (2013). Traffic routes and signals for the tonoplast. Traffic 14: 622–628. [DOI] [PubMed] [Google Scholar]
- Pourcher M., Santambrogio M., Thazar N., Thierry A.M., Fobis-Loisy I., Miège C., Jaillais Y., Gaude T. (2010). Analyses of sorting nexins reveal distinct retromer-subcomplex functions in development and protein sorting in Arabidopsis thaliana. Plant Cell 22: 3980–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M.L., Schmitz A.J., Thole J.M., Bonner H.K., Otegui M.S., Nielsen E. (2006). A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. J. Cell Biol. 172: 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M.L., Serna J., Falbel T.G., Bednarek S.Y., Nielsen E. (2004). The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16: 1589–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera M., Bassil E., Tajima H., Wimmer M., Chanoca A., Otegui M.S., Paris N., Blumwald E. (2015). pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell 27: 1200–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., et al. (2014). GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell 26: 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes F.C., Buono R., Otegui M.S. (2011). Plant endosomal trafficking pathways. Curr. Opin. Plant Biol. 14: 666–673. [DOI] [PubMed] [Google Scholar]
- Saito C.and Ueda T. (2009). Functions of RAB and SNARE proteins in plant life. Int. Rev. Cell Mol. Biol. 274: 183–233. [DOI] [PubMed] [Google Scholar]
- Scheuring D., Viotti C., Krüger F., Künzl F., Sturm S., Bubeck J., Hillmer S., Frigerio L., Robinson D.G., Pimpl P., Schumacher K. (2011). Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnatwinkel C., Christoforidis S., Lindsay M.R., Uttenweiler-Joseph S., Wilm M., Parton R.G., Zerial M. (2004). The Rab5 effector Rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2: E261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabra M.C., Wasmeier C. (2004). Controlling the location and activation of Rab GTPases. Curr. Opin. Cell Biol. 16: 451–457. [DOI] [PubMed] [Google Scholar]
- Shimada T., et al. (2003a). Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J. Biol. Chem. 278: 32292–32299. [DOI] [PubMed] [Google Scholar]
- Shimada T., Koumoto Y., Li L., Yamazaki M., Kondo M., Nishimura M., Hara-Nishimura I. (2006). AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47: 1187–1194. [DOI] [PubMed] [Google Scholar]
- Shimada T., Fuji K., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2003b). Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 16095–16100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.W., et al. (2005). An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170: 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M.L., Platre M.P., Assil S., van Wijk R., Chen W.Y., Chory J., Dreux M., Munnik T., Jaillais Y. (2014). A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J. 77: 322–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Lippé R., Christoforidis S., Gaullier J.M., Brech A., Callaghan J., Toh B.H., Murphy C., Zerial M., Stenmark H. (1998). EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394: 494–498. [DOI] [PubMed] [Google Scholar]
- Singh M.K., Krüger F., Beckmann H., Brumm S., Vermeer J.E.M., Munnik T., Mayer U., Stierhof Y.-D., Grefen C., Schumacher K., Jürgens G. (2014). Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr. Biol. 24: 1383–1389. [DOI] [PubMed] [Google Scholar]
- Shirakawa M., Ueda H., Koumoto Y., Fuji K., Nishiyama C., Kohchi T., Hara-Nishimura I., Shimada T. (2014). CONTINUOUS VASCULAR RING (COV1) is a trans-Golgi network-localized membrane protein required for Golgi morphology and vacuolar protein sorting. Plant Cell Physiol. 55: 764–772. [DOI] [PubMed] [Google Scholar]
- Sohn E.J., Kim E.S., Zhao M., Kim S.J., Kim H., Kim Y.W., Lee Y.J., Hillmer S., Sohn U., Jiang L., Hwang I. (2003). Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15: 1057–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somsel Rodman J., Wandinger-Ness A. (2000). Rab GTPases coordinate endocytosis. J. Cell Sci. 113: 183–192. [DOI] [PubMed] [Google Scholar]
- Stenmark H. (2009). Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10: 513–525. [DOI] [PubMed] [Google Scholar]
- Stenmark H., Vitale G., Ullrich O., Zerial M. (1995). Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell 83: 423–432. [DOI] [PubMed] [Google Scholar]
- Stierhof Y.D., El Kasmi F. (2010). Strategies to improve the antigenicity, ultrastructure preservation and visibility of trafficking compartments in Arabidopsis tissue. Eur. J. Cell Biol. 89: 285–297. [DOI] [PubMed] [Google Scholar]
- Sunada M., Goh T., Ueda T., Nakano A. (2016). Functional analyses of the plant-specific C-terminal region of VPS9a: the activating factor for RAB5 in Arabidopsis thaliana. J. Plant Res. 129: 93–102. [DOI] [PubMed] [Google Scholar]
- Szumlanski A.L., Nielsen E. (2009). The Rab GTPase RabA4d regulates pollen tube tip growth in Arabidopsis thaliana. Plant Cell 21: 526–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Tamura K., Takagi J., Koumoto Y., Hara-Nishimura I., Shimada T. (2010). MAG4/Atp115 is a golgi-localized tethering factor that mediates efficient anterograde transport in Arabidopsis. Plant Cell Physiol. 51: 1777–1787. [DOI] [PubMed] [Google Scholar]
- Tamura K., Takahashi H., Kunieda T., Fuji K., Shimada T., Hara-Nishimura I. (2007). Arabidopsis KAM2/GRV2 is required for proper endosome formation and functions in vacuolar sorting and determination of the embryo growth axis. Plant Cell 19: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh O.K., Hatsugai N., Tamura K., Fuji K., Tabata R., Yamaguchi K., Shingenobu S., Yamada M., Hasebe M., Sawa S., Shimada T., Hara-Nishimura I. (2015). BEACH-domain proteins act together in a cascade to mediate vacuolar protein trafficking and disease resistance in Arabidopsis. Mol. Plant 8: 389–398. [DOI] [PubMed] [Google Scholar]
- Tian G.W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Yamaguchi M., Uchimiya H., Nakano A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20: 4730–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Uemura T., Sato M.H., Nakano A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40: 783–789. [DOI] [PubMed] [Google Scholar]
- Uejima T., Ihara K., Goh T., Ito E., Sunada M., Ueda T., Nakano A., Wakatsuki S. (2010). GDP-bound and nucleotide-free intermediates of the guanine nucleotide exchange in the Rab5·Vps9 system. J. Biol. Chem. 285: 36689–36697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uejima T., Ihara K., Sunada M., Kawasaki M., Ueda T., Kato R., Nakano A., Wakatsuki S. (2013). Direct metal recognition by guanine nucleotide-exchange factor in the initial step of the exchange reaction. Acta Crystallogr. D Biol. Crystallogr. 69: 345–351. [DOI] [PubMed] [Google Scholar]
- van Leeuwen W., Okrész L., Bögre L., Munnik T. (2004). Learning the lipid language of plant signalling. Trends Plant Sci. 9: 378–384. [DOI] [PubMed] [Google Scholar]
- Vermeer J.E.M., van Leeuwen W., Tobeña-Santamaria R., Laxalt A.M., Jones D.R., Divecha N., Gadella T.W.J. Jr., Munnik T. (2006). Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47: 687–700. [DOI] [PubMed] [Google Scholar]
- Viotti C., et al. (2010). Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Hinz G. (2005). Sorting of proteins to storage vacuoles: how many mechanisms? Trends Plant Sci. 10: 316–323. [DOI] [PubMed] [Google Scholar]
- Wen L., Fukuda M., Sunada M., Ishino S., Ishino Y., Okita T.W., Ogawa M., Ueda T., Kumamaru T. (2015). Guanine nucleotide exchange factor 2 for Rab5 proteins coordinated with GLUP6/GEF regulates the intracellular transport of the proglutelin from the Golgi apparatus to the protein storage vacuole in rice endosperm. J. Exp. Bot. 66: 6137–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard A.A.D., Moore I. (2008). The functions of Rab GTPases in plant membrane traffic. Curr. Opin. Plant Biol. 11: 610–619. [DOI] [PubMed] [Google Scholar]
- Yamazaki M., Shimada T., Takahashi H., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2008). Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol. 49: 142–156. [DOI] [PubMed] [Google Scholar]