Abstract
Zinc finger proteins are the largest transcription factor family in human genome. The diverse combinations and functions of zinc finger motifs make zinc finger proteins versatile in biological processes, including development, differentiation, metabolism and autophagy. Over the last few decades, increasing evidence reveals the potential roles of zinc finger proteins in cancer progression. However, the underlying mechanisms of zinc finger proteins in cancer progression vary in different cancer types and even in the same cancer type under different types of stress. Here, we discuss general mechanisms of zinc finger proteins in transcription regulation and summarize recent studies on zinc finger proteins in cancer progression. In this review, we also emphasize the importance of further investigations in elucidating the underlying mechanisms of zinc finger proteins in cancer progression.
Keywords: Zinc finger protein, Transcription factor, Cancer progression
Background
Transcription factors play a central role in regulating gene expression, and therefore coordinate a plethora of biological processes, including differentiation, development, metabolism, apoptosis, autophagy and stemness maintenance [1–5]. Based on different DNA binding motifs, transcription factors can be majorly categorized into classical zinc fingers [6], homeodomains [7], and basic helix-loop-helix [8]. Among these, classical zinc finger containing proteins (ZNFs) form the largest family of sequence-specific DNA binding protein, which are encoded by 2 % of human genes [9, 10]. To date, 8 different classes of zinc finger motifs have been reported, including Cys2His2 (C2H2) like, Gag knuckle, Treble clef, Zinc ribbon, Zn2/Cys6, TAZ2 domain like, Zinc binding loops and Metallothionein [11]. Different types of zinc finger motifs show great diversity of biological functions. Notably, in addition to DNA binding, studies have recently revealed the RNA, protein and lipids interacting abilities of zinc finger motifs [12–15]. Therefore, with different combinations of multiple zinc finger motifs, ZNFs can greatly expand their diverse role in gene regulations under different cell contexts or stimuli. The general mechanism of gene regulation by ZNFs and their great variety of roles in cancer progression will be discussed in this review.
The transcription regulation of ZNFs
C2H2-type zinc finger motif is the largest group of all zinc finger motif classes. According to the InterPro database (updated on April 14th 2016), there are 5,926 members in the C2H2-type ZNF family. C2H2-type zinc finger motif is composed of CX2CX3FX5LX2HX3H, and its two cysteine and two histidine residues fold into a finger-like structure of a two-stranded antiparallel β-sheet and an α-helix after interacting with zinc ions [16, 17]. Studies have demonstrated that two to three successive C2H2-type zinc finger motifs are the most suitable unit for DNA binding [6]. In addition, GC-rich or GT-rich sequences serve as C2H2-type ZNF cis-regulatory elements. For example, CTGGCAGCGC has been revealed as SP1 consensus binding element to transcriptionally activate BRK1 expression, while (T/A)(G/A)CAGAA(T/G/C) is the consensus element for ZNF217 to suppress E-cadherin expression [18, 19].
In addition to tandem zinc finger motifs, C2H2-type ZNF also contains other functional domains, such as BTB (Broad-Complex, Tramtrack, and Bric-a-brac)/POZ (poxvirus and zinc finger), the Krüppel-associated box (KRAB), and SCAN (SRE-ZBP, CTfin51, AW-1 and Number 18 cDNA) domain. These functional domains may control subcellular localization, DNA binding and gene expression by regulating selective binding of the transcription factors with each other or with other cellular component. For instance, zinc finger protein GATA-1 has been reported to interact with different partners, including Fli-1, Sp1, EKLF and PU.1 [20–22].
ZNF proteins can carry out different functions with different partners and even elicit opposing actions on different partners. For example, physical interaction between GATA-1 and Fli-1, a member of Ets family of transcriptional activator, cooperatively activate the expression of megakaryocyte-specific genes, including GPIX and GPIbalpha, at transcriptional level [20]. In contrast, interacting with PU.1, another Ets family member, blocks GATA-1 DNA binding ability and therefore inhibits erythroid differentiation [23]. Recent study also shows that ZEB1, a transcription repressor of differentiation-associated genes, turns its function into a transcriptional co-activator of a common ZEB1/YAP target genes through interacting with YAP and therefore leading to aggressive cancer phenotype [24].
Studies have demonstrated that ZNF proteins show diverse regulation mechanisms on a wide variety of downstream genes through recruiting different chromatin modifiers. Some ZNF proteins work as transcriptional repressors by recruiting co-repressors [25–27]. For example, ZNF217 has been found to suppress downstream gene expression by interacting with co-repressors, including CoREST, lysine demethylase 1, histone deacetylase 2 and C-terminal binding protein [25]. Some ZNF proteins, on the other hand, work as transcriptional activators by interacting with co-activators, including CBP/p300 and C/EBP [28, 29]. These studies clearly indicate that trans-acting proteins play important roles in determining ZNFs as transcription activators or repressors.
Post-translational modifications on ZNFs
The post-translational modifications (PTMs) of ZNFs, especially acetylation and phosphorylation, add another layer of regulation for ZNFs in which transcription may be activated or repressed. GATA1, a transcription factor that contains 2 highly conserved zinc finger motifs, is found acetylated at the lysine residues adjacent to the C terminal zinc finger by acetyltransferase CBP and p300. Acetylation of GATA1 shows stable association with chromatin probably by facilitating protein interactions, such as bromodomain-containing protein Brd3 [30–32]. Erythroid Krüppel-like factor, also known as EKLF, is acetylated at lysine residues 288 and 302 near its zinc finger motif mediated by CBP and p300 [33]. The acetylated EKLF at lysine residue 288 can transactivate β-globin expression through recruiting the large erythroid complex (ERC-1) that contains SWI/SNF chromatin-remodeling proteins and histone 3.3 [33, 34]. Another C2H2 zinc finger protein, YY1, is acetylated by p300/CBP associated factor (PCAF) at its zinc finger motif and inhibits its DNA binding capacity. Acetylation mediated by p300 and PCAF at the central glycine-lysine rich domain of YY1, however, does not affect DNA binding affinity but fully suppresses target gene transcription [35].
Phosphorylation on serine or threonine residues of the ZNFs linker peptide has been reported [36]. ZNFs, including Ikaros, Sp1 and YY1, are found to be highly phosphorylated on threonine/serine residues of their linker peptide during mitosis and therefore abrogated their DNA binding ability [37, 38]. Rizkallah et al. generated an antibody raised against phosphorylated linker peptide TGEKP to show that about 50 % of all linkers in 80 % of C2H2-type ZNFs are phosphorylated, indicating that phosphorylation is a highly coordinated mechanism to keep ZNFs away from DNA during mitosis [39].
The oncogenic ZNFs in cancer progression
Recent studies revealed that aberrant expression of C2H2 ZNF proteins contributes to tumorigenesis in different aspects (summarized in Table 1). For example, amplification and overexpression of ZKSCAN3, also known as ZNF306 or ZNF309, was first reported in invasive colorectal cancers. The authors showed that ZKSCAN3 knockdown in colorectal cancer cells inhibited anchorage-independent growth and orthotopic tumor growth, while ZKSCAN3 overexpression exerted opposite effects [40]. To identify ZKSCAN3 downstream genes, these authors further conducted expression array and identified candidate target genes enriched in growth, cell migration, angiogenesis and proteolysis [41]. Studies confirm that ZKSCAN3 transcriptionally activates integrin β4 and vascular endothelial growth factor, which are involved in ZKSCAN3-mediated colorectal tumorigenesis [41]. In addition, ZKSCAN3 is also found to be amplified and overexpressed in multiple myeloma and prostate cancer [42, 43]. The overexpression of ZKSCAN3 enhances cell proliferation through transcriptionally activating cyclin D2 expression [42]. Interestingly, a recent study reveals a novel role of ZKSCAN3 in autophagy using cervical cancer, colon cancer, neuroblastoma, and ovarian cancer models [4]. Chauhan et al., show that ZKSCAN3 translocates into the nucleus and acts as a master transcriptional repressor of a large set of genes involved in autophagy and lysosome biogenesis, including Map1lC3b and Wipi2, under serum stimulation [4].
Table 1.
Summary of differential roles of ZNF proteins in cancer progression
| ZNFs | Aliases | Role | Cancer models | Target genes | Mechanism in tumorigenesis | References |
|---|---|---|---|---|---|---|
| ZKSCAN3 | ZNF306, ZNF309 | Oncogene | Colorectal cancer | Integrin β4 ↑, VEGF ↑ | Promotes cancer cell growth, migration, angiogenesis, proteolysis | [40, 41] |
| Multiple myeloma | Cyclin D2 ↑ | Enhances cell proliferation | [42] | |||
| Prostate cancer | - | Promotes cell migration | [43] | |||
| Cervical, colon, ovarian cancer, neuroblastoma | MAP1LC3B ↓, WIPI2 ↓ | Suppresses autophagy and lysosome biogenesis | [4] | |||
| ZNF322A | ZNF388, ZNF489 | Oncogene | Lung cancer | - | Chromosome locus 6p22.1 is amplified | [44] |
| ADD1 ↑, CCND1 ↑, p53 ↓ | Promotes cell growth, migration and invasion | [45] | ||||
| ZNF304 | - | Oncogene | Colorectal | p14ARF ↓, p15INK4B ↓, p16INK4A ↓ | Suppresses tumor suppressor genes through recruiting a co-repressor complex, including DNMT1 | [48] |
| Ovarian cancer | Integrin β1 ↑ | Activates Src/focal adhesion kinase and paxillin and therefore prevents anoikis | [49] | |||
| ZNF139 | ZKSCAN1, ZNF36, ZSCAN33, KOX18 | Oncogene | Gastric cancer | - | Serves an independent prognostic factor for gastric cancer patients | [50] |
| Survivin ↑, x-IAP ↑, Bcl2 ↑, Caspase-3 ↓, Bax ↓ | Promotes cell proliferation and inhibits apoptosis | [51] | ||||
| MMP-2 ↑, MMP-9 ↑, ICAM-1 ↑, TIMP-1 ↓ | Promotes cell migration and invasion | [52] | ||||
| MDR-1/P-gp ↑, MRP1 ↑, Bcl-2 ↑, Bax ↓ | Contributes to multi-drug resistance | [53] | ||||
| ZFX | ZNF926 | Oncogene | Hepatocellular carcinoma | Nanog ↑, SOX2 ↑ | Confers self-renewal properties and chemoresistance | [5] |
| Nasopharyngeal carcinoma | E-cadherin ↓ | May be involved in EMT | [54] | |||
| Glioma, lung, oral, breast cancer | - | Promotes cell proliferation and survival | [55, 56, 58, 60] | |||
| Gastric cancer | - | Promotes cell growth through up-regulating ERK-MAPK pathway | [57] | |||
| Gallbladder cancer | - | Promotes proliferation, migration and invasion potentially through activation of PI3K/AKT pathway | [59] | |||
| Glioblastoma | c-Myc ↑ | Promotes glioma stem cell maintenance | [61] | |||
| ZEB1 | ZFHX1A, DELTAEF1 | Oncogene | Breast cancer | ESRP2 ↓ | Promotes TGF-β-induced EMT | [68] |
| Glioma | - | SHP-2 up-regulates ZEB1 expression to mediate EMT, invasion and growth | [69] | |||
| Cervix, breast cancer, osteosarcoma, adrenal carcinoma | E-cadherin ↓ | Down-regulates E-cadherin and cell polarity factors by recruiting co-repressor CtBP or BRG1 | [70, 71] | |||
| Cervix, colorectal cancer | - | Activates genes involved in TGF-β/BMP signaling by recruiting p300 and P/CAF | [72, 73] | |||
| Lung cancer | E-cadhein ↑, ST14 ↑, Vimentin ↑ | Confers EMT-related acquired resistance to EGFR-TKI | [74] | |||
| Breast cancer | VEGFA ↑ | Promotes angiogenesis | [75] | |||
| ZNF545 | ZFP82 | TSG | Nasopharyngeal, esophageal, lung, gastric, colon, breast cancer | - | Induces cell apoptosis by repressing ribosome biogenesis and NF-kB and AP-1 signaling | [76] |
| ZNF331 | ZNF361, ZNF463 | TSG | Gastric cancer | DSTN ↓, EIF5A ↓, GARS ↓, DDX5 ↓, STAM ↓, UQCRFS1 ↓, SET ↓, ACTR3 ↓ | Inhibits cell growth, migration and invasion | [78] |
| Gastrointestinal, esophageal cancer | - | Promoter hypermethylation is found in various cancer types | [79, 80] | |||
| ZNF24 | ZNF191, Kox17 | TSG | Breast cancer | VEGF ↓ | Inhibits angiogenesis | [82, 83] |
| Gastric cancer | - | miR940 promotes cancer migration and invasion by targeting ZNF24 | [84] | |||
| ZNF668 | - | TSG | Breast cancer | - | Suppresses cell proliferation by promoting MDM2 autoubiquitination and therefore p53 stabilization | [85] |
| Osteosarcoma | - | Involved in DNA repair by regulating chromatin relaxation and recruiting repair proteins to DNA lesions | [86] | |||
| ZHX1 | - | TSG | Gastric cancer | - | miR-199a-3p promotes cell proliferation and suppresses apoptosis by targeting to ZHX1 | [88] |
| CCND1 ↓, CCNE ↓, Bcl2 ↓, Bax ↑, cleaved Caspase-3 ↑ | Induce G1/S arrest and apoptosis | [89] | ||||
| ZNF395 | PBF, HDBP2 | Oncogene | Ewing’s sarcoma, osteosarcoma, renal cell carcinoma | - | Overexpressed in various cancers | [90–92] |
| Glioblastoma | - | Induced under hypoxia stress | [93] | |||
| Skin and cervix cancer, glioblastoma | IFIT1/ISG56 ↑, IFI44 ↑, IFI16 ↑ | Supports inflammation and cancer progression | [94] | |||
| TSG | Liver cancer | - | miR-525-3p promotes cell migration and invasion by targeting ZNF395 | [95] | ||
| Kaiso | ZNF348, ZBTB33 | TSG | Breast and colon cancer | CCND1 ↓ | Suppresses cell proliferation | [99] |
| Oncogene | Breast cancer | Vimentin ↑, Slug ↑, ZEB1 ↑ | Involved in TGF-β-mediated metastasis | [100] | ||
| Prostate cancer | miR-31 ↓ | Promotes cell migration and invasion | [101] | |||
| Breast and colorectal cancer | HIF-1α ↓ | - | [102] |
-, target not-determined
ZNF322A, also known as ZNF388 or ZNF489, consists of 11 tandem repeats of C2H2 zinc finger motif. ZNF322A was first identified as oncogene by Lo et al., showing that ZNF322A residing region is amplified in both Asian and Caucasian lung cancer patients [44]. Further study reveals that ZNF322A promotes cell proliferation, migration and invasion through transcriptionally activating cyclin D1 and alpha-adducin but suppressing p53 in lung cancers [45]. Multivariate Cox regression analysis indicates ZNF322A is an independent risk factor of poor outcome in lung cancer patients [45]. Notably, ZNF322A mouse ortholog, Zfp322a, is reported as a novel essential component of the transcription network, which maintains the self-renewal and pluripotency of mouse embryonic stem (mES) cells [46]. Zfp322a promotes OKSM (Oct4, Klf4, Sox2, c-Myc)-induced mouse embryonic fibroblast reprogramming to mES cells by transcriptionally activating Oct4 and Nanog expression [46]. The study on Zfp322a implies a potential role of human ZNF322A in maintaining the pluripotency of embryonic stem cells or cancer stem cells.
ZNF304, which contains a KRAB domain and 13 C2H2 zinc finger motifs, was first identified by AU-motif directed display and RACE in 2002 [47]. ZNF304 plays a pivotal role in silencing tumor suppressors, including p14ARF, p15INK4B and p16INK4A, through recruiting a co-repressor complex that includes DNA methyltransferase DNMT1 [48]. In addition, an integrative bioinformatic analysis of The Cancer Genome Atlas ovarian cancer dataset and experimental validation reveals the association between ZNF304 and ovarian cancer metastasis [49]. The authors show that ZNF304 transcriptionally activates integrin β1 expression, which subsequently activates Src/focal adhesion kinase and paxillin and eventually prevents anoikis [49]. Using delivery of ZNF304 siRNA by a dual assembly nanoparticle, these authors successfully conducted a sustained ZNF304 silencing which increased anoikis and reduced ovarian tumor growth in orthotopic mouse models [49].
ZNF139 is significantly overexpressed in gastric cancer patients. Cox survival analysis reveals ZNF139 overexpression as an independent prognostic factor for gastric cancer patients [50]. ZNF139 has been reported to promote proliferation and inhibit apoptosis through up-regulating the expression of Survivin, x-IAP and Bcl-2, and down-regulating Caspase-3 and Bax [51]. In addition, ZNF139 promotes cancer migration and invasion in gastric cancer by increasing the expression of MMP-2, MMP-9 and ICAM-1, and decreasing the expression of TIMP-1 [52]. ZNF139 also contributes to multi-drug resistance by enhancing the expression of MDR-1/P-gp, MRP1, Bcl-2 while inhibiting Bax expression [53].
Overexpression of zinc finger protein, X-linked (ZFX) has been shown to promote cell growth and metastasis in laryngeal squamous cell carcinoma, glioma, non-small cell lung cancer, gastric cancer, oral squamous cell carcinoma, gallbladder cancer and breast cancer [5, 54–60]. In addition, ZFX is found to confer self-renewal properties and chemoresistance in hepatocellular carcinoma through transcriptional activation of Nanog and SOX2 expression [5]. Fang et al. also showed that ZFX transcriptionally up-regulates c-Myc expression leading to glioma stem cell maintenance [61]. Inhibition of ZFX using siRNA oligo or drug treatment suppresses cancer progression, indicating the potential of oncogenic ZNFs as therapeutic targets [62, 63].
Zinc finger E-box-binding homeobox, ZEB1, is a well-studied transcription factor involved in Epithelial-Mesenchymal Transition (EMT) in several cancer types, including breast cancer, lung cancer, pancreatic cancer and prostate cancer [64–67]. ZEB1 expression in cancer cells is elevated upon signaling induction, including TGF-β and platelet-driven growth factor receptor-α signaling [68, 69]. As an activator of EMT, increased ZEB1 binds to E-boxes containing downstream targets, including E-cadherin and cell polarity factors, and represses their transcription by recruiting co-repressors CtBP or SWI/SNF chromatin-remodeling protein BRG1 [70, 71]. Notably, studies also reveal that ZEB1 can transcriptionally activate genes involved in TGF-β/BMP signaling through recruiting co-activators, p300 and P/CAF [72, 73]. In addition to its role in EMT, ZEB1 overexpression further contributes to EMT-related acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKI) in non-small cell lung cancer through transcriptionally up-regulating E-cadherin, ST14 and vimentin [74]. Moreover, Yoshida et al., show that silencing ZEB1 expression restores sensitivity to EGFR-TKI, suggesting targeting ZEB1 could be a potential therapy to resensitize TKI-resistant tumors [74]. A recent study also reveals a novel role of ZEB1 in promoting angiogenesis in breast cancer [75]. The authors show that ZEB1 overexpression in breast cancer cells recruits Sp1 to VEGFA promoter region and activates VEGFA expression and secretion, therefore promoting angiogenesis in vitro and in vivo [75].
The tumor suppressor ZNFs in cancer progression
In addition to cancer promotion, several ZNFs have been found to function as tumor suppressors. For example, ZNF545, which is down-regulated in cancer cells as a consequence of promoter methylation, acts as a tumor suppressor by inducing cell apoptosis, repressing ribosome biogenesis and suppressing NF-kB and AP-1 signaling in nasopharyngeal, esophageal, lung, gastric, colon and breast cancer [76]. Notably, methylated degrees of five CpG sites (-232, -214, -176, -144 and -116) discriminate gastric cancer patients’ survival outcome with higher CpG methylation predicting poorer overall survival [77]. Another ZNF known to be inactivated by promoter hypermethylation is ZNF331, also known as ZNF361 or ZNF463 [78–80]. Overexpression of ZNF331 inhibits cell growth by down-regulating genes, including DSTN, EIF5A, GARS, DDX5, STAM, UQCRFS1 and SET, and inhibits cell migration/invasion by down-regulating genes, including DSTN and ACTR3 [78].
ZNF24, also known as ZNF191 or Kox17, contains 4 Krüppel-like C2H2 zinc finger domains on C-terminus that function as DNA binding domains [81]. ZNF24 suppresses VEGF expression by binding to the proximal VEGF promoter, and negatively regulates tumor growth by inhibiting angiogenesis in breast cancer [82, 83]. Using transgenic zebra fish model, Jia et al., demonstrate that expression of human ZNF24 induces vascular defects, which can be recovered by VEGF overexpression [83]. Clinical studies of human breast cancer confirm the inverse correlation between ZNF24 and VEGF, indicating the tumor suppressor role of ZNF24 in breast cancer tumorigenesis by inhibiting angiogenesis [83]. Interestingly, a recent study shows that miR940 is up-regulated in gastric cancer and promotes gastric cancer migration and invasion by targeting tumor suppressor ZNF24 [84].
ZNF668 is a member of Krüppel C2H2 zinc finger protein family, which possesses 16 C2H2-type zinc fingers. ZNF668 facilitates p53 stabilization and activity by disrupting MDM2-mediated ubiquitination and degradation in breast cancer [85]. In addition, ZNF668 interacts with Tip60 to enhance H2AX hyperacetylation in response to ionizing radiation and promote RPA phosphorylation and recruitment to DNA damage foci upon UV damage, therefore leading to chromatin relaxation and loading of DNA repair proteins [86].
Zinc-fingers and homeoboxes-1 (ZHX1), which contains two C2H2 zinc finger motifs and five homeodomains, has been reported to be down-regulated in hepatocellular carcinoma and gastric cancer [87–89]. ZHX1 induces G1/S arrest through down-regulating cyclin D1 and cyclin E expression, and enhances apoptosis through down-regulating Bcl2 and up-regulating Bax and cleaved Caspase-3 [89]. Of note, Wang et al., demonstrate that miRNA, miR-199a-3p, targets ZHX1 for RNA degradation to promote cell proliferation and suppresses apoptosis in gastric cancer. Reconstitution of ZHX1 expression abrogates gastric cancer oncogenicity [88].
ZNFs: double-edged sword in tumorigenesis
Some ZNFs have been shown to play different roles in different cancer types and stimuli. For example, ZNF395 is overexpressed in various cancers, including Ewing sarcomas, osteosarcomas and renal cells carcinomas [90–92]. Moreover, ZNF395 expression is induced under hypoxic stress in glioblastoma, neuroblastoma and skin cancer [90, 93, 94]. Hypoxia-induced ZNF395 can transcriptionally up-regulate cancer-related genes and interferon-stimulated genes, such as IFIT1/ISG56, IFI44 and IFI16, in an IKK signaling-dependent manner [94]. These results implicate ZNF395 as a novel transcription factor which supports inflammation and cancer progression. However, a recent study reveals the tumor suppressor role of ZNF395 in liver cancer. The authors show that miR-525-3p, which is overexpressed in liver cancer, promotes liver cancer cell migration and invasion by targeting and down-regulating ZNF395 expression [95]. The clinical analysis indeed confirms the inverse correlation of miR-525-3p and ZNF395 in liver cancer [95]. These studies collectively show that ZNF395 may play different roles in different cancer types.
Kaiso, also known as ZNF348 or ZBTB33, belongs to the BTB/POZ subfamily of ZNFs. Kaiso can bind to sequence-specific or methyl-CpG DNA using its zinc finger motifs, while its N-terminus POZ domain helps homodimerization or heterodimerization with chromatin co-repressors, including nuclear receptor co-repressor I [96–98]. By recruiting chromatin co-repressors, Kaiso transcriptionally suppresses downstream gene expression. Kaiso was first identified as a tumor suppressor that transcriptionally suppressed oncogenic genes in sequence- or methyl-CpG-specific manner. For example, Kaiso represses cyclin D1 expression by binding to CCND1 promoter in a sequence- and methyl-CpG-specific manner in breast and colon cancer [99]. Since then, more and more studies have demonstrated the oncogene role of Kaiso in various cancers. For example, Kaiso is found highly expressed in triple negative breast cancers and involved in TGF-β-mediated metastasis by up-regulating several EMT genes, including Vimentin, Slug and ZEB1 [100]. High expression of Kaiso in prostate cancer promotes cell migration and invasion through transcriptional suppression of miR-31 expression in methyl CpG-specific manner [101]. In addition, Pierre et al., show that Kaiso transcriptionally suppresses HIF-1α expression by targeting to methylated HIF1A promoter in breast and colorectal cancer [102]. Kaiso is a versatile ZNF, which exerts different functions in different cell types in respond to different stimuli.
Conclusion
Recent studies show that C2H2 ZNF proteins play important roles in cancer progression through regulating transcription of downstream genes, which are involved in proliferation, apoptosis, migration and invasion. Although more and more studies have been focused on the underlying mechanism of C2H2 ZNF transcription regulation, results remain conflicting. It is now understood that different layers of regulations lead C2H2 ZNF proteins to different roles in tumorigenesis. In this review, we summarize various levels of ZNF proteins regulation in tumorigenesis (Fig 1). First, differential expression levels of ZNF proteins in different cancer types are regulated by cancer-related miRNA, including miR-199a-3p, miR-525-3p, miR-940 and miR-31. Second, different environmental stimuli activate signaling cascades and therefore fine-tune ZNF protein functions through various PTMs, including phosphorylation and acetylation. PTMs regulation affects DNA binding abilities and interacting proteins recruitments of ZNF proteins. Third, ZNF proteins at different protein domains or with various PTMs recruit different interacting proteins, including transcription co-activators/co-repressors, chromatin modifiers and other transcription factors. Therefore, ZNF proteins can activate or suppress downstream genes by recruiting different interacting partners. Fourth, ZNF proteins show diverse sequence-specific DNA binding abilities with different combinations of zinc finger motifs. Knowing the complexities and diversities of ZNF proteins, it is important to elucidate the underlying mechanisms of C2H2 ZNF proteins in different cancers under different environmental stimuli. Therefore, drugs targeting specific C2H2 ZNF protein expression or activity can be developed for therapeutic strategy against tumors in a specific stage of cancer progression.
Fig. 1.
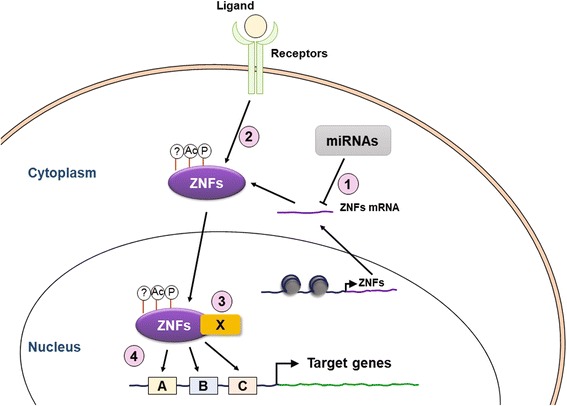
Various regulations of ZNF proteins’ functions in cancer progression. The versatile roles of ZNF proteins in cancer progression can be regulated at different levels. Differential expression of ZNF proteins in different cancer types can be regulated by 1) cancer-related miRNAs, including miR-199a-3p, miR-525-3p, miR-940 and miR-31, or 2) different environmental stimuli, which activate signaling cascades and therefore fine-tune ZNF protein functions through various of PTMs, including phosphorylation (P) and acetylation (Ac). 3) ZNF proteins at different protein domains or with various PTMs recruit different interacting proteins namely X, including transcription co-activators/co-repressors, chromatin modifiers and other transcription factors, to activate or suppress downstream genes. 4) ZNF proteins show diverse sequence-specific DNA binding abilities due to different combinations of zinc finger motifs shown as boxes
Abbreviations
BTB, Broad-Complex, Tramtrack, and Bric-a-brac; C2H2, Cys2His2; EGFR-TKI, epidermal growth factor receptor-tyrosine kinase inhibitors; EMT, Epithelial-Mesenchymal Transition; ERC-1, erythroid complex; KRAB, Krüppel-associated box; mES, mouse embryonic stem; PCAF, p300/CBP associated factor; POZ, poxvirus and zinc finger; PTMs, post-translational modifications; SCAN, SRE-ZBP, CTfin51, AW-1 and Number 18 cDNA; TGF-β, transforming growth factor-β; ZFX, zinc finger protein, X-linked; ZHX1, Zinc-fingers and homeoboxes-1; ZNF, Zinc finger
Acknowledgments
This work was supported by Taiwan Ministry of Science grant 104-2627-B-006-001, the Aim for the Top University Project grant D105-35A07, and Taiwan Ministry of Health and Welfare grant 105-TDU-B-211-124-003.
Funding
This work was supported by Taiwan Ministry of Science grant 104-2627-B-006-001, the Aim for the Top University Project grant D105-35A07, and Taiwan Ministry of Health and Welfare grant 105-TDU-B-211-124-003 to YCW.
Availability of data and materials
Data and materials related to this work are available upon request.
Authors’ contribution
JJ and YCW wrote the review. Both authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
All authors approve the manuscript for publication.
Ethics approval and consent to participate
Not applicable.
Contributor Information
Jayu Jen, Phone: +886-6-2353535 ext.5476, Email: jayujen1021@gmail.com.
Yi-Ching Wang, Phone: +886-6-2353535 ext.5502, Email: ycw5798@mail.ncku.edu.tw.
References
- 1.Arenzana TL, Schjerven H, Smale ST. Regulation of gene expression dynamics during developmental transitions by the Ikaros transcription factor. Genes Dev. 2015;29:1801–16. doi: 10.1101/gad.266999.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krebs CJ, Zhang D, Yin L, Robins DM. The KRAB zinc finger protein RSL1 modulates sex-biased gene expression in liver and adipose tissue to maintain metabolic homeostasis. Mol Cell Biol. 2014;34:221–32. doi: 10.1128/MCB.00875-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma X, Huang M, Wang Z, Liu B, Zhu Z, Li C. ZHX1 inhibits gastric cancer cell growth through inducing cell-cycle arrest and apoptosis. J Cancer. 2016;7:60–8. doi: 10.7150/jca.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50:16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai KP, Chen J, He M, Ching AK, Lau C, Lai PB, et al. Overexpression of ZFX confers self-renewal and chemoresistance properties in hepatocellular carcinoma. Int J Cancer. 2014;135:1790–9. doi: 10.1002/ijc.28819. [DOI] [PubMed] [Google Scholar]
- 6.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 7.Scott MP, Tamkun JW, Hartzell GW., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 8.Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 10.Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–3. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- 11.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–50. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konieczny P, Stepniak-Konieczna E, Sobczak K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014;42:10873–87. doi: 10.1093/nar/gku767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Font J, Mackay JP. Beyond DNA: zinc finger domains as RNA-binding modules. Methods Mol Biol. 2010;649:479–91. doi: 10.1007/978-1-60761-753-2_29. [DOI] [PubMed] [Google Scholar]
- 14.Brayer KJ, Kulshreshtha S, Segal DJ. The protein-binding potential of C2H2 zinc finger domains. Cell Biochem Biophys. 2008;51:9–19. doi: 10.1007/s12013-008-9007-6. [DOI] [PubMed] [Google Scholar]
- 15.Matthews JM, Sunde M. Zinc fingers--folds for many occasions. IUBMB Life. 2002;54:351–5. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- 16.Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 17.McCarty AS, Kleiger G, Eisenberg D, Smale ST. Selective dimerization of a C2H2 zinc finger subfamily. Mol Cell. 2003;11:459–70. doi: 10.1016/S1097-2765(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 18.Li M, Ling B, Xiao T, Tan J, An N, Han N, et al. Sp1 transcriptionally regulates BRK1 expression in non-small cell lung cancer cells. Gene. 2014;542:134–40. doi: 10.1016/j.gene.2014.03.043. [DOI] [PubMed] [Google Scholar]
- 19.Nunez N, Clifton MM, Funnell AP, Artuz C, Hallal S, Quinlan KG, et al. The multi-zinc finger protein ZNF217 contacts DNA through a two-finger domain. J Biol Chem. 2011;286:38190–201. doi: 10.1074/jbc.M111.301234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisbacher M, Holmes ML, Newton A, Hogg PJ, Khachigian LM, Crossley M, et al. Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding. Mol Cell Biol. 2003;23:3427–41. doi: 10.1128/MCB.23.10.3427-3441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory RC, Taxman DJ, Seshasayee D, Kensinger MH, Bieker JJ, Wojchowski DM. Functional interaction of GATA1 with erythroid Krüppel-like factor and Sp1 at defined erythroid promoters. Blood. 1996;87:1793–801. [PubMed] [Google Scholar]
- 22.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, et al. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–8. [PubMed] [Google Scholar]
- 24.Lehmann W, Mossmann D, Kleemann J, Mock K, Meisinger C, Brummer T, et al. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat Commun. 2016;7:10498. doi: 10.1038/ncomms10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowger JJ, Zhao Q, Isovic M, Torchia J. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene. 2007;26:3378–86. doi: 10.1038/sj.onc.1210126. [DOI] [PubMed] [Google Scholar]
- 26.Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3’ ends of ZNF genes. PLoS One. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gocke CB, Yu H. ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PLoS One. 2008;3:e3255. doi: 10.1371/journal.pone.0003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeon BN, Kim MK, Yoon JH, Kim MY, An H, Noh HJ, et al. Two ZNF509 (ZBTB49) isoforms induce cell-cycle arrest by activating transcription of p21/CDKN1A and RB upon exposure to genotoxic stress. Nucleic Acids Res. 2014;42:11447–61. doi: 10.1093/nar/gku857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meruvu S, Hugendubler L, Mueller E. Regulation of adipocyte differentiation by the zinc finger protein ZNF638. J Biol Chem. 2011;286:26516–23. doi: 10.1074/jbc.M110.212506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamonica JM, Vakoc CR, Blobel GA. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 2006;108:3736–8. doi: 10.1182/blood-2006-07-032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamonica JM, Deng W, Kadauke S, Campbell AE, Gamsjaeger R, Wang H, et al. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci U S A. 2011;108:E159–68. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamsjaeger R, Webb SR, Lamonica JM, Billin A, Blobel GA, Mackay JP. Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3. Mol Cell Biol. 2011;31:2632–40. doi: 10.1128/MCB.05413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Kadam S, Emerson BM, Bieker JJ. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Krüppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol Cell Biol. 2001;21:2413–22. doi: 10.1128/MCB.21.7.2413-2422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengupta T, Chen K, Milot E, Bieker JJ. Acetylation of EKLF is essential for epigenetic modification and transcriptional activation of the beta-globin locus. Mol Cell Biol. 2008;28:6160–70. doi: 10.1128/MCB.00919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–91. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jantz D, Berg JM. Reduction in DNA-binding affinity of Cys2His2 zinc finger proteins by linker phosphorylation. Proc Natl Acad Sci U S A. 2004;101:7589–93. doi: 10.1073/pnas.0402191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dovat S, Ronni T, Russell D, Ferrini R, Cobb BS, Smale ST. A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev. 2002;16:2985–90. doi: 10.1101/gad.1040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizkallah R, Hurt MM. Regulation of the transcription factor YY1 in mitosis through phosphorylation of its DNA-binding domain. Mol Biol Cell. 2009;20:4766–76. doi: 10.1091/mbc.E09-04-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rizkallah R, Alexander KE, Hurt MM. Global mitotic phosphorylation of C2H2 zinc finger protein linker peptides. Cell Cycle. 2011;10:3327–36. doi: 10.4161/cc.10.19.17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L, Hamilton SR, Sood A, Kuwai T, Ellis L, Sanguino A, et al. The previously undescribed ZKSCAN3 (ZNF306) is a novel “driver” of colorectal cancer progression. Cancer Res. 2008;68:4321–30. doi: 10.1158/0008-5472.CAN-08-0407. [DOI] [PubMed] [Google Scholar]
- 41.Yang L, Zhang L, Wu Q, Boyd DD. Unbiased screening for transcriptional targets of ZKSCAN3 identifies integrin beta 4 and vascular endothelial growth factor as downstream targets. J Biol Chem. 2008;283:35295–304. doi: 10.1074/jbc.M806965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang L, Wang H, Kornblau SM, Graber DA, Zhang N, Matthews JA, et al. Evidence of a role for the novel zinc-finger transcription factor ZKSCAN3 in modulating Cyclin D2 expression in multiple myeloma. Oncogene. 2011;30:1329–40. doi: 10.1038/onc.2010.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Jing Y, Qin Y, Hunsucker S, Meng H, Sui J, et al. The zinc finger transcription factor ZKSCAN3 promotes prostate cancer cell migration. Int J Biochem Cell Biol. 2012;44:1166–73. doi: 10.1016/j.biocel.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo FY, Chang JW, Chang IS, Chen YJ, Hsu HS, Huang SF, et al. The database of chromosome imbalance regions and genes resided in lung cancer from Asian and Caucasian identified by array-comparative genomic hybridization. BMC Cancer. 2012;12:235. doi: 10.1186/1471-2407-12-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jen J, Lin LL, Chen HT, Liao SY, Lo FY, Tang YA, et al. Oncoprotein ZNF322A transcriptionally deregulates alpha-adducin, cyclin D1 and p53 to promote tumor growth and metastasis in lung cancer. Oncogene. 2015;35(18):2357-69. [DOI] [PMC free article] [PubMed]
- 46.Ma H, Ng HM, Teh X, Li H, Lee YH, Chong YM, et al. Zfp322a regulates mouse ES cell pluripotency and enhances reprogramming efficiency. PLoS Genet. 2014;10:e1004038. doi: 10.1371/journal.pgen.1004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabater L, Ashhab Y, Caro P, Kolkowski EC, Pujol-Borrell R, Domínguez O. Identification of a KRAB-containing zinc finger protein, ZNF304, by AU-motif-directed display method and initial characterization in lymphocyte activation. Biochem Biophys Res Commun. 2002;293:1066–72. doi: 10.1016/S0006-291X(02)00344-3. [DOI] [PubMed] [Google Scholar]
- 48.Serra RW, Fang M, Park SM, Hutchinson L, Green MR. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. Elife. 2014;3:e02313. doi: 10.7554/eLife.02313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aslan B, Monroig P, Hsu MC, Pena GA, Rodriguez-Aguayo C, Gonzalez-Villasana V, et al. The ZNF304-integrin axis protects against anoikis in cancer. Nat Commun. 2015;6:7351. doi: 10.1038/ncomms8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Zhao Q, Fan LQ, Wang LL, Tan BB, Leng YL, et al. Zinc finger protein 139 expression in gastric cancer and its clinical significance. World J Gastroenterol. 2014;20:18346–53. doi: 10.3748/wjg.v20.i48.18346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan L, Tan B, Li Y, Zhao Q, Liu Y, Wang D, et al. Silencing of ZNF139-siRNA induces apoptosis in human gastric cancer cell line BGC823. Int J Clin Exp Pathol. 2015;8:12428–36. [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Tan BB, Zhao Q, Fan LQ, Wang D, Liu Y. ZNF139 promotes tumor metastasis by increasing migration and invasion in human gastric cancer cells. Neoplasma. 2014;61:291–8. doi: 10.4149/neo_2014_037. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Tan BB, Zhao Q, Fan LQ, Liu Y, Wang D. Regulatory mechanism of ZNF139 in multi-drug resistance of gastric cancer cells. Mol Biol Rep. 2014;41:3603–10. doi: 10.1007/s11033-014-3224-4. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Yan X, Yan L, Shan Z, Liu S, Chen X, et al. High expression of Zinc-finger protein X-linked is associated with reduced E-cadherin expression and unfavorable prognosis in nasopharyngeal carcinoma. Int J Clin Exp Pathol. 2015;8:3919–27. [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Z, Li K, Xu D, Liu Y, Tang H, Xie Q, et al. ZFX regulates glioma cell proliferation and survival in vitro and in vivo. J Neurooncol. 2013;112:17–25. doi: 10.1007/s11060-012-1032-z. [DOI] [PubMed] [Google Scholar]
- 56.Jiang M, Xu S, Yue W, Zhao X, Zhang L, Zhang C, et al. The role of ZFX in non-small cell lung cancer development. Oncol Res. 2012;20:171–8. doi: 10.3727/096504012X13548165987493. [DOI] [PubMed] [Google Scholar]
- 57.Wu S, Lao XY, Sun TT, Ren LL, Kong X, Wang JL, et al. Knockdown of ZFX inhibits gastric cancer cell growth in vitro and in vivo via downregulating the ERK-MAPK pathway. Cancer Lett. 2013;337:293–300. doi: 10.1016/j.canlet.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Ma H, Yang F, Lian M, Wang R, Wang H, Feng L, et al. Dysregulation of zinc finger protein, X-linked (ZFX) impairs cell proliferation and induces apoptosis in human oral squamous cell carcinorma. Tumour Biol. 2015;36:6103–12. doi: 10.1007/s13277-015-3292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weng H, Wang X, Li M, Wu X, Wang Z, Wu W, et al. Zinc finger X-chromosomal protein (ZFX) is a significant prognostic indicator and promotes cellular malignant potential in gallbladder cancer. Cancer Biol Ther. 2015;16:1462–70. doi: 10.1080/15384047.2015.1070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang H, Lu Y, Zheng Y, Yu X, Xia X, He X, et al. shRNA-mediated silencing of ZFX attenuated the proliferation of breast cancer cells. Cancer Chemother Pharmacol. 2014;73:569–76. doi: 10.1007/s00280-014-2379-y. [DOI] [PubMed] [Google Scholar]
- 61.Fang X, Huang Z, Zhou W, Wu Q, Sloan AE, Ouyang G, et al. The zinc finger transcription factor ZFX is required for maintaining the tumorigenic potential of glioblastoma stem cells. Stem Cells. 2014;32:2033–47. doi: 10.1002/stem.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li K, Zhu ZC, Liu YJ, Liu JW, Wang HT, Xiong ZQ, et al. ZFX knockdown inhibits growth and migration of non-small cell lung carcinoma cell line H1299. Int J Clin Exp Pathol. 2013;6:2460–7. [PMC free article] [PubMed] [Google Scholar]
- 63.Liu TY, Gong W, Tan ZJ, Lu W, Wu XS, Weng H, et al. Baicalein inhibits progression of gallbladder cancer cells by downregulating ZFX. PLoS One. 2015;10:e0114851. doi: 10.1371/journal.pone.0114851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karihtala P, Auvinen P, Kauppila S, Haapasaari KM, Jukkola-Vuorinen A, Soini Y. Vimentin, zeb1 and Sip1 are up-regulated in triple-negative and basal-like breast cancers: association with an aggressive tumour phenotype. Breast Cancer Res Treat. 2013;138:81–90. doi: 10.1007/s10549-013-2442-0. [DOI] [PubMed] [Google Scholar]
- 65.Liu Y, Zhang N, Wang Y, Xu M, Liu N, Pang X, et al. Zinc finger E-box binding homeobox 1 promotes invasion and bone metastasis of small cell lung cancer in vitro and in vivo. Cancer Sci. 2012;103:1420–8. doi: 10.1111/j.1349-7006.2012.02347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bronsert P, Kohler I, Timme S, Kiefer S, Werner M, Schilling O, et al. Prognostic significance of Zinc finger E-box binding homeobox 1 (ZEB1) expression in cancer cells and cancer-associated fibroblasts in pancreatic head cancer. Surgery. 2014;156:97–108. doi: 10.1016/j.surg.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 67.Hou L, Li Q, Yu Y, Li M, Zhang D. SET8 induces epithelial-mesenchymal transition and enhances prostate cancer cell metastasis by cooperating with ZEB1. Mol Med Rep. 2016;13:1681–8. doi: 10.3892/mmr.2015.4733. [DOI] [PubMed] [Google Scholar]
- 68.Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S, et al. TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene. 2012;31:3190–201. doi: 10.1038/onc.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang L, Zhang W, Li Y, Alvarez A, Li Z, Wang Y, et al. SHP-2-upregulated ZEB1 is important for PDGFRα-driven glioma epithelial-mesenchymal transition and invasion in mice and humans. Oncogene. 2016; doi:10.1038/onc.2016.100 [DOI] [PMC free article] [PubMed]
- 70.Shi Y, Sawada J, Sui G, el Affar B, Whetstine JR, Lan F, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–8. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 71.Sánchez-Tilló E, Lázaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490–500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 72.Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J. 2003;22:2443–52. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22:2453–62. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoshida T, Song L, Bai Y, Kinose F, Li J, Ohaegbulam KC, et al. ZEB1 mediates acquired resistance to the epidermal growth factor receptor-tyrosine kinase inhibitors in non-small cell lung cancer. PLoS One. 2016;11:e0147344. doi: 10.1371/journal.pone.0147344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu L, Tong Q, Liu S, Cui J, Zhang Q, Sun W, et al. ZEB1 upregulates VEGF expression and stimulates angiogenesis in breast cancer. PLoS One. 2016;11:e0148774. doi: 10.1371/journal.pone.0148774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng Y, Liang P, Geng H, Wang Z, Li L, Cheng SH, et al. A novel 19q13 nucleolar zinc finger protein suppresses tumor cell growth through inhibiting ribosome biogenesis and inducing apoptosis but is frequently silenced in multiple carcinomas. Mol Cancer Res. 2012;10:925–36. doi: 10.1158/1541-7786.MCR-11-0594. [DOI] [PubMed] [Google Scholar]
- 77.Deng J, Liang H, Ying G, Dong Q, Zhang R, Yu J, et al. Poor survival is associated with the methylated degree of zinc-finger protein 545 (ZNF545) DNA promoter in gastric cancer. Oncotarget. 2015;6:4482–95. doi: 10.18632/oncotarget.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu J, Liang QY, Wang J, Cheng Y, Wang S, Poon TC, et al. Zinc-finger protein 331, a novel putative tumor suppressor, suppresses growth and invasiveness of gastric cancer. Oncogene. 2013;32:307–17. doi: 10.1038/onc.2012.54. [DOI] [PubMed] [Google Scholar]
- 79.Vedeld HM, Andresen K, Eilertsen IA, Nesbakken A, Seruca R, Gladhaug IP, et al. The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers. Int J Cancer. 2015;136:844–53. doi: 10.1002/ijc.29039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang S, Linghu E, Zhan Q, Han W, Guo M. Methylation of ZNF331 promotes cell invasion and migration in human esophageal cancer. Curr Protein Pept Sci. 2015;16:322–8. doi: 10.2174/138920371604150429155255. [DOI] [PubMed] [Google Scholar]
- 81.Wang H, Sun R, Liu G, Yao M, Fei J, Shen H. Characterization of the target DNA sequence for the DNA-binding domain of zinc finger protein 191. Acta Biochim Biophys Sin Shanghai. 2008;40:704–10. doi: 10.1093/abbs/40.8.704. [DOI] [PubMed] [Google Scholar]
- 82.Harper J, Yan L, Loureiro RM, Wu I, Fang J, D’Amore PA, et al. Repression of vascular endothelial growth factor expression by the zinc finger transcription factor ZNF24. Cancer Res. 2007;67:8736–41. doi: 10.1158/0008-5472.CAN-07-1617. [DOI] [PubMed] [Google Scholar]
- 83.Jia D, Hasso SM, Chan J, Filingeri D, D’Amore PA, Rice L, et al. Transcriptional repression of VEGF by ZNF24: mechanistic studies and vascular consequences in vivo. Blood. 2013;121:707–15. doi: 10.1182/blood-2012-05-433045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu X, Ge X, Zhang Z, Zhang X, Chang J, Wu Z, et al. MicroRNA-940 promotes tumor cell invasion and metastasis by downregulating ZNF24 in gastric cancer. Oncotarget. 2015;6:25418–28. doi: 10.18632/oncotarget.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu R, Peng G, Dai H, Breuer EK, Stemke-Hale K, Li K, et al. ZNF668 functions as a tumor suppressor by regulating p53 stability and function in breast cancer. Cancer Res. 2011;71:6524–34. doi: 10.1158/0008-5472.CAN-11-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu R, Wang E, Peng G, Dai H, Lin SY. Zinc finger protein 668 interacts with Tip60 to promote H2AX acetylation after DNA damage. Cell Cycle. 2013;12:2033–41. doi: 10.4161/cc.25064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang J, Liu D, Liang X, Gao L, Yue X, Yang Y, et al. Construction of a recombinant eukaryotic human ZHX1 gene expression plasmid and the role of ZHX1 in hepatocellular carcinoma. Mol Med Rep. 2013;8:1531–6. doi: 10.3892/mmr.2013.1700. [DOI] [PubMed] [Google Scholar]
- 88.Wang Z, Ma X, Cai Q, Wang X, Yu B, Cai Q, et al. MiR-199a-3p promotes gastric cancer progression by targeting ZHX1. FEBS Lett. 2014;588:4504–12. doi: 10.1016/j.febslet.2014.09.047. [DOI] [PubMed] [Google Scholar]
- 89.Ma X, Huang M, Wang Z, Liu B, Zhu Z, Li C. ZHX1 inhibits gastric cancer cell growth through inducing cell-cycle arrest and apoptosis. J Cancer. 2016;7:60–8. doi: 10.7150/jca.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yabe H, Tsukahara T, Kawaguchi S, Wada T, Sato N, Morioka H, et al. Overexpression of papillomavirus binding factor in Ewing's sarcoma family of tumors conferring poor prognosis. Oncol Rep. 2008;19:129–34. [PubMed] [Google Scholar]
- 91.Tsukahara T, Nabeta Y, Kawaguchi S, Ikeda H, Sato Y, Shimozawa K, et al. Identification of human autologous cytotoxic T-lymphocyte-defined osteosarcoma gene that encodes a transcriptional regulator, papillomavirus binding factor. Cancer Res. 2004;64:5442–8. doi: 10.1158/0008-5472.CAN-04-0522. [DOI] [PubMed] [Google Scholar]
- 92.Dalgin GS, Holloway DT, Liou LS, DeLisi C. Identification and characterization of renal cell carcinoma gene markers. Cancer Inform. 2007;3:65–92. [PMC free article] [PubMed] [Google Scholar]
- 93.Murat A, Migliavacca E, Hussain SF, Heimberger AB, Desbaillets I, Hamou MF, et al. Modulation of angiogenic and inflammatory response in glioblastoma by hypoxia. PLoS One. 2009;4:e5947. doi: 10.1371/journal.pone.0005947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jordanovski D, Herwartz C, Pawlowski A, Taute S, Frommolt P, Steger G. The hypoxia-inducible transcription factor ZNF395 is controlled by IĸB kinase-signaling and activates genes involved in the innate immune response and cancer. PLoS One. 2013;8:e74911. doi: 10.1371/journal.pone.0074911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pang F, Zha R, Zhao Y, Wang Q, Chen D, Zhang Z, et al. MiR-525-3p enhances the migration and invasion of liver cancer cells by downregulating ZNF395. PLoS One. 2014;9:e90867. doi: 10.1371/journal.pone.0090867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Daniel JM, Spring CM, Crawford HC, Reynolds AB, Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30:2911–9. doi: 10.1093/nar/gkf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12:723–34. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 98.Park JI, Kim SW, Lyons JP, Ji H, Nguyen TT, Cho K, et al. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell. 2005;8:843–54. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 99.Donaldson NS, Pierre CC, Anstey MI, Robinson SC, Weerawardane SM, Daniel JM. Kaiso represses the cell cycle gene cyclin D1 via sequence-specific and methyl-CpG-dependent mechanisms. PLoS One. 2012;7:e50398. doi: 10.1371/journal.pone.0050398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bassey-Archibong BI, Kwiecien JM, Milosavljevic SB, Hallett RM, Rayner LG, Erb MJ, et al. Kaiso depletion attenuates transforming growth factor-β signaling and metastatic activity of triple-negative breast cancer cells. Oncogenesis. 2016;5 doi: 10.1038/oncsis.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang H, Liu W, Black S, Turner O, Daniel JM, Dean-Colomb W, et al. Kaiso, a transcriptional repressor, promotes cell migration and invasion of prostate cancer cells through regulation of miR-31 expression. Oncotarget. 2016;7:5677–89. doi: 10.18632/oncotarget.6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pierre CC, Longo J, Bassey-Archibong BI, Hallett RM, Milosavljevic S, Beatty L, et al. Methylation-dependent regulation of hypoxia inducible factor-1 alpha gene expression by the transcription factor Kaiso. Biochim Biophys Acta. 2015;1849:1432–41. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data and materials related to this work are available upon request.


