Abstract
Campylobacter jejuni is the major cause of human bacterial diarrhea worldwide. Its pathogenic mechanism remains poorly understood. cj0371 is a novel gene that was uncovered using immunoscreening. There have been no previous reports regarding its function. In this study, we constructed an insertion mutant and complement of this gene in C. jejuni and examined changes in virulence. We observed that the cj0371 mutant showed significantly increased invasion and colonization ability. We also investigated the role of cj0371 in motility, chemotaxis, and growth kinetics to further study its function. We found that the cj0371 mutant displays hypermotility, enhanced chemotaxis, and enhanced growth kinetics. In addition, we localized the Cj0371 protein at the poles of C. jejuni by fluorescence microscopy. We present data that collectively significantly proves our hypothesis that cj0371 is a new virulence-associated gene and through the influence of chemotaxis plays a negative role in C. jejuni pathogenicity.
Keywords: virulence-associated gene, adhesion and invasion, chemotaxis, motility, growth kinetics
Introduction
The food-borne pathogen Campylobacter jejuni is responsible for campylobacteriosis, the most frequently reported food-borne illness in the European Union with ~200,000 human cases reported every year (Aliouane et al., 2016). Its typical clinical signs and symptoms are fever, inflammation, severe abdominal cramping, watery diarrhea and bloody stools, post-infectious sequelae including Guillain Barre Syndrome and other neurological disorders, with a limited morbidity but high mortality (Young et al., 2007; Dash et al., 2015). Poultry is a major source of C. jejuni, and only a small proportion of campylobacteriosis cases can be attributed to other animals or environmental sources (Chen and Jiang, 2014). However, the factors and mechanisms that contribute to successful colonization in poultry and virulence in humans remains poorly understood (Chandrashekhar et al., 2015). Despite the availability of both genomic information of different C. jejuni strains and genetic tools, the complete understanding of the virulence of C. jejuni is still an ongoing effort. In contrast to other diarrhea-causing bacteria, C. jejuni does not express a large number of classical virulence factors in human campylobacteriosis (Havelaar et al., 2009). Even so, many atypical virulence factors have been discovered.
With the development of whole genome sequencing technology and bioinformatics, numerous genome sequences of many C. jejuni are now available and comparisons identified several new open reading frames that may represent species-specific proteins (Aliouane et al., 2016). Using in vivo-induced antigen technology (IVIAT), an immunoscreening method, our group has identified virulence-associated genes expressed during human and chicken infection or invasion by C. jejuni (Hu et al., 2014a,b). We successfully identified 48 unique genes expressed in vivo, including one novel gene open reading frame. Through sequence blast in NCBI, we confirmed that this novel gene is cj0371 (Gene ID: 904694, updated on 3-Feb-2016). We speculated it is a novel virulence-associated gene. Because there have not been any reports regarding this gene until now, we were strongly interested in its function.
!!!!! Bioformatics analysis indicated that cj0371 is highly conserved in C. jejuni. This study was carried out to evaluate the ability of a cj0371 mutant to invade Caco-2 cells, the level of colonization in infant rabbits, and a functional exploration of the cj0371 gene. A motility plate assay, TEM imaging and subcellular localization of the Cj0371 protein were carried out to confirm whether cj0371 plays a role in the flagella system. This report provides important information for understanding C. jejuni virulence and brings unique insight into the evolution and function of this remarkable bacteria.
Materials and methods
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. C. jejuni 11168 was used to generate the cj0371 deletion mutant and complement strain. C. jejuni were routinely grown on Campy blood-free selective medium (CCDA; Oxid Ltd., UK) or Mueller-Hinton agar (MH; BD Ltd., USA) microaerobically [85% N2(v/v), 10% CO2(v/v), and 5% O2(v/v)] in a jar at 42°C. E. coli DH5α that was used for cloning purposes was routinely cultured in Luria-Bertani (LB) medium at 37°C overnight. When necessary, culture media were supplemented with appropriate antibiotics, chloramphenicol (20 μg/ml), kanamycin (50 μg/ml), or C. jejuni supplement (SR0204E, Oxid Ltd., Basingtoke, UK).
Table 1.
Bacterial strains and plasmids used in this study.
| Strains or plasmids | Genotype | Source or reference |
|---|---|---|
| STRAINS | ||
| E.coli | ||
| DH5ɑ | endA1 hsdR17 (r−K m+K) supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR [Φ80dlac (lacZ ΔM15)] | Invitrogen |
| DH5ɑ-pRK2013 | Conjugation helper strain, DH5ɑ containing plasmid PRK2013 | Alkemade et al., 2003 |
| C. jejuni | ||
| C. jejuni 11168 | C. jejuni NCTC11168 | National Collection of Type Cultures |
| C. jejuni Δ cj0371 | C. jejuni NCTC11168 cj0371::cmr | This study |
| C. jejuni Δ cj0371+ | C. jejuni NCTC11168 cj0371::cmr (pOUA18-PmetK-cj0371) | This study |
| PLASMIDS | ||
| pMD-19T | T clone vector, Aampr | Takara |
| pMD-20T | T clone vector, Aampr | Takara |
| pUOA18 | E. coli-Campylobacter shuttle vector, Cmr | |
| pUOA18-PmetK | E. coli-Campylobacter shuttle vector with metK gene promoter | This study |
| pRY107 | Kanr resistance plasmid, E. coli-Campylobacter shuttle vector | |
| pRK2013 | E. coli-Campylobacter shuttle helper plasmid | Alkemade et al., 2003 |
| pRY107-egfp | E. coli-Campylobacter shuttle vector with egfp gene | This study |
Bioinformatics analysis
ProtParam was used to analyze the physicochemical properties of the Cj0371 protein (http://web.expasy.org/protparam/). Cj0371 has one signaling peptide and one transmembrane structure predicted by CBS Prediction Servers (http://www.cbs.dtu.dk/services/). EMBL String was used to find homologs of Cj0371 and investigate cj0371 genetic structure. All BLAST searches were conducted within BioEdit and a BLAST database was also made using BioEdit (Reuter et al., 2015).
Generation of C. jejuni 11168 Δcj0371 and C. jejuni 11168 Δcj0371+
Deletion of the cj0371 gene was achieved by homologous recombination using a suicide vector containing a homologous sequence on either side of the cj0371 gene as described previously (Miller et al., 1988; Guo et al., 2008; Javed et al., 2012; Handley et al., 2015). Briefly, the cj0371 open reading frame, including 200 bp of upstream sequence and 180 bp of downstream sequence, was amplified by PCR from C. jejuni 11168 using primers cj0371-F1 and cj0371-R1 (Table 2). Then, the fragments were cloned into pMD-19T to generate pMD-19T-cj0371. Pst I was the only restriction site in 338~393 bp of cj0371 open reading frame. The Cmr cassette was amplified from pUOA18 using Cmr-F and Cmr-R primers and ligated to pMD-19T to generate pMD-19T-Cmr. Using Pst I to digest pMD-19T-cj0371, pMD-19T-Cmr, and the Cmr fragment was ligated into pMD-19T-cj0371 to generate the suicide plasmid pMD-19T-cj0371-Cmr. Transferring the suicide plasmid into DH5α cells, transformants were selected on plates containing Ampicillin by blue-white colonies selection. After screening, one clone containing the cassette inserted in the same orientation as cj0371 was used to mutate C. jejuni 11168 by electroporation and allelic exchange. One Cmr mutant was selected on MH agar containing 20 μg/ml of chloramphenicol. The mutation was confirmed by PCR analysis and nucleotide sequencing.
Table 2.
Primers used in this study.
| Primers | Primer sequences(5′–3′) | Amplicon size | Restriction sites |
|---|---|---|---|
| cj0371-F1 | GTGCAAGCTTCGCAAAAAACAAAAAATT | 1.0 kp | Hind III |
| cj0371-R1 | GCGCGAATTCACTTCTTGCGCTGCAGCA | EcoR I | |
| cmr-F | GATCTGCAGTGGAGCGGACAACGAGTAAA | 1.1 kp | Pst I |
| cmr-R | GATCTGCAGTCAGTGCGACAAACTGGGATT | Pst I | |
| cj0371-F2 | ATAGGATCCATGAAAAAAATCAAAAAA | 606 bp | BamH I |
| cj0371-R2 | GTAGAGCTCTTAAGAGCCAAAAGAAGA | Sac I | |
| cj0371-F3 | CGCGGATCCATGAAAAAAATCAAAAAA | 603 bp | BamH I |
| cj0371-R3 | ATCCGGATCCAGAGCCAAAAGAAGAAC | BamH I | |
| PmetK-F | GCGTCTAGATAATTTCCGCTTGAAAGAGCA | 592 bp | Xba I |
| PmetK-R | CGCGGATCCTCCTTTCATTTAAAATGAACC | BamH I |
Complementation of the cj0371 mutation in C. jejuni Δcj0371 was carried out as descried previously (Zeng et al., 2009; Chandrashekhar et al., 2015). The cj0371 coding sequence along with its promoter region was amplified by PCR using specific primers (cj0371-F2 and cj0371-R2 which contain BamH I and Sac I site at 5′ ends, respectively) and cloned into pMD-20T to generate pMD-20T-cj0371. Using BamH I and Sac I to digest pMD-20T-cj0371, the fragments were cloned into pUOA18-PmetK. The sequence of pUOA18-PmetK-cj0371 was confirmed by PCR. Then, following digestion by Xba I and Sac I, PmetK-cj0371 was cloned into pRY107, which was then digested by the same enzymes. PmetK is the promoter region of metK gene. It was utilized as a strong promoter to ensure cj0371 overexpression. The recombinant plasmids were transformed into E. coli DH5ɑ, and subjected to restriction analysis to confirm that they carried the desired sequence. The plasmids were introduced into C. jejuni Δcj0371 by biparental conjugation as described previously (Guerry et al., 1994). Transconjugants were selected on MH agar plates containing chloramphenicol and Campylobacter supplement (SR0204E Oxid Ltd., UK.). One transconjugant was selected and further confirmed by PCR to verify the presence of the wild-type copy of cj0371 (Yao et al., 1993; Akiba et al., 2006).
In vitro adhesion and invasion assay
The adhesion and invasion potentials of all strains were determined using in vitro adhesion and invasion assays as described earlier (Everest et al., 1992; Tareen et al., 2013). Caco-2 cells were seeded into 24-well plates at semiconfluency (~1 × 105 per well) ~24 h prior to infection. When Caco-2 cells were grown to ~100% confluence, they were washed with PBS and inoculated with 1.0 × 107 C. jejuni, a multiplicity of infection of 100. To investigate adhesion, the infected monolayer cells were incubated for 2 h to allow invasion to occur. Following the invasion period, wells for assaying adhesion were washed 3 times with PBS. Cells were lysed using 0.1% (v/v) Triton X-100 for 7 min at room temperature, serially diluted (10-fold) in PBS and 100 μl of each dilution was spread on a CCDA plate. The plates were incubated for 48 h at 42°C under microaerobic conditions after which colony forming-units (CFUs) were counted. To assess invasion, the C. jejuni suspension was removed after 2 h and the cells were washed 3 times with PBS before further incubation with culture medium supplemented with 100 μg/ml gentamicin. Subsequently, infected cells were rinsed with PBS and lysed with 0.1% Triton X-100; plating of the bacteria was performed as described above.
Infant rabbit colonization assay
Litters of 1-day old New Zealand White infant rabbits were acquired from a commercial breeder (Jinlin infant rabbit farm, Nanjing, Jiangsu, China). The animal experimental design and protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Yangzhou University. The rabbit model was used as previously described (Ritchie et al., 2010, 2012). The infant rabbits were injected with cimetidine (50 mg/kg intraperitoneal injection) 3 h before orogastric inoculation with either 1 × 109 cfu wild type C. jejuni 11168 or Δcj0371, using a size 4 French catheter (Arrow International, Reading, PA). The C. jejuni to be tested were grown on MH agar plates for 24 h at 42°C and were harvested in sodium bicarbonate solution (2.5 g in 100 ml; PH 9), and the final concentration of inocula was adjusted by centrifugation (8 min,1000 rpm) to 5 × 109 CFU/ml. After inoculation, rabbits were euthanized at fixed times after infection, 24 and 48 h. To count the number of C. jejuni CFUs in the cecum and colon, C. jejuni was plated on selective media. Cecum and colon samples were weighed and homogenized in 1 ml PBS, serially diluted, and spotted 20 μl on CCDA plates containing Cefoperazone (32 μg/ml) and Amphotericin (10 μg/ml) for enumeration of CFU per gram of tissue. The plates were incubated at 42°C miroaerobically. Some rabbits were not colonized by C. jejuni and these rabbits were excluded from all further analyses. The limit of detection was 50 CFU/ml or 50 CFU/g.
Subcellular localization of Cj0371 protein
To determine where the Cj0371 carries out its function in C. jejuni, we used GFP tagged Cj0371 to investigate its subcellular localization by fluorescence microscopy. First, we constructed C. jejuni Δcj0371 expressing a functional green fluorescent protein of Cj0371. Using primers cj0371-F3 and cj0371-R3 (Table 2), we amplified cj0371 without its termination codon from C. jejuni 11168, and cloned it into pMD-19T to generate pMD-19T-cj0371. BamH I was used to digest pMD-19T-cj0371 and pUOA18-PmetK, then the cj0371 fragment was cloned into pUOA18-PmetK to generate pUOA18-PmetK-cj0371, following by cloning the PmetK-cj0371 fragment into pRY107-egfp. The recombinant plasmids pRY107-PmetK-cj0371-egfp were then transformed into E. coli DH5ɑ and subjected to restriction analysis to confirm that they carried the desired sequence. The plasmids were introduced into C. jejuni Δcj0371 by biparental conjugation. Transconjugants were selected and confirmed as described above. The resulting strains were grown in MH liquid medium to OD600 of 0.4 and washed with PBS once. Ten microliters of the culture volume was loaded on slides, then spotted with 10 μl 4% paraformaldehyde in the bacterial suspension and observed on a Leica SP8 STED 3X confocal microscope.
Motility plate assay and TEM imaging
The motility of mutant and complement strains was examined as previously described (Sommerlad and Hendrixson, 2007). Briefly, C. jejuni strains were grown on CCDA agar for 48 h in microaerobic conditions at 42°C. Then, the bacteria were resuspended in pre-warmed phosphate-buffered saline (PBS), and the optial density at 600 nm (OD600) of the bacterial solution to be tested was adjusted to 1.0. Each tested strain was spotted on the centers of duplicate MH plates and test tubes containing 0.4% agar. The plates were then incubated under microaerobic conditions at 42°C. After culturing for 48 h, the swarming diameter of the tested strains were compared to the wild-type C. jejuni 11168.
To examine the flagellar architecture of the mutants, 1%(wt/vol) ammonium molybdate-stained grids were used as previously described (Kalmokoff et al., 2006). C. jejuni strains were grown on CCDA agar plates, and diluted to an OD600 of 0.4. The bacteria were pelleted and resuspended in 2% glutaraldehyde (PH 7.0) for 1 h at room temperature, and samples were stained with 1% (wt/vol) ammonium molybdate-stained grids and visualized with a Tecnai 12 transmission electron microscope at an accelerating voltage of 120 kV.
Assessment of growth
All C. jejujni strains were cultured on CCDA agar for 24 h in microaerobic conditions at 42°C. Bacteria was harvested from plates and transferred to MH broth. The OD600 of the bacterial cultures to be tested was adjusted to 1.0 and diluted 10-fold. The bacterial suspensions were then subpackaged into 8 anaerobic tubes with 4 ml in each tube and cultured at 37°C with shaking (180 rpm). Every 3 h, a tube of each strain was removed to observe the OD600.
Chemotaxis assay
Using a modified hard-agar plug (HAP) procedure, we tested the chemotaxis of wild type and mutant C. jejuni (Hugdahl et al., 1988; Li et al., 2014). Each strain was grown on MH agar plates with 5% (v/v) sheep blood for 24 h at 42°C microaerobically. The bacteria were harvested with PBS, and the concentration was spectrophotometrically adjusted to 1 × 109 CFU/ml. To prepare the HAPs, we made a 4% agar solution by dissolving 20 g of agar powder in 500 ml of PBS, autoclaving the solution, and added the test chemical in the 4% agar solution tempered at 60°C. Test chemicals were dissolved in PBS at 0.2 M and then filter sterilized (0.22-μm pore-size). The test chemical solution of 10 ml was added to 10 ml of the 4.0% agar solution, mixed, and poured into petri dishes (90 × 15 mm). After cooling, the agar was cut into 8 mm-diameter plugs (HAPs) with sterile, hollow, bullet casings (8-mm inner diameter). Five milliliters bacterial suspension was mixed with the same volume of heat-melted 0.8% agar in PBS at 42°C, and then poured into petri dishes (90 × 15 mm). HAPs containing a test chemical were placed with sterile toothpicks in soft agar. The plates were incubated microaerobically at 42°C for 6 h, and the diameters of accumulative bacterial rings toward each of the attractants in the plates were measured.
Statistical analysis
Data analysis was performed using GraphPad. Statistical significance of the data was determined using Student′s t-test in cases, where only two data sets were compared. The values of P < 0.05 was considered statistically significant.
Results
Characterization of the cj0371 mutant and complement
According to the results of PCR and sequencing, cj0371 was disrupted by the insertion of the Cmr sequence. The PCR and sequencing data also confirmed that the disruption of cj0371 in C. jejuni Δcj0371 was complemented with the correct ORF compared to the wild type strain using the complementation vector. In this study, the sequences of the primers for the gene-disrupted or complemented mutant were derived from chromosomal DNA of C. jejuni 11168. Furthermore, we used an anti-Cj0371 monoclonal antibody (our group had made) to confirm that the Cj0371 protein in the mutant was undetectable. The complement mutant presented the Cj0371 protein at a higher level than the wild type strain (see the Supplementary Material).
cj0371 is a virulence-associated gene
The invasion ability of C. jejuni is an important pathogenicity-associated factor (Dasti et al., 2010). Many previous studies concluded that early mucosal damage is a result of invasion of C. jejuni into the epithelial cells (Field et al., 1986; Babakhani et al., 1993). First, we carried out an adhesion and invasion assay to examine the ability of C. jejuni Δcj0371 to infect Caco-2 cells compared to C. jejuni 11168. Importantly, we did not observe significant differences (P > 0.05) in adhesion ability between C. jejuni 11168 and C. jejuni Δcj0371 (Figure 1A), but C. jejuni Δcj0371 showed significantly increased invasion ability (P < 0.01) compared to C. jejuni 11168 (Figure 1B). Similarly, C. jejuni Δcj0371 exhibited slightly increased colonization ability (though without statistically significant differences) in infant rabbits 24 h after infection, and at 48 h post-infection, the colonization level of C. jejuni 11168 was significantly (P < 0.05) less than C. jejuni Δcj0371 (Figure 2). These results indicate that cj0371 is a virulence-associated gene.
Figure 1.
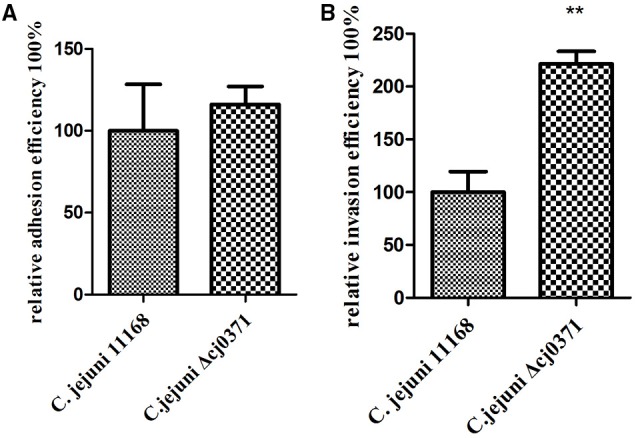
In vitro adherence and invasion assay of C. jejuni 11168 and C. jejuni Δcj0371. The adhesion phenotype of C. jejuni 11168 and C. jejuni Δcj0371 (A) and the invasion phenotype (B) were determined using Caco-2 cells. The adhesion ability of the cj0371 mutant is similar to the wild type (P > 0.05), but compared to the wild type, C. jejuni Δcj0371 showed significantly increased invasion ability (P < 0.01). *P ≤ 0.01; **P ≤ 0.05; ***P ≤ 0.001.
Figure 2.
Infant rabbit colonization assay of C. jejuni 11168 and C. jejuni Δcj0371. At 24 h PI, the colonization level of C. jejuni 11168 and C. jejuni Δcj0371 has no significant differences (P > 0.05). However, 48 h PI, the colonization level of C. jejuni Δcj0371 is significantly high than C. jejuni 11168 (P < 0.05). *P ≤ 0.01; **P ≤ 0.05; ***P ≤ 0.001.
cj0371 localizes at C. jejuni cellular poles
We constructed C. jejuni Δcj0371 expressing functional Cj0371-GFP and GFP, and observed its fluorescence using confocal microscopy. We found that Cj0371-GFP localized at the two C. jejuni poles (Figure 3). Given the polar localization of flagella in C. jejuni, we reasoned that this gene may play a direct or indirect role in the flagellar system. This observation led us to question a role for cj0371 in flagellar biosynthesis and function.
Figure 3.
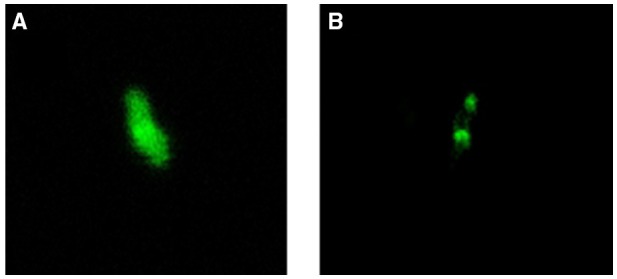
Subcellular localization of the Cj0371 protein. (A) C. jejuni Δcj0371 expressing GFP. (B) C. jejuni Δcj0371 expressing Cj0371-GFP proteins.
cj0371 doesn't directly influence C. jejuni flagellar biosynthesis
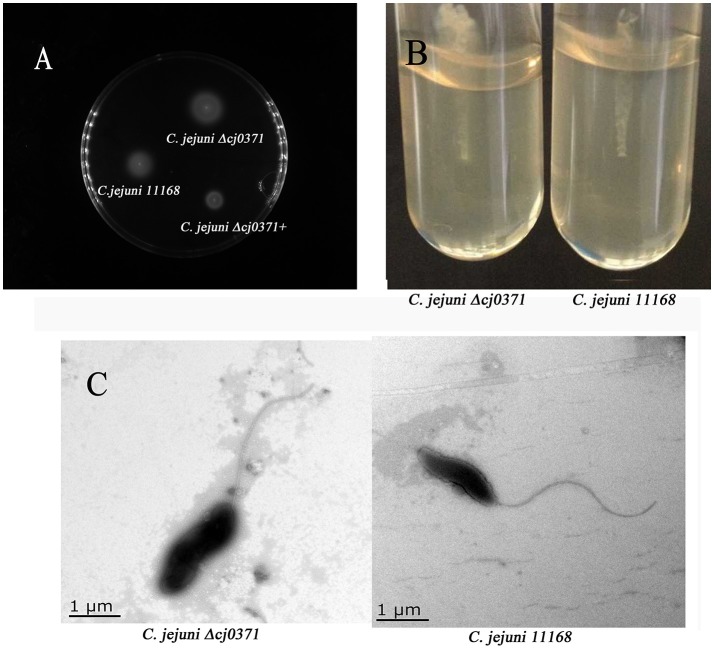
We used a soft-agar motility plate assay to compare the motility of C. jejuni Δcj0371, C. jejuni 11168, and C. jejuni Δcj0371+ (Figures 4A,B). Surprisingly, we found that C. jejuni Δcj0371 exhibited hypermotility and C. jejuni Δcj0371+ showed a motility defect. Potentially, a motility defect phenotype could not be effectively shown by a soft-agar motility plate assay (Gao et al., 2014). So we observed the flagella of C. jejuni Δcj0371 and wild type C. jejuni by transmission electron microscopy (TEM; Figure 4C). However, we did not find any difference on the bacterial surface; C. jejuni Δcj0371 exhibited apparently normal flagella at its poles, indicating that cj0371 is not directly associated with flagellar synthesis and assembly. We additionally quantified the proportion of bacteria exhibiting one or both poles with no polar flagella, but there was no difference between the mutant and wild type strain (data not shown).
Figure 4.
Motility plate assay and TEM imaging. (A) Motility analysis of C. jejuni 11168, the cj0371 mutant and the complemented strain on soft agar. WT, wild type C. jejuni 11168-, C. jejuni Δcj0371+, C. jejuni Δcj0371+. (B) C. jejuni 11168 and C. jejuni Δcj0371 were cultured in a soft agar tube for 48 h to analyze motility. (C) Transmission electron microscopy analysis of C. jejuni 11168 and the cj0371 mutant strain.
cj0371 influences chemotactic behaviors of C. jejuni
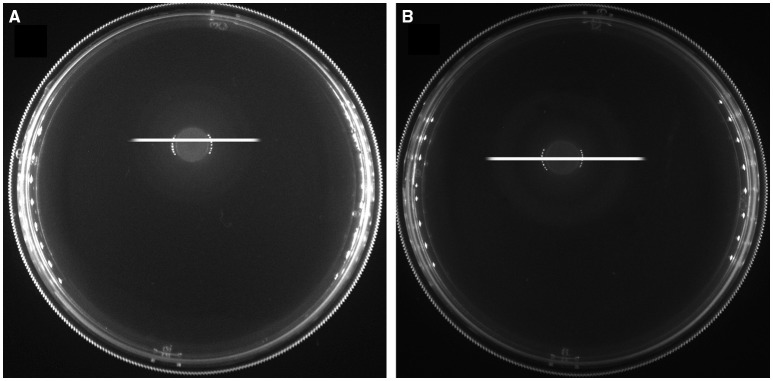
Previously, studies have speculated that C. jejuni use chemotaxis to reach particular milieu (Chang and Miller, 2006). We selected DL-malic acid, ketoglutaric acid and succinic acid, which are confirmed chemoattractants (Hugdahl et al., 1988), and utilized HAPs procedures to assay the chemotaxis of C. jejuni. We found the chemotaxis of C. jejuni Δcj0371 is stronger than C. jejuni 11168 (Figure 5 Table 3). Thus, we concluded that cj0371 is associated with C. jejuni chemotaxis.
Figure 5.
Chemotaxis of the mutant and wild-type strain. The zones of accumulation (chemoattractant) of C. jejuni Δcj0371 and C. jejuni 11168 after 6 h of incubation at 42°C. The plug contains 0.1 M DL-malic acid and the length of white lines are C. jejuni 11168 (A) and C. jejuni Δcj0371 (B) diameters of chemotaxis rings.
Table 3.
The diameters of chemotactic rings with different attractants.
| Attractant (0.1 M) | C. jejuni 11168 diameter of chemotactic ring (mm) | C. jejuniΔcj0371 diameter of chemotactic ring (mm) |
|---|---|---|
| α-ketoglutarate | 27 | 38 |
| succinate | 19 | 21 |
| DL-malic acid | 27 | 35 |
Mutation of cj0371 influences C. jejuni growth
To estimate C. jejuni Δcj0371 growth, we observed its growth curve compared to the wild-type C. jejuni 11168. We used anaerobic tubes to culture C. jejuni. According to the growth curves (Figure 6), C. jejuni Δcj0371 displays a growth increase compared with C. jejuni 11168 in MH liquid culture. C. jejuni Δcj0371 grows significantly faster than C. jejuni 11168 (6 h, P < 0.05; 12 h, P < 0.001; 18 h, P < 0.001; 24 h, P < 0.001; 30 h, P < 0.001; 36 h, P < 0.01). C. jejuni Δcj0371+ grows slower than mutant, and C. jejuni 11168 has the slowest growth rate.
Figure 6.
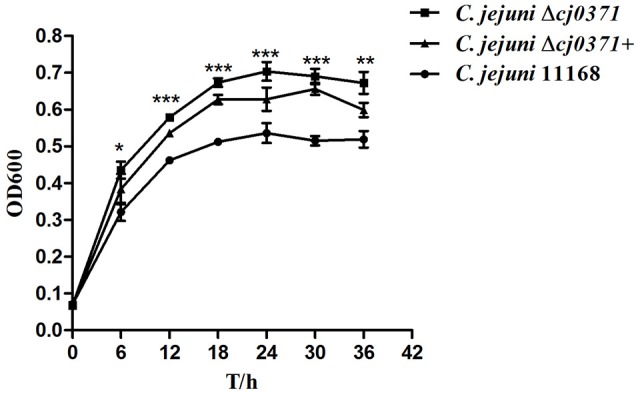
Growth kinetics of C. jejuni 11168 and the cj0371 mutant in MH broth. The strains were cultured under microaerobic conditions at 42°C. At indicated time points, bacterial suspensions in anaerobic tubes were taken to measure OD600. The experiment was repeated three times, and the results of one representative experiment are shown. *P ≤ 0.01; **P ≤ 0.05; ***P ≤ 0.001.
Discussion
To our knowledge, cj0371 was an unknown gene prior to this report. According to previous studies that investigated novel genes, we deleted the target gene from the wild-type and used a vector with a strong promoter to complement it. By observing the biological characteristics of the mutant and complement strain compared with the wild type, we expected to find a unique insight into the function of cj0371.
Adhesion and colonization of animal tissue by C. jejuni is an important step in establishing infection (Morooka et al., 1985). Mutation of cj0371 was able to significantly increase invasion of C. jejuni into Caco-2 cells compared to wild-type C. jejuni (Figure 1). In addition, mutant strain colonized in distal gastrointestinal tract of infant rabbits better than wild type C. jejuni. These results suggested that cj0371 is associated with C. jejuni virulence and that it is a negative control factor. To further prove our conclusion, we tested the expression level of cj0371 when C. jejuni 11168 infected HD-11 cell, we found that cj0371 showed up-regulated expression compared with the strain cultured in vitro (see the Supplementary Material). It is worth stressing that we selected infant rabbits as the experimental animals rather than poultry (such as chicken). One reason is that the infant rabbit colonization model has been successfully used for this pathogenic bacteria (publication pending), and another is that while poultry is the natural repository of C. jejuni, we could not successfully isolate C. jejuni from chickens after oral challenge. This could be explained by the presence of protective maternal antibodies in chick sera in the first week (Meunier et al., 2015).
Now, the recognized virulence factors of C. jejuni include motility, chemotaxis, colonization, adherence and invasion, cytolethal distending toxin (Cdt A,B,C; Dasti et al., 2010). cj0371 was selected by IVIAT, so we speculated it was a virulence-associated gene. Its amino acid sequence is homologous to Cj8486_0361 (100%), which is defined as a putative flagellar motility protein [C. jejuni subsp. jejuni CG8486] (GenBank: EDK21732.1). Flagellar systerm and is energy burden associated with motility, so many mutations affecting unrelated physiological processes can indirectly affect motility (Nielubowicz et al., 2010; Neal-McKinney and Konkel, 2012). Because Cj0371 localizes at the C. jejuni cellular poles (Figure 3) and C. jejuni has a single flagellum at both poles, meanwhile Cj0371 has a signaling peptide and transmembrane domains, we speculated that cj0371 may influence flagellar assembly and/or function. However, neither a soft-agar plate assay to observe the motility of the cj0371 mutant and complemented strain, nor observation of the flagella on the surface of C. jejuni using TEM revealed significant evidence to confirm that cj0371 is associated with flagellar synthesis and assembly.
cj0371 does not influence flagellar assembly and biosynthesis, but it can increase the virulence of C. jejuni, so we speculated that it may control other factors such as chemotaxis, growth and metabolism. Another important finding of our work is that cj0371 influences C. jejuni chemotaxis. Chemotaxis allows motile bacteria to travel toward a favorable niche or away from unfavorable conditions (Marchant et al., 2002). Cellular motility and chemotaxis have been implicated in the colonization and virulence of pathogenic bacteria and have a role in invasion and colonization of the host intestinal tract (Hartley-Tassell et al., 2010). To examine the chemotactic behaviors of cheB, cheR, and cheBR mutants, Kanungpean et al. carried out a semisolid agar motility assay (Kanungpean et al., 2011). In other worlds, motility is not only regulated by flagella but also controlled by chemotactic factors. The bacteria use a signaling cascade of protein phosphorylation and dephosphorylation reactions to control bacterial motors in response to environmental chemical changes (Miller et al., 2009). The direct association of the chemotaxis system with the flagellar apparatus affects the bacterial motility of C. jejuni (Zautner et al., 2012). In our study, the result of the motility assay is consistent with chemotaxis assay. Hypermotility and increased chemotaxis resulting in enhanced invasion and colonization seems reasonable. Including the data from the growth kinetic assay, hyperchemotaxis may allow C. jejuni Δcj0371 to travel toward a favorable niche, which benefits its growth. As for the cj0371 complement growing faster than the wild-type C. jejuni, we propose that despite driving expression of the gene with a strong promotor, this may not result in overexpression in liquid medium. Of course, these conclusions are still inferences that need to be corroborated with further research.
Based on the results of invasion and colonization experiments, we discerned cj0371 is a virulence-associated gene. However, the cj0371 mutant has an increased capacity for invasion and colonization, so we conclude that it plays a negative role in pathogenicity, which is expected to be suppressed usually during infection. But cj0371 was identified by IVIAT screening, suggesting this gene was induced during host infection. Actually, the genes screened by IVIAT are virulence-associated genes, including classical virulence genes and other virulence-associated genes (Rollins et al., 2005). We present data that collectively is not enough to prove cj0371 is a typical virulence gene. So we temporarily call it virulence-association gene. This study presents an insight into the pathogenic mechanisms of C. jejuni and provides a keen interest to pursue in further study. In future studies, we intend to use co-immunoprecipitation to discover the protein that interact with Cj0371, followed by identification of interacting protein by liquid chromatography-mass spectrometry analysis. Through uncovering protein interactions we hope to further elucidate the function of Cj0371.
Author contributions
XD as the first author, she participate to do all of the work including statistical analysis and this manuscript also was written by her. NW made the mutant strain and complement strain. FR and HT also participate to do part of the work. XJ and JH are tutor.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the All-Purpose Laboratory of the college of veterinary medicine in Yangzhou University for their support with the laser scanning confocal microscope (Leica SP8 STED 3X). We would also like to thank the testing central of Yangzhou University for their support of the Tecnai 12 transmission electron microscope. This work was supported by National Natural Science Foundation of China grant (31372449), National Key Technology R&D Program (2014BAD13B02), Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Six talent peaks project in Jiangsu Province (2015-SWYY-02).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01094
References
- Akiba M., Lin J., Barton Y. W., Zhang Q. (2006). Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J. Antimicrob. Chemother. 57, 52–60. 10.1093/jac/dki419 [DOI] [PubMed] [Google Scholar]
- Aliouane S., Pages J. M., Bolla J. M. (2016). Cloning, expression, purification, regulation, and subcellular localization of a mini-protein from Campylobacter jejuni. Curr. Microbiol. 72, 511–517. 10.1007/s00284-015-0980-x [DOI] [PubMed] [Google Scholar]
- Babakhani F. K., Bradley G. A., Joens L. A. (1993). Newborn piglet model for campylobacteriosis. Infect. Immun. 61, 3466–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekhar K., Gangaiah D., Pina-Mimbela R., Kassem I. I., Jeon B. H., Rajashekara G. (2015). Transducer like proteins of Campylobacter jejuni 81-176: role in chemotaxis and colonization of the chicken gastrointestinal tract. Front. Cell. Infect. Microbiol. 5:46. 10.3389/fcimb.2015.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Miller J. F. (2006). Campylobacter jejuni colonization of mice with limited enteric flora. Infect. Immun. 74, 5261–5671. 10.1128/IAI.01094-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Jiang X. (2014). Microbiological safety of chicken litter or chicken litter-based organic fertilizers: a review. Agriculture 4, 1–29. 10.3390/agriculture4010001 [DOI] [Google Scholar]
- Dash S., Pai A. R., Kamath U., Rao P. (2015). Pathophysiology and diagnosis of Guillain-Barre syndrome - challenges and needs. Int. J. Neurosci. 125, 235–240. 10.3109/00207454.2014.913588 [DOI] [PubMed] [Google Scholar]
- Dasti J. I., Tareen A. M., Lugert R., Zautner A. E., Gross U. (2010). Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int. J. Med. Microbiol. 300, 205–211. 10.1016/j.ijmm.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Everest P. H., Goossens H., Butzler J. P., Lloyd D., Knutton S., Ketley J. M., et al. (1992). Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J. Med. Microbiol. 37, 319–325. 10.1099/00222615-37-5-319 [DOI] [PubMed] [Google Scholar]
- Field L. H., Headley V. L., Underwood J. L., Payne S. M., Berry L. J. (1986). The chicken embryo as a model for campylobacter invasion: comparative virulence of human isolates of Campylobacter jejuni and Campylobacter coli. Infect. Immun. 54, 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B., Lara-Tejero M., Lefebre M., Goodman A. L., Galan J. E. (2014). Novel components of the flagellar system in epsilonproteobacteria. MBio 5, e01349–e01314. 10.1128/mBio.01349-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., Yao R., Alm R. A., Burr D. H., Trust T. J. (1994). Systems of experimental genetics for Campylobacter species. Methods Enzymol. 235, 474–481. 10.1016/0076-6879(94)35163-5 [DOI] [PubMed] [Google Scholar]
- Guo B. Q., Wang Y., Shi F., Barton Y. W., Plummer P., Reynolds D. L., et al. (2008). CmeR functions as a pleiotropic regulator and is required for optimal colonization of Campylobacter jejuni in vivo. J. Bacteriol. 190, 1879–1890. 10.1128/JB.01796-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley R. A., Mulholland F., Reuter M., Ramachandran V. K., Musk H., Clissold L., et al. (2015). PerR controls oxidative stress defence and aerotolerance but not motility-associated phenotypes of Campylobacter jejuni. Microbiology 161, 1524–1536. 10.1099/mic.0.000109 [DOI] [PubMed] [Google Scholar]
- Hartley-Tassell L. E., Shewell L. K., Day C. J., Wilson J. C., Sandhu R., Ketley J. M., et al. (2010). Identification and characterization of the aspartate chemosensory receptor of Campylobacter jejuni. Mol. Microbiol. 75, 710–730. 10.1111/j.1365-2958.2009.07010.x [DOI] [PubMed] [Google Scholar]
- Havelaar A. H., van Pelt W., Ang C. W., Wagenaar J. A., van Putten J. P. M., Gross U., et al. (2009). Immunity to Campylobacter: its role in risk assessment and epidemiology. Crit. Rev. Microbiol. 35, 1–22. 10.1080/10408410802636017 [DOI] [PubMed] [Google Scholar]
- Hu Y. Q., Huang J. L., Jiao X. A. (2014a). Screening of genes expressed in vivo during interaction between chicken and Campylobacter jejuni. J. Microbiol. Biotechnol. 24, 217–224. 10.4014/jmb.1308.08092 [DOI] [PubMed] [Google Scholar]
- Hu Y. Q., Huang J. L., Li Q. C., Shang Y. W., Ren F. Z., Jiao Y., et al. (2014b). Use of in vivo-induced antigen technology to identify in vivo-expressed genes of Campylobacter jejuni during human infection. J. Microbiol. Biotechnol. 24, 363–370. 10.4014/jmb.1311.11019 [DOI] [PubMed] [Google Scholar]
- Hugdahl M. B., Beery J. T., Doyle M. P. (1988). Chemotactic behavior of Campylobacter jejuni. Infect. Immun. 56, 1560–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed M. A., Cawthraw S. A., Baig A., Li J. J., McNally A., Oldfield N. J., et al. (2012). Cj1136 is required for lipooligosaccharide biosynthesis, hyperinvasion, and chick colonization by Campylobacter jejuni. Infect. Immun. 80, 2361–2370. 10.1128/IAI.00151-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmokoff M., Lanthier P., Tremblay T. L., Foss M., Lau P. C., Sanders G., et al. (2006). Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 188, 4312–4320. 10.1128/JB.01975-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanungpean D., Kakuda T., Takai S. (2011). Participation of CheR and CheB in the chemosensory response of Campylobacter jejuni. Microbiology 157, 1279–1289. 10.1099/mic.0.047399-0 [DOI] [PubMed] [Google Scholar]
- Li Z., Lou H., Ojcius D. M., Sun A., Sun D., Zhao J., et al. (2014). Methyl-accepting chemotaxis proteins 3 and 4 are responsible for Campylobacter jejuni chemotaxis and jejuna colonization in mice in response to sodium deoxycholate. J. Med. Microbiol. 63, 343–354. 10.1099/jmm.0.068023-0 [DOI] [PubMed] [Google Scholar]
- Marchant J., Wren B., Ketley J. (2002). Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol. 10, 155–159. 10.1016/S0966-842X(02)02323-5 [DOI] [PubMed] [Google Scholar]
- Meunier M., Guyard-Nicodeme M., Dory D., Chemaly M. (2015). Control strategies against Campylobacter at the poultry production level: biosecurity measures, feed additives and vaccination. J. Appl. Microbiol. 120, 1139–1173. 10.1111/jam.12986 [DOI] [PubMed] [Google Scholar]
- Miller J. F., Dower W. J., Tompkins L. S. (1988). High-voltage electroporation of bacteria: genetic transformation of Campylobacter jejuni with plasmid DNA. Proc. Natl. Acad. Sci. U.S.A. 85, 856–860. 10.1073/pnas.85.3.856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. D., Russell M. H., Alexandre G. (2009). Diversity in bacterial chemotactic responses and niche adaptation. Adv. Appl. Microbiol. 66, 53–75. 10.1016/S0065-2164(08)00803-4 [DOI] [PubMed] [Google Scholar]
- Morooka T., Umeda A., Amako K. (1985). Motility as an intestinal colonization factor for Campylobacter jejuni. Microbiology 131, 1973–1980. 10.1099/00221287-131-8-1973 [DOI] [PubMed] [Google Scholar]
- Neal-McKinney J. M., Konkel M. E. (2012). The Campylobacter jejuni CiaC virulence protein is secreted from the flagellum and delivered to the cytosol of host cells. Front. Cell. Infect. Microbiol. 2:31. 10.3389/fcimb.2012.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielubowicz G. R., Smith S. N., Mobley H. L. T. (2010). Zinc uptake contributes to motility and provides a competitive advantage to proteus mirabilis during experimental urinary tract infection. Infect. Immun. 78, 2823–2833. 10.1128/IAI.01220-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Periago P. M., Mulholland F., Brown H. L., van Vliet A. H. (2015). A PAS domain-containing regulator controls flagella-flagella interactions in Campylobacter jejuni. Front. Microbiol. 6:770. 10.3389/fmicb.2015.00770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie J. M., Rui H. P., Bronson R. T., Waldor M. K. (2010). Back to the future: studying cholera pathogenesis using infant rabbits. MBio 1, e00047–e00010. 10.1128/mBio.00047-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie J. M., Rui H. P., Zhou X. H., Iida T., Kodoma T., Ito S., et al. (2012). Inflammation and disintegration of intestinal villi in an experimental model for vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 8:e1002593. 10.1371/journal.ppat.1002593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins S. M., Peppercorn A., Hang L., Hillman J. D., Calderwood S. B., Handfield M., et al. (2005). In vivo induced antigen technology (IVIAT). Cell. Microbiol. 7, 1–9. 10.1111/j.1462-5822.2004.00477.x [DOI] [PubMed] [Google Scholar]
- Sommerlad S. M., Hendrixson D. R. (2007). Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J. Bacteriol. 189, 179–186. 10.1128/JB.01199-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareen A. M., Luder C. G. K., Zautner A. E., Gross U., Heimesaat M. M., Bereswill S., et al. (2013). The Campylobacter jejuni Cj0268c protein is required for adhesion and invasion in vitro. PLoS ONE 8:e81069. 10.1371/journal.pone.0081069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Taylor D. E. (1990). Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94, 23–28. 10.1016/0378-1119(90)90463-2 [DOI] [PubMed] [Google Scholar]
- Yao R., Alm R. A., Trust T. J., Guerry P. (1993). Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130, 127–130. 10.1016/0378-1119(93)90355-7 [DOI] [PubMed] [Google Scholar]
- Young K. T., Davis L. M., Dirita V. J. (2007). Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5, 665–679. 10.1038/nrmicro1718 [DOI] [PubMed] [Google Scholar]
- Zautner A. E., Tareen A. M., Gross U., Lugert R. (2012). Chemotaxis in Campylobacter jejuni. Eur. J. Microbiol. Immunol. 2, 24–31. 10.1556/EuJMI.2.2012.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Xu F., Lin J. (2009). Molecular, antigenic, and functional characteristics of ferric enterobactin receptor CfrA in Campylobacter jejuni. Infect. Immun. 77, 5437–5448. 10.1128/IAI.00666-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.