Abstract
Background
To improve current treatment strategies for patients with aggressive colorectal cancer (CRC), the molecular understanding of subgroups of CRC with poor prognosis is of vast importance. SOX2 positive tumors have been associated with a poor patient outcome, but the functional role of SOX2 in CRC patient prognosis is still unclear.
Methods
An in vitro cell culture model expressing SOX2 was used to investigate the functional role of SOX2 in CRC. In vitro findings were verified using RNA from fresh frozen tumor tissue or immunohistochemistry on formalin fixed paraffin embedded (FFPE) tumor tissue from a cohort of 445 CRC patients.
Results
Using our in vitro model, we found that SOX2 expressing cells displayed several characteristics of cancer stem cells; such as a decreased proliferative rate, a spheroid growth pattern, and increased expression of stem cell markers CD24 and CD44. Cells expressing SOX2 also showed down-regulated expression of the intestinal epithelial marker CDX2. We next evaluated CDX2 expression in our patient cohort. CDX2 down-regulation was more often found in right sided tumors of high grade and high stage. Furthermore, a decreased expression of CDX2 was closely linked to MSI, CIMP-high as well as BRAF mutated tumors. A decreased expression of CDX2 was also, in a stepwise manner, strongly correlated to a poor patient prognosis. When looking at SOX2 expression in relation to CDX2, we found that SOX2 expressing tumors more often displayed a down-regulated expression of CDX2. In addition, SOX2 expressing tumors with a down-regulated CDX2 expression had a worse patient prognosis compared to those with retained CDX2 expression.
Conclusions
Our results indicate that SOX2 expression induces a cellular stem cell state in human CRC with a decreased expression of CDX2. Furthermore, a down-regulated expression of CDX2 results in a poor patient prognosis in CRC and at least part of the prognostic importance of SOX2 is mediated through CDX2 down-regulation.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-016-2509-5) contains supplementary material, which is available to authorized users.
Keywords: SOX2, CDX2, Colorectal cancer, Prognosis, Cancer stem cell
Background
Colorectal cancer (CRC) is a common malignancy worldwide and the second leading cause of cancer deaths in the western world [1]. CRC is often detected at late stages contributing to the high mortality rate seen in this disease. Today, most patients receive a similar stage specific treatment strategy, however not all benefit from it. In future treatment of CRC patients, personalized therapy will be of vast importance, but this will also place higher demands on the molecular subclassification of CRC.
The SOX2 gene encodes for a transcription factor and is a member of the SRY-related HMG-box (SOX) gene family. It is known that SOX2 plays essential roles in cell fate determination, thereby regulating developmental processes [2]. In recent years, aberrant expression of SOX2 has been reported in CRC as well as several other types of cancers [3–6]. According to our previous study, SOX2 expression was found to be correlated to high tumor grade, mutated BRAF and a poor patient prognosis [7]. We further found that the expression of SOX2 was partly regulated by BRAF [7]. Expression of SOX2 has also been associated with distant metastases in right-sided colon cancer [8], suggesting that SOX2 expressing tumors represent a subgroup with poor patient outcome. In CRC, SOX2 has previously been suggested to regulate epithelial-mesenchymal transition (EMT) and increased tumor migration and invasion [9]. However, the functional role of SOX2 in CRC patient prognosis is still unclear.
Recent research has revealed that a small subgroup of tumor cells possesses characteristics associated with stem cells and have therefore been called cancer stem cells (CSCs). CSCs have the ability of self-renewal and multi-lineage differentiation, features that can cause both tumor growth and emergence of new tumors [10–12]. SOX2 expression has been associated with a stem cell state in human ovarian, cervical, pancreatic, head and neck squamous cell, and breast carcinoma [3, 13–16], but so far this has not been shown in CRC. SOX2 expression has been associated with tumors of high grade (poorly differentiated) in different cancers [7, 17–20]. The transcription factor, Caudal type homeobox 2 (CDX2), is a major regulator of the expression of intestine-specific genes involved in cell differentiation [21, 22]. CDX2 is expressed at high levels in the normal colorectal epithelium, but loss or decrease of expression is seen in a subset of CRCs [23, 24]. Previous studies have also reported that loss of CDX2 is associated with poor patient prognosis in CRC [25–27].
In this study we investigated the functional role of SOX2 in CRC using an in vitro cell culture model. We found no evidence that SOX2 was involved in regulation of EMT or cellular migration. However, SOX2 positive cells were found to display several characteristics of cancer stem cells, as well as a decreased expression of the intestinal epithelial marker CDX2. In a cohort of CRC patients, we further demonstrate that SOX2 expression is significantly associated with down-regulated expression of CDX2 and at least part of the prognostic importance of SOX2 is mediated through CDX2 down-regulation. In conclusion, we suggest that CDX2 down-regulation is partly regulated by SOX2 and contributes to a poor prognosis in this patient group.
Methods
Cell culture and cell lines
In this study, the human colon cancer cell lines Caco2, SW480 and SW620 (American Type Culture Collection, Manassas, VA, USA) were grown in Dulbecco’s modified Eagle’s medium (DMEM) with glutamax supplemented with 10 % fetal bovine serum (FBS) (Gibco, Life Technologies, Stockholm, Sweden) and maintained at 37 °C and 5 % CO2. The stable transfectant expressing increased levels of SOX2 has been previously described [7].
Migration assay
Cell migration was analyzed using transwell cell culture inserts with a pore size of 8 μm (BD Biosciences, Stockholm, Sweden) in 24-well plates. Caco2 or Caco2-SOX2 were seeded at a density of 1x105 per insert in cell culture medium with 10 % FBS for 2–3 h. Media was subsequently exchanged to serum-free DMEM and cells were allowed to migrate towards either culture media supplemented with 10 % FBS or serum-free medium for 20 h at 37 °C and 5 % CO2. Cells remaining on the inside of the insert were removed with cotton swabs and the cells that migrated through the membrane were fixed and stained with Coomassie blue (Bio-Rad Laboratories, Solna, Sweden). For quantification, three fields were chosen randomly and migrating cells were counted at x10 magnification using a light microscope. The experiment was repeated three times.
Proliferation assay
Cell proliferation was assessed by the XTT assay (Roche Diagnostics, Bromma, Sweden) according to the manufacturer’s instructions. In brief, Caco2 or Caco2-SOX2 cells were cultured in a 96-well plate at a density of 5x103 per well in cell culture media supplemented with 10 % FBS for 72 h at 37 °C and 5 % CO2. The cells were then incubated with XTT labeling for 4 h at 37 °C before the absorbance was measured with an ELISA reader at a wavelength of 490 nm. A reference wavelength at 650 nm was also measured. Quadruplicates of each sample were analyzed and the experiment was repeated three times.
Real time PCR
The NucleoSpin RNA kit (Macherey-Nagel, Duren, Germany) was used for isolation of total RNA from cultured cells, and cDNA was synthesized with the SuperScript II Reverse Transcriptase (Invitrogen, Life Technologies, Stockholm, Sweden). Fresh frozen human tumor tissue was homogenized using the gentleMACS Dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany) before total RNA was isolated with the High Pure RNA Paraffin Kit (Roche Diagnostics, Stockholm, Sweden) and then converted into cDNA using the SuperScript VILO cDNA Synthesis Kit (Invitrogen, Life Technologies, Stockholm, Sweden). All steps were performed according to manufacturer’s protocols.
In the present study, primers for GAPDH, RPL13A, SOX2, MMP3, MMP11, E-cadherin, Snail and Fibronectin were from DNA Technology A/S (Aarhus, Denmark) and their sequences are listed in Additional file 1. For the remaining genes, Quantitect Primer Assays (Qiagen, Sollentuna, Sweden) were used. Quantitative RT-PCR reactions were performed on an ABI 7900HT instrument (Applied Biosystems, Life Technologies, Stockholm, Sweden) with the following thermal cycling conditions used: 50 °C for 2 min and then an initial denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Gene expression was normalized to GAPDH for cultured cells or RPL13A for fresh frozen tumor specimens. Standard deviations were calculated for the mean of triplicate reactions.
Clinical samples
CRC specimens included in this study were from the Colorectal Cancer in Umeå Study (CRUMS) [28]. Tumor tissue samples were collected from patients with primary CRC that underwent tumor-resective surgery between 1995 and 2003 at Umeå University Hospital, Sweden. Formalin-fixed paraffin-embedded (FFPE) tissue was sampled from all patients and fresh frozen tumor tissue was collected from a subgroup of the patients. One pathologist did all histopathological classifications by reviewing routinely stained tumor sections, as previously described [28]. Clinical data were obtained from the patient records, and survival data was collected during autumn 2012. 445 cases were included in this study, but due to unavailable or insufficient tumor sample or negative staining in adjacent normal colon epithelium (n = 14), 431 of the tumors could be successfully analyzed for CDX2 expression.
Analyses of microsatellite instability (MSI) screening status, CpG island methylator phenotype (CIMP) status and mutational status of BRAF and KRAS have previously been described [29, 30]. In brief, MSI screening status was determined in FFPE tissue samples by immunohistochemical analyses of the expression of four mismatch repair proteins (MLH1, MSH2, MSH6 and PMS2). Tumors lacking nuclear staining for at least one of the four proteins were considered to have a positive MSI screening status, compared to those with a negative MSI screening status, referred to as microsatellite stable (MSS). CIMP status was determined by the MethyLight method with previously described primer and probe sequences. An eight gene panel (CDKN2A, MLH1, CACNA1G, NEUROG1, RUNX3, SOCS1, IGF2, and CRABP1) was used for evaluation of the hypermethylation status: CIMP-negative tumors (no promoter hypermethylation), CIMP-low tumors (one to five genes methylated) or CIMP-high tumors (six to eight genes methylated). BRAFV600E mutation was detected by the Taqman allelic discrimination assay [31] (reagents from Applied Biosystems, Life Technologies, Stockholm, Sweden). KRAS mutational status was determined by sequencing using Big Dye v. 3.1 (Applied Biosystems, Life Technologies, Stockholm, Sweden). The expression of SOX2 has previously been evaluated in this patient cohort [7], where nuclear staining was assessed as either negative or positive.
Immunohistochemistry
FFPE CRC specimens were cut at 4-μm and then dried, deparaffinized and rehydrated. CDX2 mouse monoclonal antibody (clone CDX2-88, Biogenex, Fremont, CA, USA) was used at a dilution of 1:50 and visualized by the iVIEW DAB Detection kit on an Ventana Benchmark Ultra staining machine (Ventana Medical Systems, Tucson, AZ, USA), with the CC1 standard pretreatment. Normal colon mucosa was used as positive control. The slides were counterstained with hematoxylin. Immunohistochemical staining of CDX2 was evaluated under light microscopy by one observer two times under supervision of an experienced pathologist. In cases with discrepant scoring, a third final evaluation was made. Nuclear CDX2 staining was scored as: <5 % positive tumor cells, 5-50 % positive tumor cells or >50 % positive tumor cells. Normal colon mucosa, if included in the sample, was used as an internal positive control.
Statistical analyses
IBM SPSS Statistics software version 21 (SPSS Inc., Chicago, Illinois, USA) was used for statistical analyses. The nonparametric Mann–Whitney U-test was performed in order to compare the differences in gene expression levels between two groups. Cross-tabulations for associations between CDX2 expression and different clinicopathological and molecular variables were analyzed with χ2 tests. To estimate cancer-specific survival, Kaplan-Meier survival analysis was used, and the log-rank test was used for comparisons between groups. Cancer-specific survival was defined as death with known disseminated or recurrent disease. Patients lacking survival data or patients who died with postoperative complications within one month after surgery (n = 34) were excluded from the survival analyses. For multivariable analyses, Cox proportional hazard models were used. P < 0.05 was considered statistically significant for all analyses.
Results
To gain mechanistic insights to the prognostic importance of SOX2 in CRC, we created a stable transfectant of the CRC cell line Caco2 expressing increased levels of SOX2 (Caco2-SOX2) as previously described [7]. The Caco2 cell line was chosen to represent one of the largest subgroups of sporadic CRCs; CIMP negative, MSS and wild-type in KRAS and BRAF [32].
SOX2 is not a major regulator of EMT and cellular migration in CRC cells
In CRC, SOX2 has previously been linked to EMT and increased migration and invasion [9]. EMT is linked to changes in expression of several transcription factors and cellular adhesion molecules. To investigate these events in our in vitro model, we compared the expression of EMT related factors in Caco2 and Caco2-SOX2 cells. Even though the epithelial marker E-cadherin (CDH1) was found to be significantly decreased by SOX2, the expression of the transcription factors Snail (SNAI1), Slug (SNAI2) and Twist1, controlling E-cadherin expression were unaltered or decreased (Fig. 1a). Furthermore, expression of Fibronectin, N-cadherin and Vimentin, associated with a mesenchymal phenotype, were not increased but instead severely decreased in SOX2 expressing cells (Fig. 1a). We also investigated the expression of several matrix metalloproteinases (MMPs) to examine possible effects on the extracellular matrix, caused by SOX2 expression. Most of the investigated MMPs were found to be down-regulated in Caco2-SOX2 cells (Fig. 1b). We further compared the migratory ability of Caco2 and Caco2-SOX2 cells using Boyden transwell migration experiments. Cellular migration was found to be significantly decreased in Caco2 cells expressing SOX2 compared to Caco2 wild type cells (Fig. 1c). Together, these results incline that SOX2 is not a major regulator of EMT or cellular migration and invasion in our in vitro model.
Fig. 1.
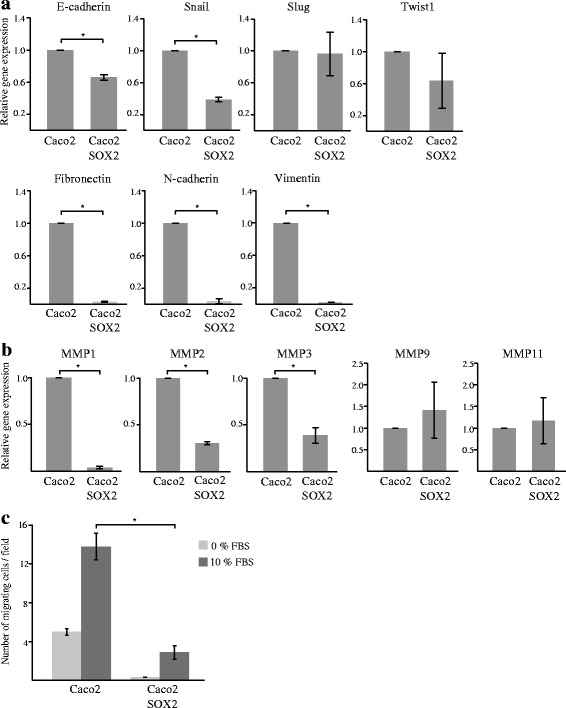
Evaluation of EMT and cellular migration in response to SOX2 expression. Caco2 cells and Caco2 cells stably transfected with SOX2 (Caco2-SOX2) was analyzed by RT-PCR for the expression of a EMT related genes, or b MMPs. Shown is relative gene expression from three or more independent experiments ± SD with Caco2 levels set as 1. c Cellular migration was evaluated using Boyden chamber experiments. Shown is mean number of migrating cells ± SD from three independent experiments. Significant differences are indicated by * (P < 0.05)
SOX2 induces a stem cell state in CRC cells
Further investigations revealed that Caco2 cells expressing high levels of SOX2 had a lower proliferative rate (Fig. 2a), were less adherent and displayed a spheroid growth pattern compared to Caco2 wild type cells that were more confluent (Fig. 2b). Accordingly, Caco2-SOX2 cells showed decreased expression of several important adhesion molecules (Fig. 2c). Decreased proliferation and adhesion are events indicative of a cancer stem cell state. We further analyzed cancer stem cell markers, CD44, CD24 and CD133, associated with aggressive cancer types and poor prognosis in CRC [33] (Fig. 2d). Expression of CD44 and CD24 was found to be significantly increased in Caco2 cells expressing SOX2. The levels of CD133 were instead found to be decreased. Together, the phenotype seen in Caco2-SOX2 cells suggests that SOX2 might induce a cancer stem cell state in CRC leading to increased aggressiveness and poorer patient prognosis.
Fig. 2.
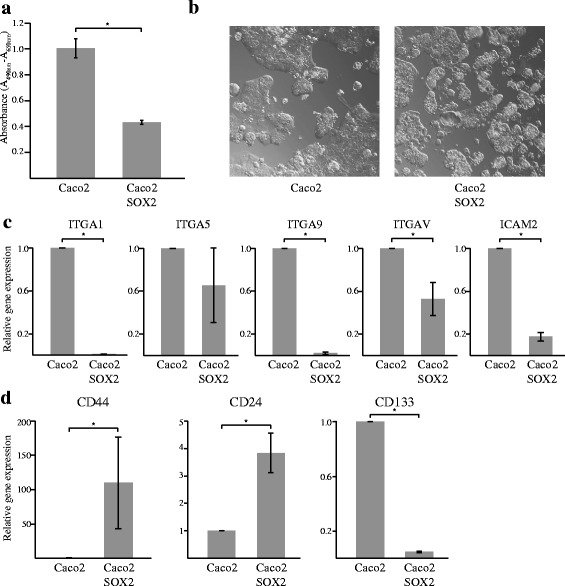
SOX2 induces a CSC state in CRC cells. Factors associated with a CSC state was evaluated in Caco2 cells and Caco2 cells stably transfected with SOX2 (Caco2-SOX2). a Proliferation of cells as measured by XTT cell proliferation assay. b Morphological evaluation of cells. c Expression of cellular adhesion molecules as evaluated by RT-PCR. Shown is relative expression with Caco2 cells set as 1. d Expression of cancer stem cell markers as evaluated by RT-PCR. Shown is relative expression with Caco2 cells set as 1. Gene expression analyses were reproduced three times and mean values ± SD is shown. Significant differences are indicated by * (P < 0.05)
SOX2 is inversely associated with expression of the intestinal epithelial cell marker CDX2
We previously found that SOX2 positive CRC tumors more often are poorly differentiated [7]. A poor cell differentiation is also found in tumors that loose the expression of CDX2, an intestine-specific transcription factor essential for intestinal homeostasis and for the maintenance of an intestinal epithelial phenotype [34]. Furthermore, loss of CDX2 has been linked to more aggressive tumors and a poor outcome in CRC [25–27]. We compared the expression of SOX2 and CDX2 in Caco2 and Caco2-SOX2 cells. We found that Caco2-SOX2 cells showed significantly decreased levels of CDX2, compared to control Caco2 cells (Fig. 3a). Furthermore, we compared SW480 and SW620 CRC cell lines, derived from a primary and metastatic tumor, respectively, resected from a single patient. A high expression of SOX2 was found in the metastatic cell line, and correlated to a significantly decreased expression of CDX2 (Fig. 3a). We further analyzed fresh frozen tumor tissue from 25 CRC patients by RT-PCR for the expression of SOX2 and CDX2. Generally, in tumors with a high expression of SOX2 the levels of CDX2 were either low or absent (Fig. 3b). Likewise, in tumors with a high expression of CDX2, the levels of SOX2 were low or absent (Fig. 3b). These findings suggest that SOX2 expression is associated with a down-regulated expression of CDX2.
Fig. 3.
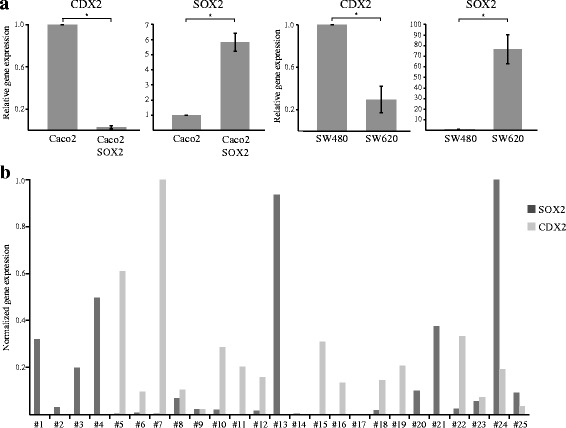
SOX2 is associated with down-regulated expression of CDX2. a Caco2, Caco2-SOX2, SW480 and SW620 cells were analyzed by RT-PCR for the expression of CDX2 and SOX2. Shown is relative gene expression from three independent experiments ± SD with Caco2 or SW480 levels set as 1. Significant differences are indicated by * (P < 0.05). b RNA from fresh frozen tumor tissue from 25 CRC patients was analyzed by RT-PCR for the expression of SOX2 and CDX2. Shown is normalized gene expression
SOX2 is associated with a down-regulated expression of CDX2 in CRC patients
The expression of CDX2 was evaluated by immunohistochemistry in a large cohort of 445 CRC patients. Nuclear CDX2 expression was scored in tumor tissue as; less than 5 % positive cells, 5–50 % positive cells or, more than 50 % positive cells. Representative images of the immunohistochemical stainings of CDX2 can be found in Fig. 4a. In total, 43.4 % of patients showed less than 50 % CDX2 positive tumor nuclei, and of those 14.4 % showed a close to complete lack of CDX2 expression (<5 % positive cells) (Table 1). A down-regulated expression of CDX2 was more often found in right sided tumors (P < 0.001) and tumors of higher stage (P < 0.001) (Table 1). Furthermore, loss of CDX2 expression was significantly associated with poorly differentiated tumors (P < 0.001) (Table 1). In survival analyses, a down-regulated expression of CDX2 correlated, in a stepwise manner, to a poor patient survival (Log-rank P < 0.001) (Fig. 4b). The prognostic importance of CDX2 down-regulation stayed significant in multivariable Cox regression analyses adjusting for stage, age, sex, localization and grade (for patients with 5–50 % CDX2 (HR = 1.54, 95 % CI 1.04–2.28, P = 0.031) and for patients with <5 % CDX2 (HR = 2.45, 95 % CI 1.50–4.01, P < 0.001)).
Fig. 4.
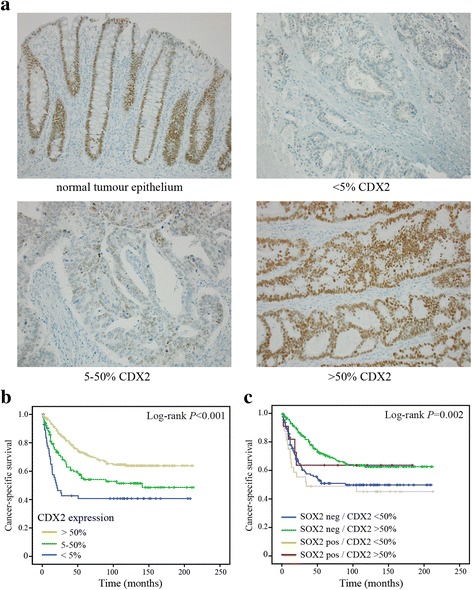
Evaluation of CDX2 expression in CRC. a Representative images of immunohistological stainings of CDX2 in human CRC tissue specimens; normal colon epithelium and CRC with <5 % expression, 5–50 % expression and >50 % expression of CDX2. b Kaplan-Meier survival analyses of CDX2 expression in CRC. c Kaplan-Meier survival analyses in subgroups of CRC defined as SOX2 positive or negative, and CDX2 < 50 % expression and >50 % expression. Log-rank tests were used to calculate P-values
Table 1.
CDX2 expression in relation to clinicopathological characteristics in CRC
| CDX2 expression | ||||
|---|---|---|---|---|
| <5 % | 5–50 % | >50 % | P valuea | |
| Frequencies, n (%) | 62 (14.4) | 125 (29.0) | 244 (56.6) | |
| Sex, n (%) | 0.667 | |||
| Male | 33 (14.3) | 63 (27.3) | 135 (58.4) | |
| Female | 29 (14.5) | 62 (31.0) | 109 (54.5) | |
| Age, n (%) | 0.275 | |||
| ≤59 years | 16 (21.1) | 18 (23.7) | 42 (55.3) | |
| 60–69 years | 16 (14.7) | 29 (26.6) | 64 (58.7) | |
| 70–79 years | 20 (12.9) | 54 (34.8) | 81 (52.3) | |
| ≥80 years | 10 (11.0) | 24 (26.4) | 57 (62.6) | |
| TNM stage, n (%)b | <0.001 | |||
| I | 2 (3.0) | 21 (31.8) | 43 (65.2) | |
| II | 19 (11.2) | 36 (21.3) | 114 (67.5) | |
| III | 17 (19.3) | 32 (36.4) | 39 (44.3) | |
| IV | 24 (24.2) | 33 (33.3) | 42 (42.4) | |
| Localization, n (%)b | <0.001 | |||
| Right colon | 41 (29.5) | 27 (19.4) | 71 (51.1) | |
| Left colon | 9 (7.1) | 39 (30.7) | 79 (62.2) | |
| Rectum | 12 (7.5) | 59 (36.6) | 90 (55.9) | |
| Grade, n (%)b | <0.001 | |||
| Highly to moderately differentiated | 13 (6.3) | 45 (22.0) | 147 (71.7) | |
| Moderately to poorly differentiated | 49 (22.5) | 80 (36.7) | 89 (40.8) | |
aχ2 test
bThe following numbers of missing cases were present: TNM stage, 9; localization, 4; grade, 8
When looking at molecular subgroups of CRC, a down-regulated expression of CDX2 was closely linked to CIMP-high (P < 0.001), MSI (P < 0.001) and BRAF mutated tumors (P < 0.001) (Table 2). Furthermore, a down-regulated expression of CDX2 was more often found in SOX2 positive tumors (P < 0.001) (Table 2). Of the SOX2 positive tumors, 73.9 % had less than 50 % CDX2 positive tumor cells, and of these 32.6 % had less than 5 % positive cells (Table 2).
Table 2.
CDX2 expression in relation to molecular characteristics in CRC
| CDX2 expression | ||||
|---|---|---|---|---|
| <5 % | 5–50 % | >50 % | P valuea | |
| Frequencies, n (%) | 62 (14.4) | 125 (29.0) | 244 (56.6) | |
| MSI screening status, n (%)b | <0.001 | |||
| MSI | 27 (41.5) | 14 (21.5) | 24 (36.9) | |
| MSS | 33 (9.4) | 108 (30.8) | 210 (59.8) | |
| CIMP status, n (%)b | <0.001 | |||
| CIMP-negativec | 8 (3.8) | 66 (31.1) | 138 (65.1) | |
| CIMP-lowc | 25 (15.6) | 46 (28.8) | 89 (55.6) | |
| CIMP-highc | 29 (52.7) | 10 (18.2) | 16 (29.1) | |
| BRAF V600E, n (%)b | <0.001 | |||
| wild type | 27 (7.4) | 110 (30.3) | 226 (62.3) | |
| mutated | 35 (58.3) | 11 (18.3) | 14 (23.3) | |
| KRAS (codon 12, 13), n (%)b | 0.027 | |||
| wild type | 55 (16.2) | 88 (26.0) | 196 (57.8) | |
| mutated | 7 (8.2) | 33 (38.8) | 45 (52.9) | |
| SOX2 expression, n (%)b | <0.001 | |||
| SOX2 negative | 44 (12.3) | 100 (27.9) | 215 (59.9) | |
| SOX2 positive | 15 (32.6) | 19 (41.3) | 12 (26.1) | |
Abbreviations: MSI microsatellite instability, MSS, microsatellite stable, CIMP CpG island methylator phenotype (according to an eight-gene CIMP panel)
aχ2 test
bThe following numbers of missing cases were present: MSI screening status, 15; CIMP status, 4; BRAF V600E, 8; KRAS, 7; SOX2 expression, 26
cCIMP negative, no promoter hypermethylation; CIMP low, one to five genes methylated; CIMP high, six to eight genes methylated
SOX2 positive tumors with a down-regulated expression of CDX2 (<50 % positive cells) had a worse prognosis than SOX2 positive tumors with retained CDX2 expression (>50 % positive cells) (Fig. 4c), suggesting that a part of the negative role of SOX2 on prognosis might be through down-regulation of CDX2. 40.2 % of the SOX2 negative tumors also showed a down-regulated expression of CDX2 (less than 50 % positive cells) (Table 2). In SOX2 negative tumors, CDX2 down-regulation also resulted in a poorer patient prognosis (Fig. 4c), suggesting that CDX2 expression in itself is a strong prognostic factor and likely can be regulated also independently of SOX2.
Discussion
Expression of SOX2 is associated with a poor patient prognosis in CRC, but so far the molecular mechanisms have not been fully elucidated. Here we have studied the functional role of SOX2 in CRC. Using an in vitro cell culture model, we could show that expression of SOX2 was associated with a cellular stem cell state and decreased expression of the intestinal epithelial marker CDX2. The correlation of SOX2 expression and a decreased expression of CDX2 could further be verified in our patient cohort. We also found that decreased expression of CDX2 correlated to a poor patient survival, and that SOX2 expressing tumors with a down-regulated CDX2 expression had a worse patient prognosis compared to those with a retained CDX2 expression, suggesting that a part of the negative role of SOX2 on prognosis might be through down-regulation of CDX2.
In a previous report by Han et al., a role for SOX2 in EMT and increased migration and invasion in CRC was presented [9]. In our in vitro model expressing SOX2, we found that SOX2 did not increase the migratory effect of tumor cells and was not a major regulator of EMT. Instead, our cells expressing SOX2 showed a lower proliferative rate, were less adherent and displayed a more spheroid growth pattern compared to wild type cells. These are all characteristics of CSCs. CSCs possess stem cell-like features, like the ability of self-renewal and multi-lineage differentiation and they have been proposed to retain their tumorigenic capacity and to be the cells responsible for initiation, maintenance and spreading of the tumor [35, 36]. Sphere-forming CSCs have been shown to be more aggressive (metastatic) in vivo than adherent cells [37]. Stem cells divide more slowly than differentiated cells, and the quiescent slow-cycling phenotype seen in CSCs probably plays a role in tumor recurrence as well as resistance to treatment [38, 39]. A possible SOX2 induced CSC state in our in vitro model could thereby be one explanation to the decreased survival seen in patients with SOX2 positive tumors. In line with this hypothesis, cancer cells expressing SOX2 showed an increased expression of the stem cell markers CD24 and CD44. The expression of the stem cell marker CD133 was instead decreased. CD24 has previously been shown to be regulated by SOX2 [5]. CD44 has been identified as a potential CSC marker in CRC [40] and has also been shown to be a more selective colon CSC marker than CD133 since decreased expression of CD44, but not CD133, has been shown to reduce both clonal formation and tumor formation [41]. Other studies have also indicated that CD133 might not be a good CSC marker in CRC, since knocking-down the gene expression of CD133 does not induce cellular differentiation in CRC [42], and both CD133 positive and CD133 negative CRC subpopulations are capable of tumor initiation [43]. CD44 is also the main receptor of the ECM component hyaluronan [44], and it has been shown that expression of CD44 on tumor cells correlate with cancer cell adhesion to endothelial cells and also with metastasis [45]. In a previous study, a cluster of stem-like factors, including SOX2 and CD44, identified patients with a worse prognosis [46]. We plan to further investigate the role of SOX2 in cancer stem cell differentiation and tumor progression. In few previous studies, the suggested role of SOX2 in cancer stem cell differentiation has, in difference to our study, at least partly been linked to EMT related factors [14, 15]. Further studies of the functional role of SOX2 in human cancers are required to clarify these differences.
In our previous study of SOX2 expression in CRC, we have shown that poorly differentiated tumors more often are SOX2 positive [7]. Poorly differentiated tumors have also been associated with decreased expression of the intestinal epithelial marker CDX2 (reviewed in [47]). Therefore, we were interested in studying the expression of CDX2 in correlation to expression of SOX2. Two different cell lines expressing SOX2 showed a decreased expression of CDX2 compared to the wild type cell lines. RT-PCR analyses of fresh frozen tumor tissue verified that tumors with a high expression of SOX2 had low or absent expression of CDX2. When analyzing the expression of CDX2 by immunohistochemistry in our patient cohort, we found that SOX2 positive tumors were highly associated with a decreased expression of CDX2; 73.9 % of the SOX2 positive tumors had less than 50 % CDX2 positive tumor cells, and of these 32.6 % had less than 5 % positive cells. Together these results suggest that expression of SOX2 is correlated to a down-regulated expression of CDX2.
A down-regulated expression of CDX2 in CRC has previously been linked to subgroups of tumors defined as CIMP-high, MSI, and BRAF mutated although results are still inconclusive (reviewed in [47]). In our cohort, a down-regulated expression of CDX2 was closely linked to CIMP-high (P < 0.001), MSI (P < 0.001) and BRAF mutated tumors (P < 0.001) (Table 2), strengthening previously published findings. This finding is also in line with our previous results that SOX2 is partly regulated by BRAF [7]. Down-regulated expression of CDX2 was also correlated to a poor patient prognosis in our cohort, similar to other reports in CRC [25–27]. In our study, the prognostic importance of CDX2 remained significant in multivariable analyses adjusted for stage and other confounders, suggesting that CDX2 is a powerful prognostic factor in CRC. When combining the expression of CDX2 and SOX2 we found that SOX2 positive tumors with a decreased expression of CDX2 had a worse patient prognosis compared to those with a retained CDX2 expression, indicating that at least a part of the negative role of SOX2 on prognosis might be through down-regulation of CDX2. This subgroup, showing SOX2 expression but a down-regulated expression of CDX2, has previously been shown to predict a worse patient outcome in gastric cancer [48]. Since decreased expression of CDX2 also could be seen in SOX2 negative tumors, we speculate that CDX2 can be down-regulated both by SOX2 dependent and SOX2 independent mechanisms. SOX2 expression is often seen only in a part of the tumor, and since SOX2 expression is analyzed in just one tissue section per tumor [7], the number of SOX2 positive tumors in our patient cohort may be underestimated and some of the SOX2 negative tumors with down-regulated expression of CDX2 might actually be SOX2 positive. However, likely there are also other events involved.
CDX2 has been suggested to be a target gene of the Hippo pathway in CRC [49]. The Hippo pathway normally plays critical roles in cell proliferation, growth and apoptosis, but when deregulated it is instead involved in initiation and progression of tumors [50, 51]. Studies have also suggested that SOX2 is involved in the deregulation of the Hippo pathway [52, 53], and therefore we speculate that the SOX2 mediated down-regulation of CDX2 might partly be through this pathway. Kuzmichev et al. have further shown that SOX21 can repress the expression of CDX2 in CRC, and that SOX21 is induced by SOX2 [54]. SOX21 might therefore be another possible pathway that SOX2 utilizes to regulate the expression CDX2. The mechanisms behind the regulation of CDX2 expression by SOX2 will be further investigated.
Conclusions
Using an in vitro cell model, we found that SOX2 in CRC induces a CSC state with down-regulated expression of the intestinal epithelial transcription factor CDX2. In our patient cohort, the expression of SOX2 was highly and significantly associated with a down-regulated expression of CDX2. Furthermore, SOX2 expressing tumors with down-regulated expression of CDX2 had a particularly poor prognosis. We suggest that the poor prognosis seen in patients with SOX2 positive tumors is at least partly mediated through down-regulated expression of CDX2.
Abbreviations
CDX2, caudal type homeobox 2; CIMP, CpG island methylator phenotype; CRC, colorectal cancer; CRUMS, Colorectal Cancer in Umeå Study; CSC, cancer stem cell; DMEM, Dulbecco’s modified Eagle’s medium; EMT, epithelial-mesenchymal transition; FBS, fetal bovine serum; FFPE, formalin-fixed paraffin-embedded; MMP, matrix metalloproteinase; MSI, microsatellite instability; MSS, microsatellite stable; SOX, SRY-related HMG-box
Acknowledgements
We are grateful to Kerstin Näslund for expert technical assistance and Dr. Anna M. Dahlin and Anna Löfgren-Burström for previous molecular evaluations in CRUMS.
Funding
This study was supported by grants from The Cancer Research Foundation in Northern Sweden, OE and Edla Johanssons foundation, Petrus and Augusta Hedlunds Foundation, Magn. Bergvall. Foundation, The Swedish Cancer Society, The Swedish Research Council and Umeå University.
Availability of data and materials
The relevant data supporting the conclusions of this article are included within the article and its additional files.
Authors’ contributions
IVL, SE, RP and MLW participated in the conception and design of the study. IVL, SE, VE, ÅÖ and RP collected data. IVL, SE, RP and MLW analyzed and interpreted data. IVL, SE and RP were involved in writing the draft manuscript. All authors reviewed and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The handling of tissue samples and patient data in the present study was approved by the Research Ethics Committee at Umeå University, Umeå, Sweden (dnr 99–330), including the procedure whereby patients verbally gave their informed consent. This consent was documented in each patient record, and considered by the ethical committee to be sufficient.
Additional file
Sequences of primers used for RT-PCR. (DOCX 17 kb)
Contributor Information
Ida V. Lundberg, Email: ida.lundberg@umu.se
Sofia Edin, Phone: +46 (0)907854431, Email: sofia.edin@umu.se.
Vincy Eklöf, Email: vincy.eklof@umu.se.
Åke Öberg, Email: ake_oberg@yahoo.se.
Richard Palmqvist, Email: richard.palmqvist@umu.se.
Maria L. Wikberg, Email: maria.wikberg@umu.se
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer Stat. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12(1):15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, Pandiella A, Rezola R, Martin AG. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31(11):1354–65. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 4.Sanada Y, Yoshida K, Ohara M, Oeda M, Konishi K, Tsutani Y. Histopathologic evaluation of stepwise progression of pancreatic carcinoma with immunohistochemical analysis of gastric epithelial transcription factor SOX2: comparison of expression patterns between invasive components and cancerous or nonneoplastic intraductal components. Pancreas. 2006;32(2):164–70. doi: 10.1097/01.mpa.0000202947.80117.a0. [DOI] [PubMed] [Google Scholar]
- 5.Fang X, Yu W, Li L, Shao J, Zhao N, Chen Q, Ye Z, Lin SC, Zheng S, Lin B. ChIP-seq and functional analysis of the SOX2 gene in colorectal cancers. Omics. 2010;14(4):369–84. doi: 10.1089/omi.2010.0053. [DOI] [PubMed] [Google Scholar]
- 6.Li XL, Eishi Y, Bai YQ, Sakai H, Akiyama Y, Tani M, Takizawa T, Koike M, Yuasa Y. Expression of the SRY-related HMG box protein SOX2 in human gastric carcinoma. Int J Oncol. 2004;24(2):257–63. [PubMed] [Google Scholar]
- 7.Lundberg IV, Lofgren Burstrom A, Edin S, Eklof V, Oberg A, Stenling R, Palmqvist R, Wikberg ML. SOX2 expression is regulated by BRAF and contributes to poor patient prognosis in colorectal cancer. PLoS One. 2014;9(7):e101957. doi: 10.1371/journal.pone.0101957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumann J, Bahr F, Horst D, Kriegl L, Engel J, Luque RM, Gerhard M, Kirchner T, Jung A. SOX2 expression correlates with lymph-node metastases and distant spread in right-sided colon cancer. BMC Cancer. 2011;11:518. doi: 10.1186/1471-2407-11-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X, Fang X, Lou X, Hua D, Ding W, Foltz G, Hood L, Yuan Y, Lin B. Silencing SOX2 induced mesenchymal-epithelial transition and its expression predicts liver and lymph node metastasis of CRC patients. PLoS One. 2012;7(8):e41335. doi: 10.1371/journal.pone.0041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–84. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 11.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10(6):717–28. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 12.O’Connor ML, Xiang D, Shigdar S, Macdonald J, Li Y, Wang T, Pu C, Wang Z, Qiao L, Duan W. Cancer stem cells: a contentious hypothesis now moving forward. Cancer Lett. 2014;344(2):180–7. doi: 10.1016/j.canlet.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Bareiss PM, Paczulla A, Wang H, Schairer R, Wiehr S, Kohlhofer U, Rothfuss OC, Fischer A, Perner S, Staebler A, et al. SOX2 expression associates with stem cell state in human ovarian carcinoma. Cancer Res. 2013;73(17):5544–55. doi: 10.1158/0008-5472.CAN-12-4177. [DOI] [PubMed] [Google Scholar]
- 14.Liu XF, Yang WT, Xu R, Liu JT, Zheng PS. Cervical cancer cells with positive Sox2 expression exhibit the properties of cancer stem cells. PLoS One. 2014;9(1):e87092. doi: 10.1371/journal.pone.0087092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herreros-Villanueva M, Zhang JS, Koenig A, Abel EV, Smyrk TC, Bamlet WR, de Narvajas AA, Gomez TS, Simeone DM, Bujanda L, et al. SOX2 promotes dedifferentiation and imparts stem cell-like features to pancreatic cancer cells. Oncogenesis. 2013;2:e61. doi: 10.1038/oncsis.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SH, Oh SY, Do SI, Lee HJ, Kang HJ, Rho YS, Bae WJ, Lim YC. SOX2 regulates self-renewal and tumorigenicity of stem-like cells of head and neck squamous cell carcinoma. Br J Cancer. 2014;111(11):2122–30. doi: 10.1038/bjc.2014.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pham DL, Scheble V, Bareiss P, Fischer A, Beschorner C, Adam A, Bachmann C, Neubauer H, Boesmueller H, Kanz L, et al. SOX2 expression and prognostic significance in ovarian carcinoma. Int J Gynecol Pathol. 2013;32(4):358–67. doi: 10.1097/PGP.0b013e31826a642b. [DOI] [PubMed] [Google Scholar]
- 18.Ruan J, Wei B, Xu Z, Yang S, Zhou Y, Yu M, Liang J, Jin K, Huang X, Lu P, et al. Predictive value of Sox2 expression in transurethral resection specimens in patients with T1 bladder cancer. Med Oncol. 2013;30(1):445. doi: 10.1007/s12032-012-0445-z. [DOI] [PubMed] [Google Scholar]
- 19.Sholl LM, Long KB, Hornick JL. Sox2 expression in pulmonary non-small cell and neuroendocrine carcinomas. Appl Immunohistochem Mol Morphol. 2010;18(1):55–61. doi: 10.1097/PAI.0b013e3181b16b88. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, He W, Lu C, Wang Z, Wang J, Giercksky KE, Nesland JM, Suo Z. Oct3/4 and Sox2 are significantly associated with an unfavorable clinical outcome in human esophageal squamous cell carcinoma. Anticancer Res. 2009;29(4):1233–41. [PubMed] [Google Scholar]
- 21.Suh E, Chen L, Taylor J, Traber PG. A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol Cell Biol. 1994;14(11):7340–51. doi: 10.1128/MCB.14.11.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suh E, Traber PG. An intestine-specific homeobox gene regulates proliferation and differentiation. Mol Cell Biol. 1996;16(2):619–25. doi: 10.1128/MCB.16.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moskaluk CA, Zhang H, Powell SM, Cerilli LA, Hampton GM, Frierson HF., Jr Cdx2 protein expression in normal and malignant human tissues: an immunohistochemical survey using tissue microarrays. Mod Pathol. 2003;16(9):913–9. doi: 10.1097/01.MP.0000086073.92773.55. [DOI] [PubMed] [Google Scholar]
- 24.Kaimaktchiev V, Terracciano L, Tornillo L, Spichtin H, Stoios D, Bundi M, Korcheva V, Mirlacher M, Loda M, Sauter G, et al. The homeobox intestinal differentiation factor CDX2 is selectively expressed in gastrointestinal adenocarcinomas. Mod Pathol. 2004;17(11):1392–9. doi: 10.1038/modpathol.3800205. [DOI] [PubMed] [Google Scholar]
- 25.Dawson H, Koelzer VH, Lukesch AC, Mallaev M, Inderbitzin D, Lugli A, Zlobec I. Loss of Cdx2 expression in primary tumors and lymph node metastases is specific for mismatch repair-deficiency in colorectal cancer. Frontiers in oncology. 2013;3:265. doi: 10.3389/fonc.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong KD, Lee D, Lee Y, Lee SI, Moon HY. Reduced CDX2 expression predicts poor overall survival in patients with colorectal cancer. Am Surg. 2013;79(4):353–60. [PubMed] [Google Scholar]
- 27.Bae JM, Lee TH, Cho NY, Kim TY, Kang GH. Loss of CDX2 expression is associated with poor prognosis in colorectal cancer patients. World J Gastroenterol. 2015;21(5):1457–67. doi: 10.3748/wjg.v21.i5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahlin AM, Henriksson ML, Van Guelpen B, Stenling R, Oberg A, Rutegard J, Palmqvist R. Colorectal cancer prognosis depends on T-cell infiltration and molecular characteristics of the tumor. Mod Pathol. 2011;24(5):671–82. doi: 10.1038/modpathol.2010.234. [DOI] [PubMed] [Google Scholar]
- 29.Dahlin AM, Palmqvist R, Henriksson ML, Jacobsson M, Eklof V, Rutegard J, Oberg A, Van Guelpen BR. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin Cancer Res. 2010;16(6):1845–55. doi: 10.1158/1078-0432.CCR-09-2594. [DOI] [PubMed] [Google Scholar]
- 30.Eklof V, Wikberg ML, Edin S, Dahlin AM, Jonsson BA, Oberg A, Rutegard J, Palmqvist R. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br J Cancer. 2013;108(10):2153–63. doi: 10.1038/bjc.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benlloch S, Paya A, Alenda C, Bessa X, Andreu M, Jover R, Castells A, Llor X, Aranda FI, Massuti B. Detection of BRAF V600E mutation in colorectal cancer: comparison of automatic sequencing and real-time chemistry methodology. J Mol Diagn. 2006;8(5):540–3. doi: 10.2353/jmoldx.2006.060070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, Arango D, Strausberg RL, Buchanan D, Wormald S, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74(12):3238–47. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 33.Sahlberg SH, Spiegelberg D, Glimelius B, Stenerlow B, Nestor M. Evaluation of cancer stem cell markers CD133, CD44, CD24: association with AKT isoforms and radiation resistance in colon cancer cells. PLoS One. 2014;9(4):e94621. doi: 10.1371/journal.pone.0094621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coskun M, Troelsen JT, Nielsen OH. The role of CDX2 in intestinal homeostasis and inflammation. Biochim Biophys Acta. 2011;1812(3):283–9. doi: 10.1016/j.bbadis.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–99. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 36.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66(19):9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 37.Gaviraghi M, Tunici P, Valensin S, Rossi M, Giordano C, Magnoni L, Dandrea M, Montagna L, Ritelli R, Scarpa A, et al. Pancreatic cancer spheres are more than just aggregates of stem marker-positive cells. Biosci Rep. 2011;31(1):45–55. doi: 10.1042/BSR20100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore N, Lyle S. Quiescent, slow-cycling stem cell populations in cancer: a review of the evidence and discussion of significance. J Oncol. 2011;2011.doi:10.1155/2011/396076. [DOI] [PMC free article] [PubMed]
- 39.Pannuti A, Foreman K, Rizzo P, Osipo C, Golde T, Osborne B, Miele L. Targeting Notch to target cancer stem cells. Clin Cancer Res. 2010;16(12):3141–52. doi: 10.1158/1078-0432.CCR-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104(24):10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du L, Wang H, He L, Zhang J, Ni B, Wang X, Jin H, Cahuzac N, Mehrpour M, Lu Y, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14(21):6751–60. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 42.Feng HL, Liu YQ, Yang LJ, Bian XC, Yang ZL, Gu B, Zhang H, Wang CJ, Su XL, Zhao XM. Expression of CD133 correlates with differentiation of human colon cancer cells. Cancer Biol Ther. 2010;9(3):216–23. doi: 10.4161/cbt.9.3.10664. [DOI] [PubMed] [Google Scholar]
- 43.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118(6):2111–20. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61(7):1303–13. doi: 10.1016/0092-8674(90)90694-A. [DOI] [PubMed] [Google Scholar]
- 45.Orian-Rousseau V. CD44, a therapeutic target for metastasising tumours. Eur J Cancer. 2010;46(7):1271–7. doi: 10.1016/j.ejca.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 46.Ong CW, Chong PY, McArt DG, Chan JY, Tan HT, Kumar AP, Chung MC, Clement MV, Soong R, Van Schaeybroeck S, et al. The prognostic value of the stem-like group in colorectal cancer using a panel of immunohistochemistry markers. Oncotarget. 2015;6(14):12763–73. doi: 10.18632/oncotarget.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olsen J, Espersen ML, Jess P, Kirkeby LT, Troelsen JT. The clinical perspectives of CDX2 expression in colorectal cancer: a qualitative systematic review. Surg Oncol. 2014;23(3):167–76. doi: 10.1016/j.suronc.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Camilo V, Barros R, Celestino R, Castro P, Vieira J, Teixeira MR, Carneiro F, Pinto-de-Sousa J, David L, Almeida R. Immunohistochemical molecular phenotypes of gastric cancer based on SOX2 and CDX2 predict patient outcome. BMC Cancer. 2014;14:753. doi: 10.1186/1471-2407-14-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang K, Zhou G, Zhang Q, Li J, Zhang C. Expression of hippo pathway in colorectal cancer. Saudi J Gastroenterol. 2014;20(3):188–94. doi: 10.4103/1319-3767.133025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H, Jiang D, Chi F, Zhao B. The Hippo pathway regulates stem cell proliferation, self-renewal, and differentiation. Protein Cell. 2012;3(4):291–304. doi: 10.1007/s13238-012-2919-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–57. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 52.Basu-Roy U, Bayin NS, Rattanakorn K, Han E, Placantonakis DG, Mansukhani A, Basilico C. Sox2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat Commun. 2015;6:6411. doi: 10.1038/ncomms7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bora-Singhal N, Nguyen J, Schaal C, Perumal D, Singh S, Coppola D, Chellappan S. YAP1 regulates OCT4 activity and SOX2 expression to facilitate self-renewal and vascular mimicry of stem-like cells. Stem Cells. 2015;33(6):1705–18. doi: 10.1002/stem.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuzmichev AN, Kim SK, D’Alessio AC, Chenoweth JG, Wittko IM, Campanati L, McKay RD. Sox2 acts through Sox21 to regulate transcription in pluripotent and differentiated cells. Curr Biol. 2012;22(18):1705–10. doi: 10.1016/j.cub.2012.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The relevant data supporting the conclusions of this article are included within the article and its additional files.


