Abstract
This study aimed to elucidate aspects of the epidemiology of Blastocystis in Nigerian school children, including the distribution of subtypes (STs) and ST alleles. A total of 199 genomic DNAs extracted from fecal samples from 199 Nigerian children aged 2–14 years were tested by real-time polymerase chain reaction for Blastocystis. Positive DNAs were submitted to barcoding by PCR and sequencing to obtain information on STs and ST alleles. A total of 167 (84%) samples were positive for Blastocystis, with prevalence increasing by age. No association between Blastocystis colonization and gender (P = 0.51) or type/presence of toilet facilities (P = 0.21) was observed. Blastocystis carriers were more prone to using water collected from wells than from sachets (P = 0.0044). Moreover, Blastocystis positivity was associated with positivity for fecal-orally transmitted protozoa (P = 0.018) and helminths (P < 0.0001). A clear inverse association of Blastocystis colonization and malaria infection was observed (P < 0.0001); however, malaria-positive children being younger than malaria-negative children, this finding was attributed to the age effect of Blastocystis colonization. ST data were available for 127/167 (76%) samples. Fifty-one children were positive for ST1, while 42 and 33 children were colonized with ST2 and ST3, respectively; a single case of ST7 was observed. By and large, the ST alleles identified for ST1 and ST2 did not differ from those observed in humans in other regions of the world; meanwhile, the distribution of ST3 alleles was remarkably distinct and potentially specific to humans in sub-Saharan Africa.
Introduction
Blastocystis is a common single-celled intestinal parasite of humans and animals.1–3 Its role in human health and disease remains unclear despite the fact that it has been known for more than a century. Although tools for differentiating colonization from infection are still not available, the use of DNA-based technologies for detection of the organism and genetic characterization of the genus has recently resulted in a few remarkable findings: Blastocystis 1) comprises at least 17 ribosomal lineages, eight of which are shared between human and non-human hosts,4 2) is a common and stable colonizer of the intestine,5 3) is associated with particular intestinal bacterial communities,6 4) appears to be associated to low body mass index,6 5) is more common in healthy individuals than in patients with infectious, functional, and inflammatory bowel disease,7 and 6) is probably much more common than previously anticipated. With regard to the latter, a prevalence of 50% was found in an Irish community of elderly, healthy individuals,5 and a prevalence of 100% was recently identified in Senegalese children,8 just to mention a couple of examples.
Cost-effectively increasing test sensitivity and specificity, the use of real-time polymerase chain reaction (PCR) for detection of Blastocystis is now considered state of the art, omitting the need for highly experienced personnel for microscopic identification.9,10 However, real-time PCR for surveying Blastocystis in sub-Saharan African cohorts has so far been included only by El Safadi and others.8 DNAs screened by real-time PCR and found positive can be processed by barcoding as previously described, enabling both subtype (ST) and allele identification relevant to studies on the molecular epidemiology of Blastocystis.11,12 To date, a total of at least 140 different ST alleles have been recorded in the Blastocystis Subtype (18S) and Sequence Typing (MLST) Databases, available at www.pubmlst.org/blastocystis.
Blastocystis might be the most common intestinal parasite in humans and has a global distribution. It is clear that DNA-based analysis of Blastocystis has recently taken us a long way in terms of increasing our understanding of parasite transmission, including host specificity, but also our understanding of its overall public health impact. To further this understanding, knowledge on the prevalence and distribution of STs in the background population in different parts of the world is important for reference. In this study, we surveyed a cohort of Nigerian children13 for Blastocystis with a view to elucidating aspects of its epidemiology, including prevalence and distribution of STs and ST alleles in a cohort of individuals representing the background population.
MATERIALS AND METHODS
Study individuals and accompanying data.
Study site, study population, acquisition of sociodemographic data, and DNA extraction is described in the recent articles by Efunshile and others.13,14 In brief, samples were collected in Ilero, Nigeria, from preschool nursery children and schoolchildren between 2 and 14 years of age. Demographic data available for analysis included data on gender, religion, toilet facilities, and water sources. Information on coinfection with other intestinal parasites and malaria infection status was also available.
Samples, DNA extraction, PCR, sequencing, and data analysis.
In this study, 199 genomic DNAs, previously extracted directly from stool from 199 unique children by use of the QIAamp DNA Stool Mini Kit (Qiagen GmBH, Hilden, Germany) and screened for Entamoeba histolytica, Entamoeba dispar, Giardia intestinalis, and Cryptosporidium spp. by real-time PCR,13 were all screened for Blastocystis using the real-time PCR method previously described, and cycle threshold (Ct) values were recorded for each positive sample.9 Subsequently, all PCR-positive samples were subtyped by barcoding according to Scicluna and others12 and Stensvold.11 In brief, PCR products were sequenced unilaterally using the BhRDr primer by Eurofins Genomics (MWG-Biotech, Ebersberg, Germany). Resulting ab1 sequence files were manually edited using FinchTV (Geospiza, Seattle, WA), and identification of STs and ST alleles was performed using the Blastocystis Subtype (18S) and Sequence Typing (MLST) Databases, available at http://pubmlst.org/Blastocystis.11,15
Statistical analysis.
Utilizing the SAS Analytics, Business Intelligence and Data Management Software (Cary, NC) categorical variables were cross-tabulated, and χ2 tests were used to test for associations between Blastocystis infection and epidemiological parameters. The Mantel–Haenszel test, a nonparametric method, was used specifically for the variables “water source” and “toilet facility.” A t test was used to compare the Ct values of the Blastocystis-positive samples that we were able to ST using conventional PCR with Ct values of the Blastocystis-positive samples that we were unable to ST. The Ct values of ST1, ST2, and ST3 were compared pairwise using the analysis of variance test, and confidence intervals for the difference between STs were generated using the Bonferroni correction method. A probability (P) value < 0.05 was considered statistically significant.
Ethical considerations.
The study was ethically approved by the research and ethical committee of the Federal Teaching Hospital, Abakaliki, Nigeria. Written informed consent was also obtained from all the parents or guardians of the participating children.
RESULTS
Blastocystis was detected in 167 (84%) of the samples, of which 127 could be processed successfully with regard to ST identification. The Ct values of positive samples that could be subtyped (27.6, standard deviation [SD] = 4.5) and not subtyped (32.9, SD = 5.5) were compared using the t test, and significantly higher Ct values were observed in the samples that we were unable to ST (P < 0.0001). The majority of the children (N = 51) were colonized with ST1; however, the proportion of children colonized with ST2 (N = 42) or ST3 (N = 33) was also substantial. Other than these three STs, a single case of ST7 was identified (Table 1 ). Among ST1 strains, allele 3 was observed in only one case, while allele 4 was detected in 39 cases; we were unable to assign an allele to 11 ST1-positive specimens. Among ST2-positive samples, allele 11 was seen in one case, allele 12 in 30 cases, and allele 15 in eight cases, while we were unable to assign an allele to three ST2-positive specimens. ST3 strains included 13 cases of allele 36, 10 cases of allele 37, one case of allele 38, three cases of allele 39, and one case of allele 49; we were unable to assign an allele to five ST3-positive specimens. No allele information was assigned to the single positive ST7 specimen. Our inability to assign allele information to a total of 20 samples was mainly due to ambiguous nucleotide sequence chromatograms, which precluded distinctive allele calling. However, in none of these cases were the sequence traces so ambiguous that they did not enable ST calling.
Table 1.
Prevalence of Blastocystis and distribution of STs, ST alleles, and corresponding average real-time PCR Ct values
| Prevalence npositive/nsamples (%) | Average Ct value (SD) | |
|---|---|---|
| Blastocystis positive | 167/199 (84) | 28.9 (5.3) |
| Samples subtyped | 127/167 (76) | 27.6 (4.5) |
| Samples not subtyped | 40/167 (24) | 32.9 (5.5) |
| ST1 | 51/199 (26) | 27.3 (4.2) |
| Allele 3 | 1/199 (0.5) | 23.4 (–†) |
| Allele 4 | 39/199 (20) | 26.9 (4.0) |
| NA* | 11/51 | 28.9 (4.8) |
| ST2 | 42/199 (21) | 26.4 (3.4) |
| Allele 11 | 1/199 (0.5) | 26.6 (–) |
| Allele 12 | 30/199 (15) | 26.5 (3.7) |
| Allele 15 | 8/199 (4.0) | 26.2 (3.4) |
| NA | 3/42 | 25.8 (1.1) |
| ST3 | 33/199 (17) | 29.7 (5.4) |
| Allele 36 | 13/199 (6.5) | 30.2 (6.8) |
| Allele 37 | 10/199 (5.0) | 28.2 (4.3) |
| Allele 38 | 1/199 (0.5) | 26.0 (–) |
| Allele 39 | 3/199 (1.5) | 32.2 (3.1) |
| Allele 49 | 1/199 (0.5) | 37.2 (–) |
| NA | 5/33 | 29.1 (4.7) |
| ST7 | 1/199 (0.50) | 22.7 (–) |
| NA | 1/1 | 22.7 (–) |
Ct = cycle threshold; PCR = polymerase chain reaction; SD = standard deviation; ST = subtype.
Not applicable (unsuccessful subtyping).
No SD specified since only one positive sample.
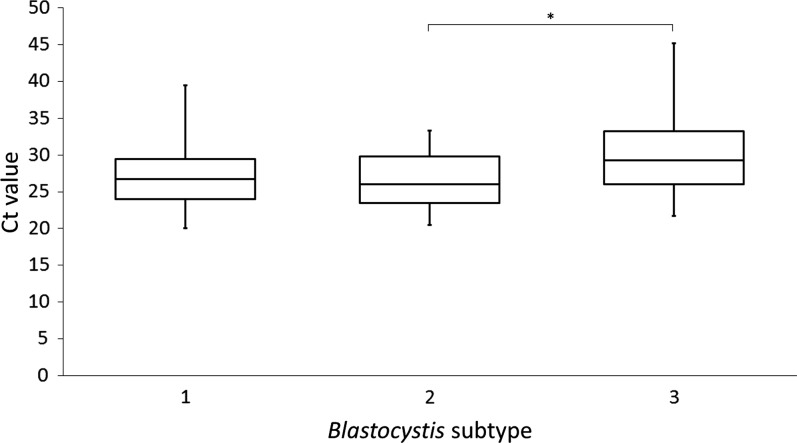
There was a significant difference between Ct values pertaining to ST1, ST2, and ST3 (P = 0.005). A pairwise comparison of the confidence intervals for ST1 (27.3, SD = 4.2), ST2 (26.4, SD = 3.4), and ST3 (29.7, SD = 5.4) showed that Ct values differed significantly only between ST2 and ST3 (P = 0.006), with differences remaining nonsignificant between ST1 and ST2 (P = 0.89) and ST1 and ST3 (P = 0.07) (Figure 1).
Figure 1.
Box plot Ct values for positive samples stratified by subtype (ST). Whiskers (minimum and maximum), box (from the 25th to the 75th percentile), and line (median). * Statistical significance, P = 0.006.
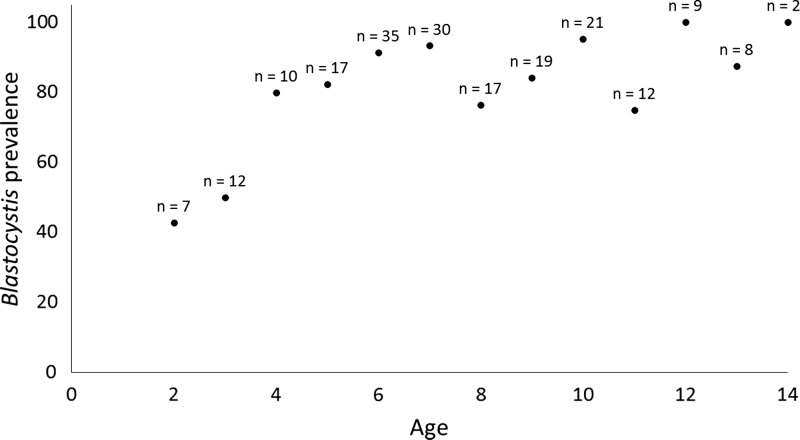
The prevalence of Blastocystis increased by age, eventually reaching an upper plateau, which included most of the tested age groups (Figure 2). The mean age of the children positive for ST1, ST2, and ST3 was 7.6 (SD = 2.8), 8.5 (SD = 2.3), and 7.8 (SD = 2.4), respectively.
Figure 2.
Prevalence of Blastocystis according to age (n indicates the number of children in each age group).
There was no association between Blastocystis colonization and gender (P = 0.51) or type/presence of toilet facilities (P = 0.21). Blastocystis carriers were more likely to use water collected from wells rather than from sachets (P = 0.0044). Moreover, Blastocystis positivity was associated with positivity for fecal-orally transmitted protozoa (P = 0.018) and helminths (P < 0.0001). A clear inverse association of Blastocystis colonization and malaria infection was observed (P < 0.0001) (Table 2 ). The mean age of malaria-positive (N = 50) and malaria-negative children (N = 149) was 5.6 (SD = 2.8) and 8.1 (SD = 2.6), respectively (P < 0.0001).
Table 2.
Distribution of Blastocystis in relation to selected sociodemographic variables and other parasites
| Variable | Blastocystis positive (N = 167) | Blastocystis negative (N = 32) | χ2 test (P) |
|---|---|---|---|
| Gender | |||
| Male (N = 110) | 94 | 16 | 0.51 |
| Female (N = 89) | 73 | 16 | |
| Religion | |||
| Christianity (N = 114) | 95 | 19 | 0.79 |
| Islam (N = 85) | 72 | 13 | |
| Toilet facility* | |||
| Bush (N = 162) | 134 | 28 | 0.21† |
| Pit latrine (N = 30) | 26 | 4 | |
| Water source‡ | |||
| Well (N = 181) | 155 | 26 | 0.0044† |
| Sachet (N = 14) | 8 | 6 | |
| Other intestinal protozoa§ | |||
| Positive (N = 94) | 85 | 9 | 0.018 |
| Negative (N = 105) | 82 | 23 | |
| Helminths∥ | |||
| Positive (N = 157) | 141 | 16 | < 0.0001 |
| Negative (N = 42) | 26 | 16 | |
| Malaria | |||
| Positive (N = 50) | 32 | 18 | < 0.0001 |
| Negative (N = 149) | 135 | 14 | |
Did not include data on “water closet” or “other facilities” due to too small numbers.
Mantel–Haenszel χ2 test.
Did not include “other source” due to too small numbers.
Includes Giardia sp. (no. of test-positive children: 74), Entamoeba dispar (no. of test-positive children: 37), and Cryptosporidium sp. (number of test-positive children: 2).13
Includes Ascaris (no. of test-positive children: 94), hookworm (no. of test-positive children: 57), Trichuris (no. of test-positive children: 5), Enterobius (no. of test-positive children: 5), Taenia (no. of test-positive children: 1), and unknown helminths (no. of test-positive children: 4).
DISCUSSION
To date, only few studies have included real-time PCR-based screening for Blastocystis.8,10,16 In the study by El Safadi and others,8 who used both conventional PCR and real-time PCR, a prevalence of 100% was observed in Senegalese children aged 6 to 10 years. In this study, we found a somewhat lower prevalence, namely, 84%; however, there is growing evidence that colonization by Blastocystis is positively associated with increasing age.17,18 In this study, only half or less of the children aged 3 or less were positive; meanwhile, in children aged 6 years or more, Blastocystis was more or less an obligate finding. Hence, given the trend toward Blastocystis being more common in older children (Figure 2), the data presented in this study appear in line with the data produced by El Safadi and others.8 In the future, studies including more children in the lower age groups (infants and toddlers) should be included to verify the lower prevalence of Blastocystis in these age groups.
Blastocystis is expected to be passed on primarily by fecal–oral transmission, which is also supported by data from this study showing that Blastocystis positivity was associated with positivity for fecal-orally transmitted intestinal parasites. Although we find no reason to believe that toddlers are less exposed to fecal–oral transmission than children who are 4 years old or older, older children have been exposed for a longer period, and since long-lasting Blastocystis colonization is not unusual,5 this finding may simply reflect an accumulation of cases with age. Meanwhile, it is also possible that the composition of the gut microbiota in small children does not favor Blastocystis colonization. For instance, if diarrhea is more common in toddlers than in older children, the dysbiosis potentially linked to episodes of diarrhea might account for the lower prevalence of Blastocystis in younger children; indeed there is an increasing body of evidence suggesting that Blastocystis colonization is inversely linked to intestinal microbiota dysbiosis.6,7,19
Given the studies showing that real-time PCR is much more sensitive for detection than other methods,9,10 the high rate of Blastocystis carrier status identified in this study is unsurprising.
The distribution of STs identified in this study confirmed previous trends from studies carried out outside of Europe.20 We confirmed the conspicuously virtual absence of ST4 in extra-European regions,8,20–24 and apart from one case of ST7, only ST1, ST2, and ST3 were found. With regard to the latter three STs, ST1 was most common, although not statistically so, and, interestingly, ST3 was the least common of the three. Even more interestingly, allele 34, which reflects the most commonly reported ST allele of ST3 in other parts of the world including Ireland,5 United Arab Emirates,22 India,21 and Colombia,24 was absent, while five other ST alleles (36, 37, 38, 39, and 49) were identified, most of which have also been found in Old World monkeys, although rarely.25 More than 80% of the ST3 strains for which allele information could be obtained reflected either allele 36 or 37, which appear to be very infrequent in humans in other regions. Hence, our data appear to suggest that while ST1 and ST2 strains found in humans do not appear to differ across the globe, the distribution of ST3 strains is clearly subject to geographical variability, with humans in sub-Saharan Africa potentially representing a cohort of people colonized by ST3 strains that are only rarely found in humans outside sub-Saharan Africa. Certainly, it appears that differences in exposure and/or intra-host specificity not reflected at ST level can be identified at allele level. Unfortunately, no previous studies on Blastocystis carried out in sub-Saharan African countries have included data on ST alleles, and so confirmation of these data is warranted.
A number of samples contained so little Blastocystis DNA (as evidenced by high Ct values) that subtyping using the original method published by Scicluna and others12 was unsuccessful. However, the modified barcoding method recently developed by Scanlan and others26 and which used a nested PCR approach might be useful for subtyping those samples that cannot be subtyped by the original setup.
Based on the differences observed in Ct values for the three STs ST1, ST2, and ST3, individuals colonized by ST2 appeared to have higher loads of Blastocystis in stool than those with ST3, while no differences were seen in Ct values between ST1 and ST2 or between ST1 and ST3.
The pronounced negative association between Blastocystis colonization and the presence of malaria is interesting, although difficult to interpret. Nevertheless, malaria-negative children were significantly older than malaria-positive children; hence, the aforementioned age effect on Blastocystis could, at least in part, explain this finding.
Blastocystis has been found in water on multiple occasions.27,28 The fact that Blastocystis carriers were more likely to use water collected from wells rather than from sachets might indicate that the water in the wells contribute to Blastocystis transmission, either by being contaminated already within the well, or by being contaminated during the subsequent handling of the water collected from the well.
In conclusion, we investigated the prevalence and ST distribution of Blastocystis in a cohort of Nigerian children aged 2–14 years, finding an overall prevalence of 84% with older children being more frequently colonized than toddlers. The distribution of STs (mainly ST1, ST2, and ST3) was unsurprising. However, the fact that ST1 and ST2 alleles were similar to those found in humans in other regions while ST3 alleles differed from ST3 alleles found in humans in non-African countries should stimulate interest in the field of parasite evolution (including host specificity and parasite phylogeography), host–parasite interactions, and public health.
ACKNOWLEDGMENTS
We thank Lis Lykke Wassmann and Gitte Jensen for their excellent technical support.
Footnotes
Financial support: Akinwale M. Efunshile's work is based on a scholarship provided by the German Academic Exchange Services (DAAD grant A/10/98666). Christen R. Stensvold's work is partly funded by the Marie Curie Actions (Call: FP7-PEOPLE-2012-CIG; grant ID: 321614).
Authors' addresses: Casper S. Poulsen, National Food Institute, Danish Technical University, Kongens Lyngby, Denmark, E-mail: csapou@food.dtu.dk. Akinwale M. Efunshile, Department of Medical Microbiology, Ebonyi State University, Abakaliki, Nigeria, E-mail: drefunshile@yahoo.com. Jenna A. Nelson, Elon University, Elon, NC, E-mail: jnelson19@elon.edu. Christen R. Stensvold, Department of Microbiology and Infection Control, Statens Serum Institut, Copenhagen, Denmark, E-mail: run@ssi.dk.
References
- 1.Clark CG, van der Giezen M, Alfellani MA, Stensvold CR. Recent developments in Blastocystis research. Adv Parasitol. 2013;82:1–32. doi: 10.1016/B978-0-12-407706-5.00001-0. [DOI] [PubMed] [Google Scholar]
- 2.Tan KS, Mirza H, Teo JD, Wu B, Macary PA. Current views on the clinical relevance of Blastocystis spp. Curr Infect Dis Rep. 2010;12:28–35. doi: 10.1007/s11908-009-0073-8. [DOI] [PubMed] [Google Scholar]
- 3.Stensvold CR. Xiao L, Ryan U, Feng Y. Biology of Foodborne Parasites. CRC Press; 2015. Blastocystis. Boca Raton, FL. [Google Scholar]
- 4.Alfellani MA, Taner-Mulla D, Jacob AS, Imeede CA, Yoshikawa H, Stensvold CR, Clark CG. Genetic diversity of Blastocystis in livestock and zoo animals. Protist. 2013;164:497–509. doi: 10.1016/j.protis.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Scanlan PD, Stensvold CR, Rajilić-Stojanović M, Heilig HG, De Vos WM, O'Toole PW, Cotter PD. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol Ecol. 2014;90:326–330. doi: 10.1111/1574-6941.12396. [DOI] [PubMed] [Google Scholar]
- 6.Andersen LO, Bonde I, Nielsen HB, Stensvold CR. A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiol Ecol. 2015;91 doi: 10.1093/femsec/fiv072. doi: 10.1093/femsec/fiv072. [DOI] [PubMed] [Google Scholar]
- 7.Krogsgaard LR, Engsbro AL, Stensvold CR, Nielsen HV, Bytzer P. The prevalence of intestinal parasites is not greater among individuals with irritable bowel syndrome: a population-based case-control study. Clin Gastroenterol Hepatol. 2015;13:507–513. doi: 10.1016/j.cgh.2014.07.065. e2. [DOI] [PubMed] [Google Scholar]
- 8.El Safadi D, Gaayeb L, Meloni D, Cian A, Poirier P, Wawrzyniak I, Delbac F, Dabboussi F, Delhaes L, Seck M, Hamze M, Riveau G, Viscogliosi E. Children of Senegal River Basin show the highest prevalence of Blastocystis sp. ever observed worldwide. BMC Infect Dis. 2014;14 doi: 10.1186/1471-2334-14-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stensvold CR, Ahmed UN, Andersen LO, Nielsen HV. Development and evaluation of a genus-specific, probe-based, internal-process-controlled real-time PCR assay for sensitive and specific detection of Blastocystis spp. J Clin Microbiol. 2012;50:1847–1851. doi: 10.1128/JCM.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirier P, Wawrzyniak I, Albert A, El Alaoui H, Delbac F, Livrelli V. Development and evaluation of a real-time PCR assay for detection and quantification of Blastocystis parasites in human stool samples: prospective study of patients with hematological malignancies. J Clin Microbiol. 2011;49:975–983. doi: 10.1128/JCM.01392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stensvold CR. Comparison of sequencing (barcode region) and sequence-tagged-site PCR for Blastocystis subtyping. J Clin Microbiol. 2013;51:190–194. doi: 10.1128/JCM.02541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scicluna SM, Tawari B, Clark CG. DNA barcoding of Blastocystis. Protist. 2006;157:77–85. doi: 10.1016/j.protis.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Efunshile MA, Ngwu BA, Kurtzhals JA, Sahar S, König B, Stensvold CR. Molecular detection of the carriage rate of four intestinal protozoa with real-time polymerase chain reaction: possible overdiagnosis of Entamoeba histolytica in Nigeria. Am J Trop Med Hyg. 2015;93:257–262. doi: 10.4269/ajtmh.14-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Efunshile AM, Olawale T, Stensvold CR, Kurtzhals JAL, Konig B. Epidemiological study of the association between malaria and helminth infections in Nigeria. Am J Trop Med Hyg. 2015;92:578–582. doi: 10.4269/ajtmh.14-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stensvold CR, Alfellani M, Clark CG. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect Genet Evol. 2012;12:263–273. doi: 10.1016/j.meegid.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Jones MS, Ganac RD, Hiser G, Hudson NR, Le A, Whipps CM. Detection of Blastocystis from stool samples using real-time PCR. Parasitol Res. 2008;103:551–557. doi: 10.1007/s00436-008-1006-4. [DOI] [PubMed] [Google Scholar]
- 17.Abdulsalam AM, Ithoi I, Al-Mekhlafi HM, Khan AH, Ahmed A, Surin J, Mak JW. Prevalence, predictors and clinical significance of Blastocystis sp. in Sebha, Libya. Parasit Vectors. 2013;6:86. doi: 10.1186/1756-3305-6-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabrine-Santos M, Cintra Eo N, Carmo RA, Nascentes GA, Pedrosa AL, Correia D, Oliveira-Silva MB. Occurrence of Blastocystis spp. in Uberaba, Minas Gerais, Brazil. Rev Inst Med Trop Sao Paulo. 2015;57:211–214. doi: 10.1590/S0036-46652015000300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen AM, Stensvold CR, Mirsepasi H, Engberg J, Friis-Moller A, Porsbo LJ, Hammerum AM, Nordgaard-Lassen I, Nielsen HV, Krogfelt KA. Active ulcerative colitis associated with low prevalence of Blastocystis and Dientamoeba fragilis infection. Scand J Gastroenterol. 2013;48:638–639. doi: 10.3109/00365521.2013.780094. [DOI] [PubMed] [Google Scholar]
- 20.Alfellani MA, Stensvold CR, Vidal-Lapiedra A, Onuoha ES, Fagbenro-Beyioku AF, Clark CG. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013;126:11–18. doi: 10.1016/j.actatropica.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 21.Pandey PK, Verma P, Marathe N, Shetty S, Bavdekar A, Patole MS, Stensvold CR, Shouche YS. Prevalence and subtype analysis of Blastocystis in healthy Indian individuals. Infect Genet Evol. 2015;31:296–299. doi: 10.1016/j.meegid.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 22.AbuOdeh R, Ezzedine S, Samie A, Stensvold CR, ElBakri A. Prevalence and subtype distribution of Blastocystis in healthy individuals in Sharjah, United Arab Emirates. Infect Genet Evol. 2016;37:158–162. doi: 10.1016/j.meegid.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 23.Abu-Madi M, Aly M, Behnke JM, Clark CG, Balkhy H. The distribution of Blastocystis subtypes in isolates from Qatar. Parasit Vectors. 2015;8:465. doi: 10.1186/s13071-015-1071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez JD, Sanchez LV, Bautista DC, Corredor AF, Florez AC, Stensvold CR. Blastocystis subtypes detected in humans and animals from Colombia. Infect Genet Evol. 2014;22:223–228. doi: 10.1016/j.meegid.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 25.Alfellani MA, Jacob AS, Perea NO, Krecek RC, Taner-Mulla D, Verweij JJ, Levecke B, Tannich E, Clark CG, Stensvold CR. Diversity and distribution of Blastocystis sp. subtypes in non-human primates. Parasitology. 2013;140:966–971. doi: 10.1017/S0031182013000255. [DOI] [PubMed] [Google Scholar]
- 26.Scanlan PD, Stensvold CR, Cotter PD. Development and application of a Blastocystis subtype-specific PCR assay reveals that mixed-subtype infections are common in a healthy human population. Appl Environ Microbiol. 2015;81:4071–4076. doi: 10.1128/AEM.00520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee LI, Chye TT, Karmacharya BM, Govind SK. Blastocystis sp.: waterborne zoonotic organism, a possibility? Parasit Vectors. 2012;5:130. doi: 10.1186/1756-3305-5-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leelayoova S, Rangsin R, Taamasri P, Naaglor T, Thathaisong U, Mungthin M. Evidence of waterborne transmission of Blastocystis hominis. Am J Trop Med Hyg. 2004;70:658–662. [PubMed] [Google Scholar]