Summary
The rigorous characterization of distinct induced pluripotent stem cells (iPSC) derived from multiple reprogramming technologies, somatic sources, and donors is required to understand potential sources of variability and downstream potential. To achieve this goal, the Progenitor Cell Biology Consortium performed comprehensive experimental and genomic analyses of 58 iPSC from ten laboratories generated using a variety of reprogramming genes, vectors, and cells. Associated global molecular characterization studies identified functionally informative correlations in gene expression, DNA methylation, and/or copy-number variation among key developmental and oncogenic regulators as a result of donor, sex, line stability, reprogramming technology, and cell of origin. Furthermore, X-chromosome inactivation in PSC produced highly correlated differences in teratoma-lineage staining and regulator expression upon differentiation. All experimental results, and raw, processed, and metadata from these analyses, including powerful tools, are interactively accessible from a new online portal at https://www.synapse.org to serve as a reusable resource for the stem cell community.
Highlights
-
•
Comprehensive portal for diverse iPSC, protocols, metadata, and genomic assays
-
•
Recurrent CNV occur during reprogramming, impact oncogenes and tumor suppressors
-
•
DNA methylation is influenced by cell of origin in iPSC
-
•
PSC X-chromosome inactivation impacts lineage differentiation outcomes
Inherent differences exist among induced pluripotent stem cells (iPSC) from different laboratories, donor of origin, reprogramming methods, and culturing conditions. To carefully evaluate these differences, Lutzko and colleagues from the Progenitor Cell Biology Consortium characterize dozens of iPSC generated with diverse methods to identify methodological and donor-specific differences.
Introduction
Pluripotent stem cells (PSC) have been used to study human development, model disease, and generate cellular tools for regenerative medicine. Human embryonic stem cells (hESC) have been considered the functional, genetic, and epigenetic gold standard in the field (Thomson et al., 1998). Methods of somatic cell reprogramming to generate induced PSC (iPSC) (Takahashi and Yamanaka, 2006) are continually being improved and have enabled the generation of iPSC using a variety of somatic cell sources, gene combinations, and methodologies. However, due to the intensive resources required for iPSC generation and characterization, direct comparisons of iPSC generated using a wide range of technologies and cell sources from multiple independent laboratories have rarely been performed, making it unclear whether all methodologies produce iPSC with a similar quality and stability.
A variety of studies have compared the expression profiles, pluripotentiality, and genetic and epigenetic stability of hESC and iPSC including lines generated using different strategies, distinct parental somatic cell types, or reprogramming methods (Bock et al., 2011, International Stem Cell Initiative et al., 2007, Müller et al., 2011, Rouhani et al., 2014, Schlaeger et al., 2015). However, these have been limited to a few variables, have multiple methods or laboratories collecting and processing samples, and typically employ a single genomics platform. “Multi-omics” analyses have proved to be essential in deciphering complex gene regulatory programs, as demonstrated by analyses of iPSC reprogramming transitional states (Clancy et al., 2014, Lee et al., 2014, Tonge et al., 2014).
The Progenitor Cell Biology Consortium (PCBC) of the National Heart, Lung and Blood Institute was founded to study iPSC reprogramming and differentiation and develop strategies to address the challenges presented by the transplantation of these cells. These questions include, but are not limited to: (1) Do iPSC consistently generate all three germ layers? (2) How prevalent is copy-number variation (CNV) in iPSC generated using different reprogramming methodologies? (3) Do different reprogramming methods affect global methylation, gene, splicing and microRNA (miRNA) expression profiles? (4) Can aberrant PSC gene regulation be identified on a global basis? (5) How do variables such as X-chromosome inactivation (XCI) affect iPSC quality, stability, and differentiation potential? To advance these goals, the PCBC developed a Central Cell Characterization Core and Bioinformatics Core to perform standardized and comprehensive characterization of iPSC generated using different somatic cell sources, methodologies, and vectors. The characterized iPSC are being made available through WiCell Research Institute.
Using integrative analyses across genomic analysis platforms, we present comparative results on phenotype, genetics, epigenetics, and gene regulation for a diverse panel of iPSC and hESC. Standardized methods and strict control of reagents during cell culture, sample collection, and assay performance were used to evaluate the innate potential and limitations of these cells with fewer confounding factors. Our use of this uniform analytical methodology allowed us to discover candidate regulators of the fate of reprogrammed cells. To maximize the utility of this resource, we developed an interactive open data portal for access to the raw data, metadata, results, and protocols from these experiments for further analysis (https://www.synapse.org/PCBC).
Results
Study Design and Synapse Analysis Portal
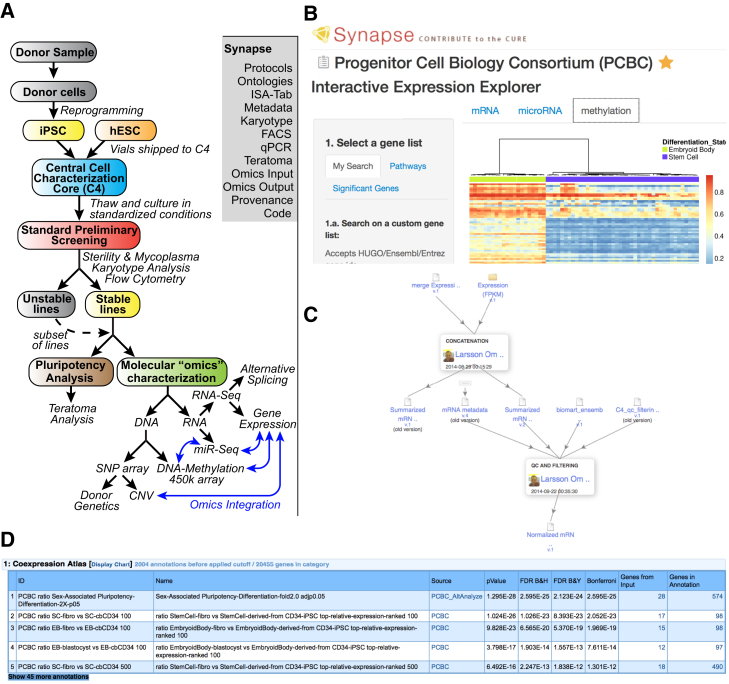
An overview of the study is presented in Figure 1. The evaluation of iPSC from multiple laboratories and methodologies required highly structured cell-line annotations and well-documented protocols to make comprehensive comparisons possible. Metadata standards were developed to capture the origin of each line, starting cell type, donor demographics, and reprogramming parameters (derivation method, vector type, reprogramming genes, culture conditions). These metadata were provided by the originating laboratory and confirmed and augmented with in vitro genetic and experimental characterization of the line. RNA sequencing (RNA-seq) was performed at an acceptable depth to facilitate accurate gene-expression quantification (Supplemental Experimental Procedures). To facilitate use of the protocols, genomic analyses, and metadata produced through this effort, we developed a sophisticated interactive data portal, the interface of which is exemplified in Figure 1. In addition to integrated provenance annotations for every raw data file, script, or processed result file, data can be queried through an interactive heatmap viewer that displays and inter-relates gene expression, DNA methylation, and miRNA expression for queried genes, pathways, and gene signatures produced in the analyses described here. These signatures have been further propagated into ToppGene (Chen et al., 2009) for interactive queries. Synapse IDs are included to access the resources, data, metadata, ontologies, and other information through the Synapse online repository.
Figure 1.
Study Design and Synapse Results Portal
(A) Overview of study design and data deposited online in Synapse. FACS, fluorescence-activated cell sorting; ISA-Tab, investigation/study/assay tab-delimited format.
(B) Synapse heatmap viewer displaying probes corresponding to EB-induced transcription factors and associated DNA-methylation levels to visually detected outliers in PSC.
(C) Provenance for the creation of normalized gene-expression values, associated scripts, quality control, metadata, and annotations.
(D) Gene-enrichment analysis in ToppGene displaying top-ranking PCBC stem cell signatures.
Screening of Lines
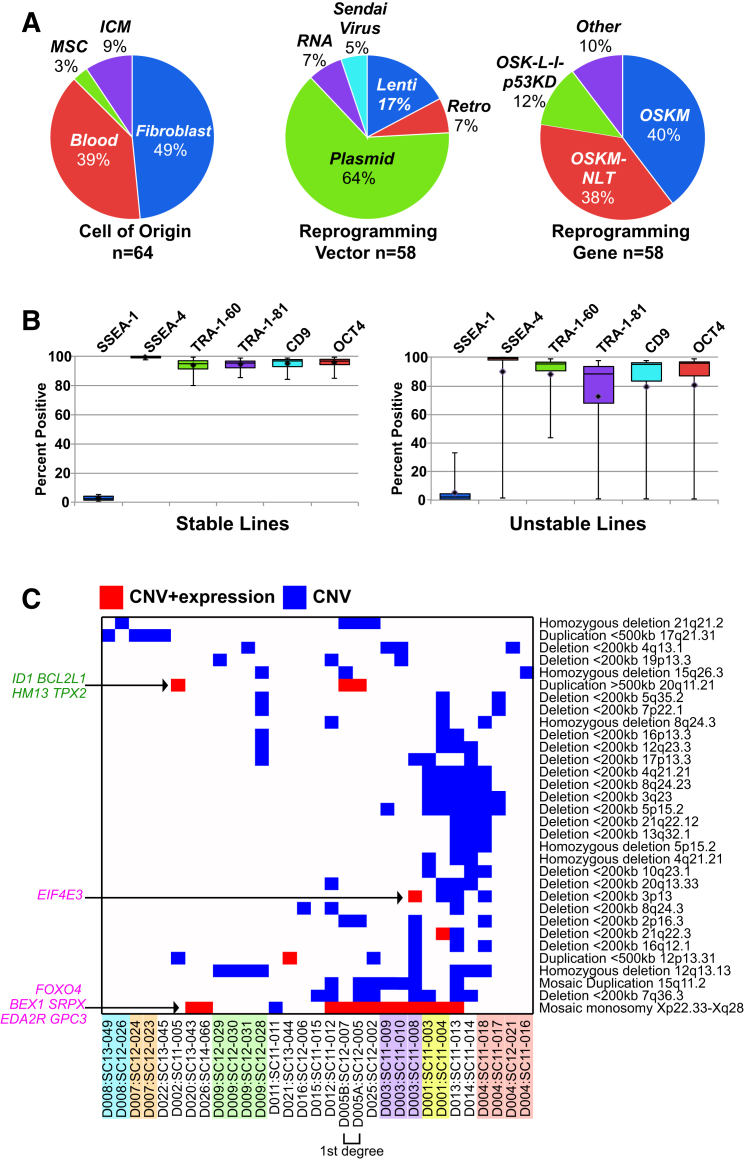
The data from the first 64 lines (58 iPSC and 6 hESC) enrolled in the study are presented here with their characteristics outlined in Figure 2A (details in syn2767694). All lines completed a standardized screen to ensure they met a basic set of criteria. This included self-renewal in defined feeder-free conditions, expression of markers of pluripotency and a lack of expression of markers of differentiation, a normal karyotype, and the ability to grow sufficient quantities of cells for the analyses (Tables S2 and S3; Figure S1). Overall, 6 hESC and 35 iPSC (64%) met these criteria and 23 iPSC did not (36%) (Table S4). Abnormal karyotypes were observed in seven lines (Table S5), with karyotypes for all lines available (syn2679104). The most consistent flow cytometry anomalies were TRA-1-81 and TRA-1-60 below 90% or an increase in SSEA-1 above 5% (Figure 2B). Due to contamination, difficulty in expanding cells, and/or abnormal karyotype, not all lines were included in functional pluripotency assays.
Figure 2.
iPSC Line Characteristics, Flow Cytometry Analysis, and CNV Accumulation
(A) Reprogramming variables for: originating cell type (left), reprogramming vector (middle), and gene combination (right). Reprogramming gene combinations: OSKM is composed of POU5F1 (also known as OCT4), SOX2, KLF4, and c-MYC; OSK-L-l-p53KD includes LIN28A and TP53 knockdown and l-MYC instead of c-MYC; OSKM-NLT includes NANOG, LIN28A, and SV40 large T antigen. ICM, inner cell mass; MSC, mesenchymal stem cell.
(B) Flow cytometry analysis classified iPSC as stable (n = 41) or unstable (n = 11). Boxes represent the first and third quartiles, whiskers show the complete range, and the horizontal line is the median.
(C) Commonly observed CNV predicted as non-benign or clinically significant and observed from at least three independent genetic donors are listed on the right and shown as a heatmap (blue). Red cells indicate that CNV overlaps genes with concordant expression differences. Concordant genes with known function are labeled on the left, with previously identified tumor-suppressor genes in purple and cell-growth-promoting and oncogenesis-promoting genes in green. Lines from the same donor are highlighted in the same color.
Pluripotency Analysis
Pluripotency was evaluated in a teratoma assay on 49 lines. Forty-six of the lines met the screening criteria outlined in Table S3 and 45 of these lines generated teratomas. Three lines did not meet the PSC screening criteria with decreased expression of self-renewal markers and increased differentiation in culture (SC12-021, SC12-023, and SC14-082), and all three successfully generated teratomas. All teratomas were scored by a clinical pathologist, and representatives of all three embryonic germ layers were identified in all tumors (detailed information is available at Synapse syn2882785). We performed immunostaining analysis on teratomas from a subset of lines to confirm pluripotency (muscle-specific actin [MSA], neurofilament, and α-fetoprotein) and OCT4 to evaluate the presence of undifferentiated PSC (Figure S1). This included two lines that did not meet the screening criteria and independent iPSC from the same donor as controls (Table S7), and three teratomas that had regions histopathologically classified as poorly differentiated as well as independent teratomas generated from the same lines (Table S8). The immunostaining confirmed pluripotency in all tumors (Figure S1). OCT4 staining was observed in one teratoma with a poorly differentiated region (SC12-034), although other teratomas from this line were fully differentiated and did not have OCT-4-stained regions. Two teratomas from other lines (SC11-014 and SC11-0013) with poorly differentiated regions did not have OCT4 immunoreactivity, although we did not have adjacent sections for staining (Table S8).
Evaluation of CNV Changes in iPSC
Genetic stability was evaluated between independent lines with common donors by CNV SNP microarrays. Although two SNP genotyping arrays were used, all lines derived from a single donor were run on the same platform (see Experimental Procedures). Variations were observed in all lines and on all chromosomes (Figure S2). Excluding human leukocyte antigen-associated regions, 724 non-benign or clinically significant CNV from 529 unique genomic loci were identified (syn3105726). Although not significant, lines reprogrammed with integrating vectors trended toward a higher frequency of clinically significant CNV (58%) compared with non-integrating vectors (41%).
Our study included different iPSC generated from the same donor sample and reprogramming methods, thereby enabling the direct evaluation of the CNV present in the donor versus those induced during reprogramming and culture. We observed CNV that were specific to the donor, and others present among multiple genetically distinct iPSC (Figure S2). We identified lines generated from the same donor samples that had variable CNV (Table S6), with some donors having higher frequencies of CNV than others (such as D001, 2, 3, 4, and 9).
We discovered 102 non-benign CNV shared by at least two distinct donors, with 83 of these CNV variably present in two or more distinct samples from a common donor. Two donors (D004 and D003) were solely responsible for 46 of these CNV, while 26 were recurrent among multiple donors (Figure S2C). A more stringent analysis considering CNV shared among at least three donors identified a set of 31 frequently affected genomic loci, suggesting that they occurred during iPSC reprogramming or that the starting samples were mosaic (Abyzov et al., 2012, McConnell et al., 2013, Young et al., 2012) (Figure 2).
Comparison of the CNV and RNA-seq data identified 19 non-benign and clinically significant CNV that overlap with differentially expressed genes in a manner consistent with the detected duplication or deletion (syn2731183) (Figure 2C). This included 88 downregulated genes in deleted regions, 79 of which correspond to the frequently observed X-chromosome mosaic monosomy. Among 26 upregulated genes in duplicated regions, a duplication of 20q11.21 corresponded to the upregulation of nine overlapping genes, including four (ID1, BCL2L1, HM13, and TPX2) previously shown to promote hESC survival or oncogenic potential (Nguyen et al., 2014). We also found compatible regulation of the cancer susceptibility genes FYN (6q21 duplication), ERCC2 (19q13.32 duplication), and NIN (14q22.1 duplication), as well as the tumor-suppressor genes FOXO4, BEX1, SRPX, EDA2R, GPC3 (X monosomy), ING2 (4q35.1 deletion), and EIF4E3 (3p13 deletion) (Osborne et al., 2013). These results are consistent with these CNV conferring a survival or proliferative advantage.
Global Expression and Methylation Analysis of PSC
To understand the molecular determinants of PSC quality as a function of reprogramming method and somatic cell origin, we performed mRNA, miRNA, and methylation profiling on iPSC and hESC with profiles from hESC-derived embryoid bodies (EB) as a control.
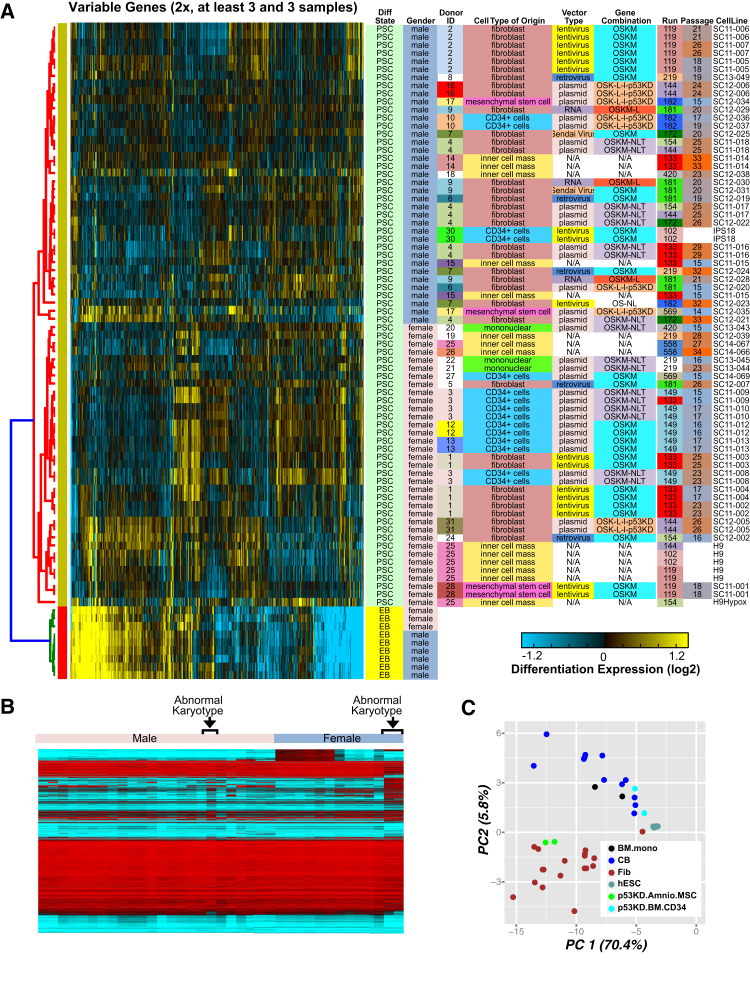
Relative to EB, hESC and iPSC were largely indistinguishable from each other at the global gene-expression level by both hierarchical clustering and principal component analysis (PCA) (syn3107554). Greater variability was observed from analogous DNA methylation and miRNA profiles (Figure S3). However, restricting the analysis to genes with varying expression only in PSC identifies donor, sex, reprogramming technology, and originating laboratory as the major driving covariates by hierarchical clustering (Figure 3A). These differences did not clearly associate with the passage number of the profiled PSC. Of interest, H9 cells (D025) analyzed greater than ten passages apart displayed a highly variable signature with the higher passage more similar to EB. Likewise, one of two mesenchymal stem cell-derived iPSC from the same donor and laboratory (D017) exhibited a similar EB-like signature. Neither the D017 nor the H9 samples displayed apparent global DNA methylation differences, demonstrating the utility of distinct genomic platforms in assessing PSC quality (Figure S3).
Figure 3.
Global Gene Expression and Methylation Variation between iPSC
(A) Hierarchical clustering of the most variable genes observed among iPSC and hESC (n = 1,031). These genes were chosen by selecting the reliably expressed genes (n = 9,670) that varied at least 2-fold between six or more samples and correlated (ρ > 0.5) to the expression of at least ten other genes (AltAnalyze, Predict Groups analysis). Yellow indicates upregulated and blue indicates downregulated genes. Expression is shown relative to day 17 EB derived from multiple hESC (median-based normalization applied to preferentially identify PSC variance). Selected metadata associated with each cell line are shown on the right, with identical terms in each column sharing a color.
(B) Expression clustering of all CpG methylation probes on the X chromosome, with blue indicating hypo- and red hypermethylation. Lines with an abnormal karyotype are indicated.
(C) Principal component analysis of all differentially methylated probes for all evaluated PSC lines, colored according to cell of origin. BM.mono, bone marrow-derived monocytes; CB, umbilical cord blood; Fib, fibroblasts; Amnio.MSC, amniotic fluid-derived mesenchymal stem cells; BM.CD34, bone marrow-derived CD-34+ cells; p53KD, OSKL-l-p53KD reprogramming vector.
To identify differences associated with major cell-line variables, we performed all possible pairwise comparisons from each metadata category for gene expression, splicing, miRNA, and DNA methylation (syn3094629). We identified 355 differentially expressed genes from these comparisons and 3,451 differential methylated DNA probes. As expected, laboratory of origin accounted for the largest number of differences, likely because several iPSC derivation protocols and cell types of origin were largely unique to a single laboratory (e.g., RNA-based reprogramming, stromal priming) and could therefore mask handling or other technical differences between laboratories. The major distinguishing reprogramming variables from the methylation analyses were cell type of origin (1,427 probes), method of reprogramming (1,346 probes), and sex (520 probes). Clustering of these methylation profiles readily distinguished lines based on both sex and abnormal karyotypes (Figure 3B), while PCA segregated samples based on cell of origin (Figure 3C). Although these samples consistently segregated by cell of origin independently of the donor sex, these differences could not be directly attributed to blood and fibroblast somatic methylation profile differences (data not shown).
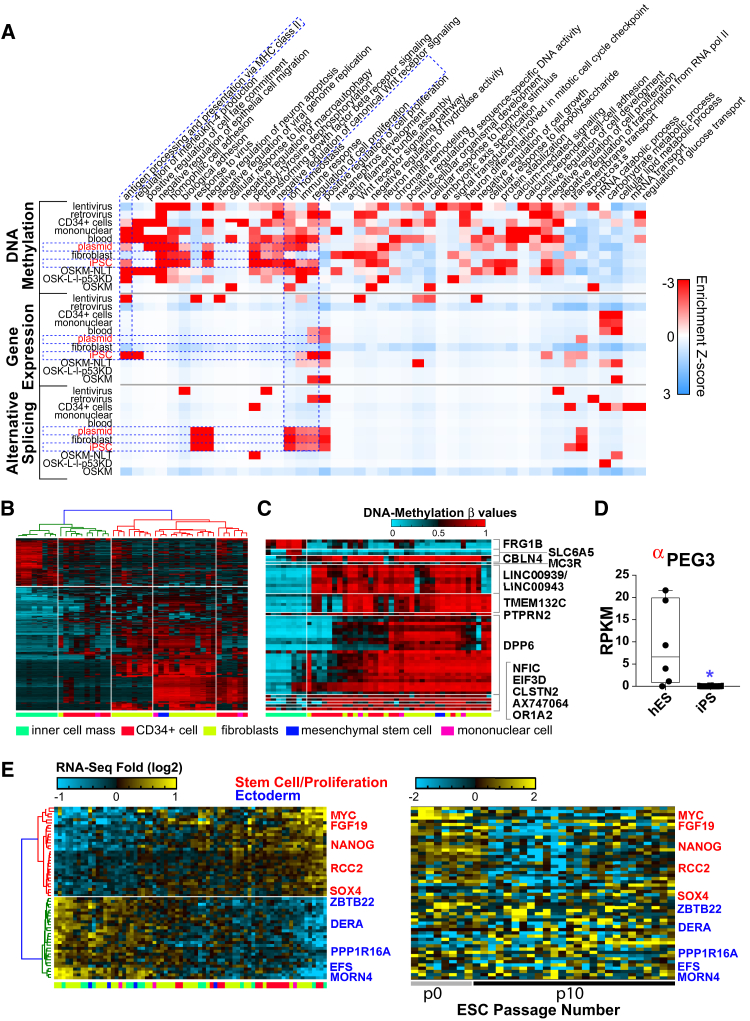
To examine the impact on possible pathways, we looked at the enrichment of our discovered reprogramming regulated genes among gene ontology (GO) terms for each of the different measurement platforms (Figure 4A). The most prominent pathway level effects were found in the methylation comparisons with hESC for a wide array of biological comparisons and tested reprogramming variables. We observed consistent regulation of inflammatory and immune response, ion homeostasis, and regulation of cell proliferation gene sets, particularly among all iPSC compared with hESC, among the different profiling technologies.
Figure 4.
Global Reprogramming Impact on Pathways in PSC
(A) Pathway-level impact of reprogramming methodology or initiating cell type as compared with hESC, based on statistically enriched GO terms for each comparison and profiling technology. Red indicates higher Z scores, corresponding to lower GO-Elite enrichment p values. Dashed blue lines indicate common regulated pathways in the different applied profiling methods and in the same reprogramming comparisons.
(B) DNA-methylation profiles for probes significantly differing between hESC and iPSC (non-adjusted p < 0.05). Colored bars below each cluster indicate cell of origin.
(C) Hierarchically clustered subset of the DNA-methylation probes with the lowest p values (adjusted p < 0.05). Associated genes for each probe cluster are indicated on the right.
(D) PEG3 expression in hESC and iPSC lines. Box and whiskers plot of unique donor PSC values are represented. Anti-correlated gene expression and DNA methylation are denoted by a red alpha. A blue asterisk indicates significant differential expression.
(E) Expression clustering (HOPACH) of genes correlated and anti-correlated with NANOG and MYC gene expression in all PSC. To the right of this cluster, early- (P0) and late-passage (P10) single-cell hESC (Yan et al., 2013) are shown in the same gene order. Genes with red names are negative regulators of differentiation, stem cell maintenance genes, or positive regulators of cell proliferation, while genes with blue names are associated with ectoderm differentiation, based on ToppGene-associated annotations from multiple sources.
To determine whether differential methylation might be a source of observed gene-expression differences, we compared the expression profiles of these differentially regulated genes and probes, based on common gene annotations (e.g., promoter, body, or UTR location of the probe). This analysis indicates that ∼21% of all differentially methylated probes correspond to gene-expression changes in the PSC, while ∼43% of all differentially expressed genes appear to be due to underlying differential DNA methylation (Pearson ρ < −0.5). Only negative correlations were considered from these analyses. Taken together, these data suggest that while iPSC are largely similar to hESC at the level of gene expression, observed differences are frequently correlated with changes in DNA methylation.
Comparison of hESC and iPSC
Among DNA-methylation profiles, comparison of all iPSC to hESC yielded 180 differentially methylated sites, with 52% of these anti-correlated with gene expression (n = 93). A more relaxed analysis (non-adjusted moderated t test p < 0.05) of unique donor samples indicated that methylation probes largely segregated by donor and cell of origin when subjected to hierarchical clustering (Figure 4B). In agreement with previously published studies, DPP6, TMEM132C, and PTPRT were among the most differentially methylated loci between iPSC and hESC (Figure 4C). In addition, we found that several interesting gene loci were hypomethylated (FRG1B, SLC6A5) and hypermethylated (PTPRN2, LINC00939, CBLN4, MC3R, NFIC, EIF3D, CLSTN2, AX747064, and OR1A2) in iPSC. Genes hypermethylated in iPSC were associated with neuronal differentiation and genomic targets of the polycomb repressive complex 2 (PRC2) (ToppGene). The most highly differentially expressed iPSC versus hESC gene, the paternally imprinted PEG3, was also anti-correlated with DNA-methylation probes (Pearson ρ < −0.98) (Figure 4D).
A close examination of the expression of core pluripotency factors across all PSC identified a large number of genes correlated and anti-correlated with NANOG and MYC (Figure 4E). Genes coexpressed with NANOG and MYC were enriched in negative regulators of differentiation, stem cell maintenance, and positive regulation of cell proliferation, while anti-correlated genes were enriched in experimentally observed ectoderm differentiation upregulated genes (ToppGene). To test whether these differences could be related to PSC quality and increased passaging, we compared the expression of these same genes with hESC from a previously described single-cell RNA-seq dataset (Figure 4E) (Yan et al., 2013). Clustering of both early (passage 0) and late (passage 10) single-cell hESC confirmed that NANOG and MYC high lines were most similar to early-passage hESC.
Genomic Impact of Reprogramming Technology, Cell of Origin, and iPSC Stability
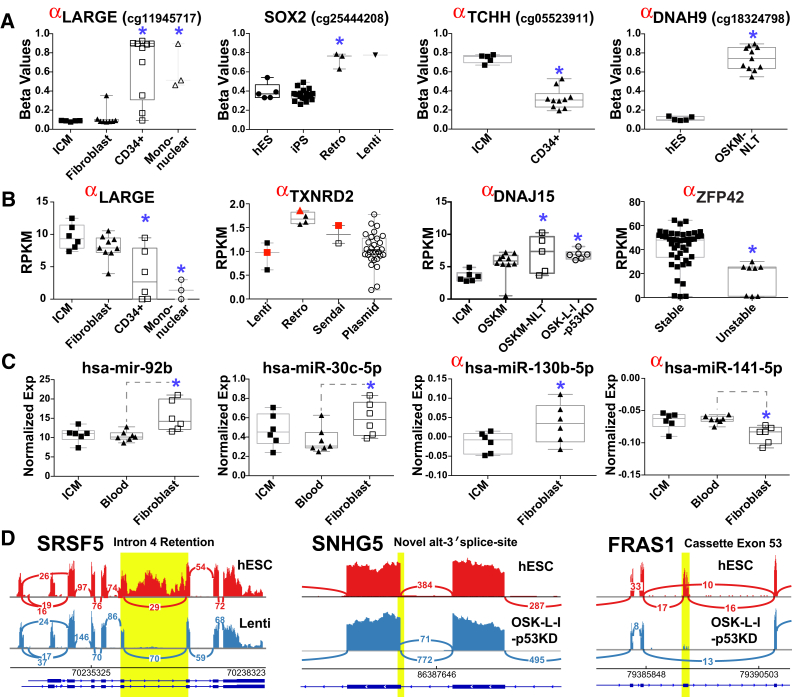
Among the 64 lines evaluated, 41 underwent genomics characterization, with five unstable lines included as controls. These 46 lines comprised five cell-of-origin groups, five reprogramming vector types, and five distinct gene combinations. Comprehensive pairwise comparisons of all metadata categories across each genomics platform highlighted a large number of genes (syn3106206), splicing variants (syn3106266), methylation probes (syn3106255), and miRNAs (syn3106244) strongly associated with one or more of these variables (Figure 5). To our knowledge, few of these molecular differences have previously been reported. Many of the most significant differences were observed among differentially methylated probes (Figures 5A and S4A). For example, SOX2 was hypermethylated in retroviral lines relative to all hESC and nearly all iPSC. Reciprocal differences in gene expression were frequently observed for these and many other differentially methylated genes.
Figure 5.
Candidate Factors Associated with iPSC Derivation Method
(A–C) The top differentially regulated (A) DNA methylation, (B) mRNA gene expression, and (C) normalized miRNA expression profiles associated with each indicated comparison. Box and whiskers plot of unique donor PSC values are represented. For TXNRD2, sample expression values for the same genetic donor (D007) are indicated for the three indicated reprogramming methods in red. Anti-correlated gene expression and DNA methylation are denoted by a red alpha. Blue asterisks indicate significantly differentially expressed genes (adjusted p < 0.05) versus hESC or the indicated comparator. Retro, retroviral; Lenti, lentiviral; RPKM, reads per kilobase per million mapped reads.
(D) Examples of splicing events visualized in the software IGV (Broad Institute), with associated genomic read-alignment depth and junction read counts indicated for a single representative sample.
For all genomic analyses, the small number of unique donors available for certain reprogramming methods limited the power of our analysis. However, the availability of a small number of iPSC derived from the same donor with different methods provides additional confirmation of our findings. For example, differentially expressed retroviral and lentiviral associated genes (e.g., TXNRD2, JUN, UCP2, and HIST1H2BF; Figures 5B and S4B) were consistently observed from uniquely reprogrammed lines from a single donor (D007). Notably, these genes are involved in multiple pathways related to oxidative stress. Differential expression of multiple genes affecting cell growth and differentiation (ID2, ID4, JAG1, IGFBP5, and GLT1D1) were observed with OSK-L-l-p53KD, relative to other gene reprogramming combinations or hESC. In unstable lines, decreased expression of crucial PSC genes (ZFP42 and TRIM6) was associated with increased promoter and gene methylation of these genes. Using a 96-gene qPCR panel we verified differential expression for multiple genes where corresponding probes were present (e.g., ZFP42; Figure S4C).
In total, 41 miRNAs were statistically associated with at least one reprogramming variable. Among these, we observed three miRNAs (miR-92b, miR-30c-1, miR-30c-2) with predicted mRNA targets that were differentially expressed in a reciprocal manner (GO-Elite) (Figure 5C). Five of the 41 regulated miRNAs were also anti-correlated with methylation probes (miR-141, miR-130b, miR-191, miR-660, miR-548f-1), suggesting regulation in part by DNA methylation.
Alternative splicing and promoter usage was evaluated in our RNA-seq data. Comparison of hESC and hESC-derived EBs identified 129 alternative exon events with a false discovery rate p < 0.05 (syn3106284), including many well-validated events (in MBD2, DNMT3B, SLK, ADD3, MARK3, FYN, NUMB, NAV2, and NFYA) (Gopalakrishna-Pillai and Iverson, 2011, Lu et al., 2014, Salomonis et al., 2009) (Figure S5A), suggesting that these data are reliable for more in-depth evaluation. A total of 77 alternative exons were significant in a pairwise comparison of all major reprogramming or cell-of-origin variables in PSC. Manual examination of highly differential but non-significant splicing events suggest that many are valid, but detected with lower sensitivity due to reduced sequencing depth (Figures 5D and S5B).
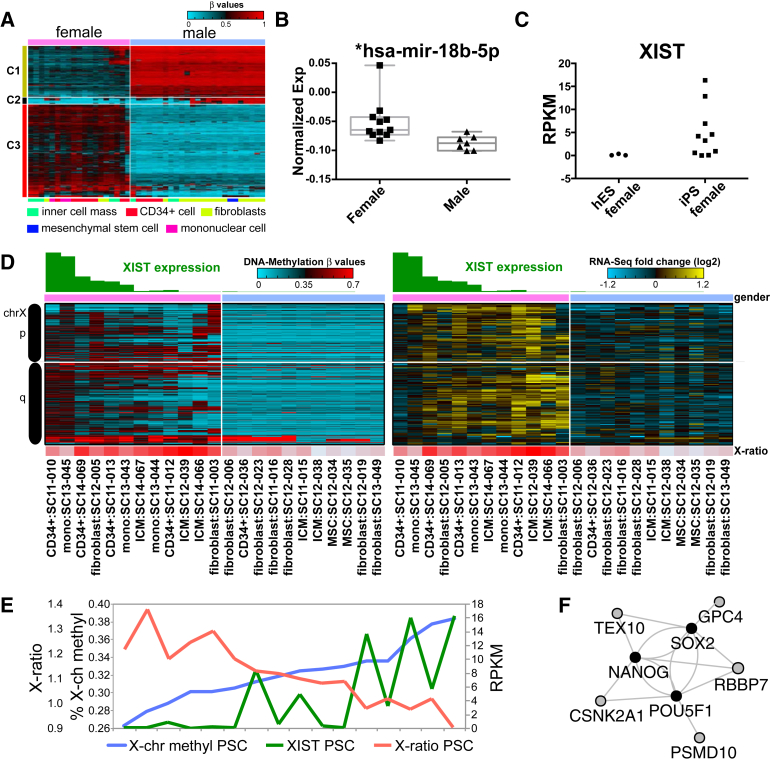
Effect of XCI and Donor Sex
A significant potential confounder in this dataset is donor sex difference. A total of 520 probes were differentially methylated between male and female donors, the majority of which were localized to allosomes (457 probes) (Figure 6A). Similarly, most differentially expressed genes between male and females were also localized to allosomes (43 out of 60), as were differentially expressed miRNA (4 out of 7). Predicted mRNA targets (GO-Elite) of one X-chromosomal miRNA (miR-18b) were enriched among male versus female RNA upregulated genes (Figure 6B). This miRNA was also found to be anti-correlated to its own DNA-methylation probes, suggesting that it is regulated by DNA methylation.
Figure 6.
Impact of X-Chromosome Inactivation and Sex on iPSC
(A) Segregation of PSC based on differentially regulated sex-associated DNA-methylated probes (HOPACH).
(B) Normalized miRNA expression values in male and female lines from distinct donors. Box and whiskers plot of unique donor PSC values are represented.
(C) XIST expression in independent female hESC and iPSC samples from unique donors.
(D) Heatmaps of all anti-correlated (Pearson ρ < −0.6) methylation probes (left) and genes (right) on the X chromosome, ordered by genomic location. XIST expression values are in green and X-to-autosome expression ratios are below the heatmaps.
(E) Comparison of distinct measures of XCI within female PSC as determined by RNA-seq (X-to-autosome ratio, XIST expression) and DNA-methylation array (% X-chr methylation).
(F) Protein-protein interactions (BioGRID database) between genes anti-correlated with XIST (gray) and core pluripotency factors (black).
Genes associated with autosomal differential DNA methylation were enriched for PRC2 factors and targets of the PRC2 transcription factor Suz12 (Figures S6B and S6C). Only one DNA-methylation-regulating gene, MECP2, was itself differentially methylated between females and males. This is consistent with prior studies that have identified MECP2 as a target of X inactivation (Vallot et al., 2015).
In mouse ESC and human somatic cells, aberrant loss of XIST expression and corresponding breakdown of normal XCI has been associated with reduced developmental and increased oncogenic potential. In human PSC, XIST expression is required for the initiation of XCI but not for XCI maintenance. Multiple classes of female PSC have been described including those which only undergo XCI upon differentiation (class I), those that already have undergone XCI (class II and III), and PSC that have lost XIST during culture and have undergone eroded XCI (class III) (Hall et al., 2008, Silva et al., 2008, Vallot et al., 2015). Six of ten iPSC from distinct female donors show little to no XIST expression by RNA-seq, with no expression in any of the three hESC (Figure 6C). Restricted analysis of probes on the X chromosome found 1,118 methylation probes anti-correlated for the same PSC with gene expression (syn3107536). These largely overlapped with a prior set of described XCI-associated probes, 619 out of 3,279 (Nazor et al., 2012) (syn3107535). From these 1,118 probes, we find that lines without XIST expression have a decrease in X-chromosome methylation and increased X-to-autosome gene-expression ratio (Figures 6D and S6C). Each of these three measures of XCI were correlated to each other (ρ > 0.6 or ρ < −0.6) (Figure 6E). The observed continuum of predicted XCI among PSC lines supports prior proposed models of variable or precocious XCI among cells within each PSC line, rather than 100% conformity (Hall et al., 2008, Silva et al., 2008). Although spontaneous differentiation in some cultures could account for the increased XCI, both XIST expression and X-to-autosome ratios were largely consistent in biological RNA-seq replicates from the same PSC line (data not shown). Among 116 genes anti-correlated (Pearson ρ < −0.6) with XIST expression, five (RBBP7, CSNK2A1, PSMD10, GPC4, and TEX10) shared protein interactions with at least one core pluripotency factor (POU5F1, SOX2, or NANOG) (BioGRID database). The X-chromosome localized nucleosome remodeling factor RBBP7 was the most anti-correlated with XIST expression and interacts with all three pluripotency factors at the protein level (Figure 6E).
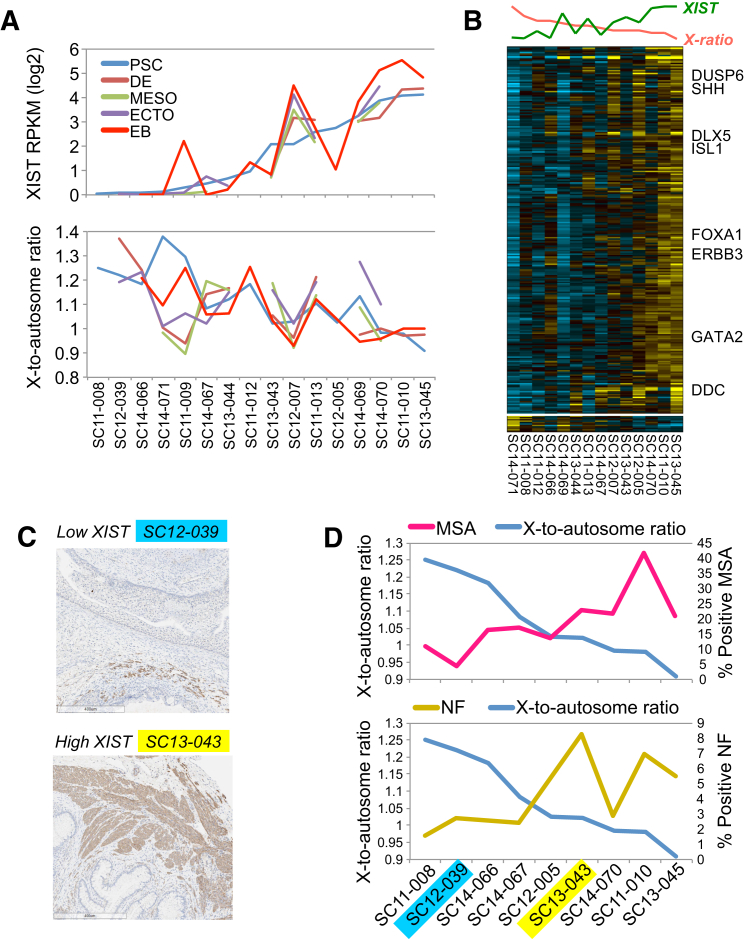
In addition to these expression differences, 646 autosome and allosome probes were differentially methylated in XIST-high versus XIST-low female lines (unique donors, all probes considered). Among the 236 known associated genes, eight transcription factors were hypermethylated (Brachyury, ZNF628, CUX1) or hypomethylated (MZF1, SCRT2, SCML1, TFCP2, ZNF148) with high XIST expression. Several of these factors have important roles in lineage differentiation (Brachyury, SCRT2, TFCP2, CUX1) or proliferation (MZF1, ZNF148, CUX1) (Figure S6D). As these genes promote distinct differentiation pathways, we subjected a set of female PSC (n = 16) to short-term directed differentiation assays for definitive endoderm, mesoderm, ectoderm, and EB and performed RNA-seq. Although XIST expression in these lines changed upon differentiation, high XIST lines generally remained high (average 2-fold increase versus PSC) and XIST-low remained low (Figure 7A). While most of these lines retained similar X-to-autosome ratios during differentiation as well, notable variance among a few lines was observed (SC11-009, SC14-069, SC14-071) (Figure 7A). Comparison of the number of passages in these two sets of lines revealed that low XIST PSC had undergone significantly more passages in culture (Student’s t test p < 0.05, ∼6 passages more on average). At least one prior study has demonstrated that failure to induce to XIST expression upon PSC differentiation is a hallmark of XCI erosion (Mekhoubad et al., 2012). Taken together, the low XIST female PSC in this study appear to be most consistent with class III or eroded XCI. An exception to this rule was SC11-009. This line was found to transition from low to high XIST expression from the PSC to the EB. This same trend was observed in all lineage and EB differentiations, suggesting induction of XCI (class I).
Figure 7.
EB Differentiation Outcomes Correlate with Differential X-Chromosome Inactivation among Female Stem Cell Lines
(A) Comparison of XIST expression and X-to-autosome ratios in PSC and EB for the same lines (PSC line name indicated below the plots).
(B) HOPACH clustered heatmap of differentially expressed genes in EB differentiated from female PSC lines. Developmental regulators are shown on the right of the plot. PSC X-to-autosome ratio and XIST expression are displayed above the plot.
(C) Immunohistochemistry of two teratomas derived from a low and high XIST female PSC using a muscle-specific actin (MSA) antibody.
(D) Quantification of the percentage of positive MSA or neurofilament (NF) staining in adjacent teratoma sections relative to PSC X-to-autosome ratio.
To analyze the differentiation gene-expression results in the context of XCI, we focused on genes with expression correlated (ρ > 0.6 or ρ < −0.6) to multiple measures of PSC XCI in each differentiation state (PSC XIST expression, X-to-autosome ratio, and degree of X-chromosome methylation). In EB, we found 267 genes correlated to multiple measures of XCI and only 12 anti-correlated (Figure 7B). These correlated genes were most enriched (ToppGene) in proteins associated with methylation (e.g., TRMT11, COMTD1, HENMT1, CAMKMT), neuron-fate commitment (GATA2 ISL1, FOXA1, SHH), heart development (ERBB3, ISL1, SSH), and other broad developmental processes. Analysis of the early germ-layer differentiations revealed possible precocious induction of genes anti-correlated with measures of XCI in each of the differentiations, such as myocardin and SMAD3 in definitive endoderm (Figure S6E, syn5565603). As an improved means to evaluate the differentiation potential of these lines, we performed immunohistochemistry on teratoma sections from female PSC (Figures 7C, 7D, and S6F). On average, we detected ∼18% positive MSA staining and ∼4% neurofilament (NF) staining in adjacent histological sections. Strikingly, both differentiation markers were correlated to multiple measures of PSC XCI (>0.6), with MSA most highly correlated to XIST expression (ρ = 0.71) and NF to X-to-autosome ratio (ρ = 0.67). These results are in agreement with prior studies indicating that PSC with increased XIST and XCI results in improved differentiations relative to PSC undergoing XCI erosion (Mekhoubad et al., 2012).
Discussion
The large-scale profiling of dozens of iPSC and previously characterized hESC represents an important analytical reference for the stem cell research community. Evaluating these lines using the same post-reprogramming culture conditions and profiling technologies has allowed us to carefully examine many possible variables. The creation of metadata standards and associated ontologies was essential to make informed comparisons and identify confounders in our study. All metadata, raw genomic files, protocols, processed results, and analyses are provided in Synapse (Omberg et al., 2013).
Our studies identified 23 iPSC lines with adverse characteristics such as contamination, karyotypic abnormalities, flow cytometry, or culture morphology consistent with differentiation. Surprisingly, teratomas generated from 45 of 46 lines, including three with characteristics of differentiation, were pluripotent as they contained cells from all three embryonic germ layers. Notably, three pluripotent teratomas also contained undifferentiated cells identified by histological or immunostaining analyses, although independent tumors from the same lines were fully differentiated and did not. Given that the teratoma assay is commonly used to confirm PSC pluripotency and quality (Muller et al., 2010), these results suggest that teratoma analysis should be considered within the context of other analyses and results to determine the quality of the PSC line and not as a stand-alone quality measure.
Evaluation of deleterious CNV provided strong evidence that the same genetic abnormalities can occur in distinct iPSC lines and that such abnormalities can arise during the reprogramming process. As described in other studies, we were unable to exclude the possibility that there was heterogeneity in the starting cell population (Ma et al., 2014). CNV that were coincident with differential expression frequently resulted in the deletion of known tumor suppressors or duplication of cell growth/oncogenic factors. Such genetic abnormalities could result in clonal selection advantages that are undesirable for clinical applications (Cunningham et al., 2012).
The DNA-methylation, gene-expression, miRNA, and splicing differences observed in these studies represent intriguing differences in PSC that could result in differences in pluripotentiality, cell growth, or potential tumorigenicity in vivo. The existence of consistent patterns between DNA methylation and mRNA or miRNA expression provides an additional layer of confidence in these observations. While methylation profiles were highly and consistently different among iPSC and hESC, fewer differences were observed for mRNA and miRNA. Although many DNA-methylation probes were identified that were highly distinct between iPSC and hESC, including a number of probes anti-correlated with gene expression (FRG1B, CCL28, CR1L, PEG3), none could perfectly distinguish between these cell types.
At a global level, the analysis of DNA-methylation profiles provides important insights into molecular and cell growth characteristics of PSC that would otherwise be difficult to identify. Reprogramming-associated variations in X-linked CpG methylation is of particular interest because of the complex variations in degrees of XCI and X-chromosome reactivation between various hierarchical states of pluripotency. Our analysis highlighted two distinct populations of XCI female cells, with the most hypermethylated X-chromosomal PSC split between very high and absent XIST expression. Interestingly, none of the female hESC lines in this study expressed XIST, whereas many of the iPSC do. Our analyses of differentiated female PSC identified correlates between XCI and pluripotentiality that substantiate prior proposed models and provide additional candidate molecular regulators for investigation (Mekhoubad et al., 2012, Silva et al., 2008).
Genes anti-correlated with XIST, principally RBBP7, share protein interactions with core pluripotency regulators, and these differences persist in the EB. This result is particularly intriguing given that RBBP7, a partner of PRC2 implicated in nucleosome binding, and SUZ12, a component of PRC2 required for stability of the complex and EZH1/2 mediated catalytic activity, were highly enriched factors in our analysis of differentially methylated sex-associated autosomal genes. In addition to RBBP7, predicted regulation by PRC2 was recurrent in a number of our covariate analyses, including iPSC versus hESC differentially methylated probes. A growing body of literature now supports an important role for PRC2 in pluripotency, XCI, and differentiation as a recruitment tool of PRC1 (Cheng et al., 2014). Although likely not relevant in vivo, such physical protein and epigenetic interactions could be undesirable in iPSC for programmed lineage differentiation.
The resource presented here provides a standardized and annotated dataset for the characterization of hESC and iPSC that can be applied to the discovery of molecular determinants underlying specific biological properties of iPSC and their use for future clinical applications. It provides information on the most relevant, informative, and efficient assays to use for iPSC characterization. Finally, the new data repository encodes a standard for the biological, genomic, and epigenomic characteristics of high-quality, stable iPSC that will serve as a valuable resource as iPSC technology moves into clinical translation. The many observed reprogramming and cell-of-origin gene-expression and splicing differences provide intriguing starting hypotheses to fuel new research.
We aim to improve the breadth and utility of this new resource by adding additional pluripotent lines and differentiated products. Integration of previously published and new datasets will further facilitate advanced cross-comparison analyses, many of which can be achieved using the online data analysis and exploratory tools provided within the Synapse programmatic and web interface. By providing consistent cell-line descriptions, protocols, and associated data in an easy-to-access online repository, we hope that these observations will fuel future research into the role of these gene signatures in resulting progenitor populations.
Experimental Procedures
Methods and Data Availability
All data and methods described herein are available at https://www.synapse.org/PCBC (http://dx.doi.org/10.7303/syn1773109) and/or Supplemental Experimental Procedures. Accessions for specific methods are provided in methods sections and Table S9. For interactive analyses, customized data exploration options have been integrated into Synapse to facilitate gene-level, cluster, and ToppGene functional enrichment analyses.
Cell Lines
The lines brought into the study included commonly used but distinct variables from multiple laboratories (Figure 2A). The line identifiers, originating laboratory, and key contributing scientists for each line are provided in Table S1.
Genomic and Epigenetic Molecular Characterization
mRNA-Seq libraries were prepared with the Illumina TruSeq kit RNA V2. Single-end libraries were sequenced at a depth of between 10 and 30 million 50-nt reads on an Illumina HiSeq 2000. A small number (n = 3) of ESC and iPSC were also sequenced at a depth of ∼50 million paired-end, stranded reads, for comparison. miRNA libraries were prepared with the Illumina TruSeq Small RNA kit and sequenced to 1–4 million reads. Methylation was assessed with the Illumina HumanMethylation450 BeadChip with annotations provided by ENCODE (Encode Project Consortium, 2012). Two different assays were used for CNV analysis. 21 cell lines were assayed with the Illumina CytoSNP-850K BeadChip, and 29 cell lines with the Illumina HD HumanOMNI-Quad BeadChip platform. Thirty-seven lines were assayed using a TaqMan Low Density Array (Life Technologies, 4385344) containing a panel of stem cell and pluripotency marker genes (syn3107327).
Data Processing
FASTQ files were aligned to the human genome build GRCh37 and University of California Santa Cruz transcriptome reference (Rosenbloom et al., 2014) using TopHat 2.0.9 (Kim et al., 2013) (syn1773110). Gene-level RPKM (reads per kilobase per million mapped reads) and alternative splicing estimates were obtained from AltAnalyze (Emig et al., 2010). miRNA expression was quantified with mirExpress v2.1.4 (Wang et al., 2009) using the human miRBase 20.0 reference (syn2247097). Methylation arrays were normalized with the minfi R package (Aryee et al., 2014) (syn2677441). Raw data and processing scripts with exact parameters used are available and are linked together by provenance in Synapse (Table S9).
Author Contributions
J.W., W.H., A.K.H., P.M., A.M., J.A.C., B.A., and C.L. conceived of and designed the study. N.S., P.J.D., L.O., R.S., S.B., L.S., S.H.S., M.K., C.M., K.D., and S.H. generated and assembled the data. N.S., P.J.D., L.O., R.S., S.B., J.H., M.K., S.K.S., K.D., Y.W., E.Z., P.M., J.A.C., B.A., and C.L. analyzed and interpreted the data. N.S., P.J.D., L.O., L.S., S.H.S., J.H., and C.L. wrote the manuscript.
Acknowledgments
The authors thank the PCBC line contributors in Table S1; Dr. Michael Terrin, Ling Tang, Andrea Lefever, and Liz Casher from the Administrative Coordinating Center; and Dr. Elke Grassman and Diana Nordling from the CCHMC Translational Core Laboratories for support. This work was supported by the NHLBI Progenitor Cell Biology Consortium, Administrative Coordinating Center (U01HL099997), Cell Characterization Core, Bioinformatics Core, and PCBC2012Pilot_01. Other support was provided by the National Heart, Lung, and Blood Institute (NHLBI) (U01HL099775), the National Institute for Child Health and Human Development (NICHD) (R01HD082098), the National Institute General Medical Sciences (NIGMS) (R01GM110628), and the National Eye Institute (NEI) (R01EY023962). Under a licensing agreement between Life Technologies Corporation and the Johns Hopkins University, E.Z. is entitled to a share of royalty received by the University on sales of human induced pluripotent stem cell lines. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its Conflict of Interest policies.
Published: June 9, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, Supplemental Results, six figures, and nine tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.05.006.
Supplemental Information
References
- Abyzov A., Mariani J., Palejev D., Zhang Y., Haney M.S., Tomasini L., Ferrandino A.F., Rosenberg Belmaker L.A., Szekely A., Wilson M. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z.D., Ziller M., Croft G.F., Amoroso M.W., Oakley D.H. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B., Ren X., Kerppola T.K. KAP1 represses differentiation-inducible genes in embryonic stem cells through cooperative binding with PRC1 and derepresses pluripotency-associated genes. Mol. Cell. Biol. 2014;34:2075–2091. doi: 10.1128/MCB.01729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy J.L., Patel H.R., Hussein S.M., Tonge P.D., Cloonan N., Corso A.J., Li M., Lee D.S., Shin J.Y., Wong J.J. Small RNA changes en route to distinct cellular states of induced pluripotency. Nat. Commun. 2014;5:5522. doi: 10.1038/ncomms6522. [DOI] [PubMed] [Google Scholar]
- Cunningham J.J., Ulbright T.M., Pera M.F., Looijenga L.H. Lessons from human teratomas to guide development of safe stem cell therapies. Nat. Biotechnol. 2012;30:849–857. doi: 10.1038/nbt.2329. [DOI] [PubMed] [Google Scholar]
- Emig D., Salomonis N., Baumbach J., Lengauer T., Conklin B.R., Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010;38:W755–W762. doi: 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishna-Pillai S., Iverson L.E. A DNMT3B alternatively spliced exon and encoded peptide are novel biomarkers of human pluripotent stem cells. PLoS One. 2011;6:e20663. doi: 10.1371/journal.pone.0020663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L.L., Byron M., Butler J., Becker K.A., Nelson A., Amit M., Itskovitz-Eldor J., Stein J., Stein G., Ware C. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J. Cell. Physiol. 2008;216:445–452. doi: 10.1002/jcp.21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Stem Cell Initiative, Adewumi O., Aflatoonian B., Ahrlund-Richter L., Amit M., Andrews P.W., Beighton G., Bello P.A., Benvenisty N., Berry L.S. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.S., Shin J.Y., Tonge P.D., Puri M.C., Lee S., Park H., Lee W.C., Hussein S.M., Bleazard T., Yun J.Y. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat. Commun. 2014;5:5619. doi: 10.1038/ncomms6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Loh Y.H., Li H., Cesana M., Ficarro S.B., Parikh J.R., Salomonis N., Toh C.X., Andreadis S.T., Luckey C.J. Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell. 2014;15:92–101. doi: 10.1016/j.stem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Morey R., O'Neil R.C., He Y., Daughtry B., Schultz M.D., Hariharan M., Nery J.R., Castanon R., Sabatini K. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–183. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell M.J., Lindberg M.R., Brennand K.J., Piper J.C., Voet T., Cowing-Zitron C., Shumilina S., Lasken R.S., Vermeesch J.R., Hall I.M. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S., Bock C., de Boer A.S., Kiskinis E., Meissner A., Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F.J., Goldmann J., Loser P., Loring J.F. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell. 2010;6:412–414. doi: 10.1016/j.stem.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Müller F.J., Schuldt B.M., Williams R., Mason D., Altun G., Papapetrou E.P., Danner S., Goldmann J.E., Herbst A., Schmidt N.O. A bioinformatic assay for pluripotency in human cells. Nat. Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazor K.L., Altun G., Lynch C., Tran H., Harness J.V., Slavin I., Garitaonandia I., Muller F.J., Wang Y.C., Boscolo F.S. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.T., Geens M., Mertzanidou A., Jacobs K., Heirman C., Breckpot K., Spits C. Gain of 20q11.21 in human embryonic stem cells improves cell survival by increased expression of Bcl-xL. Mol. Hum. Reprod. 2014;20:168–177. doi: 10.1093/molehr/gat077. [DOI] [PubMed] [Google Scholar]
- Omberg L., Ellrott K., Yuan Y., Kandoth C., Wong C., Kellen M.R., Friend S.H., Stuart J., Liang H., Margolin A.A. Enabling transparent and collaborative computational analysis of 12 tumor types within the Cancer Genome Atlas. Nat. Genet. 2013;45:1121–1126. doi: 10.1038/ng.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne M.J., Volpon L., Kornblatt J.A., Culjkovic-Kraljacic B., Baguet A., Borden K.L. eIF4E3 acts as a tumor suppressor by utilizing an atypical mode of methyl-7-guanosine cap recognition. Proc. Natl. Acad. Sci. USA. 2013;110:3877–3882. doi: 10.1073/pnas.1216862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2014;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhani F., Kumasaka N., de Brito M.C., Bradley A., Vallier L., Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014;10:e1004432. doi: 10.1371/journal.pgen.1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N., Nelson B., Vranizan K., Pico A.R., Hanspers K., Kuchinsky A., Ta L., Mercola M., Conklin B.R. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput. Biol. 2009;5:e1000553. doi: 10.1371/journal.pcbi.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeger T.M., Daheron L., Brickler T.R., Entwisle S., Chan K., Cianci A., DeVine A., Ettenger A., Fitzgerald K., Godfrey M. A comparison of non-integrating reprogramming methods. Nat. Biotechnol. 2015;33:58–63. doi: 10.1038/nbt.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S.S., Rowntree R.K., Mekhoubad S., Lee J.T. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tonge P.D., Corso A.J., Monetti C., Hussein S.M., Puri M.C., Michael I.P., Li M., Lee D.S., Mar J.C., Cloonan N. Divergent reprogramming routes lead to alternative stem-cell states. Nature. 2014;516:192–197. doi: 10.1038/nature14047. [DOI] [PubMed] [Google Scholar]
- Vallot C., Ouimette J.F., Makhlouf M., Feraud O., Pontis J., Come J., Martinat C., Bennaceur-Griscelli A., Lalande M., Rougeulle C. Erosion of X chromosome inactivation in human pluripotent cells initiates with XACT coating and depends on a specific heterochromatin landscape. Cell Stem Cell. 2015;16:533–546. doi: 10.1016/j.stem.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Wang W.C., Lin F.M., Chang W.C., Lin K.Y., Huang H.D., Lin N.S. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics. 2009;10:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. Single-cell RNA-seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Young M.A., Larson D.E., Sun C.W., George D.R., Ding L., Miller C.A., Lin L., Pawlik K.M., Chen K., Fan X. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.