Abstract
Transcriptional and post-transcriptional regulation of gene expression defines the neurobiological mechanisms that bridge genetic and environmental risk factors with neurobehavioral dysfunction underlying the addictions. More than 1000 genes in the eukaryotic genome code for multifunctional RNA binding proteins (RBPs) that can regulate all levels of RNA biogenesis. More than 50% of these RBPs are expressed in the brain where they regulate alternative splicing, transport, localization, stability, and translation of RNAs during development and adulthood. RBP dysfunction can exert global effects on their targetomes that underlie neurodegenerative disorders such as Alzheimer’s and Parkinson’s disease as well as neurodevelopmental disorders, including autism and schizophrenia. Here, we consider the evidence that RBPs influence key molecular targets, neurodevelopment, synaptic plasticity, and neurobehavioral dysfunction underlying the addictions. Increasingly well-powered genome-wide association studies in humans and mammalian model organisms combined with ever more precise transcriptomic and proteomic approaches will continue to uncover novel and possibly selective roles for RBPs in the addictions. Key challenges include identifying the biological functions of the dynamic RBP targetomes from specific cell types throughout subcellular space (e.g., the nuclear spliceome versus the synaptic translatome) and time and manipulating RBP programs through post-transcriptional modifications to prevent or reverse aberrant neurodevelopment and plasticity underlying the addictions.
Introduction
RNA binding proteins (RBPs) bind both RNAs and proteins to regulate all aspects of mRNA biogenesis and metabolism, “from the cradle (transcription) to the grave (decay)” (Doyle & Kiebler, 2012). RBPs bind and package specific pre-mRNAs and proteins into unique and highly dynamic ribonucleoprotein (RNP) complexes to regulate splicing, editing, polyadenylation, nuclear export, localization, translation, and stability (Glisovic et al., 2008). More than 1000 mammalian genes code for RBPs and 20% of all protein products represent RBPs (Gerstberger, Hafner & Tuschl, 2014), reflecting the extensive splicing and diversity of RBP function. RBPs possess modular RNA binding motifs that cooperatively determine target specificity as well as auxiliary domains that mediate protein-protein interactions and post-translational modifications that can modify RNA binding, transport, and localization of RBPs (Glisovic et al., 2008). Importantly, post-translational modification of RBP signaling is potentially a useful strategy to prevent and treat a variety of disease states (Wang et al., 2009; Kim et al., 2014).
Dozens of RBPs have established roles in neurodevelopment and synaptic plasticity (Doxakis, 2014) and a large literature documents the contribution of RBPs to neurodegenerative disorders (Romano & Buratti, 2013) and neurodevelopmental disorders such as autism and schizophrenia (Bill et al., 2013; Fernandez, Rajan & Bagni, 2013). However, much less is known regarding RBPs and neuropsychiatric disorders such as the addictions. There are several reasons to suspect that RBPs play a crucial role in the addictions. First, alternative splicing – one of the key nuclear functions of RBPs – is highly prevalent within the CNS and is associated with several psychiatric disorders (Glatt et al., 2011). Human genome-wide association studies (GWAS) have yet to uncover statistically significant associations between RBPs and the addictions; however, several genome-wide significant intronic variants for psychiatric disorders have been identified within RBP targets that affect RBP binding and splicing (Glatt et al., 2011). Second, several RBPs play a critical role in neurodevelopment and thus, could mediate transcriptomic programs that are activated following encounter with stressors during critical developmental periods and increase risk for the addictions (Andersen & Teicher, 2009). Third, drug-induced synaptic plasticity is an important component throughout all stages of addiction and dozens of RBPs have been identified that exhibit cytoplasmic function in transporting, localizing, and translating mRNAs in synaptic plasticity (Thomas et al., 2014; Tolino, Kohrmann & Kiebler, 2012).
Considering the addictions as neuropsychiatric disorders that have a neurodevelopmental component
Genetic and fluctuating environmental risk factors affect neurodevelopment and the later neurobiological responses to external stimuli (van Loo & Martens, 2007). The addictions are gene x environment disorders that require drug exposure to manifest. Both genetic and environmental risk factors likely interact to affect neurodevelopment and neuropharmacological sensitivity to reinforcing stimuli, including drugs and associated cues (Leyton & Vezina, 2014; Andersen & Teicher, 2009). Severe childhood adversity is a key environmental risk factor that greatly increases susceptibility to the addictions (Kendler et al., 2000) and early life stress causes structural changes that affect mesolimbic reward function, including reduced hippocampal and prefrontal cortical development that may heighten dopamine release in the nucleus accumbens (Andersen & Teicher, 2009). We posit that a subset of RBPs orchestrate neurodevelopmental plasticity induced by environmental risk factors that increases susceptibility to the addictions. The mesocorticolimbic dopaminergic circuitry is involved in reward/aversion processing and positive/negative reinforcement learning in the addictions and includes midbrain dopaminergic neuron projections from the ventral tegmental area to the medial prefrontal cortex and nucleus accumbens and glutamatergic projections from the prefrontal cortex to the nucleus accumbens (Volman et al., 2013). Mesocorticolimbic dysfunction is common in numerous neurodevelopmental and neuropsychiatric disorders, especially the addictions (Dichter, Damiano & Allen, 2012). Understanding the potential contribution of RBPs in mesocorticolimbic development, environmental risk factor-induced plasticity, and drug-induced plasticity could improve our understanding of the heritable basis of addictive disorders, especially within the context of gene x environment (G x E) interactions (Wermter et al., 2010).
As an example of a hypothesized role for RBPs in G x E interactions in neurodevelopment and plasticity underlying psychiatric traits, a genetic variant in the gene coding for brain-derived neurotrophic factor (BDNF; the Val66/Met allele) that decreases activity-dependent secretion of BDNF is associated with phenotypic variation in psychiatric endophenotypes (e.g., fear/aversion learning) during childhood versus adolescence (Casey et al., 2009). Furthermore, individuals carrying the Val66/Met BDNF variant that also underwent early childhood adversity (institutionalization) showed a decrease in cortical volume, an increase in amygdala volume, an increase in behavioral anxiety, and an increase in cortisol stress response (Casey et al., 2009). Accumulating evidence indicates that RBPs can regulate translation of BDNF (Allen et al., 2013; Vanevski & Xu, 2015) and that an increase in BDNF signaling can increase translation and synaptic localization of RBPs (Castren et al., 2002). In this review, we summarize recent examples of RBPs that affect primary molecular targets and cellular, neurodevelopmental, and neurobehavioral function relevant to the addictions. We begin by discussing examples that contain the most direct experimental and circumstantial evidence that draws a link to the addictions (FMRP, hnRNPs). Toward the end, we discuss those RBPs for which there is less, yet accumulating evidence (RBFOX and CELF proteins). Finally, we include a section on RNA editing and the addictions, as this exciting new area of research comprises distinct molecular mechanisms from the remainder of the review. Because multiple RBPs frequently coordinate in large RNP complexes to coordinate post-transcriptional regulation of mRNAs, we have attempted to draw links between various discussed RBPs and their families whenever relevant, while keeping in mind that it would be beyond the scope of the review to document all possible connections between the large number of RBPs that are discussed.
FMRP
Fragile X mental retardation protein (FMRP) is a polyribosome-associated neuronal RBP that targets and translationally represses mRNAs associated with synaptic plasticity and has been implicated in autism, affective disorders, ADHD, bipolar disorder, schizophrenia and the addictions (Fernandez, Rajan & Bagni, 2013; Smith et al., 2014). FMRP is expressed throughout neurodevelopment and is necessary for proper differentiation, migration, axon formation, refinement and stabilization, synapse formation and circuit wiring of neocortical layers (Till, 2010). FMRP inhibits ribosomal translocation of mRNAs that is relieved following activity-dependent signaling to permit cytoskeletal remodeling that underlies synaptic plasticity (Darnell et al., 2011; Darnell & Klann, 2013). The N-terminal region contains two Tudor domains that bind noncoding RNAs followed by a nuclear localization signal NLS. The middle of the protein contains two hnRNP K homolology (KH) domains that further specify RNA and protein interactions. The C-terminal region contains a nuclear export signal followed by an RGG box that directly binds to mRNA targets (Fernandez, Rajan & Bagni, 2013).
Signaling by several different receptor types modulate FMRP activity, including mGluRs, AMPA, GABA, NMDA, TrkB, dopamine, and cannabinoid receptors (Fernandez, Rajan & Bagni, 2013). Accordingly, FMRP targets hundreds of neurodevelopmental and neuroplasticity proteins involved in cytoskeletal remodeling; many of these targets have been associated with the addictions, including mGluR1, mGluR5, PSD-95, CYFIP1/2, GABA-A receptor subunits, NR2A/2B NMDA receptor subunits, Homer1, neuroligins, CREB binding protein and D1 dopamine receptor-coupled GRK2 (Wang et al., 2008; Darnell & Klann, 2013). FMRP also targets elongation factor-1 and −2 (EF-1, EF-2), argonaute 1/2 (Ago1/2), and Dicer which are ubiquitously involved in protein translation and miRNA processing, respectively (Darnell & Klann, 2013). Thus, FMRP can also exert widespread, indirect regulation of the translatome.
With regard to drugs of abuse, deletion of FMRP perturbs midbrain dopaminergic neuron development, amphetamine-induced dopamine release in the striatum and prefrontal cortex, psychostimulant-induced locomotor activity, stereotypy, and drug reward (Fish et al., 2013; Fulks et al., 2010). FMRP deletion also disrupts cocaine-induced neurobehavioral plasticity in the nucleus accumbens, including decreased locomotor sensitization, enhanced stereotypy, perturbed dendritic morphology, changes in AMPA/NMDA receptor ratios and glutamatergic transmission, and reduced cocaine reward that is associated with increased mGluR5 activation (Smith et al., 2014). In prefrontal cortical neurons, D1 dopamine receptor activation is sufficient to induce FMRP phosphorylation and synthesis of synaptic proteins involved in glutamate receptor trafficking and plasticity (Wang et al., 2008; Wang, Kim & Zhuo, 2010). In another example that indirectly implicates FMRP in the addictions, cytoplasmic FMRP-interacting protein 1 (CYFIP1) inhibits elf4E–mediated cap-dependent mRNA translation of proteins involved in actin cytoskeleton remodeling and dendritic spine maturation (De Rubeis et al., 2013; Napoli et al., 2008) and mutations in the closely related Cyfip2 gene in mice modulates psychostimulant-induced locomotor activity and sensitization, dendritic morphology, and AMPA receptor neurotransmission (Kumar et al., 2013). In addition to an mGluR5 mechanisms underlying neurobehavioral dysfunction in the absence of FMRP (Smith et al., 2014; Bear, Huber & Warren, 2004), neuronal activity and BDNF/TrkB signaling can regulate expression of FMRP in the hippocampus (Castren et al., 2002) and FMRP deletion perturbs both BDNF and TrkB spatiotemporal expression and signaling in neurodevelopment and neuroplasticity (Castren & Castren, 2014). Thus, both BDNF/TrkB and mGluR5 signaling could converge on FMRP mechanisms of psychostimulant neurobehavioral plasticity (Smith et al., 2014; Kumar et al., 2013).
Because FMRP deletion increases protein translation and disrupts structural and synaptic plasticity induced by glutamate receptor signaling (e.g., mGluR-LTD), an important question is whether or not fine tuning glutamatergic signaling by the use of pharmaceuticals targeting NMDA receptors and mGluR can improve the outcome of neurodevelopmental and neuropsychiatric disorders associated with FMRP dysfunction (Michalon et al., 2012; Bear, Huber & Warren, 2004), including the addictions (Cleva et al., 2010). FMRP also undergoes post-translational modifications, including mGluR-mediated dephosphorylation and relief from translational repression as well as mTOR-mediated dephosphorylation by protein phosphatase 2A (PP2A) and re-establishment of translational repression via PP2A suppression (Narayanan et al., 2008; Ceman et al., 2003), presenting additional opportunities for perturbing FMRP mechanisms of neurodevelopmental and neuropsychiatric dysfunction.
hnRNPs
Heterogeneous nuclear ribonucleoproteins (hnRNPs) are a large and diverse group of nucleoplasmic-localized multifunctional RBPs that form RNP complexes and can regulate splicing, export, localization, translation, and stability (Han, Tang & Smith, 2010; Dreyfuss, Kim & Kataoka, 2002). Rapidly accumulating studies are identifying new contributions of hnRNPs to neurodevelopment (Sinnamon et al., 2012; Liu & Szaro, 2011) and synaptic plasticity (Zhang, Neubert & Jordan, 2012; Leal, Afonso & Duarte, 2014; Folci et al., 2014; Sinnamon & Czaplinski, 2011). hnRNPs contain highly conserved, modular RNA recognition motifs (RRMs) that determine sequence binding specificity and affinity and intevening regions that localize and permit generalized and specific roles in post-transcriptional processing. The hnRNPs were originally classified based on their coimmunopurification in RNA complexes with monoclonal antibodies against the founding member hnRNP C, thus identifying hnRNP A-U (Dreyfuss et al., 1993). Although hnRNPs can be highly similar in protein structure and function, their nomenclature is not consistent with their sequence homology, suggesting independent evolution and expansion of several RRMs. As examples, some hnRNPs share greater sequence similarity with other RBP classes such as hnRNP A1 versus ELAVL4/CELF1 or hnRNP C versus the SR protein transformer-2β (Tra2β) (Tang et al., 2012). Furthermore, other hnRNPs such as E/K, I/L, U, I (PTBP-1), and hnRNP F/H contain separate, quasi-RRMs that do not share sequence homology with canonical RRMs of hnRNPs (Tang et al., 2012). Thus, discussing hnRNPs as a separate class is somewhat arbitrary but for ease of reference, here we will discuss addiction-relevant RBPs that have been named hnRNPs and in the immediately following sections, we will discuss hnRNPs that have alias names.
The mu opioid receptor (MOR) is a primary molecular target for the addictive properties of opioids and other drugs of abuse (Contet, Kieffer & Befort, 2004) and undergoes alternative splicing by hnRNP H1 binding to the intronic AGGG sequence and recruiting hnRNP A1, A2B1, AB, C, H3, and U which results in exclusion of exon 2 and decreased expression of Oprm1 (mu opioid receptor; MOR) (Xu et al., 2014). The MOR intronic SNP rs9479757 decreased binding of hnRNP H1, resulting in exon 2 skipping and increased MOR expression that was associated with increased severity of heroin dependence (Xu et al., 2014). hnRNP H1 and hnRNP F can also post-transcriptionally regulate MOR expression by repressing translation at the 5’ UTR (Song et al., 2012), providing a second level of MOR regulation by hnRNP H1.
We used fine-scale gene mapping with interval-specific congenic mouse lines to identify a 206 kb region on chromosome 11 containing Hnrnph1 (the gene coding for hnRNP H1) and Rufy1 that was necessary for reduced sensitivity methamphetamine-induced locomotor activity. Replicate mouse lines harboring transcription activator-like effector nucleases (TALENs)-induced frameshift deletions in Hnrnph1 recapitulated the QTL phenotype, thus identifying Hnrnph1 as the quantitative trait gene (Yazdani et al., 2015). Inheritance of this caused a decrease in expression of Bdnf, Elavl2, Elavl4, and Nurr1 (nuclear receptor related 1 protein) (Yazdani et al., 2015), a transcription factor that is crucial for the development and function of midbrain dopaminergic neurons (Campos-Melo et al., 2013). hnRNP H1 (along with hnRNP A1, K, and M) has been shown to be co-regulated with Nurr1 expression across neuronal cell lines (Johnson et al., 2011) and a recent review of proteomic studies of neuronal and brain expression with drugs of abuse identified psychostimulant- and opioid-induced changes in hnRNP H2 and A1 expression that could regulate gene networks enriched for protein modification/degradation, synaptic/neuronal function, transmission, and signaling (Wang, Yuan & Li, 2011). We are currently evaluating the potential role of Hnrnph1 in the rewarding and reinforcing properties of psychostimulants and opioids and changes in the transcriptome and spliceome associated with Hnrnph1 dysfunction.
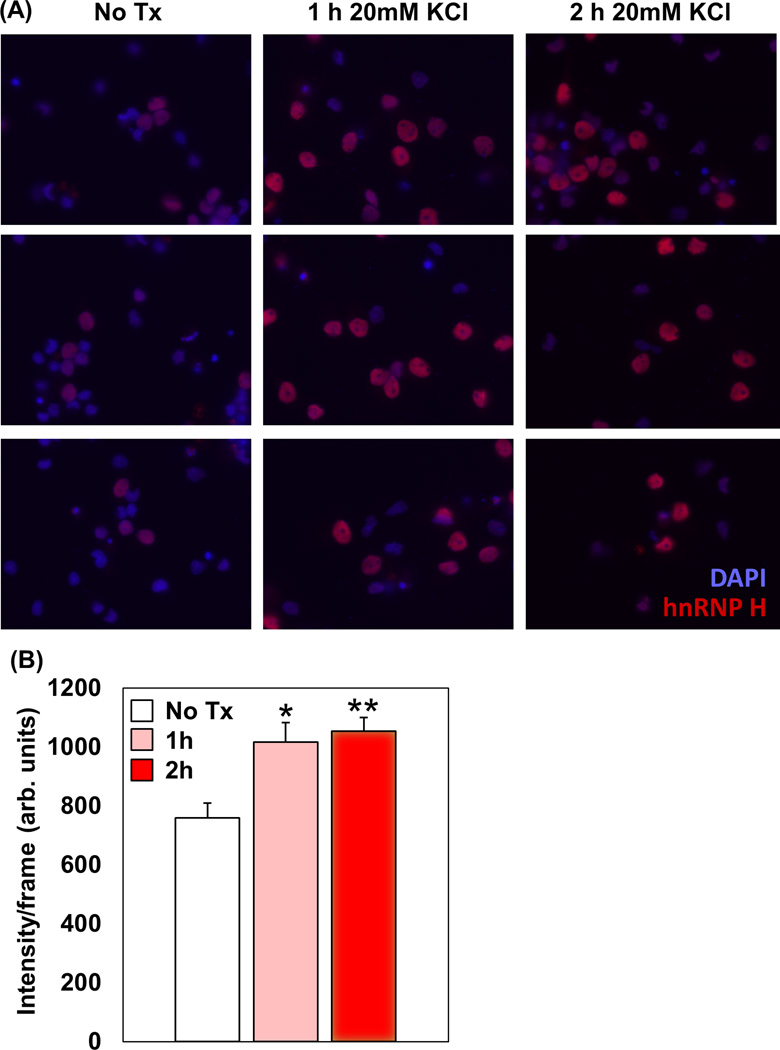
Hnrnph1 is highly and ubiquitously expressed throughout the mouse brain (Lein et al., 2007). It contains three quasi-RRMs that mediate binding to poly-(G) tracts to either enhance or silence splicing (Han, Tang & Smith, 2010). hnRNP H protein is primarily localized to the nucleus in primary rat cortical neurons and depolarization via KCl application increased the intensity of nuclear immunocytochemical staining (Fig. 1), suggesting activity-dependent nuclear function that is consistent with its role in alternative splicing and polyadenylation (Katz et al., 2010). Nevertheless, hnRNP H1 also contains three, intervening glycine-rich domains and the central domain contains a NLS that permits bidirectional transport between the nucleus and cytoplasm (Van Dusen et al., 2010). These modular glycine-rich domains are also necessary for regulation of splicing (Wang et al., 2012) and likely mediate interaction with other proteins, including complexing and cooperating with hnRNP A1, RBFOX2, and hnRNP F to enhance the suppress exon inclusion (Mauger, Lin & Garcia-Blanco, 2008; Fisette et al., 2010). hnRNP H1 can also cooperate with hnRNP I (see PCBP-1 below) to form a a splicer enhancer complex in neurons (Chou et al., 1999).
Figure 1. Immunocytochemical staining of hnRNP H in rat cortical neurons following KCl-induced depolarization.
(A): Primary neocortical neurons were dissected from E18 Sprague-Dawley rat embryos (Charles River Laboratories). Dissociated neurons were cultured neurons for 1 week. For the control, no treatment (No Tx) group, 1 ml of conditioned media was replaced with 1 ml of neurobasal media. For the 1 h and 2 h Tx groups, 1 mL of conditioned media was replaced with 1ml of 20 mM KCl-enriched neurobasal media. Treated neurons were then washed, fixed, permeabilized, blocked, and incubated with primary hnRNP H antibody (1:500 Rabbit polyclonal, Bethyl Labs) in 1% BSA overnight at 4° C. 12 h later, neurons were washed and incubated with an Alexa Fluor 594 antibody (1:500 Donkey anti-Rabbit, Life Technologies) in 1% BSA. Processed coverslips were then stained with DAPI (blue) and mounted onto glass slides. Images were collected using a Zeiss AxioObserver microscope under uniform settings for all three groups. 20 serial images (frames) were captured per condition and fluorescence was quantified using ImageJ under a uniform threshold range. Note both an increase in the number of H1 stained neurons following 1–2 h of KCl Tx as well as an increase in the fluorescent staining intensity after KCl treatment. (B): Semi-quantification of fluorescence staining intensity. One-way ANOVA indicated a main effect of genotype (F2,57 = 8.4; P = 0.0006). *P = 0.01; **P <0.001 (unpaired t-tests versus No Tx).
To summarize, increasing evidence indicates that hnRNPs such as hnRNP H1 may be involved in establishing multiple addictions. The role of hnRNP H1 in activity-dependent synaptic plasticity is not known although intriguingly, Hnrnph1 was identified as one of the most significantly upregulated transcripts in transcriptome analysis of cortical brain tissue following experimental traumatic brain injury (Kobori, Clifton & Dash, 2002) which could indicate a role in regenerative plasticity. Available evidence from our laboratory indicates that neuronal stimulation induces an increase in hnRNP H staining that is localized to the nucleus (Fig. 1), suggesting that the contribution of hnRNP H1 to neuroplasticity is either indirect (e.g., via nuclear splicing or nuclear polyadenylation of synaptic targets) or that dendritic localization of hnRNP H requires signaling through specific receptors. We are currently testing the latter hypothesis in the context of the addictions by treating neurons with dopamine receptor agonists and examining changes in hnRNP H staining. Because hnRNP H1 can undergo several post-translational modifications such as phosphorylation, methylation, and sumoylation (Chaudhury, Chander & Howe, 2010), there is potential to fine tune hnRNP H1 signaling to prevent or normalize plasticity associated with the addictions.
PCBPs (including hnRNP K)
Polycytosine-binding proteins (PCBPs) 1–4 (hnRNP E1-E4) and hnRNP K, bind with high affinity to poly(C) DNA and RNA sequences to regulate transcription and post-transcriptional processing via three, modular hnRNP K homolology (KH) RNA binding domains (Han, Tang & Smith, 2010). PCBPs also contain an intervening sequence between the second and third KH domains with nuclear localization signals (PCBP1 and 2) or an hnRNP K-specific nuclear shuttling domain to permit bidirectional transport. hnRNP K also has a SH-3 (KI) binding domain and both hnRNP K and PCBP4 contain an NLS in the third KH domain (Choi et al., 2009). hnRNP K, PCBP1, and PCBP2 are located primarily in the nucleus whereas PCBP3 and 4 are located primarily in the cytoplasm (Chaudhury, Chander & Howe, 2010).
In neurodevelopment, PCBPs regulate axonogenesis (Thyagarajan & Szaro, 2004) and corticogenesis (Pilaz & Silver, 2015). PCBP1–3 are expressed in the adrenal medulla, midbrain and locus coeruleus, respectively where they bind the 3’ UTR of tyrosine hydroxylase (TH, the enzyme necessary for synthesis of dopamine and norepinephrine) in the cytoplasm to increase stability and protein translation (Boschi et al., 2015; Czyzyk-Krzeska & Beresh, 1996). In addition to regulating catecholamine synthesis, hnRNP K, PCBP1, and PCBP2 can all bind to the MOR promoter to activate MOR transcription whereas PCBP3 acts as a transcriptional repressor (Choi et al., 2008; Choi et al., 2007; Choi et al., 2009).
hnRNP K interacts with ELAVL2 to control neuronal differentiation (Yano, Okano & Okano, 2005) and regulates axonogenesis via post-transcriptional interaction with genes involved in neurodevelopment, transport, localization, and cytoskeleton, including GAP-43 (Liu & Szaro, 2011; Liu, Gervasi & Szaro, 2008). hnRNP K exhibits early developmental expression in several components of the mesocorticolimbic circuitry and subsequently a more restricted expression to the hippocampus (Blanchette, Fuentes Medel & Gardner, 2006). hnRNP K can bind to the TH promoter to activate transcription (Banerjee et al., 2014) and co-localizes with TH in the ventral midbrain. Thus, hnRNP K could regulate catecholamine synthesis during midbrain dopaminergic neuron development, maintenance, and synaptic plasticity in adulthood (Folci et al., 2014). hnRNPK also binds to the polyadenylation sequence within the 3’UTR of the serotonin transporter (SERT), a primary molecular target for cocaine reward (Sora et al., 2001), to increase SERT protein levels by preventing miR-16-mediated inhibition of translation (Yoon et al., 2013). In addition to its link to catecholamines and monoamines, hnRNP K can bind to the MOR promoter and to the promoter of the beta-2 subunit of the neuronal nicotinic acetylcholine receptor to activate transcription (Choi et al., 2008; Du, Melnikova & Gardner, 1998). Finally, morphine can stimulate hnRNP K translation in multiple brain regions independent from transcription (Lee et al., 2014).
To summarize, PCBPs and hnRNP K are associated with neurodevelopment, catecholaminergic, monoaminergic, and opioidergic, and cholinergic signaling and thus, are prime suspects in regulating neurotransmitter signaling underlying the addictions. Importantly, post-translational modifications can also significantly impact cell function of PCBPs. A notable example is ERK-induced phosphorylation of hnRNP K which translocates hnRNP K to the cytoplasm where it can then bind to the 3’ UTR of mRNAs to regulate protein translation (Habelhah et al., 2001). Therefore, therapeutic modulation of hnRNP K signaling could be a future treatment avenue in the addictions.
PTBPs (including PTBP-1; a.k.a. hnRNP I)
Polypyrimidine tract-binding protein (PTBPs) bind to both intronic and exonic polypyrimidine tract sequences to repress exon inclusion. PTBPs contain four RRMs, a nuclear localization signal and a nuclear export signal in the N-terminus that permit alternative splicing, shuttling, polyadenylation, transport, localization, stabilization, and translation (Romanelli, Diani & Lievens, 2013; Keppetipola et al., 2012). RRM-1 and RRM2 are separated by flexible linkers that permit independent conformations whereas RRM-3 and −4 form a globular structure and binds to nearby, intervening pyrimidine tracts to induce RNA looping (Romanelli, Diani & Lievens, 2013). Alternatively spliced isoforms of PTBP1 can contain longer intervening domains or isoforms that lack RRM-1 and −2, thus diversifying PTBP targets and function within the context of an RNP complex. Splicing repression by PTBP can depend on its proximity to the splice site, its antagonism by other RBP splicing co-factors such as CELF proteins (Spellman et al., 2005; Gromak et al., 2003) and neuro-oncological ventral antigen1 (Nova1) (Polydorides et al., 2000), and its interaction with co-repressors such as the Raver proteins (Henneberg et al., 2010). In addition to splicing, PTBP1 can bind directly to 3’ UTRs to regulate cleavage in polyadenylation, 3’ UTR exon inclusion, and mRNA stability (Sawicka et al., 2008). Finally, PTBPs interact with PCBPs act at the internal ribosomal entry site (IRES) to initiate translation (Bushell et al., 2006).
During neurodevelopment, reciprocal changes in PTBP1 and 2 expression coordinate differential splicing of PSD-95 to control the timing of neuronal differentiation (Zheng et al., 2012). PTBP1 is expressed in neural progenitors and decreases during differentiation and is restricted to glia during adulthood. In contrast, expression of the gene paralog PTBP2 increases during neuronal differentiation, decreases during cortical maturation, and shows moderate neuronal expression during adulthood (Keppetipola et al., 2012). Notably, PTBP1 inhibits the expression of PSD-95 in neural progenitors whereby it represses the inclusion of a coding exon which results in nonsense-mediated decay and delays excitatory synapse stabilization and maturation (Zheng et al., 2012; Keppetipola et al., 2012). A similar mechanism is used by PTBPs to inhibit their own expression (Boutz et al., 2007). PTBP2 is essential for postnatal survival and genome-wide RNA target analysis of the developing mouse brain revealed that PTBP2 inhibits a splicing program involved in neuronal cell cycle, proliferation, actin cyotoskeleton, and neuronal differentiation (Licatalosi et al., 2012).
With regard to dopamine receptors and the addictions, the short and long splice forms of the D2 dopamine receptor (D2L and D2S) are highly conserved and have different signaling properties and physiological function (Picetti et al., 1997) that have been associated with the addictions (Smith et al., 2002; Levran et al., 2015). Both PTBP1 and Nova1 promote exon 6 inclusion whereas hnRNP M inhibits exon 6 inclusion, thus bi-directionally regulating D2L versus D2S expression (Sasabe, Futai & Ishiura, 2011; Park et al., 2011). Overexpression of PTBP1 in vitro was associated with a decrease in transcription of ΔfosB, a stable splice variant of the transcription factor fosB that accumulates in the nucleus accumbens following chronic administration of drugs of abuse and is associated with sustained drug-induced synaptic plasticity (Alibhai et al., 2007; Nestler, Barrot & Self, 2001). Thus, perhaps post-translationally modifying PTBP1, e.g., via phosphorylation and cytoplasmic translocation (Xie et al., 2003), could modulate ΔfosB-mediated neuroplasticity associated with the addictions.
ELAVL
The neuronal-specific mammalian embryonic lethal, abnormal vision-like (ELAVL)2, 3, and 4 RBPs are an RBP family based on homology to ELAV protein in Drosophila (Robinow et al., 1988) and regulate the transport, stabilization, localization and translation of mRNAs. They contain three RRMs that mediate binding to intronic targets for splicing as well as binding to highly conserved AU-rich element (ARE) sequences of 3’UTR targets to regulate stability (Colombrita, Silani & Ratti, 2013), including Nova1 (Ratti et al., 2008). ELAVL4 can also enhance cap-dependent translation via structural unwinding of 5’ UTRs (Fukao et al., 2009). ELAVL RBPs contribute to all stages of neuronal differentiation, maintenance, synaptogenesis, and activity-dependent synaptic plasticity (Perrone-Bizzozero & Bolognani, 2002). The localization of ELAVL RBPs in the neocortex and hippocampus (Okano & Darnell, 1997) suggest potential involvement in neurodevelopmental and neuroanatomical risk for the addictions (Andersen & Teicher, 2009). RBP target analysis of ELAVL of mouse forebrain tissue identified a spliceome enriched for axonal and synaptic cytoskeleton dynamics and 3’UTR-regulated genes involved in amino acid synthesis. Of note, ELAVL regulates the splicing and half-life of glutaminase, indicating an essential role in excitatory neurotransmission (Ince-Dunn et al., 2012).
Acute cocaine treatment caused a decrease in whole brain FMRP expression and an increase in ELAVL expression that was associated with an increase in expression of genes enriched for dendritic synaptic plasticity (Tiruchinapalli, Caron & Keene, 2008). Combining repeated cocaine administration with swim stress increased ELAVL4 phosphorylation and translation in the hippocampus as well as expression of its canonical cytoskeletal target, Growth Associated Protein (GAP)-43 (Pascale et al., 2011), which could contribute to structural plasticity and drug associative learning (Bolognani et al., 2007). Importantly, ELAVL4 selectively binds to the unique ARE sequence of the long 3’ UTR but not the short 3’ UTR of BDNF mRNA to increase BDNF stability and activity-dependent translation (Allen et al., 2013). The increase in BDNF translation is mediated by PKC-induced phosphorylation of ELAVL4 which disinhibits 3’UTR-mediated translational repression of the long 3’UTR BDNF mRNA in hippocampal neuronal dendrites (Vanevski & Xu, 2015). Selective regulation of the long 3’ UTR of BDNF is important because it is responsible for activity-dependent neuronal translation of BDNF (Lau et al., 2010) and synaptic maturation in the dendrites (An et al., 2008). Psychostimulant administration increases the expression of BDNF (Russo et al., 2009) and in turn, BDNF can regulate neuronal expression of FMRP and synaptic protein translation (Castren & Castren, 2014). Thus, an ELAVL-mediated increase in activity-dependent translation of BDNF and other mRNA targets may work in parallel with FMRP to induce neurobehavioral plasticity in response to drugs of abuse (Smith et al., 2014). Interestingly, an increase in BDNF expression may serve as a biomarker for severity of psychostimulant addiction and vulnerability to relapse in recently abstinent individuals (Sinha, 2011).
ZBP1
Zipcode Binding Protein-1 (ZBP1) is a cytoplasmic protein that shuttles between the nucleus and cytoplasm via its NES and NLS sequence. ZBP2 is a second, homologous RBP that is localized to the nucleus and cooperates with ZBP1 to shuttle beta-actin mRNA into the cytoplasm (Gu et al., 2002; Pan et al., 2007). ZBPs share significant homology with PCBPs and contain two RRMs and four KH-type RNA binding motifs. KH3 and KH4 domains of ZBP1 form a pseudodimer that recognizes a 54 nucleotide zipcode sequence on the 3’ UTR of beta-actin mRNA for transport and translation in dendrites (Doyle & Kiebler, 2012; Gu et al., 2002; Huttelmaier et al., 2005; Ross et al., 1997; Farina et al., 2003).
ZBP1 represses beta-actin translation in the cytoplasm that is relieved upon Src phosphorylation at Tyr396 (Huttelmaier et al., 2005). Both BDNF and netrin-1 stimulate phosphorylation of ZBP1 at Tyr396 and increased protein synthesis of beta actin in a model of axon guidance in cortical neurons (Sasaki et al., 2010; Welshhans & Bassell, 2011). Neurodevelopmental studies demonstrate ZBP1 transport and localization of beta-actin in synaptogenesis of dendritic filopodia (Eom et al., 2003) and neurotrophin-induced growth cone motility (Zhang et al., 2001). Furthermore, ZBP1 is necessary for NMDA receptor-dependent targeting of beta-actin mRNA to the hippocampal dendrites (Tiruchinapalli et al., 2003) as well as dendritic arborization induced by Src-induced ZBP1 phosphorylation and relief of translational repression (Perycz et al., 2011). In addition to dendritic plasticity, axonal regeneration in severed sensory neurons involves ZBP1-mediated transport of both beta-actin and GAP-43 mRNAs to localize protein synthesis and increase axonal growth and branching, respectively (Donnelly et al., 2011; Donnelly et al., 2013).
Similar to ELAVL-associated increased expression of GAP-43 following in vivo administration of cocaine and stress (Pascale et al., 2011), in vivo axonal injury induced an increased in expression of GAP-43 that interacts with ELAVL4 as well as ZBP1 in a complex to induce axonal outgrowth and branching (Yoo et al., 2013). It was recently shown that although both ZBP1 and ELAVL4 interact with beta-actin, ELAVL4 binds specifically to the ARE sequence in the 3’ UTR whereas ZBP1 requires a specific secondary structure (Kim et al., 2015) that could permit their ability to form a EVAL/ZBP complex in neuroplasticity underlying axonal regeneration and possibly cocaine neuroplasticity. In further support of a link between ZBP1 and cocaine, transgenic, ectopic expression of ZBP1 in the striatum during adulthood blocked cocaine-induced conditioned place preference that was rescued by eliminating ZBP1 expression. Direct, experimental target analysis identified nearly 200 transcripts involved in synaptic plasticity that could be responsible for the effect of ZBP1 on cocaine reward, including the scaffolding gene Homer1 that negatively regulates cocaine reward (Szumlinski et al., 2004) as well as cadherins, transcription factors, kinases, ion channels, and Ras members (Lapidus et al., 2012). Interestingly, we previously identified both a behavioral QTL and a cis-acting eQTL from striatal tissue on chromosome 11 for Igf2bp1 (insulin growth factor 2 mRNA binding protein 1; the gene coding for ZBP1 protein) that was causally associated with reduced methamphetamine-induced locomotor activity and increased Igf2bp1 expression. In light of the recent ZBP1 findings discussed above, Igf2bp1 could represent a quantitative trait gene underlying methamphetamine stimulant sensitivity (Bryant et al., 2012).
An intriguing possibility is that native ZBP1 expression could be re-awakened in the mature brain following chronic exposure to drugs of abuse and regulate a neurodevelopmental program that underlies certain addictions such as cocaine (Dong & Nestler, 2014). Furthermore, based on the combined evidence described above, ZBP1 could promote BDNF-mediated structural recovery of damaged catecholaminergic and monoaminergic axons following administration of neurotoxic drugs such as methamphetamine and MDMA (Adori et al., 2010; Ares-Santos et al., 2014).
RBFOX1 (A2BP1)
RBFOX (RNA binding protein, fox-1 homolog) proteins are neuronal splicing factors that promote both exon inclusion and skipping that depends on the position of the canonical UGCAUG binding motif near the exon (Underwood et al., 2005; Zhang et al., 2008). RBFOX proteins contain a highly conserved, identical RRM that is responsible for binding RNAs and less conserved regions in the N-terminus and NLS-containing C-terminus that also dictate splicing of RBFOX proteins themselves and other proteins which can govern subcellular localization and increase the functional diversity of these RBPs (Kuroyanagi, 2009).
Genome-wide target analysis of RBFOX2 in human embryonic stem cells identified several RBP splicing factors as targets, including hnRNP A2/B1, H1, H2, and PTBP (Yeo et al., 2009)., which emphasizes the high degree of network connectivity in genomic coordination of RBP splicing (Huelga et al., 2012). Enrichment analysis of the predicted RBFOX spliceome revealed sets of genes involved in neuromuscular, cytoskeleton, ion channel, and phosphorylation functions (Zhang et al., 2008). Experimental identification of direct RBFOX targets in the mouse brain identified intronic splicing targets and 3’ UTR targets that could regulate alternative polyadenylation and mRNA stability. RBFOX targets were enriched for cytoskeleton anchoring, scaffolding and signaling, and neuronal projections. Increased expression of RBFOX1 and RBFOX3 and decreased expression of RBFOX2 were associated with a change in the RBFOX spliceome programs from E17 to adulthood (Weyn-Vanhentenryck et al., 2014).
RBFOX1 is a neurodevelopmental splicing RBP whose dysfunction is associated with autism, intellectual disability, attention deficit hyperactive disorder, bipolar disorder, and schizophrenia (Bill et al., 2013; Fogel et al., 2012). CNS-specific knockout of Rbfox1 in mice resulted in enhanced hippocampal neuronal excitability and susceptibility to seizures, demonstrating a role for RBFOX1 in neuronal excitation and synaptic transmission (Gehman et al., 2011). RBFOX1 is expressed throughout development in the mouse and human basal forebrain, neocortex, and hippocampus (Hammock & Levitt, 2011; Fogel et al., 2012). Spliceome and transcriptome analysis of differentiated primary human neural progenitor cells following RBFOX1 knockdown identified parallel networks of transcription factors, splicing factors, and synaptic proteins involved in neurogenesis, neurodevelopment, maintenance, cytoskeletal organization and cell adhesion, projection, proliferation, and synapse function. Notably, differential splicing was observed for several genes coding for RBPs, including HNRNPD, HNRNPA1, ELAVL2, and HNRNPH1 (Fogel et al., 2012) which in turn, was associated with a perturbation in their predicted splicing programs based on their RNA binding motifs. For example, 205 of the total 996 alternative splicing events that were identified following RBFOX1 knockdown contained the binding site for hnRNP H1, suggesting that RBFOX1 regulates the splicing of HNRNPH1 (Fogel et al., 2012). Conversely, hnRNP H1 regulates the splicing activity of RBFOX1/2 by interacting with the C-terminal domain (Sun et al., 2012). In addition to splicing HNRNPH1, RBFOX2 can form a complex with hnRNP H1 and F to silence splicing of other genes (Mauger, Lin & Garcia-Blanco, 2008). Thus, hnRNP H1 and RBFOX proteins could coordinate splicing in affecting methamphetamine stimulant behavior (Yazdani et al., 2015) and heroin addiction (Xu et al., 2014).
Recent, direct evidence implicating RBFOX in the nucleus accumbens in the addictions comes from a genome-wide trancriptomic and epigenomic study of chronic cocaine administration in mice that identified a translocation of RBFOX1 to the nucleus which was associated with an increase in splicing events that coincided with the location of histone modifications. Furthermore, site-specific Cre-mediated knockdown in the nucleus accumbens of floxed RBFOX1 mice blocked cocaine reward (Feng et al., 2014), which together suggests that that RBFOX1 coordinates an adaptive splicing program underlying cocaine dependence. Interestingly, RBFOX1 variants have been nominally associated with nicotine dependence, alcohol dependence, and cocaine reward and regulate the splicing of cell adhesion molecule genes in dopaminergic neurons that are associated with neurodevelopmental connectivity, plasticity, and genetic variation underlying the addictions (Zhong et al., 2015).
CELF4 and CELF6
CELF (CUG-BP, ELAV-like factor) is a family of highly expressed multifunctional RBPs in the brain that have both nuclear splicing and cytoplasmic functions in RNA processing. CELF and ELAV can cooperatively promote splicing in mammalian neurons and a recent study in C. elegans indicates that they co-regulate overlapping and distinct splicing networks to determine cholinergic versus GABAergic neuronal cell type (Norris et al., 2014). CELF RBPs bind to pyrimidine-rich sequences and compete with PTBPs to activate splicing (Spellman et al., 2005; Gromak et al., 2003). Notably, the high sequence similarity between hnRNP A1 versus CELF1 suggests shared functions (Tang et al., 2012). CELF RBPs contain highly conserved RNA recognition motifs (RRM)-1 and −2 at the N-terminus and a third RRM at the C-terminus (Dasgupta & Ladd, 2012). RRM-1 and −2 are separated from RRM-3 by a non-conserved divergent linker domain that differentiates CELF1–2 from CELF 3–6 in determining RNA-protein and protein-protein interactions in forming target-specific RNP complexes (Gallo & Spickett, 2010; Dasgupta & Ladd, 2012). The RRMs contain RNP motifs that typically bind to introns, 3’ UTRs, and 5’ UTRs of mRNAs to regulate splicing, poly(A)-specific ribonuclease recruitment and deadenylation [e.g., with c-fos (Moraes, Wilusz & Wilusz, 2006)], polyadenylation, mRNA stability and translation (Dasgupta & Ladd, 2012). Both the divergent domain and C-terminus contain the signals that determine nuclear versus cytosolic localization of CELF1 and CELF2 (Ladd & Cooper, 2004; Fujimura, Kano & Murata, 2008). Importantly, CELF proteins contain multiple phosphorylation sites that regulate protein and RNA interactions to influence protein stability, localization and translation of CELF targets (Dasgupta & Ladd, 2012).
CELF RBPs are linked to several neurological and neurodegenerative disorders, as well as neurodevelopmental and neuropsychiatric disorders, including social communication problems in autism (CELF4, CELF6), epilepsy (CELF4), bipolar disorder (CELF5), and schizophrenia (CELF5) (Ladd, 2013; Welter et al., 2014; Dougherty et al., 2013). Recent evidence implicates both CELF4 and CELF6 in the addictions. CELF4 is a brain-specific isoform that is expressed throughout development and is highly expressed in the hippocampus, amygdala, and cortex. CELF4 loss of function in mice causes seizures (Yang et al., 2007) and a functional deficit in excitatory synaptic transmission in cortical and hippocampal neurons (Wagnon et al., 2011). The complex seizure phenotype implicates a role for CELF4 in corticothalamic development (Wagnon et al., 2011). RNA target analysis of CELF4 in cortical and hippocampal tissue identified several 3’ UTR-targeted mRNAs involved in synaptic transmission (Wagnon et al., 2012). Of potential relevance to the addictions, genetic variation in CELF4 showed a nominal association (P =3–5 × 10−6) with the subjective amphetamine response (Hart et al., 2012) and has been linked to hyperphagia-related obesity (Comuzzie et al., 2012; Halgren et al., 2012).
Translational profiling of ribosome-bound mRNAs from mouse serotonergic neurons combined with analysis of human genetic variants identified CELF6 as a candidate gene associated with autism. Celf6 knockouts exhibit reduced brain serotonin, deficits in ultrasonic vocalizations in neonatal pups, and behavioral resistance to change (Dougherty et al., 2013). CELF6 is expressed in both the nucleus and cytoplasm throughout development and exhibits localized expression in monoaminergic and catecholaminergic cells in the basal forebrain, ventral tegmental area, substantia nigra, raphe nuclei, and locus coeruleus (Maloney, Khangura & Dougherty, 2015). Thus, Celf6 could influence neurodevelopment and function of the mesocorticolimbic reward circuitry. In support, Celf6 knockouts show a disruption of conditioned cocaine reward (Dougherty, 2015). CELF6 can promote both exon inclusion and skipping (Ladd et al., 2004). It will be important to determine the direct nuclear and cytoplasmic targets of CELF6 which will provide insight into the subcellular mechanisms of behavioral and psychiatric dysfunction.
ADAR1/2
RNA editing is yet another type of distinct and adaptive pre-mRNA processing mechanism that can increase proteome diversity to regulate neurodevelopment, plasticity, and human disease, including neurological and psychiatric disorders such as schizophrenia, bipolar disorder, and depression (Li & Church, 2013). Adenosine deaminase acting on RNA (ADAR)-1, 2, and 3 are nuclear, enzymatic RBPs that contain double-stranded RNA binding motifs and destabilize pre-mRNA to “edit” adenosine (A) nucleotides to guanosine-mimicking RNA inosine (I) nucleotides. A-I editing changes the complementary nucleotide to cytosine, thus modulating RNA base pairing and potentially splice site, transport, ribosome binding, translational efficiency, and amino acid sequence (Slotkin & Nishikura, 2013; Li & Church, 2013). Compared to extensive editing of noncoding RNA, re-coding of coding exons is rare in mammals and these sites are enriched for proteins involved in neuronal function, including neuronal excitability, vesicular release, and cytoskeleton architecture (Rosenthal, 2015). The expansion of RNA editing from rodents to non-human primates to humans suggests and important role in brain evolution and cognition (Li & Church, 2013). Although the regulation of ADAR in RNA editing is poorly understood, it is relevant to note that FMRP interacts with ADAR in Drosophila to modulate enzymatic activity and editing of mRNA transcripts that affect synaptic morphology (Bhogal et al., 2011; Bassell, 2011). This observation highlights an additional function of FMRP and suggests that FMRP and ADAR could work together in neurodevelopment and neuroplasticity in the addictions.
ADAR-2 has an established role in editing of the GluA2 subunit of the AMPA receptor at Q/R site 607 which reduces calcium permeability (Geiger et al., 1995) and may protect against neuronal excitotoxicity in vivo (Higuchi et al., 2000). With regard to the addictions, cocaine abstinence and cocaine-primed reinstatement of self-administration in rats has been associated with decreased ADAR-2 expression and decreased GluA2 editing of the Q/R site in the nucleus accumbens shell. Overexpression of ADAR-2 prevented relapse and the concomitant increase in surface receptor expression of AMPAR (Schmidt et al., 2014). Thus, unedited AMPAR receptors are associated with a model of drug relapse that could be mitigated by restoration of surface edited AMPA receptors.
In addition to AMPA receptor editing, ADAR proteins also edit the 5-HT2C receptor [a promising drug target for treating addictive disorders (Higgins & Fletcher, 2015)] at five different sites within the second intracellular loop, yielding up to 24 brain region-specific isoforms that could alter pre-mRNA splicing, ligand affinity, ligand-induced g-protein signaling, ligand-induced blockade of constitutive activity, and brain region-specific functions, including modulation of dopamine release (Werry et al., 2008; Burns et al., 1997). Interestingly, rats categorized as high versus low novelty seeking (a trait that correlates with future drug use) show differences in 5-HT2C receptor editing in the mesocorticolimbic circuitry, in particular the nucleus accumbens shell that could affect dopamine transmission (Dracheva et al., 2009). An increase in alcohol drinking in different mouse strains was associated with an increase in anxiety as well as an increase in ADAR1/2 expression and 5-HT2C editing in the nucleus accumbens and dorsal raphe nucleus (Watanabe et al., 2014). Finally, chronic nicotine decreased editing of the 5-HT2C in the hippocampus that was associated with depressive-like behavior during nicotine withdrawal (Zaniewska et al., 2015; Zaniewska et al., 2010). Thus, differential 5-HT2C receptor editing could potentially predispose individuals to addiction risk as well as mediate the negative emotional states that support addictive behaviors.
An increase in 5-HT2C receptor editing is associated with hyperphagia and obesity in patients with Prader-Willi Syndrome and in mice expressing the fully edited 5-HT2C receptor (Kawahara et al., 2008). Furthermore, mice that differentially express the wild-type or catalytically inactive version of ADAR2 show hyperphagia and obesity (Singh et al., 2007) and increased preference for high fat food over running (Akubuiro et al., 2013). These physiological and behavioral changes indicate a food “addiction” propensity that is supported by increased mRNA expression of ADAR2, D1 and D2 dopamine receptors, MOR, 5-HT2C long and short splice variants in the hypothalamus as well as increased mRNA expression of D1 dopamine receptors in the striatum. PET imaging in mice using a tail vein injection of [18F] flurodeoxyglucose indicated an increase in glucose metabolism in the mesolimbic reward circuitry, hypothalamus, and hippocampus. The combined observations implicate hyperactive food-directed reward processing in ADAR2 transgenics (Akubuiro et al., 2013; Singh et al., 2011); however, direct evidence linking RNA editing to food or drug addictive behaviors is still lacking.
To summarize, changes in AMPA receptor and 5-HT receptor editing are associated with exposure to abused substances and could contribute to susceptibility and synaptic plasticity underlying the addictions. Future studies will likely involve the use of advanced genome editing approaches to directly test the causal, spatiotemporal role of re-coded proteins as well as non-coding RNAs in the brain in the establishment and maintenance of addictive behaviors.
Summary
This review highlights the diversity of RBP functions in regulating transcription, RNA metabolism, neurodevelopment and neuroplasticity relevant to the addictions (Figure 2). Although beyond the scope of this review, RBPs also interact with noncoding RNAs and there are several new and emerging RNA modifications that could be of relevance to the addictions (Satterlee et al., 2014). Identifying the key cell type-specific splice forms and unique functions of RBP splice variants and their alternatively spliced targets will be critical to yielding novel, biologically relevant discoveries in the addictions. Do a specific set of RBPs become recruited during environmental stress exposure or drug exposure that influence addiction risk? What are the transcriptomes, spliceomes and translatomes (King & Gerber, 2014) that these RBPs govern in response to risk exposure, drug exposure, and the interactive exposure to both? How do these programs change across neurodevelopment, across repeated drug exposure as addiction progress, and across recovery during abstinence? In addition to splicing, post-translational modifications of RBPs, including phosphorylation, ubiquitination, sumoylation, and methylation can further regulate the transport, stabilization, degradation, and binding of RBPs to their RNA targets and to other proteins (Chaudhury, Chander & Howe, 2010), thus adding additional layers of regulation in RBP function. This complexity may one day be harnessed to perturb RBP signaling in preventing and treating the addictions as has recently been demonstrated in other disease models (Kim et al., 2014; Wang et al., 2009).
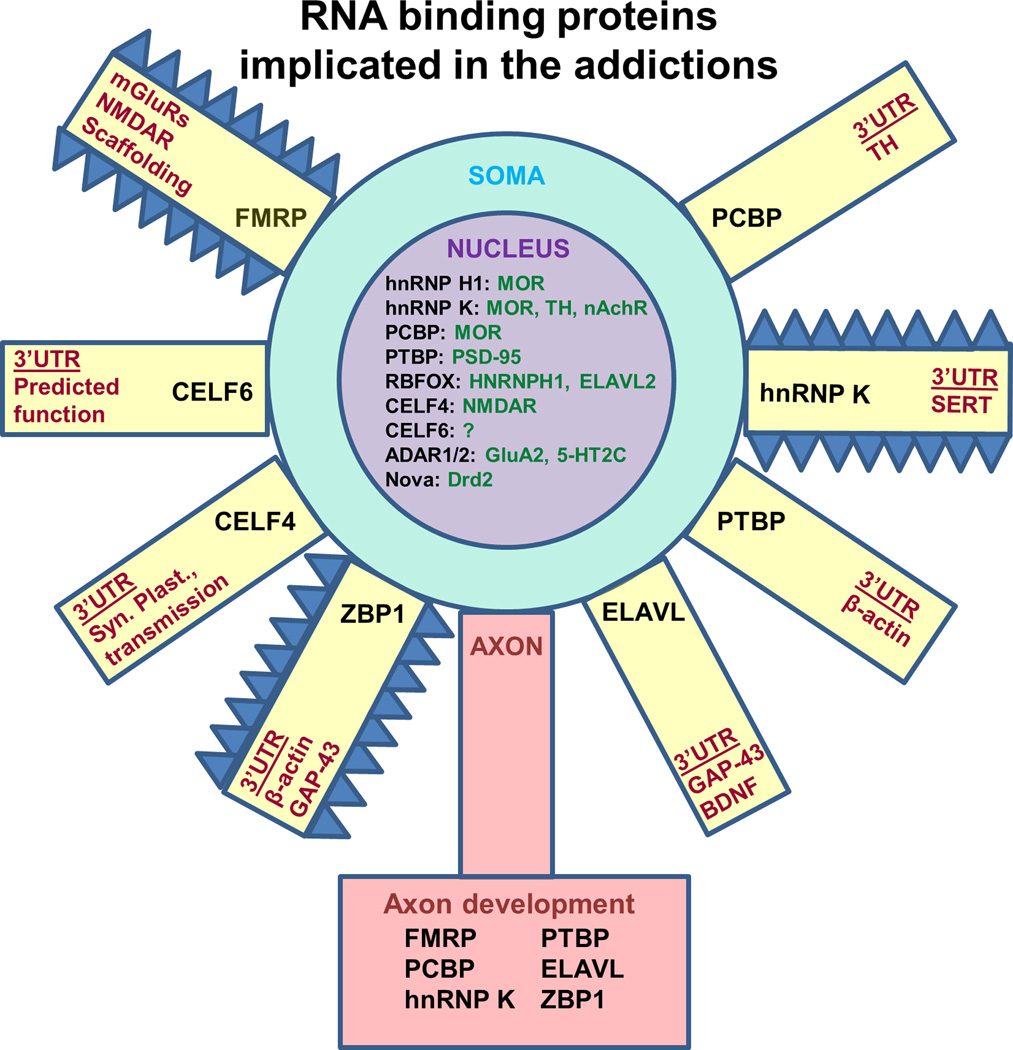
Figure 2. RNA binding proteins implicated in the addictions.
Many RBPs have the capability to shuttle between the nucleus and cytoplasm to regulate all levels of RNA post-transcriptional processing. Here, we illustrate the location of action of the main examples that are discussed and some of their well-characterized targets. For the RBPs that are illustrated, we have also indicated whether not they contribute to axon development by listing them in the axon terminal (bottom). The yellow rectangles denote dendrites. The blue spikes indicate an association with structural changes in dendritic morphology.
We limited our discussion to RBPs where multiple lines of evidence implicate a potential importance in the addictions. However, additional RBPs are certainly going to be uncovered in the addictive process from neurodevelopment to drug-induced neuroplasticity. For instance, cytoplasmic polyadenylation element binding proteins (CPEBs) plays an important role in synaptic plasticity (Ivshina, Lasko & Richter, 2014). Second, the SR protein Tra2β can be spliced by the dopamine signaling molecule DARPP-32 (Benderska et al., 2010) and can promote splicing of RGS proteins (regulators of g-protein signaling) in response to morphine (Li et al., 2013; Traynor, 2010). Third, KH-type Nova proteins also regulate splicing of the D2 dopamine receptor and neocortical synaptic proteins involved in neurotransmitter release and signaling, receptor localization, synaptogenesis, axonogenesis, actin organization, cell adhesion, and extracellular matrix organization (Park et al., 2011; Ule et al., 2005).
The topic of RBPs and the addictions is clearly a wide open and promising field of research; understanding how these remarkable proteins coordinate splicing and translation during neurodevelopment and neuroplasticity promises to yield dividends in understanding and treating neuropsychiatric diseases.
Table 1.
RBPs associated with the addictions
| RBP | Addiction- relevant targets |
RNA function | Neurodevelopment neuroplastic function |
Addiction- relevant circuitry |
Addiction- associated result |
References |
|---|---|---|---|---|---|---|
| ADAR1/2 | 5HT-2C receptor, AMPA receptor, GABA-A receptor |
RNA editing, pri-miRNA binding and miRNA biogenesis |
Neural differentiation and induction |
Ubiquitous expression, hypothalamus, striatum, mesocorticolimbic circuitry, dorsal raphe nucleus, forebrain, cortex, hippocampus, diencephalon |
Overeating, motivation to eat in a competing reward environment, novelty-induced locomotor activity, alcohol drinking, cocaine abstinence and seeking, nicotine withdrawal |
(Satterlee et al., 2014; Akubuiro et al., 2013; Dracheva et al., 2009; Watanabe et al., 2014; Schmidt et al., 2014; Zaniewska et al., 2015) |
| CELF4 | NMDA receptors, MAPT |
Splicing, 3’ UTR binding and mRNA stability, translation, localization |
Neuronal differentiation and excitation, corticothalamic development, synaptic transmission and function, predicted synaptic plasticity |
Ubiquitous expression, hippocampus, cortex |
Amphetamine response in humans, hyperactivity, hyperphagia- associated obesity, food-related obsessions |
(Hart et al., 2012; Halgren et al., 2012; Comuzzie et al., 2012) |
| CELF6 | Serotonergic neurons, MAPT |
Splice enhancement and repression |
Expressed during neurodevelopment, beginning at E14 |
Basal forebrain, ventral tegmental area, locus coeruleus, prefrontal and hippocampal cortices, hypothalamus |
Cocaine conditioned reward |
(Dougherty, 2015; Dougherty et al., 2013; Maloney, Khangura & Dougherty, 2015) |
| ELAVL | cfos, BDNF, GAP-43, Nova1, GABA-A receptor, glycine alpha-2 receptor |
Splicing, 3’ UTR binding, translation, polyadenylation , stability, protein synthesis, transport |
Neuronal differentiation, maintenance, maturation, axon growth, dendritogenesis, synaptic plasticity |
Neocortex, hippocampus |
Cocaine-induced changes in expression and phosphorylation |
(Tiruchinapalli, Caron & Keene, 2008; Pascale et al., 2011) |
| FMRP | Cyfip1/2, AMPA receptor, mGluR1, mGluR5, ADAR, PSD- 95, Homer1, CREB BP, GRK2 |
Transport, translational repression via stalling of ribosomal translocation, stability |
Cell fate, progenitor maintenance, neurite, axonal and dendritic outgrowth Glutamate receptor trafficking, dendritic spine morphology |
Mesocorticolimbic circuitry, hippocampus |
Cocaine and amphetamine neurobehavioral plasticity, synaptic and structural plasticity |
(Smith et al., 2014; Kumar et al., 2013; Pilaz & Silver, 2015; Fish et al., 2013; Fulks et al., 2010; Bhogal et al., 2011) |
| HNRNPA1 | NMDA receptors |
Splicing, spliceosome, translation, export, stability |
Neurogenesis | Ubiquitous expression |
Opioid and psychostimulant- induced changes in expression. Co- regulated with Nurr1. |
(Wang, Yuan & Li, 2011; Johnson et al., 2011) |
| HNRNPH1 | OPRM1, RBFOX, NMDA receptor, Per2 |
Splicing, regulation of splicing activity, polyadenylation and cleavage, 5’ UTR translational suppression |
Unknown | Ubiquitous Expression |
Heroin addiction, MOR splicing, methamphetamine stimulant response, association with Nurr1 expression |
(Xu et al., 2014; Yazdani et al., 2015; Johnson et al., 2011; Song et al., 2012) |
| HNRNPH2 | Splicing | Unknown | Low expression | Opioid and psychostimulant- induced change in expression |
(Wang, Yuan & Li, 2011) | |
| HNRNPK | TH, MOR, SERT, neuronal nicotinic acetylcholine receptor beta2 |
Transcription, silencing, splicing, 3’UTR binding and stability, translation |
Hippocampal synaptic plasticity, dendritic spine morphology, filopodia formation, synapse maturation, axon development |
Mesocorticolimbic circuitry, hippocampus, ventral midbrain |
Activates TH and MOR transcription, promotes SERT translation, activates transcription of beta2 subunit of the neuronal nicotinic acetylcholine receptor |
(Banerjee et al., 2014; Choi et al., 2008; Du, Melnikova & Gardner, 1998) |
| HNRNPM1 | D2 dopamine receptor |
Unknown | Unknown | Unknown | Splicing of D2 dopamine receptor |
(Park et al., 2011) |
| Nova1/2 | D2 dopamine receptor, GABA-A and GABA-B receptors, NMDA receptor, nicotinic acetylcholine receptor, GIRK2 |
Repression of splicing, polyadenylation |
Synaptogenesis, neuronal migration of cortical and purkinje neurons, neuronal inhibition |
Neocortex and midbrain (Nova1), hindbrain, motoneurons and ventral spinal cord (Nova2) |
Splicing of D2 dopamine receptor |
(Park et al., 2011) |
| PCBP 1–4 (HNRNPE1-4) |
Tyrosine hydroxylase, MOR |
Transcription, 3’UTR binding and stability, translational co- activation and repression |
Corticogenesis, axonogenesis |
TH-positive cells of the midbrain, locus coeruleus |
Nicotine-induced changes in TH mRNA |
(Boschi et al., 2015; Pilaz & Silver, 2015; Choi et al., 2007; Choi et al., 2008; Choi et al., 2009) |
| PTBP1-2 (HNRNPI) |
D2 dopamine receptor, ΔfosB, PSD- 95, PTBP, GABA-A receptor, Grip1, glycine receptor alpha2 |
Splicing, Polyadenylation , nonsense- mediated decay, transport, stability, translation initiation |
Neuronal differentiation, synapse maturation and stability, plasticity |
Cortex, hippocampus |
D2 dopamine receptor splicing |
(Sasabe, Futai & Ishiura, 2011) |
| RBFOX1 (A2BP1) |
Cell adhesion molecules, HNRNPH1, PACAP receptor, GABA-A receptor, NMDA receptor, Grip1, Csnk1d, PTBP |
Splicing, 3’ UTR binding and stability, polyadenylation |
Transmission, membrane excitability |
Ubiquitous, basal forebrain, neocortex, hippocampus, nucleus accumbens |
Cocaine conditioned reward, nicotine and alcohol dependence |
(Fogel et al., 2012; Feng et al., 2014; Zhong et al., 2015) |
| Tra2β | RGS4, glutamate receptors |
Splicing | Neuronal survival, differentiation, cortical neurogenesis |
Locus coeruleus, cerebral cortex |
Morphine-induced splicing of RGS4. DARPP-32 induces splicing of Tra2β |
(Li et al., 2013; Benderska et al., 2010) |
| ZBP1 | B-actin, GAP-43, Homer1, many other predicted targets |
3’ UTR binding and stability, translational repression, axonal mRNA transport and localization |
Axon guidance, outgrowth and branching; dendritic development and morphology, hippocampal development and plasticity, corticogenesis, axonogenesis, synaptogenesis, regeneration |
Ubiquitous expression, forebrain, hippocampus |
Cocaine conditioned reward, methamphetamine stimulant response |
(Lapidus et al., 2012; Bryant et al., 2012) |
Acknowledgments
We thank Kristen E. Hokenson and Daniel J. Appico for assistance in neuronal cell culture and imaging as well as Drs. Shelley J. Russek, Benjamin Wolozin, and Tsuneya Ikezu for use of equipment. This work is supported by NIH grants R01DA039168 (C.D.B.), R00DA029635 (C.D.B.), R03DA038287 (C.D.B.), R21DA038738 (C.D.B.), and T32GM008541 (N.Y.). Additional support comes from The Jack Spivack Excellence in Neurosciences Research Award (C.D.B.) and the Transformative Training Program in Addiction Science (Burroughs Wellcome Fund 1011479; N.Y.).
Footnotes
The authors have no conflict of interests to declare.
REFERENCES
- Adori C, Ando RD, Ferrington L, Szekeres M, Vas S, Kelly PA, Hunyady L, Bagdy G. Elevated BDNF protein level in cortex but not in hippocampus of MDMA-treated Dark Agouti rats: a potential link to the long-term recovery of serotonergic axons. Neuroscience letters. 2010;478:56–60. doi: 10.1016/j.neulet.2010.04.061. [DOI] [PubMed] [Google Scholar]
- Akubuiro A, Bridget Zimmerman M, Boles Ponto LL, Walsh SA, Sunderland J, McCormick L, Singh M. Hyperactive hypothalamus, motivated and non-distractible chronic overeating in ADAR2 transgenic mice. Genes, brain, and behavior. 2013;12:311–322. doi: 10.1111/gbb.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibhai IN, Green TA, Potashkin JA, Nestler EJ. Regulation of fosB and DeltafosB mRNA expression: in vivo and in vitro studies. Brain research. 2007;1143:22–33. doi: 10.1016/j.brainres.2007.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M, Bird C, Feng W, Liu G, Li W, Perrone-Bizzozero NI, Feng Y. HuD promotes BDNF expression in brain neurons via selective stabilization of the BDNF long 3'UTR mRNA. PloS one. 2013;8:e55718. doi: 10.1371/journal.pone.0055718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3' UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SL, Teicher MH. Desperately driven and no brakes: developmental stress exposure and subsequent risk for substance abuse. Neuroscience and biobehavioral reviews. 2009;33:516–524. doi: 10.1016/j.neubiorev.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares-Santos S, Granado N, Espadas I, Martinez-Murillo R, Moratalla R. Methamphetamine causes degeneration of dopamine cell bodies and terminals of the nigrostriatal pathway evidenced by silver staining. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2014;39:1066–1080. doi: 10.1038/npp.2013.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K, Wang M, Cai E, Fujiwara N, Baker H, Cave JW. Regulation of tyrosine hydroxylase transcription by hnRNP K and DNA secondary structure. Nature communications. 2014;5:5769. doi: 10.1038/ncomms6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ. Fragile balance: RNA editing tunes the synapse. Nature neuroscience. 2011;14:1492–1494. doi: 10.1038/nn.2982. [DOI] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends in neurosciences. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Benderska N, Becker K, Girault JA, Becker CM, Andreadis A, Stamm S. DARPP-32 binds to tra2-beta1 and influences alternative splicing. Biochimica et biophysica acta. 2010;1799:448–453. doi: 10.1016/j.bbagrm.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogal B, Jepson JE, Savva YA, Pepper AS, Reenan RA, Jongens TA. Modulation of dADAR-dependent RNA editing by the Drosophila fragile X mental retardation protein. Nature neuroscience. 2011;14:1517–1524. doi: 10.1038/nn.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill BR, Lowe JK, Dybuncio CT, Fogel BL. Orchestration of neurodevelopmental programs by RBFOX1: implications for autism spectrum disorder. International review of neurobiology. 2013;113:251–267. doi: 10.1016/B978-0-12-418700-9.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette AR, Fuentes Medel YF, Gardner PD. Cell-type-specific and developmental regulation of heterogeneous nuclear ribonucleoprotein K mRNA in the rat nervous system. Gene expression patterns : GEP. 2006;6:596–606. doi: 10.1016/j.modgep.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Qiu S, Tanner DC, Paik J, Perrone-Bizzozero NI, Weeber EJ. Associative and spatial learning and memory deficits in transgenic mice overexpressing the RNA-binding protein HuD. Neurobiology of learning and memory. 2007;87:635–643. doi: 10.1016/j.nlm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Boschi NM, Takeuchi K, Sterling C, Tank AW. Differential expression of polycytosine-binding protein isoforms in adrenal gland, locus coeruleus and midbrain. Neuroscience. 2015;286:1–12. doi: 10.1016/j.neuroscience.2014.11.038. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & development. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant CD, Kole LA, Guido MA, Sokoloff G, Palmer AA. Congenic dissection of a major QTL for methamphetamine sensitivity implicates epistasis. Genes, brain, and behavior. 2012;11:623–632. doi: 10.1111/j.1601-183X.2012.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Molecular cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Campos-Melo D, Galleguillos D, Sanchez N, Gysling K, Andres ME. Nur transcription factors in stress and addiction. Frontiers in molecular neuroscience. 2013;6:44. doi: 10.3389/fnmol.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey BJ, Glatt CE, Tottenham N, Soliman F, Bath K, Amso D, Altemus M, Pattwell S, Jones R, Levita L, McEwen B, Magarinos AM, Gunnar M, Thomas KM, Mezey J, Clark AG, Hempstead BL, Lee FS. Brain-derived neurotrophic factor as a model system for examining gene by environment interactions across development. Neuroscience. 2009;164:108–120. doi: 10.1016/j.neuroscience.2009.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castren M, Lampinen KE, Miettinen R, Koponen E, Sipola I, Bakker CE, Oostra BA, Castren E. BDNF regulates the expression of fragile X mental retardation protein mRNA in the hippocampus. Neurobiology of disease. 2002;11:221–229. doi: 10.1006/nbdi.2002.0544. [DOI] [PubMed] [Google Scholar]
- Castren ML, Castren E. BDNF in fragile X syndrome. Neuropharmacology. 2014;76 Pt C:729–736. doi: 10.1016/j.neuropharm.2013.05.018. [DOI] [PubMed] [Google Scholar]
- Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Human molecular genetics. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1's multifunctional regulatory roles. RNA (New York, N.Y.) 2010;16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Hwang CK, Song KY, Law PY, Wei LN, Loh HH. Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochemical and biophysical research communications. 2009;380:431–436. doi: 10.1016/j.bbrc.2009.01.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Kim CS, Hwang CK, Song KY, Law PY, Wei LN, Loh HH. Novel function of the poly(C)-binding protein alpha CP3 as a transcriptional repressor of the mu opioid receptor gene. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2007;21:3963–3973. doi: 10.1096/fj.07-8561com. [DOI] [PubMed] [Google Scholar]
- Choi HS, Song KY, Hwang CK, Kim CS, Law PY, Wei LN, Loh HH. A proteomics approach for identification of single strand DNA-binding proteins involved in transcriptional regulation of mouse mu opioid receptor gene. Molecular & cellular proteomics : MCP. 2008;7:1517–1529. doi: 10.1074/mcp.M800052-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MY, Rooke N, Turck CW, Black DL. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Molecular and cellular biology. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleva RM, Gass JT, Widholm JJ, Olive MF. Glutamatergic targets for enhancing extinction learning in drug addiction. Current neuropharmacology. 2010;8:394–408. doi: 10.2174/157015910793358169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C, Silani V, Ratti A. ELAV proteins along evolution: back to the nucleus? Molecular and cellular neurosciences. 2013;56:447–455. doi: 10.1016/j.mcn.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, Butte NF. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PloS one. 2012;7:e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contet C, Kieffer BL, Befort K. Mu opioid receptor: a gateway to drug addiction. Current opinion in neurobiology. 2004;14:370–378. doi: 10.1016/j.conb.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Czyzyk-Krzeska MF, Beresh JE. Characterization of the hypoxia-inducible protein binding site within the pyrimidine-rich tract in the 3'-untranslated region of the tyrosine hydroxylase mRNA. The Journal of biological chemistry. 1996;271:3293–3299. doi: 10.1074/jbc.271.6.3293. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nature neuroscience. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T, Ladd AN. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley interdisciplinary reviews.RNA. 2012;3:104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, Pasciuto E, Li KW, Fernandez E, Di Marino D, Buzzi A, Ostroff LE, Klann E, Zwartkruis FJ, Komiyama NH, Grant SG, Poujol C, Choquet D, Achsel T, Posthuma D, Smit AB, Bagni C. CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron. 2013;79:1169–1182. doi: 10.1016/j.neuron.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichter GS, Damiano CA, Allen JA. Reward circuitry dysfunction in psychiatric and neurodevelopmental disorders and genetic syndromes: animal models and clinical findings. Journal of neurodevelopmental disorders. 2012;4 doi: 10.1186/1866-1955-4-19. 19-1955-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Nestler EJ. The neural rejuvenation hypothesis of cocaine addiction. Trends in pharmacological sciences. 2014;35:374–383. doi: 10.1016/j.tips.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Park M, Spillane M, Yoo S, Pacheco A, Gomes C, Vuppalanchi D, McDonald M, Kim HH, Merianda TT, Gallo G, Twiss JL. Axonally synthesized beta-actin and GAP-43 proteins support distinct modes of axonal growth. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:3311–3322. doi: 10.1523/JNEUROSCI.1722-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, Yoon SO, Bassell GJ, Twiss JL. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. The EMBO journal. 2011;30:4665–4677. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD. Functional analyses of Celf6 – an RNA binding protein regulating early communicative behavior. Genomic and neurobiological studies of RNA binding proteins in complex traits relevant to psychiatric disorders, Winter Conference on Brain Resarch. 2015 [Google Scholar]
- Dougherty JD, Maloney SE, Wozniak DF, Rieger MA, Sonnenblick L, Coppola G, Mahieu NG, Zhang J, Cai J, Patti GJ, Abrahams BS, Geschwind DH, Heintz N. The disruption of Celf6, a gene identified by translational profiling of serotonergic neurons, results in autism-related behaviors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:2732–2753. doi: 10.1523/JNEUROSCI.4762-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxakis E. RNA binding proteins: a common denominator of neuronal function and dysfunction. Neuroscience bulletin. 2014;30:610–626. doi: 10.1007/s12264-014-1443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M, Kiebler MA. A zipcode unzipped. Genes & development. 2012;26:110–113. doi: 10.1101/gad.184945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracheva S, Lyddon R, Barley K, Marcus SM, Hurd YL, Byne WM. Editing of serotonin 2C receptor mRNA in the prefrontal cortex characterizes high-novelty locomotor response behavioral trait. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2009;34:2237–2251. doi: 10.1038/npp.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nature reviews. Molecular cell biology. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annual Review of Biochemistry. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Du Q, Melnikova IN, Gardner PD. Differential effects of heterogeneous nuclear ribonucleoprotein K on Sp1- and Sp3-mediated transcriptional activation of a neuronal nicotinic acetylcholine receptor promoter. The Journal of biological chemistry. 1998;273:19877–19883. doi: 10.1074/jbc.273.31.19877. [DOI] [PubMed] [Google Scholar]
- Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. The Journal of cell biology. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, Maze I, Shao N, Kennedy P, Koo J, Dias C, Laitman B, Stockman V, LaPlant Q, Cahill ME, Nestler EJ, Shen L. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome biology. 2014;15 doi: 10.1186/gb-2014-15-4-r65. R65-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez E, Rajan N, Bagni C. The FMRP regulon: from targets to disease convergence. Frontiers in neuroscience. 2013;7:191. doi: 10.3389/fnins.2013.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisette JF, Toutant J, Dugre-Brisson S, Desgroseillers L, Chabot B. hnRNP A1 and hnRNP H can collaborate to modulate 5' splice site selection. RNA (New York, N.Y.) 2010;16:228–238. doi: 10.1261/rna.1890310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish EW, Krouse MC, Stringfield SJ, Diberto JF, Robinson JE, Malanga CJ. Changes in sensitivity of reward and motor behavior to dopaminergic, glutamatergic, and cholinergic drugs in a mouse model of fragile X syndrome. PloS one. 2013;8:e77896. doi: 10.1371/journal.pone.0077896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, Gao F, Parikshak N, Konopka G, Geschwind DH. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Human molecular genetics. 2012;21:4171–4186. doi: 10.1093/hmg/dds240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folci A, Mapelli L, Sassone J, Prestori F, D'Angelo E, Bassani S, Passafaro M. Loss of hnRNP K impairs synaptic plasticity in hippocampal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:9088–9095. doi: 10.1523/JNEUROSCI.0303-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Kano F, Murata M. Dual localization of the RNA binding protein CUGBP-1 to stress granule and perinucleolar compartment. Experimental cell research. 2008;314:543–553. doi: 10.1016/j.yexcr.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Fukao A, Sasano Y, Imataka H, Inoue K, Sakamoto H, Sonenberg N, Thoma C, Fujiwara T. The ELAV protein HuD stimulates cap-dependent translation in a Poly(A)- and eIF4A–dependent manner. Molecular cell. 2009;36:1007–1017. doi: 10.1016/j.molcel.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Fulks JL, O'Bryhim BE, Wenzel SK, Fowler SC, Vorontsova E, Pinkston JW, Ortiz AN, Johnson MA. Dopamine Release and Uptake Impairments and Behavioral Alterations Observed in Mice that Model Fragile X Mental Retardation Syndrome. ACS chemical neuroscience. 2010;1:679–690. doi: 10.1021/cn100032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo JM, Spickett C. The role of CELF proteins in neurological disorders. RNA biology. 2010;7:474–479. doi: 10.4161/rna.7.4.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Jr, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nature genetics. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nature reviews. Genetics. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Cohen OS, Faraone SV, Tsuang MT. Dysfunctional gene splicing as a potential contributor to neuropsychiatric disorders. American journal of medical genetics.Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2011;156B:382–392. doi: 10.1002/ajmg.b.31181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS letters. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, Matlin AJ, Cooper TA, Smith CW. Antagonistic regulation of alpha-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA (New York, N.Y.) 2003;9:443–456. doi: 10.1261/rna.2191903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. The Journal of cell biology. 2002;156:41–51. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelhah H, Shah K, Huang L, Ostareck-Lederer A, Burlingame AL, Shokat KM, Hentze MW, Ronai Z. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nature cell biology. 2001;3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- Halgren C, Bache I, Bak M, Myatt MW, Anderson CM, Brondum-Nielsen K, Tommerup N. Haploinsufficiency of CELF4 at 18q12.2 is associated with developmental and behavioral disorders, seizures, eye manifestations, and obesity. European journal of human genetics : EJHG. 2012;20:1315–1319. doi: 10.1038/ejhg.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammock EA, Levitt P. Developmental expression mapping of a gene implicated in multiple neurodevelopmental disorders, A2bp1 (Fox1) Developmental neuroscience. 2011;33:64–74. doi: 10.1159/000323732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. The Biochemical journal. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- Hart AB, Engelhardt BE, Wardle MC, Sokoloff G, Stephens M, de Wit H, Palmer AA. Genome-wide association study of d-amphetamine response in healthy volunteers identifies putative associations, including cadherin 13 (CDH13) PloS one. 2012;7:e42646. doi: 10.1371/journal.pone.0042646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberg B, Swiniarski S, Sabine B, Illenberger S. A conserved peptide motif in Raver2 mediates its interaction with the polypyrimidine tract-binding protein. Experimental cell research. 2010;316:966–979. doi: 10.1016/j.yexcr.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Higgins GA, Fletcher PJ. Therapeutic Potential of 5-HT2C Receptor Agonists for Addictive Disorders. ACS chemical neuroscience. 2015;6:1071–1088. doi: 10.1021/acschemneuro.5b00025. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, Ares M, Jr, Yeo GW. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell reports. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Ince-Dunn G, Okano HJ, Jensen KB, Park WY, Zhong R, Ule J, Mele A, Fak JJ, Yang C, Zhang C, Yoo J, Herre M, Okano H, Noebels JL, Darnell RB. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron. 2012;75:1067–1080. doi: 10.1016/j.neuron.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivshina M, Lasko P, Richter JD. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annual Review of Cell and Developmental Biology. 2014;30:393–415. doi: 10.1146/annurev-cellbio-101011-155831. [DOI] [PubMed] [Google Scholar]
- Johnson MM, Michelhaugh SK, Bouhamdan M, Schmidt CJ, Bannon MJ. The Transcription Factor NURR1 Exerts Concentration-Dependent Effects on Target Genes Mediating Distinct Biological Processes. Frontiers in neuroscience. 2011;5:135. doi: 10.3389/fnins.2011.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nature methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Grimberg A, Teegarden S, Mombereau C, Liu S, Bale TL, Blendy JA, Nishikura K. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:12834–12844. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Bulik CM, Silberg J, Hettema JM, Myers J, Prescott CA. Childhood sexual abuse and adult psychiatric and substance use disorders in women: an epidemiological and cotwin control analysis. Archives of General Psychiatry. 2000;57:953–959. doi: 10.1001/archpsyc.57.10.953. [DOI] [PubMed] [Google Scholar]
- Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Critical reviews in biochemistry and molecular biology. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Lee SJ, Gardiner AS, Perrone-Bizzozero NI, Yoo S. Different motif requirements for the localization zipcode element of beta-actin mRNA binding by HuD and ZBP1. Nucleic acids research. 2015;43:7432–7446. doi: 10.1093/nar/gkv699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQ, Lee VM, Finkbeiner S, Gitler AD, Bonini NM. Therapeutic modulation of eIF2alpha phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nature genetics. 2014;46:152–160. doi: 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HA, Gerber AP. Translatome profiling: methods for genome-scale analysis of mRNA translation. Briefings in functional genomics. 2014 doi: 10.1093/bfgp/elu045. [DOI] [PubMed] [Google Scholar]
- Kobori N, Clifton GL, Dash P. Altered expression of novel genes in the cerebral cortex following experimental brain injury. Brain research.Molecular brain research. 2002;104:148–158. doi: 10.1016/s0169-328x(02)00331-5. [DOI] [PubMed] [Google Scholar]
- Kumar V, Kim K, Joseph C, Kourrich S, Yoo SH, Huang HC, Vitaterna MH, de Villena FP, Churchill G, Bonci A, Takahashi JS. C57BL/6N mutation in Cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science (New York, N.Y.) 2013;342:1508–1512. doi: 10.1126/science.1245503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cellular and molecular life sciences : CMLS. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN. CUG-BP, Elav-like family (CELF)-mediated alternative splicing regulation in the brain during health and disease. Molecular and cellular neurosciences. 2013;56:456–464. doi: 10.1016/j.mcn.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN, Cooper TA. Multiple domains control the subcellular localization and activity of ETR-3, a regulator of nuclear and cytoplasmic RNA processing events. Journal of cell science. 2004;117:3519–3529. doi: 10.1242/jcs.01194. [DOI] [PubMed] [Google Scholar]
- Ladd AN, Nguyen NH, Malhotra K, Cooper TA. CELF6, a member of the CELF family of RNA-binding proteins, regulates muscle-specific splicing enhancer-dependent alternative splicing. The Journal of biological chemistry. 2004;279:17756–17764. doi: 10.1074/jbc.M310687200. [DOI] [PubMed] [Google Scholar]
- Lapidus KA, Nwokafor C, Scott D, Baroni TE, Tenenbaum SA, Hiroi N, Singer RH, Czaplinski K. Transgenic expression of ZBP1 in neurons suppresses cocaine-associated conditioning. Learning & memory (Cold Spring Harbor, N.Y.) 2012;19:35–42. doi: 10.1101/lm.024471.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AG, Irier HA, Gu J, Tian D, Ku L, Liu G, Xia M, Fritsch B, Zheng JQ, Dingledine R, Xu B, Lu B, Feng Y. Distinct 3'UTRs differentially regulate activity-dependent translation of brain-derived neurotrophic factor (BDNF) Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15945–15950. doi: 10.1073/pnas.1002929107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal G, Afonso PM, Duarte CB. Neuronal activity induces synaptic delivery of hnRNP A2/B1 by a BDNF-dependent mechanism in cultured hippocampal neurons. PloS one. 2014;9:e108175. doi: 10.1371/journal.pone.0108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PT, Chao PK, Ou LC, Chuang JY, Lin YC, Chen SC, Chang HF, Law PY, Loh HH, Chao YS, Su TP, Yeh SH. Morphine drives internal ribosome entry site-mediated hnRNP K translation in neurons through opioid receptor-dependent signaling. Nucleic acids research. 2014;42:13012–13025. doi: 10.1093/nar/gku1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Levran O, Randesi M, da Rosa JC, Ott J, Rotrosen J, Adelson M, Kreek MJ. Overlapping dopaminergic pathway genetic susceptibility to heroin and cocaine addictions in African Americans. Annals of Human Genetics. 2015;79:188–198. doi: 10.1111/ahg.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyton M, Vezina P. Dopamine ups and downs in vulnerability to addictions: a neurodevelopmental model. Trends in pharmacological sciences. 2014;35:268–276. doi: 10.1016/j.tips.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Church GM. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nature neuroscience. 2013;16:1518–1522. doi: 10.1038/nn.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Li Y, Cui SC, Qi Y, Zhao JJ, Liu XY, Xu P, Chen XH. Splicing factor transformer-2beta (Tra2beta) regulates the expression of regulator of G protein signaling 4 (RGS4) gene and is induced by morphine. PloS one. 2013;8:e72220. doi: 10.1371/journal.pone.0072220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes & development. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gervasi C, Szaro BG. A crucial role for hnRNP K in axon development in Xenopus laevis. Development (Cambridge, England) 2008;135:3125–3135. doi: 10.1242/dev.022236. [DOI] [PubMed] [Google Scholar]
- Liu Y, Szaro BG. hnRNP K post-transcriptionally co-regulates multiple cytoskeletal genes needed for axonogenesis. Development (Cambridge, England) 2011;138:3079–3090. doi: 10.1242/dev.066993. [DOI] [PubMed] [Google Scholar]
- Maloney SE, Khangura E, Dougherty JD. The RNA-binding protein Celf6 is highly expressed in diencephalic nuclei and neuromodulatory cell populations of the mouse brain. Brain structure & function. 2015 doi: 10.1007/s00429-015-1005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger DM, Lin C, Garcia-Blanco MA. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Molecular and cellular biology. 2008;28:5403–5419. doi: 10.1128/MCB.00739-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalon A, Sidorov M, Ballard TM, Ozmen L, Spooren W, Wettstein JG, Jaeschke G, Bear MF, Lindemann L. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74:49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA (New York, N.Y.) 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E–BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. The Journal of biological chemistry. 2008;283:18478–18482. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, Self DW. DeltaFosB: a sustained molecular switch for addiction. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11042–11046. doi: 10.1073/pnas.191352698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AD, Gao S, Norris ML, Ray D, Ramani AK, Fraser AG, Morris Q, Hughes TR, Zhen M, Calarco JA. A pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Molecular cell. 2014;54:946–959. doi: 10.1016/j.molcel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Huttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Molecular and cellular biology. 2007;27:8340–8351. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Iaccarino C, Lee J, Kwon I, Baik SM, Kim M, Seong JY, Son GH, Borrelli E, Kim K. Regulatory roles of heterogeneous nuclear ribonucleoprotein M and Nova-1 protein in alternative splicing of dopamine D2 receptor pre-mRNA. The Journal of biological chemistry. 2011;286:25301–25308. doi: 10.1074/jbc.M110.206540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale A, Amadio M, Caffino L, Racagni G, Govoni S, Fumagalli F. ELAV-GAP43 pathway activation following combined exposure to cocaine and stress. Psychopharmacology. 2011;218:249–256. doi: 10.1007/s00213-010-2152-9. [DOI] [PubMed] [Google Scholar]
- Perrone-Bizzozero N, Bolognani F. Role of HuD and other RNA-binding proteins in neural development and plasticity. Journal of neuroscience research. 2002;68:121–126. doi: 10.1002/jnr.10175. [DOI] [PubMed] [Google Scholar]
- Perycz M, Urbanska AS, Krawczyk PS, Parobczak K, Jaworski J. Zipcode binding protein 1 regulates the development of dendritic arbors in hippocampal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:5271–5285. doi: 10.1523/JNEUROSCI.2387-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picetti R, Saiardi A, Abdel Samad T, Bozzi Y, Baik JH, Borrelli E. Dopamine D2 receptors in signal transduction and behavior. Critical reviews in neurobiology. 1997;11:121–142. doi: 10.1615/critrevneurobiol.v11.i2-3.20. [DOI] [PubMed] [Google Scholar]
- Pilaz LJ, Silver DL. Post-transcriptional regulation in corticogenesis: how RNA-binding proteins help build the brain. Wiley interdisciplinary reviews.RNA. 2015;6:501–515. doi: 10.1002/wrna.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratti A, Fallini C, Colombrita C, Pascale A, Laforenza U, Quattrone A, Silani V. Post-transcriptional regulation of neuro-oncological ventral antigen 1 by the neuronal RNA-binding proteins ELAV. The Journal of biological chemistry. 2008;283:7531–7541. doi: 10.1074/jbc.M706082200. [DOI] [PubMed] [Google Scholar]
- Robinow S, Campos AR, Yao KM, White K. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science (New York, N.Y.) 1988;242:1570–1572. doi: 10.1126/science.3144044. [DOI] [PubMed] [Google Scholar]
- Romanelli MG, Diani E, Lievens PM. New insights into functional roles of the polypyrimidine tract-binding protein. International journal of molecular sciences. 2013;14:22906–22932. doi: 10.3390/ijms141122906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M, Buratti E. Targeting RNA binding proteins involved in neurodegeneration. Journal of biomolecular screening. 2013;18:967–983. doi: 10.1177/1087057113497256. [DOI] [PubMed] [Google Scholar]
- Rosenthal JJ. The emerging role of RNA editing in plasticity. The Journal of experimental biology. 2015;218:1812–1821. doi: 10.1242/jeb.119065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Molecular and cellular biology. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SJ, Mazei-Robison MS, Ables JL, Nestler EJ. Neurotrophic factors and structural plasticity in addiction. Neuropharmacolog. 2009;(56 Suppl 1):73–82. doi: 10.1016/j.neuropharm.2008.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe T, Futai E, Ishiura S. Polypyrimidine tract-binding protein 1 regulates the alternative splicing of dopamine receptor D2. Journal of neurochemistry. 2011;116:76–81. doi: 10.1111/j.1471-4159.2010.07086.x. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Welshhans K, Wen Z, Yao J, Xu M, Goshima Y, Zheng JQ, Bassell GJ. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local beta-actin synthesis and growth cone turning. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:9349–9358. doi: 10.1523/JNEUROSCI.0499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterlee JS, Basanta-Sanchez M, Blanco S, Li JB, Meyer K, Pollock J, Sadri-Vakili G, Rybak-Wolf A. Novel RNA modifications in the nervous system: form and function. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:15170–15177. doi: 10.1523/JNEUROSCI.3236-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochemical Society transactions. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- Schmidt HD, McFarland KN, Darnell SB, Huizenga MN, Sangrey GR, Cha JH, Pierce RC, Sadri-Vakili G. ADAR2-dependent GluA2 editing regulates cocaine seeking. Molecular psychiatry. 2014 doi: 10.1038/mp.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Kesterson RA, Jacobs MM, Joers JM, Gore JC, Emeson RB. Hyperphagia-mediated obesity in transgenic mice misexpressing the RNA-editing enzyme ADAR2. The Journal of biological chemistry. 2007;282:22448–22459. doi: 10.1074/jbc.M700265200. [DOI] [PubMed] [Google Scholar]
- Singh M, Singh MM, Na E, Agassandian K, Zimmerman MB, Johnson AK. Altered ADAR 2 equilibrium and 5HT(2C) R editing in the prefrontal cortex of ADAR 2 transgenic mice. Genes, brain, and behavior. 2011;10:637–647. doi: 10.1111/j.1601-183X.2011.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R. New findings on biological factors predicting addiction relapse vulnerability. Current psychiatry reports. 2011;13:398–405. doi: 10.1007/s11920-011-0224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnamon JR, Czaplinski K. mRNA trafficking and local translation: the Yin and Yang of regulating mRNA localization in neurons. Acta biochimica et biophysica Sinica. 2011;43:663–670. doi: 10.1093/abbs/gmr058. [DOI] [PubMed] [Google Scholar]
- Sinnamon JR, Waddell CB, Nik S, Chen EI, Czaplinski K. Hnrpab regulates neural development and neuron cell survival after glutamate stimulation. RNA (New York, N.Y.) 2012;18:704–719. doi: 10.1261/rna.030742.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin W, Nishikura K. Adenosine-to-inosine RNA editing and human disease. Genome medicine. 2013;5:105. doi: 10.1186/gm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JW, Fetsko LA, Xu R, Wang Y. Dopamine D2L receptor knockout mice display deficits in positive and negative reinforcing properties of morphine and in avoidance learning. Neuroscience. 2002;113:755–765. doi: 10.1016/s0306-4522(02)00257-9. [DOI] [PubMed] [Google Scholar]
- Smith LN, Jedynak JP, Fontenot MR, Hale CF, Dietz KC, Taniguchi M, Thomas FS, Zirlin BC, Birnbaum SG, Huber KM, Thomas MJ, Cowan CW. Fragile X mental retardation protein regulates synaptic and behavioral plasticity to repeated cocaine administration. Neuron. 2014;82:645–658. doi: 10.1016/j.neuron.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KY, Choi HS, Law PY, Wei LN, Loh HH. Post-transcriptional regulation of mu-opioid receptor: role of the RNA-binding proteins heterogeneous nuclear ribonucleoprotein H1 and F. Cellular and molecular life sciences : CMLS. 2012;69:599–610. doi: 10.1007/s00018-011-0761-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sora I, Hall FS, Andrews AM, Itokawa M, Li XF, Wei HB, Wichems C, Lesch KP, Murphy DL, Uhl GR. Molecular mechanisms of cocaine reward: combined dopamine and serotonin transporter knockouts eliminate cocaine place preference. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5300–5305. doi: 10.1073/pnas.091039298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, McGlincy N, Grellscheid SN, Southby J, Wollerton M, Smith CW. Regulation of alternative splicing by PTB and associated factors. Biochemical Society transactions. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Fregoso O, Krainer AR. Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA (New York, N.Y.) 2012;18:274–283. doi: 10.1261/rna.030486.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumlinski KK, Dehoff MH, Kang SH, Frys KA, Lominac KD, Klugmann M, Rohrer J, Griffin W, 3rd, Toda S, Champtiaux NP, Berry T, Tu JC, Shealy SE, During MJ, Middaugh LD, Worley PF, Kalivas PW. Homer proteins regulate sensitivity to cocaine. Neuron. 2004;43:401–413. doi: 10.1016/j.neuron.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Tang YH, Han SP, Kassahn KS, Skarshewski A, Rothnagel JA, Smith R. Complex evolutionary relationships among four classes of modular RNA-binding splicing regulators in eukaryotes: the hnRNP, SR, ELAV-like and CELF proteins. Journal of Molecular Evolution. 2012;75:214–228. doi: 10.1007/s00239-012-9533-0. [DOI] [PubMed] [Google Scholar]
- Thomas MG, Pascual ML, Maschi D, Luchelli L, Boccaccio GL. Synaptic control of local translation: the plot thickens with new characters. Cellular and molecular life sciences : CMLS. 2014;71:2219–2239. doi: 10.1007/s00018-013-1506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan A, Szaro BG. Phylogenetically conserved binding of specific K homology domain proteins to the 3'-untranslated region of the vertebrate middle neurofilament mRNA. The Journal of biological chemistry. 2004;279:49680–49688. doi: 10.1074/jbc.M408915200. [DOI] [PubMed] [Google Scholar]
- Till SM. The developmental roles of FMRP. Biochemical Society transactions. 2010;38:507–510. doi: 10.1042/BST0380507. [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Caron MG, Keene JD. Activity-dependent expression of ELAV/Hu RBPs and neuronal mRNAs in seizure and cocaine brain. Journal of neurochemistry. 2008;107:1529–1543. doi: 10.1111/j.1471-4159.2008.05718.x. [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolino M, Kohrmann M, Kiebler MA. RNA-binding proteins involved in RNA localization and their implications in neuronal diseases. The European journal of neuroscience. 2012;35:1818–1836. doi: 10.1111/j.1460-9568.2012.08160.x. [DOI] [PubMed] [Google Scholar]
- Traynor J. Regulator of G protein-signaling proteins and addictive drugs. Annals of the New York Academy of Sciences. 2010;1187:341–352. doi: 10.1111/j.1749-6632.2009.05150.x. [DOI] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, Zeeberg BR, Kane D, Weinstein JN, Blume J, Darnell RB. Nova regulates brain-specific splicing to shape the synapse. Nature genetics. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Molecular and cellular biology. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dusen CM, Yee L, McNally LM, McNally MT. A glycine-rich domain of hnRNP H/F promotes nucleocytoplasmic shuttling and nuclear import through an interaction with transportin 1. Molecular and cellular biology. 2010;30:2552–2562. doi: 10.1128/MCB.00230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loo KM, Martens GJ. Genetic and environmental factors in complex neurodevelopmental disorders. Current Genomics. 2007;8:429–444. doi: 10.2174/138920207783591717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanevski F, Xu B. HuD interacts with Bdnf mRNA and is essential for activity-induced BDNF synthesis in dendrites. PloS one. 2015;10:e0117264. doi: 10.1371/journal.pone.0117264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volman SF, Lammel S, Margolis EB, Kim Y, Richard JM, Roitman MF, Lobo MK. New insights into the specificity and plasticity of reward and aversion encoding in the mesolimbic system. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:17569–17576. doi: 10.1523/JNEUROSCI.3250-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Briese M, Sun W, Mahaffey CL, Curk T, Rot G, Ule J, Frankel WN. CELF4 regulates translation and local abundance of a vast set of mRNAs, including genes associated with regulation of synaptic function. PLoS genetics. 2012;8:e1003067. doi: 10.1371/journal.pgen.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Mahaffey CL, Sun W, Yang Y, Chao HT, Frankel WN. Etiology of a genetically complex seizure disorder in Celf4 mutant mice. Genes, brain, and behavior. 2011;10:765–777. doi: 10.1111/j.1601-183X.2011.00717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Kuyumcu-Martinez MN, Sarma S, Mathur N, Wehrens XH, Cooper TA. PKC inhibition ameliorates the cardiac phenotype in a mouse model of myotonic dystrophy type 1. The Journal of clinical investigation. 2009;119:3797–3806. doi: 10.1172/JCI37976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kim SS, Zhuo M. Roles of fragile X mental retardation protein in dopaminergic stimulation-induced synapse-associated protein synthesis and subsequent alpha-amino-3-hydroxyl-5-methyl-4-isoxazole-4-propionate (AMPA) receptor internalization. The Journal of biological chemistry. 2010;285:21888–21901. doi: 10.1074/jbc.M110.116293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wu LJ, Kim SS, Lee FJ, Gong B, Toyoda H, Ren M, Shang YZ, Xu H, Liu F, Zhao MG, Zhuo M. FMRP acts as a key messenger for dopamine modulation in the forebrain. Neuron. 2008;59:634–647. doi: 10.1016/j.neuron.2008.06.027. [DOI] [PubMed] [Google Scholar]
- Wang J, Yuan W, Li MD. Genes and pathways co-associated with the exposure to multiple drugs of abuse, including alcohol, amphetamine/methamphetamine, cocaine, marijuana, morphine, and/or nicotine: a review of proteomics analyses. Molecular neurobiology. 2011;44:269–286. doi: 10.1007/s12035-011-8202-4. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ma M, Xiao X, Wang Z. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nature structural & molecular biology. 2012;19:1044–1052. doi: 10.1038/nsmb.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Yoshimoto K, Tatebe H, Kita M, Nishikura K, Kimura M, Tanaka M. Enhancement of alcohol drinking in mice depends on alterations in RNA editing of serotonin 2C receptors. The international journal of neuropsychopharmacology / official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2014;17:739–751. doi: 10.1017/S1461145713001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshhans K, Bassell GJ. Netrin-1-induced local beta-actin synthesis and growth cone guidance requires zipcode binding protein 1. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:9800–9813. doi: 10.1523/JNEUROSCI.0166-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic acids research. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wermter AK, Laucht M, Schimmelmann BG, Banaschewski T, Sonuga-Barke EJ, Rietschel M, Becker K. From nature versus nurture, via nature and nurture, to gene x environment interaction in mental disorders. European child & adolescent psychiatry. 2010;19:199–210. doi: 10.1007/s00787-009-0082-z. [DOI] [PubMed] [Google Scholar]
- Werry TD, Loiacono R, Sexton PM, Christopoulos A. RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacology & therapeutics. 2008;119:7–23. doi: 10.1016/j.pharmthera.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, Krainer AR, Darnell RB, Zhang C. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell reports. 2014;6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Lee JA, Kress TL, Mowry KL, Black DL. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lu Z, Xu M, Pan L, Deng Y, Xie X, Liu H, Ding S, Hurd YL, Pasternak GW, Klein RJ, Cartegni L, Zhou W, Pan YX. A heroin addiction severity-associated intronic single nucleotide polymorphism modulates alternative pre-mRNA splicing of the mu opioid receptor gene OPRM1 via hnRNPH interactions. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:11048–11066. doi: 10.1523/JNEUROSCI.3986-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Mahaffey CL, Berube N, Maddatu TP, Cox GA, Frankel WN. Complex seizure disorder caused by Brunol4 deficiency in mice. PLoS genetics. 2007;3:e124. doi: 10.1371/journal.pgen.0030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Okano HJ, Okano H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. The Journal of biological chemistry. 2005;280:12690–12699. doi: 10.1074/jbc.M411119200. [DOI] [PubMed] [Google Scholar]
- Yazdani N, Parker CC, Shen Y, Guido MA, Kole LA, Kirkpatrick SL, Lim JE, Sokoloff G, Cheng R, Johnson WE, Palmer AA, Bryant CD. Hnrnph1 is a quantitative trait gene for methamphetamine sensitivity. Uppsala, Sweden: International Behavioural and Neural Genetics Society; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nature structural & molecular biology. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Kim HH, Kim P, Donnelly CJ, Kalinski AL, Vuppalanchi D, Park M, Lee SJ, Merianda TT, Perrone-Bizzozero NI, Twiss JL. A HuD-ZBP1 ribonucleoprotein complex localizes GAP-43 mRNA into axons through its 3' untranslated region AU-rich regulatory element. Journal of neurochemistry. 2013;126:792–804. doi: 10.1111/jnc.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, McKenna MC, Rollins DA, Song M, Nuriel T, Gross SS, Xu G, Glatt CE. Anxiety-associated alternative polyadenylation of the serotonin transporter mRNA confers translational regulation by hnRNPK. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11624–11629. doi: 10.1073/pnas.1301485110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaniewska M, Alenina N, Wydra K, Frohler S, Kusmider M, McCreary AC, Chen W, Bader M, Filip M. Discovering the mechanisms underlying serotonin (5-HT)2A and 5-HT2C receptor regulation following nicotine withdrawal in rats. Journal of neurochemistry. 2015;134:704–716. doi: 10.1111/jnc.13192. [DOI] [PubMed] [Google Scholar]
- Zaniewska M, McCreary AC, Wydra K, Filip M. Effects of serotonin (5-HT)2 receptor ligands on depression-like behavior during nicotine withdrawal. Neuropharmacology. 2010;58:1140–1146. doi: 10.1016/j.neuropharm.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes & development. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Neubert TA, Jordan BA. RNA binding proteins accumulate at the postsynaptic density with synaptic activity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:599–609. doi: 10.1523/JNEUROSCI.2463-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- Zheng S, Gray EE, Chawla G, Porse BT, O'Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nature neuroscience. 2012;15:381–388. doi: 10.1038/nn.3026. S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Drgonova J, Li CY, Uhl GR. Human cell adhesion molecules: annotated functional subtypes and overrepresentation of addiction-associated genes. Annals of the New York Academy of Sciences. 2015 doi: 10.1111/nyas.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]