Abstract
Dengue virus (DENV) (Flavivirus, Flaviviridae) is a reemerging arthropod-borne virus with a worldwide circulation, transmitted mainly by Aedes aegypti and Aedes albopictus mosquitoes. Since the first detection of its main transmitting vector in 1992 and the invasion of DENV-1 in 1993, Costa Rica has faced dengue outbreaks yearly. In 2007 and 2013, Costa Rica experienced two of the largest outbreaks in terms of total and severe cases. To provide genetic information about the etiologic agents producing these outbreaks, we conducted phylogenetic analysis of viruses isolated from human samples. A total of 23 DENV-1 and DENV-2 sequences were characterized. These analyses signaled that DENV-1 genotype V and DENV-2 American/Asian genotype were circulating in those outbreaks. Our results suggest that the 2007 and 2013 outbreak viral strains of DENV-1 and DENV-2 originated from nearby countries and underwent in situ microevolution.
Dengue is the most important emergent and reemergent arboviral disease in tropical and subtropical regions. Dengue virus (DENV) is a member of the genus Flavivirus of the family Flaviviridae and has four antigenically distinct serotypes (DENV 1–4). Each serotype is further classified into four or five genotypes on the basis of their genetic diversity.1 Clinical presentations range from asymptomatic, undifferentiated fever, and “classic” dengue to severe forms, such as hemorrhagic fever and shock syndrome.2, ref-type="bibr">3 Given the geographical distribution of its main transmitting vector Aedes aegypti, more than half of the world population is at risk of infection. It is estimated that nearly 400 million dengue infections and 500,000 severe dengue cases occur worldwide annually.4 Interestingly, some genotypes and certain lineages within genotypes have been more frequently associated with severe disease outcomes.5,6 Therefore, to better understand dengue outbreaks in hyperendemic regions, it is important to thoroughly describe the genetic diversity of circulating serotypes.
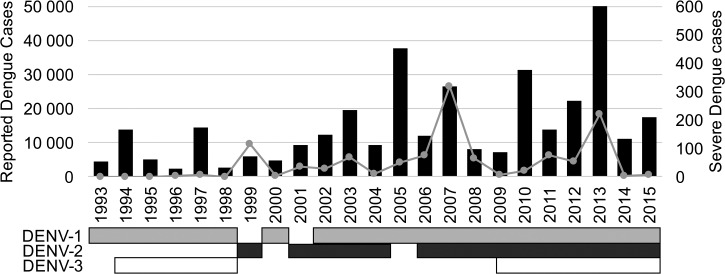
The first dengue outbreak reported in Costa Rica occurred in 1993 with DENV-1 as the only circulating serotype.7 Since then, dengue cases have been documented annually—a consequence of the endemicity of Aedes vectors that sustain local transmission.8 Epidemiological and laboratory-based surveillance suggests a pattern of serotype disappear and reintroduction during the first decade after introduction.9 In addition, sustained co-circulation of three serotypes (DENV-1, DENV-2, and DENV-3) has been reported in later outbreaks, making Costa Rica a hyperendemic country.9 Although dengue infections are always present in Costa Rica, peaks in transmission events are observed every 2–5 years, similar to other countries in the region.10 The largest dengue outbreak in Costa Rica occurred in 2013, with almost 50,000 cases suspected on the basis of laboratory testing and/or clinical symptoms (taken together with epidemiological link to previous laboratory-confirmed cases from a particular area) (Figure 1). Before 2013, there was a major outbreak in 2007. Although it was apparently smaller (26,000 total reported cases), more severe dengue cases (318 severe cases and eight deaths) was observed. The latter were classified using the National Ministry of Health criteria (based on existing World Health Organization (WHO) criteria in 2007 and then the new 2009 WHO criteria for the 2013). Despite the persistent prevalence and impact of dengue in Costa Rica for almost 18 years, there is little knowledge about DENV genetics in Costa Rica. Here, we present the molecular and phylogenetic characterization of dengue strains recovered from human samples collected in the 2007 and 2013 major outbreaks in Costa Rica.
Figure 1.
Reported dengue cases (black bars), severe dengue cases (continuous gray line), and circulating serotypes per year in Costa Rica. These results were retrieved from epidemiological and laboratory-based surveillance done by the Ministry of Health.11
To understand the genetic profile and possible origin of circulating strains, we conducted analyses of the E protein gene sequence. For this, whole blood samples were taken from patients for virus isolation no more than 6 days after fever onset. This study was approved by the Scientific Ethics Committee of the University of Costa Rica (VI-5305-2015). All patient identifiers were removed from blood samples before analysis in the laboratory. Samples were inoculated on to C6/36 cells and kept at 32–33°C until cytopathic effect was apparent. Dengue isolates with a maximum of two passages were obtained and analyzed from patients of the 2013 outbreak. Isolates from the 2007 outbreak with less than four passages were generously donated by the National Reference Center of Virology in the Costa Rican Institute of Research and Teaching in Nutrition and Health. After reverse transcription polymerase chain reaction confirmation of dengue serotype, the aforementioned clinical isolates (passage two or four, respectively from 2013 or 2017 outbreaks) were expanded and kept frozen at −70°C for other experiments. A total of 23 DENV-1 and DENV-2 isolates were characterized in this molecular study.
Viral RNA was extracted using TRIzol Reagent® (Invitrogen, Waltham, MA) from culture supernatants. Complementary DNA (cDNA) was synthesized using the RevertAid™ H Minus (ThermoFisher Scientific, Waltham, MA) following the manufacturer's instructions with random primers. A seminested PCR was performed using TopTaq Master Mix (Qiagen, Hilden, Germany) and specific primers as described elsewhere.12 In brief, PCR was implemented using cDNA and D1 and D2 primers, amplifying a product of 511 bp corresponding to a fragment of the capsid and premembrane (C/prM) genes. After serotype identification, cDNA segments between 2,474 and 2,577 nucleotides encompassing the prM and E genes of DENV were amplified by PCR using the consensus primer D1 and serotype-specific reverse primers as described elsewhere.13 PCR products were then purified using Exonuclease I and FastAP Thermosensitive Alkaline Phosphatase treatment (Thermo Fisher Scientific, Waltham, MA). Both strands of the amplicons were sequenced using Sanger sequencing (Macrogen Inc., Seoul, South Korea).
Previously described E protein gene sequence of geographically related strains of DENV-1 and DENV-2 were obtained from GenBank for analyses. Multiple sequence alignments were performed using Multiple Alignment using Fast Fourier Transform in Geneious v8.1.7 (www.geneious.com). Phylogenetic trees were assembled using the maximum likelihood statistical method based on GTR+G+I and GTR+I for DENV-1 and DENV-2, respectively. The identification of the best nucleotide substitution model and the construction of phylogenetic trees were performed using MEGA v6.0 (www.megasoftware.com). The robustness of the resulting tree was established by bootstrap analysis with 1,000 replications.
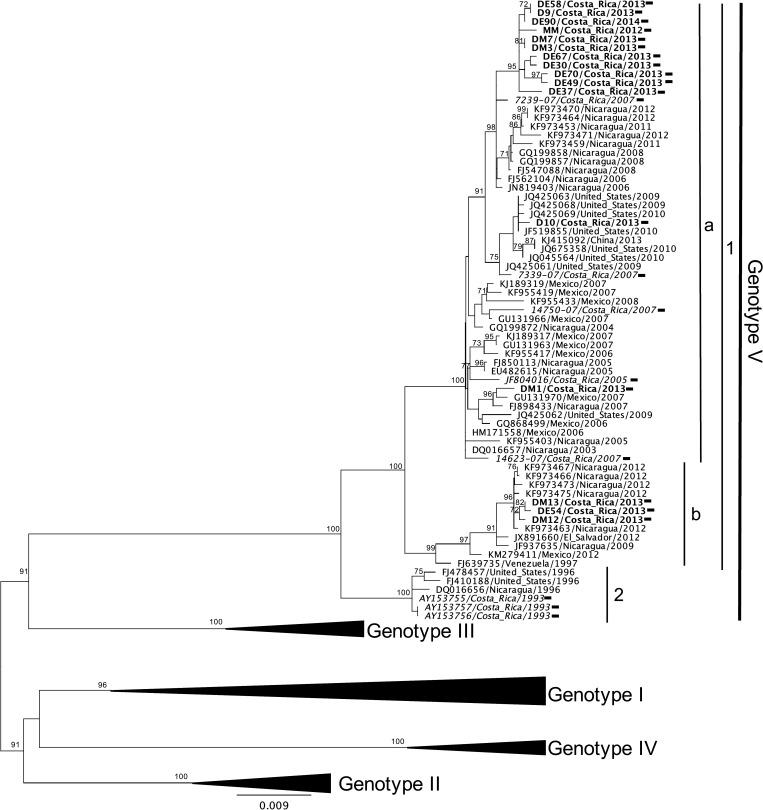
A total of 20 isolates of DENV-1 were successfully recovered from both outbreaks. The phylogenetic tree retrieved from the full E protein sequence of DENV-1 is shown in Figure 2. A total of 91 DENV-1 sequences were used for this analysis: 71 from GenBank and 20 new sequences from this study (Supplemental Table 1). As shown in Figure 2, isolates from Costa Rica are divided into two clades (1 and 2) corresponding solely to genotype V. This genotype comprises the majority of American and Caribbean strains.14 DENV-1 strains from 2013 outbreak (shown in bold) clustered in one clade, is further subdivided into two subclades (a and b). This suggests that the circulating viruses during this outbreak were from at least two distinct sources. All sequences from 2013 clustered together with strains from neighboring countries, especially from Nicaragua. Sequences from 2005 and 2007 grouped in one of these subclades (a), suggesting that some of the 2013 circulating virus originated from local transmission and in situ microevolution. The sequences of the other subclade (b) were more closely related to 2012 sequences reported from Nicaragua. This implies that some of the 2013 outbreak strains from Costa Rica most likely originated from virus circulating in neighboring Nicaragua a year before. Furthermore, the phylogenetic tree displays a pattern that suggests a replacement of the 1993 circulating clade of DENV-1 (clade 2). Lineage extinction and replacement is a well-described phenomenon in DENV evolution at local levels. ref-type="bibr">15 Nevertheless, a larger number of isolates would need to be analyzed to confirm the extinction of this clade.
Figure 2.
Maximum likelihood tree of 91 DENV-1 E gene sequences (1,485 bp) including 24 Costa Rican strains (black rectangle), 20 obtained from this study (bold font), and four obtained from GenBank (italic font). Significant bootstrap values (≥ 70) are indicated at the respecting nodes. The sequences were named according to reference number/country/year of collection or isolation. Major clades and subclades comprising Costa Rican strains are indicated with numbers and letters, respectively.
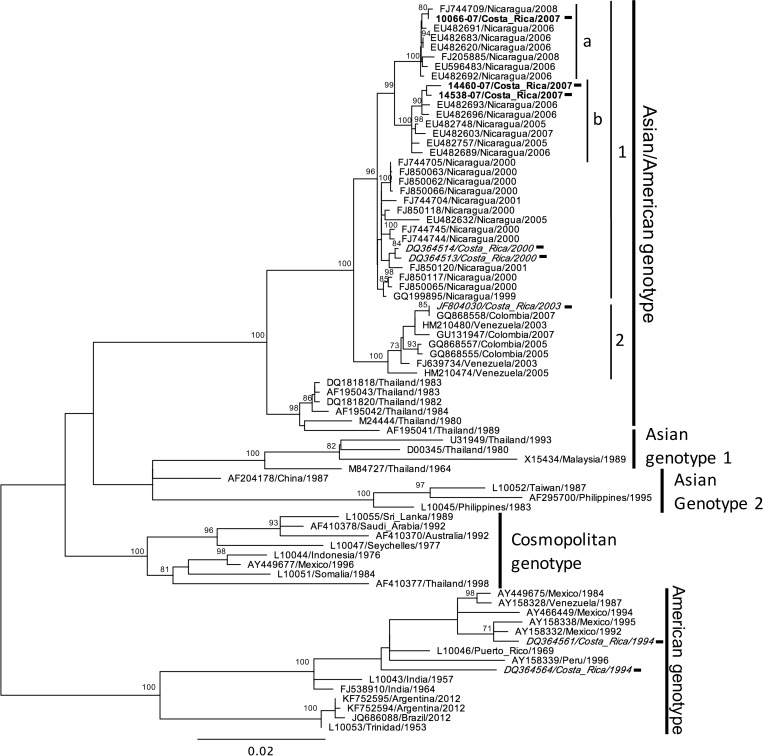
DENV-2 was not isolated from among the 2013 samples included in this study, so for the DENV-2 analysis, only the 2007 sequences generated in this study were included in the final dataset of 76 DENV-2 E protein gene sequences (73 from GenBank, three from 2007 viral strains) (Supplemental Table 2). The resulting phylogenetic tree is shown in Figure 3. The three Costa Rican sequences from 2007 (shown in bold) were grouped in the American/Asian genotype, the principal circulating DENV-2 genotype in this region.16 The 2007 isolates fell into two subclades (a and b), indicating co-circulation of at least two lineages. As seen for some DENV-1 strains of the 2013 outbreak, DENV-2 sequences from 2007 seem to be closely related to viruses reported from Nicaragua a year before that were associated with more severe cases.17 While it is tempting to speculate that this strain might be more virulent and thus explain the higher number of severe cases reported in 2007 in Costa Rica, studies of the clinical outcome and immunological backgrounds of patient would be necessary to confirm such a hypothesis. Finally two sequences belonging to the American genotype were reported from Costa Rica in 1994.18 It is plausible that the strains of this genotype did not maintain their circulation in the country, since DENV-2 was not detected by laboratory-based surveillance until 1999 (Figure 1).19
Figure 3.
Maximum likelihood tree of 76 DENV-2 E gene sequences (1,485 bp) including eight Costa Rican strains (black rectangle), three obtained from this study (bold font), and five obtained from GenBank (italic font). Significant bootstrap values (≥ 70) are indicated at the respecting nodes. The sequences were named according to reference number/country/year of collection or isolation. Major clades and subclades comprising Costa Rican strains are indicated with numbers and letters, respectively.
This study revealed that the circulating genotypes of the 2007 and 2013 outbreaks in Costa Rica were closely related to other DENVs found in the region. A larger number of dengue isolates and further analysis considering viral genetics, herd immunity, and geographical distribution are needed to thoroughly understand dengue dynamics in Costa Rica. Nevertheless, continuous monitoring of circulating strains is crucial to detect the appearance of newer strains and their role in dengue epidemiology in Costa Rica. The social and commercial activity between countries of Central America and their proximity means that exchange of virus strains is inevitable. The present study therefore complements previous dengue molecular epidemiology research by providing a more complete picture of dengue dynamics in Central America. This can inform implementation of control measures and interventions to prevent impeding epidemics. In addition, information on DENV molecular diversity is important for the establishment of immunoprophylactic approaches to control the disease in the region.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elizabeth Saenz, head of the National Reference Center of Virology in National Reference Center of Virology in the Costa Rican Institute of Research and Teaching in Nutrition and Health (INCIENSA), for providing the 2007 outbreak isolates. We also thank Francisco Vega and Carlos Vargas for valuable technical assistance and Jane Thompson for English proofreading services. We are infinitely grateful to Christine Carrington from the University of West Indies, Trinidad and Tobago for critical reading of the manuscript.
Footnotes
Financial support: This work was supported by the University of Costa Rica Project VI-803-B2-285, FEES-CONARE VI-803-B4-656 and the Graduate Studies System (SEP) of the University of Costa Rica.
Authors' addresses: Claudio Soto-Garita and Eugenia Corrales-Aguilar, Virology-CIET (Research Center for Tropical Diseases), Faculty of Microbiology, University of Costa Rica, San Jose, Costa Rica, E-mails: claudioenrique.soto@ucr.ac.cr and eugenia.corrales@ucr.ac.cr. Teresita Somogyi, Virology-CIET (Research Center for Tropical Diseases), Faculty of Microbiology, University of Costa Rica, San Jose, Costa Rica, and Molecular Diagnostics Unit, Hospital Mexico, Caja Costarricense del Seguro Social, San José, Costa Rica, E-mail: tsomogyi@ccss.sa.cr. Amanda Vicente-Santos, Virology-CIET (Research Center for Tropical Diseases), Faculty of Microbiology, University of Costa Rica, San Jose, Costa Rica, and Conservation Genetics, School of Biology, University of Costa Rica, San José, Costa Rica, E-mail: amandavic@gmail.com.
References
- 1.Weaver SC, Vasilakis N. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol. 2009;9:523–540. doi: 10.1016/j.meegid.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilder-Smith A, Ooi EE, Vasudevan SG, Gubler DJ. Update on dengue: epidemiology, virus evolution, antiviral drugs, and vaccine development. Curr Infect Dis Rep. 2010;12:157–164. doi: 10.1007/s11908-010-0102-7. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization and Special Programme for Research and Training in Tropical Diseases World Health Organization and Special Programme for Research and Training in Tropical Diseases. Dengue: Guidelines for Diagnosis, Treatment, Prevention, and Control. new edition. Geneva, Switzerland: 2009. doi:WHO/HTM/NTD/DEN/2009.1. [Google Scholar]
- 4.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yung CF, Lee KS, Thein TL, Tan LK, Gan VC, Wong JGX, Lye DC, Ng LC, Leo YS. Dengue serotype-specific differences in clinical manifestation, laboratory parameters and risk of severe disease in adults, Singapore. Am J Trop Med Hyg. 2015;92:999–1005. doi: 10.4269/ajtmh.14-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315–341. doi: 10.1016/s0065-3527(03)59009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerdas-Quesada CA, Retana-Salazar AP. Characterization and phylogenetic relationships of strains of dengue type 1 virus from Costa Rica. Rev Biol Trop. 2007;55:365–372. doi: 10.15517/rbt.v55i2.6016. [DOI] [PubMed] [Google Scholar]
- 8.Mena N, Troyo A, Bonilla-Carrión R, Rica C. Factores asociados con la incidencia de dengue en Costa Rica. Rev Panam Salud Publica. 2011;29:234–242. doi: 10.1590/s1020-49892011000400004. [DOI] [PubMed] [Google Scholar]
- 9.Iturrino-Monge R, Avila-Agüero ML, Avila-Agüero CR, Moya-Moya T, Cañas-Coto A, Camacho-Badilla K, Zambrano-Mora B. Seroprevalence of dengue virus antibodies in asymptomatic Costa Rican children, 2002–2003: a pilot study. Rev Panam Salud Publica. 2006;20:39–43. doi: 10.1590/s1020-49892006000700005. [DOI] [PubMed] [Google Scholar]
- 10.Bennett SN, Drummond J, Kapan DD, Suchard MA, Munoz-Jordán JL, Pybus OG, Holmes EC, Gubler DJ. Epidemic dynamics revealed in dengue evolution. Mol Biol Evol. 2010;27:811–818. doi: 10.1093/molbev/msp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ministerio de Salud de Costa Rica Análisis De Situación De Salud: Dengue 2015. 2015. http://www.ministeriodesalud.go.cr/index.php/vigilancia-de-la-salud/analisis-de-situacion-de-salud Available at. Accessed October 25, 2015.
- 12.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Díaz FJ, Black WC, Farfán-Ale JA, Loroño-Pino MA, Olson KE, Beaty BJ. Dengue virus circulation and evolution in Mexico: a phylogenetic perspective. Arch Med Res. 2006;37:760–773. doi: 10.1016/j.arcmed.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Villabona-Arenas CJ, Zanotto PM de A. Worldwide spread of dengue virus type 1. PLoS One. 2013;8:e62649. doi: 10.1371/journal.pone.0062649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrillo-Valenzo E, Danis-Lozano R, Velasco-Hernandez JX, Sanchez-Burgos G, Alpuche C, Lopez I, Rosales C, Baronti C, de Lamballerie X, Holmes EC, Ramos-Castaneda J. Evolution of dengue virus in Mexico is characterized by frequent lineage replacement. Arch Virol. 2010;155:1401–1412. doi: 10.1007/s00705-010-0721-1. [DOI] [PubMed] [Google Scholar]
- 16.Añez G, Morales-Betoulle ME, Rios M. Circulation of different lineages of dengue virus type 2 in Central America, their evolutionary time-scale and selection pressure analysis. PLoS One. 2011;6:e27459. doi: 10.1371/journal.pone.0027459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.OhAinle M, Balmaseda A, Macalalad AR, Tellez Y, Zody MC, Saborio S, Nunez A, Lennon NJ, Birren BW, Gordon A, Henn MR, Harris E. Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci Transl Med. 2011;3:114ra128. doi: 10.1126/scitranslmed.3003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Gubler DJ, McMillan WO. Molecular evolution of dengue 2 virus in Puerto Rico: positive selection in the viral envelope accompanies clade reintroduction. J Gen Virol. 2006;87:885–893. doi: 10.1099/vir.0.81309-0. [DOI] [PubMed] [Google Scholar]
- 19.Choudhury MA, Lott WB, Aaskov J. Distribution of fitness in populations of dengue viruses. PLoS One. 2014;9:e107264. doi: 10.1371/journal.pone.0107264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.