Abstract
Tagging of individual proteins with genetically encoded fluorescent proteins (FPs) has been used extensively to study localization and interactions in live cells. Recent developments in single-molecule localization microscopy have enabled the dynamic visualization of individual tagged proteins inside living cells. However, tagging proteins with FPs is not without problems: formation of insoluble aggregates and inhibition of native functions of the protein are well-known issues. Previously reported artifacts manifest themselves at all expression levels of the FP-tagged proteins, making the design of control experiments relatively straightforward. Here, we describe a previously uncharacterized mislocalization artifact of Entacmaea quadricolor red fluorescent protein variants that is detectable at the single-molecule level in live Escherichia coli cells.
Main Text
Biological imaging, in particular fluorescence microscopy, has revolutionized our understanding of many biological processes. In particular, the ability to endogenously tag proteins in cells using genetic fusions of fluorescent proteins (FPs) has provided important insights into subcellular localizations and structures formed by proteins in live cells as part of normal metabolism (1). While genetic fusions of fluorescent proteins represent the most convenient strategy to image biological phenomena while approximating native conditions, they are not without problems. Besides well-known problems of aggregation and modification of function, recent reports provide evidence for other types of artifacts related to the fusion protein, substantiating the observation that fluorescent proteins are not inert reporters. Notably, it has been reported that Clp proteins in Escherichia coli formed artifactual clusters when tagged with particular FPs (2). More recently, it has been demonstrated that photoswitchable FPs can influence the localization of DNA-binding proteins in E. coli (3).
Red fluorescent proteins (RFPs) constitute an important part of the fluorescent toolkit, not only because they extend the number of available channels for imaging, but also because cellular autofluorescence tends to be lower in these channels. Dimerization of RFPs is a well-known downside, but the latest generation RFPs promise to be monomeric and possess much improved photophysical properties (4, 5). To test the suitability of these RFPs for live cell imaging in E. coli, we expressed these RFPs in the absence of any tagged protein and uncovered a mislocalization artifact of RFPs that is detectable at the single molecule level.
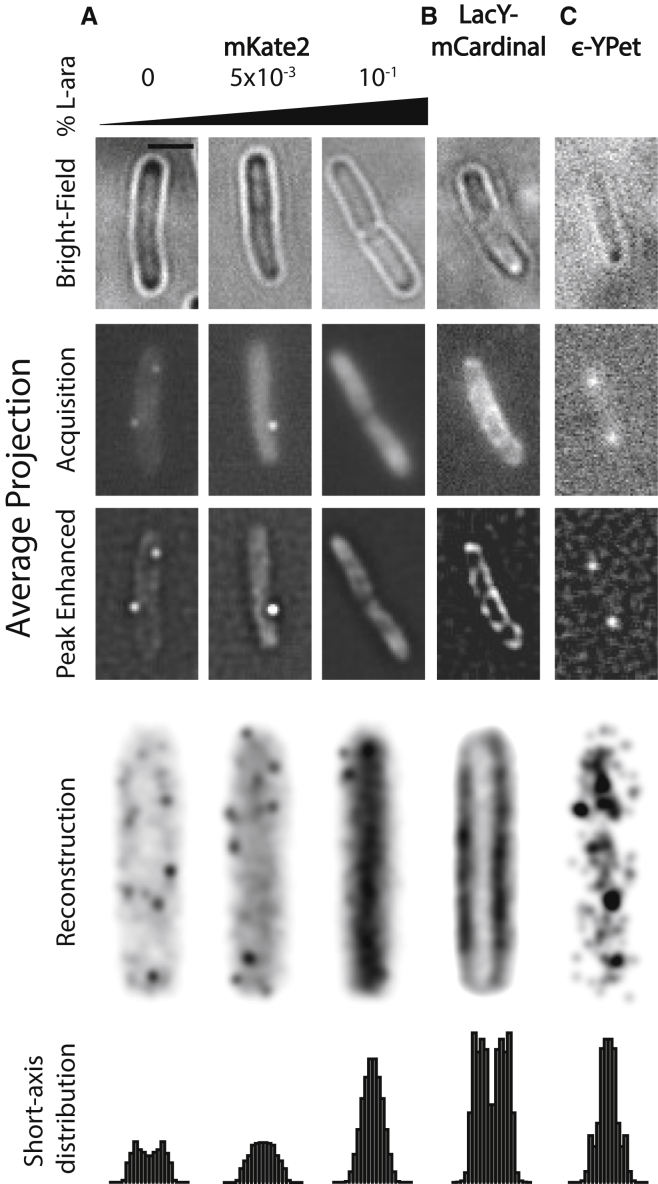
We expressed the Entacmaea quadricolor RFP derivative, mKate2 (5), from an arabinose-inducible pBAD promoter at single-molecule levels in E. coli (MG1655) cells (Fig. 1 A; see the Supporting Material). Fluorescence (and hence, copy numbers) of mKate2 could be tuned by varying the concentration of l-arabinose in the growth medium (see Fig. S1) (6). Unexpectedly, fluorescence images of individual cells revealed that mKate2 exhibits distinct, dynamic foci when expressed at very low levels. These foci localize at the cell periphery, suggestive of association with the membrane (Fig. 1 A; Movie S1). It should be noted that in the emission window corresponding to the red channel, MG1655 does exhibit cellular autofluorescence (see Fig. S2 A). However, this cellular autofluorescence does not exhibit the membrane localization observed in MG1655 expressing mKate2 (compare Fig. S2 A with Fig. S1 A). Further, when imaged and analyzed identically, a greater number of localizations are obtained in the mKate2 strain compared to the WT background (39 vs. 8 peaks/cell for the mKate2 MG1655 strain and WT MG1655, respectively) consistent with leaky expression of mKate2 from the pBAD promoter. This localization is consistent with that of the FP-tagged membrane protein LacY-mCardinal (Fig. 1 B) (4) and distinct from that of the nucleoid-associated replisome protein DnaQ-YPet (the E subunit of DNA polymerase III tagged with the YFP variant YPet; Fig. 1 C). At higher concentrations of l-arabinose (0.1%), the localization of mKate2 resembles that of a cytosolic protein (Fig. 1 A).
Figure 1.
Bright-field (top), average projections of fluorescence acquisitions (second row) and acquisitions filtered to enhance peaks (third row), reconstructed images and short-axis projections (bottom) of (A) mKate2 expressed under a pBAD promoter in the presence of varying amounts of L-arabinose: 0 (ncells = 142, npeaks = 5575), 5 × 10−3 (ncells = 137, npeaks = 8085), 10−1 (ncells = 149, npeaks = 13,283) percent; and (B) mCardinal fusion of LacY expressed under a pBAD promoter on a plasmid in fixed E. coli MG1655 (ncells = 26, npeaks = 22,954). (C) A chromosomally expressed YPet fusion of dnaQ (E subunit of polymerase III) in live E. coli MG1655 (ncells = 154, npeaks = 277). Scale bar represents 2.0 μm. Short axis distribution histograms were normalized for number of cells measured.
There are at least two possibilities for why membrane-associated foci may not be visible at higher mKate2 concentrations. The first is that at higher concentrations there is no longer sufficient contrast to distinguish individual membrane-bound mKate2 molecules as foci above the diffuse background of cytosolic molecules. A second possibility is that the distribution of mKate2 molecules between the membrane and cytosol changes as a function of mKate2 concentration, with a greater proportion of molecules bound to the membrane at low concentrations. In our previous work we demonstrated under similar conditions that we could detect a population of monomeric, membrane-bound UmuC-mKate2 molecules against a background of cytosolic mKate2 (7), indicating that our ability to detect foci is not limited by contrast in this concentration regime.
Expressed without fusion partners, YPet did not exhibit clear foci and appeared to be homogenously cytosolic (Fig. S2 B), whereas the mKate2 derivative mCardinal exhibited distinct foci at low expression levels (Fig. S2 C). The membrane-foci artifact was not detected in MG1655 exhibiting leaky expression of mCherry under the pBAD promoter (Fig. S2 D). Together, these observations suggest that artifactual membrane localization may be specific to E. quadricolor TagRFP derivatives.
As the artifact is evident when imaging at very low expression levels, analyzing the localization of low-copy-number RFP fusions should still be possible, provided that appropriate control measurements are made. For example, in our recent work on the localization of DNA polymerase V in E. coli we observed an interaction between the fusion protein UmuC-mKate2 and the cell inner membrane (7). To demonstrate that membrane interaction is a bona fide property of UmuC, as opposed to an artifact introduced by mKate2, we used an inducible plasmid to increase the concentration of mKate2 to levels where the artifact could no longer be detected and showed that foci of UmuC-mKate2 remained visible on the membrane. As an additional confirmation, electron microscopy revealed the membrane association of wild-type UmuC in fixed cells, using antibody-conjugated gold nanoparticles.
These findings support previous work indicating that fluorescent proteins and their derivatives from different organisms possess characteristics that can differentially influence the behavior of the tagged protein. Control experiments should account for the possibility that some artifacts may manifest at certain expression levels. These findings underscore the importance of verifying observations of fluorescently tagged proteins with orthogonal techniques or multiple tags wherever possible.
Author Contributions
H.G. cloned the constructs used in this study; H.G., V.E.A.C., and A.R. performed the microscopy experiments; H.G., V.E.A.C., and C.M.P. wrote the code used to analyze data with inputs from A.R.; and A.M.v.O. and A.R. supervised the work. Article was written with input from all authors.
Acknowledgments
We thank Dr. Michael Lin for kindly providing the pBAD-mCardinal and pCDNA-mCardinal constructs that were used in this study. We thank Elizabeth Wood and Michael Cox for kindly providing the DnaQ-YPet and UmuC-mKate2 constructs.
Editor: David Piston.
Footnotes
Supporting Materials and Methods, three figures, and one movie are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30394-0.
Supporting Material
References
- 1.Stracy M., Uphoff S., Kapanidis A.N. In vivo single-molecule imaging of bacterial DNA replication, transcription, and repair. FEBS Lett. 2014;588:3585–3594. doi: 10.1016/j.febslet.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 2.Landgraf D., Okumus B., Paulsson J. Segregation of molecules at cell division reveals native protein localization. Nat. Methods. 2012;9:480–482. doi: 10.1038/nmeth.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang S., Moffitt J.R., Zhuang X. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc. Natl. Acad. Sci. USA. 2014;111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu J., Haynes R.D., Lin M.Z. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat. Methods. 2014;11:572–578. doi: 10.1038/nmeth.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shcherbo D., Murphy C.S., Chudakov D.M. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guzman L.M., Belin D., Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson A., McDonald J.P., van Oijen A.M. Regulation of mutagenic DNA polymerase V activation in space and time. PLoS Genet. 2015;11:e1005482. doi: 10.1371/journal.pgen.1005482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.