Summary
Meiosis is the key step in gametogenesis. However, the mechanism of mammalian meiosis remains poorly understood due to the lack of an in vitro model. Here, we report that retinoic acid (RA) is sufficient for inducing leptotene/zygotene spermatocytes from cultured mouse spermatogonial stem cells. Multiple genes regulated by RA were identified by RNA sequencing. RA in combination with pup Sertoli cell co-culture resulted in a higher induction efficiency of 28%. Comparisons in the transcriptomic profiles of the induced spermatogenic cells and the isolated ones revealed the progressive induction of the germ cells. Using this model, we showed that Stra8, Agpat3, Fam57a, Wdr91, and Sox30 contributed to the proliferation and meiosis initiation differentially. In conclusion, we have efficiently generated spermatocytes using an RA/pup Sertoli cell-based in vitro model and provided proof-of-concept evidence for its application in identifying genes involved in mammalian meiosis.
Key words: spermatogonial stem cells, spermatocytes, Sertoli cells, meiosis, retinoic acid
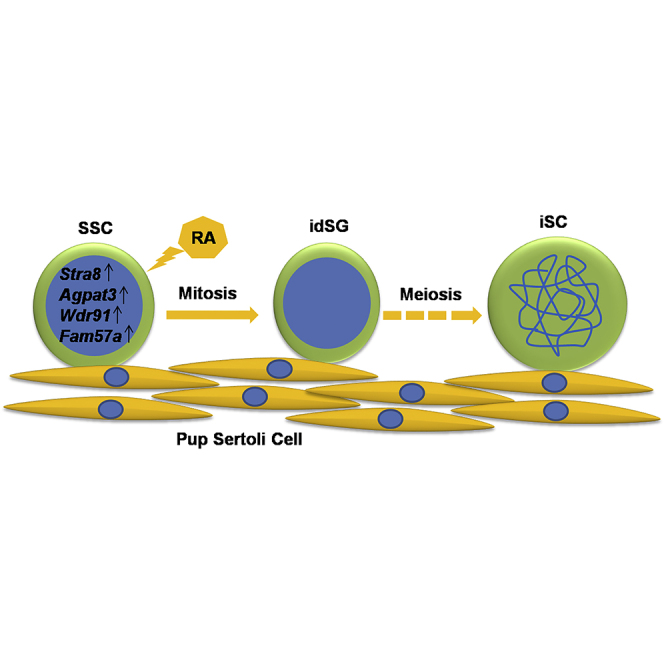
Graphical Abstract
Highlights
-
•
Retinoic acid (RA) is sufficient for the induction of spermatocytes
-
•
RA activates meiotic genes while repressing genes for SSC self-renewal
-
•
An in vitro model for meiosis based on RA and pup Sertoli cells is established
-
•
Genes for meiosis have been identified using the in vitro model
In this article, Han and colleagues show that retinoic acid (RA), which regulates a large number of genes, is sufficient for inducing spermatocytes from cultured mouse spermatogonial stem cells. RA and Sertoli cell co-culture from pup mice constitute an in vitro meiosis model of higher efficiency. The authors further show that this model can be used to identify meiotic genes.
Introduction
Meiosis is the key step of gametogenesis, which ensures the production of haploid gametes from their diploid precursors and the recombination of genetic materials from the parents. For lower eukaryotes such as yeasts, meiosis is initiated under unfavorable environmental conditions whereby the extracellular signals are integrated at the transcriptional level of master regulatory genes, which in turn activate the expression of downstream targets (van Werven and Amon, 2011). In mammals, meiosis initiation is believed to be mainly regulated by the production, storage, and metabolism of retinol and its metabolite retinoic acid (RA) (Griswold et al., 2012). RA signaling is mediated by its target genes such as Stra8, which is essential for the initiation and progression of meiosis (Anderson et al., 2008, Baltus et al., 2006, Bowles et al., 2006, Koubova et al., 2006, Mark et al., 2008). Intrinsic factors such as DAZL (deleted in azoospermia-like) prepare diploid germ cells ready for meiosis initiation when extracellular signals are received (Lin et al., 2008).
Somatic cells play important roles in mammalian meiosis initiation, partially because these cells govern the production and degradation of RA. Female germ cells initiate meiosis shortly after sex determination, while their male counterparts enter a quiescent state because their surrounding somatic cells, the Sertoli cells, express RA-metabolizing enzymes, such as CYP26b1, which effectively degrades RA before it can act upon the germ cells (Bowles et al., 2006, Koubova et al., 2006, MacLean et al., 2007). Paracrine factors such as FGF9 and intrinsic ones such as NANOS2 are also essential for meiosis inhibition and sex-specific gene expression in males (Bowles et al., 2010, Saba et al., 2013). After birth, a subpopulation of male germ cells turn into spermatogonial stem cells (SSCs), which undergo life-long self-renewal and differentiation to produce sperm.
The attempts to generate gametes in vitro can be dated back to almost a century ago. During the early phase of this long journey, the strategy of organ/tissue culture of the gonads was used and several studies reported the derivation of spermatocytes using rodent testes or human testicular biopsy (Song and Wilkinson, 2012). This strategy reached maturity when the Ogawa group reported that pup testis explants supported the derivation of functional spermatids and sperm from either autologous spermatogonia or transplanted mouse SSCs (mSSCs) (Sato et al., 2011a, Sato et al., 2011b). However, this ex vivo model is not ideal for elucidating the mechanisms of meiosis due to the complex constituents of the culture system. Another direction of the effort was to use dissociated testicular cells to derive gametes. Although several studies reported that pachytene spermatocytes or even haploid spermatogenic cells can be derived in the germ cell/Sertoli cell co-cultures, the low efficiency also limits its value for research and practical applications (Sa et al., 2008, Tres and Kierszenbaum, 1983). Moreover, none of these studies used long-term SSC cultures, which make large-scale amplification and gene modification of germ cells possible.
The third strategy for gamete generation is to induce gametes directly from pluripotent stem cells. About a decade ago, two groups showed that mouse embryonic stem cells (ESCs) could be induced to become oocytes or spermatids, respectively (Geijsen et al., 2004, Hubner et al., 2003). Several similar studies using human ESCs and induced pluripotent stem cells followed in subsequent years without functional evaluation of the induced gametes (Kee et al., 2009, Panula et al., 2010). One common problem of these studies is that the efficiency of gamete induction is unsatisfyingly low, except for one report that claimed that as much as 11% of the remaining cells after a long inducing period were haploids (West et al., 2010). Most of the other attempts to derive gametes in vitro succeeded in harvesting primordial germ cells (PGCs) but failed to generate haploid germ cells, and in many cases successful gametogenesis initiated by these induced PGCs was possible only after they were transplanted to the otherwise sterile gonads (Clark et al., 2004, Hayashi et al., 2011, Hayashi et al., 2012, Hayashi and Surani, 2009, Irie et al., 2014, Sugawa et al., 2015, Toyooka et al., 2003).
mSSCs can be maintained and genetically manipulated in vitro over extensive periods, providing a valuable resource for in vitro studies of meiosis and post-meiotic development (Kanatsu-Shinohara et al., 2003, Kubota et al., 2004). In the past decade, this culture system has been used to identify key growth factors for the proliferation of mSSCs as well as the signaling pathways and the downstream target genes (Takashima et al., 2015). However, a long-standing and vital question is whether these stem cells can be stimulated in culture to complete spermatogenesis. In the present study, we show that RA by itself is sufficient to induce mSSCs to become primary spermatocytes, and that RA together with Sertoli cells from pup mice form a more efficient in vitro model of meiosis. This model can be used to identify genes involved in meiosis. Our work builds the foundation for accomplishing gametogenesis in vitro, which in turn contributes to a better understanding of the mechanisms of gametogenesis and the development of treatment of human infertility.
Results
Retinoic Acid Is Sufficient to Induce mSSCs into Zygotene Spermatocytes
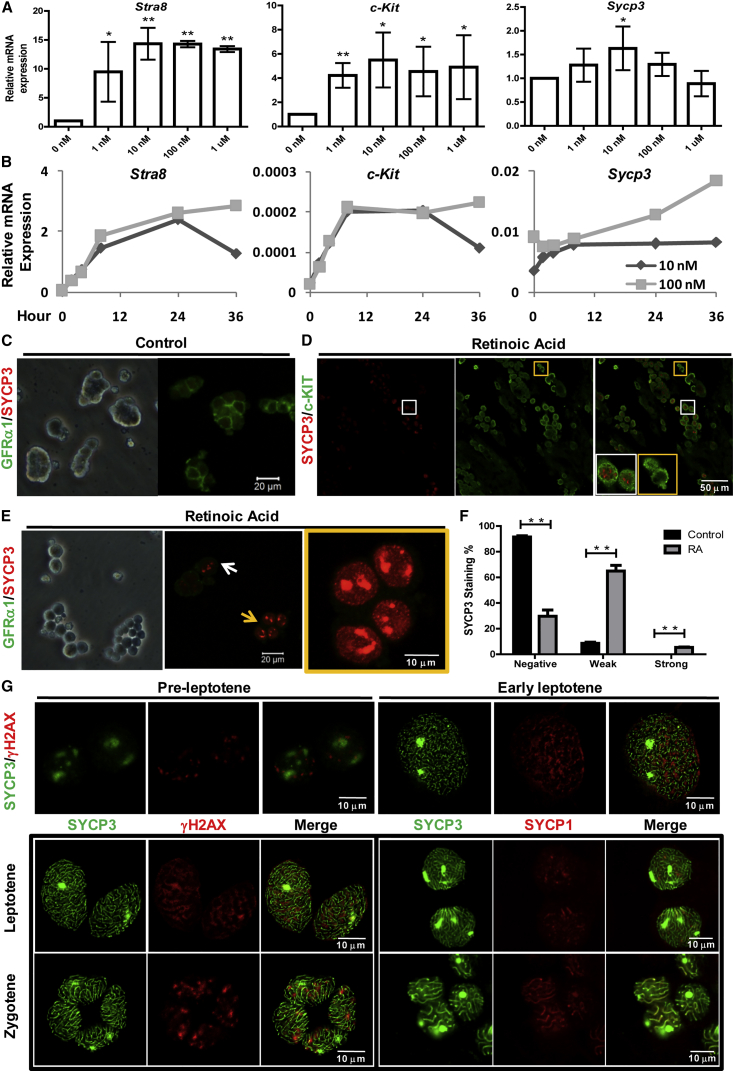
We set out to examine the effect of RA on meiosis initiation of cultured mSSCs. Normal mSSC cultures were maintained on the mouse embryonic fibroblast (MEF) feeder layer in serum-free medium and were passaged in a 1:4 ratio every 3–5 days (Figure S1). At the beginning of the experiment, mSSCs were plated onto laminin-coated feeder-free plates and treated with different concentrations of RA in serum-free medium. After 4 hr of treatment, the mRNA expression of three genes involved in spermatogonial differentiation or meiosis, Stra8, c-Kit, and Sycp3, was observed to be upregulated by RA at concentrations as low as 1 nM (Figure 1A), and were maximized by 10 nM RA. The time-course expression of these genes showed that 100 nM RA was more effective in maintaining the RNA levels for as long as 36 hr (Figure 1B). Subsequently, 100 nM and 24 hr were decided to be the optimal concentration and time span of RA treatment for meiosis induction. The feeder- and serum-free mSSCs without RA induction formed small tight clumps and expressed the stem cell marker GFRα1 but not SYCP3 (Figure 1C). On day 3 of induction, the cells turned into differentiating spermatogonia as indicated by the expression of c-KIT (Figure 1D). SYCP3 was expressed in most of these c-KIT+ cells but not in others, indicating the further differentiation of the majority of the induced cell. At this stage, the immunostaining of SYCP3 was weak and many brighter speckles were present, and we named these cells W cells for the weak staining of SYCP3. To define the identity of c-KIT+ cells and evaluate the differentiation-promoting role of RA, we labeled mSSCs with GFP by lentiviral infection (GFP-mSSCs) for the convenience of germ cell identification, and these GFP-mSSCs were treated with vehicle or RA, respectively. On day 3 of induction, the induced germ cells were collected and transplanted into the testis of busulfan-pretreated recipient mice. Based on the numbers of colonies established, RA treatment reduced the stem cell activity by about 3-fold (Figure S3D). On day 5 of induction, the induced cells were in the form of monolayer patches with clear cell boundaries (Figure 1E). In addition to the W cells, we also observed cells with strong SYCP3 immunostaining, and we named these cells S cells. Both the W and S cells at this stage did not express GFRα1, and represented about 70% and 5% of the cells on the plate, respectively (Figure 1F). Higher magnification images showed that the W cells also expressed γH2AX, a DNA-damage marker for meiosis prophase (Figure 1G), indicating they are probably pre-leptotene spermatocytes, although differentiating spermatogonia were also likely. For the S cells, high-resolution images of co-staining of either γH2AX and SYCP3 or SYCP1 and SYCP3 clearly revealed the presence of (early) leptotene and zygotene spermatocytes (Figure 1G). These results showed that RA alone was sufficient to induce mSSCs into spermatocytes, of which the most differentiated were zygotene spermatocytes.
Figure 1.
Induction of Spermatocytes from mSSCs by RA
(A) Determination of the optimal concentration of RA based on the expression of three genes involved in meiosis (Stra8, Sycp3) or in the differentiation of spermatogonia (c-Kit). Serum-free and feeder-free mSSC cultures were treated with different concentrations of RA for 4 hr, and the mRNA expression evaluated by qRT-PCR. β-Actin was used as internal control to normalize the expression of target genes. Error bars represent mean ± SD from three independent experiments. Asterisks denote statistically significant differences based on t-test (∗p < 0.05, ∗∗p < 0.01).
(B) Determination of the optimal time period of RA treatment. mSSCs were treated with 10 nM and 100 nM RA for different time periods.
(C) Co-immunostaining of mSSC marker GFRα1 and meiosis marker SYCP3 on uninduced mSSCs. Note that these cells were GFRα1+SYCP3−.
(D) Co-immunostaining of differentiating spermatogonia marker c-KIT and SYCP3 on induced germ cells on day 3 of induction. Yellow rectangle represents the c-KIT+SYCP3− cells while the white rectangle indicates the c-KIT+SYCP3+ cells. Higher-magnification pictures of the cells in the yellow or white rectangle are also shown in the corner of merge picture. Note that almost all the cells were c-KIT+ but only a fraction were SYCP3+.
(E) Co-immunostaining of GFRα1 and SYCP3 on cells on day 5 of induction. White arrow indicates cells with weak speckled staining of SYCP3 (W cells), and yellow arrow indicates cells with strong staining (S cells), of which a higher-magnification picture with a yellow frame is shown alongside.
(F) Quantification of SYCP3- cells, W cells, and S cells. Error bars represent mean ± SD from three independent experiments. Asterisks represent statistically significant differences based on t-test (∗∗p < 0.01).
(G) Co-immunostaining of SYCP3 and γH2AX, or SYCP1 and SYCP3 of cells on day 5 of induction to show different types of meiotic spermatocytes.
Gene Network Regulated by RA Signaling in mSSCs
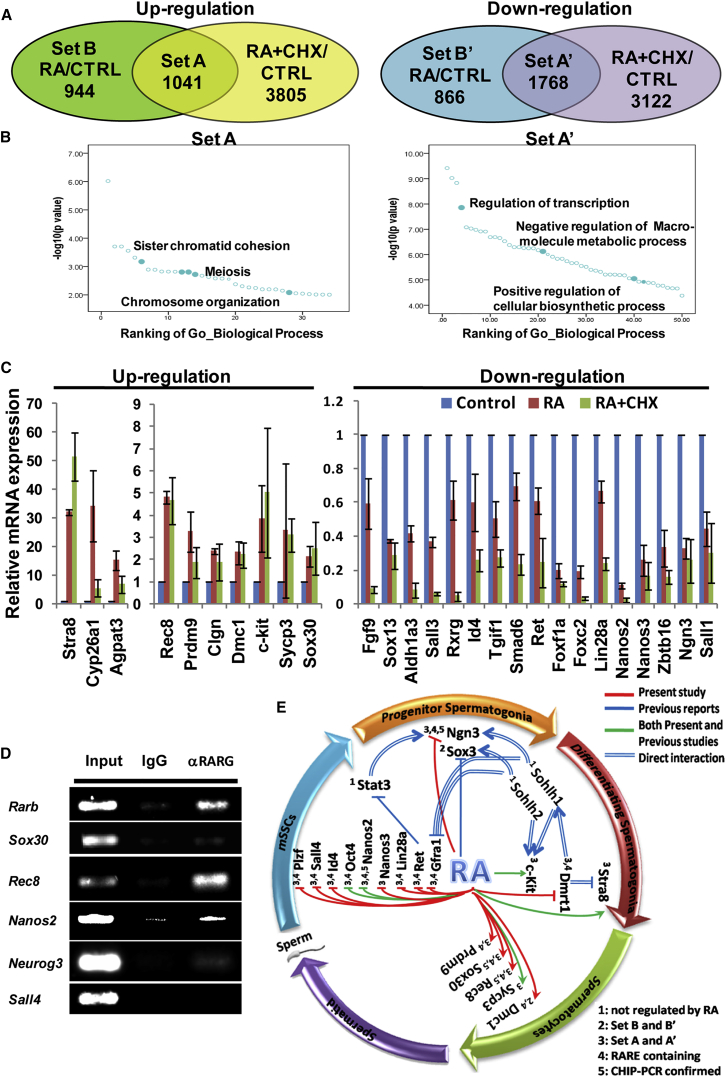
We next used RNA sequencing (RNA-seq) to identify RA-regulated genes treated by 100 nM RA for 24 hr. We also included samples treated with both RA and cycloheximide (CHX), a protein synthesis inhibitor in eukaryotic organisms, to identify potential direct target genes of RA. The optimal concentration of CHX was determined to be 0.25 μg/ml based on its concentration-dependent effects on cell growth and on the translation of Stra8 mRNA (Figures S2A and S2B).
Based on the RNA-seq data of duplicated samples of each treatment, 1,985 upregulated and 2,634 downregulated genes were identified in response to RA treatment (Figure S3B). By comparing with genes up- or downregulated by RA + CHX, 1,041 upregulated and 1,768 downregulated potential direct target genes of RA were acquired (Figure 2A and Table S1). Functional annotation term (FAT) enrichment analysis showed that RA-regulated genes (set A and set A′) were enriched with FATs related to many processes such as cell-cycle process, meiosis, signal transduction, metabolic process, development, regulation of gene expression, and reproduction (Figure 2B and Table S2).
Figure 2.
A Network of Genes Regulated by RA Signaling
(A) Up- and down-regulated genes by RA and RA + CHX and their intersections. Feeder-free and serum-free mSSCs were treated with vehicle or RA or RA plus cycloheximide (RA + CHX) for 24 hr, and then total RNA from different samples was extracted for RNA-seq analysis.
(B) Gene ontology term analysis of RA-regulated genes. Left and right panels show the significantly enriched terms for up- and down-regulated genes, respectively.
(C) qRT-PCR confirmation of a panel of RA-regulated genes. Most of these genes are well known for their roles in spermatogenesis. Error bars denote mean ± SD from three independent assays.
(D) Identification of the several potential direct target genes of RA/RAR by ChIP-PCR. Putative RARE in the promoters of Rarb, Sox30, Rec8, Nanos2, and Neurog3 were identified by motif scanning using a bioinformatics method. ChIP assays were conducted using the RARG antibody.
(E) A small network of genes regulated by RA was constructed manually based on our RNA-seq data and literature mining. Genes reported to be involved in mSSC self-renewal, progenitor spermatogonia proliferation, differentiation and meiosis initiation of differentiating spermatogonia, and meiosis of spermatocytes were placed underneath the corresponding stages of spermatogenesis. Regulatory interactions identified by the present study (red line) or previous reports (blue) or both (green) were labeled with lines of different color. The double lines indicate a direct binding of a transcription factor to the promoters of their target genes. The numbers denote different types of evidence for the regulations by RA. 1, 2, 3 indicate RNA-seq data; 4, bioinformatics scanning of the RARE; and 5, ChIP-PCR results.
See also Figures S2 and S3.
Marker genes of undifferentiated spermatogonia (mSSCs and progenitor spermatogonia) such as Gfrα1, c-Ret, Plzf/Zbtb16, Neurog3/Ngn3, Nanos2, Nanos3, Id4, Lin28a, and Pou5f1/Oct4 were all downregulated while those of differentiating spermatogonia such as Stra8, c-Kit, Dmrt1, and those of meiotic spermatocytes such as Sycp3, Rec8, Prdm9, and Dmc1 were upregulated. Interestingly, RA repressed the expression of 8 SOX family genes and 17 FOX family members. Genes involved in RA signaling or metabolism such as Rarb, Rarg, Cyp26a1, Cyp26a1, Aldh1a3, and Aldh1b1 were also regulated by RA. The expression changes of some of those genes were confirmed by qRT-PCR (Figure 2C).
We also scanned the promoter regions spanning from −10,000 bp to 5,000 bp of the transcription start site of these RA-regulated genes for the RA response element (RARE). The results revealed that their promoters were enriched with RAREs. Chromatin immunoprecipitation (ChIP)-PCR results indicated the predicted RAREs in the promoters of Rarb, Sox30, Rec8, Nanos2, and Neurog3 were indeed bound by RARG (Figure 2D and Table S3). In contrast, RARG did not bind to the examined RARE on the Sall4 promoter. Moreover, we also performed the experiments using RARA antibody whereby the results were consistent with the ones using RARG antibody (Figure S3C), indicating that the RARA may also play a role during the differentiation of mSSCs. Based on these results and those from the literature, we manually constructed a small gene-regulatory network centered on the action of RA (Figure 2E). It was obvious that RA repressed genes involved in promoting the proliferation of undifferentiated spermatogonia, which included mSSCs and progenitor spermatogonia, while it activated genes involved in spermatogonial differentiation as well as the initiation and progression of meiosis.
Meiosis Induced by Sertoli Cell Co-culture
We were interested in whether meiosis could be induced by co-culture of Sertoli cells, which are the only somatic cell type that makes physical contact with spermatogenic cells in vivo. The primary cultures of three types of Sertoli cells from pup (5–7 days post partum [dpp]), puberty (3 weeks post partum [wpp]), and adult (7–8 wpp) mice were compared for their ability to support meiosis initiation. To amplify the cell numbers and remove any contaminated germ cells, we passaged Sertoli cells once and treated them with either mitomycin (pup Sertoli cells) or Tris-HCl buffer (puberty and adult Sertoli cells) before use. The Sertoli cell cultures were more than 90% pure and free of germ cell contaminations based on the immunostainings of the Sertoli cell marker WT1 and N-CADHERIN, and the germ cell marker MVH and SYCP3 (Figures S4A–S4C).
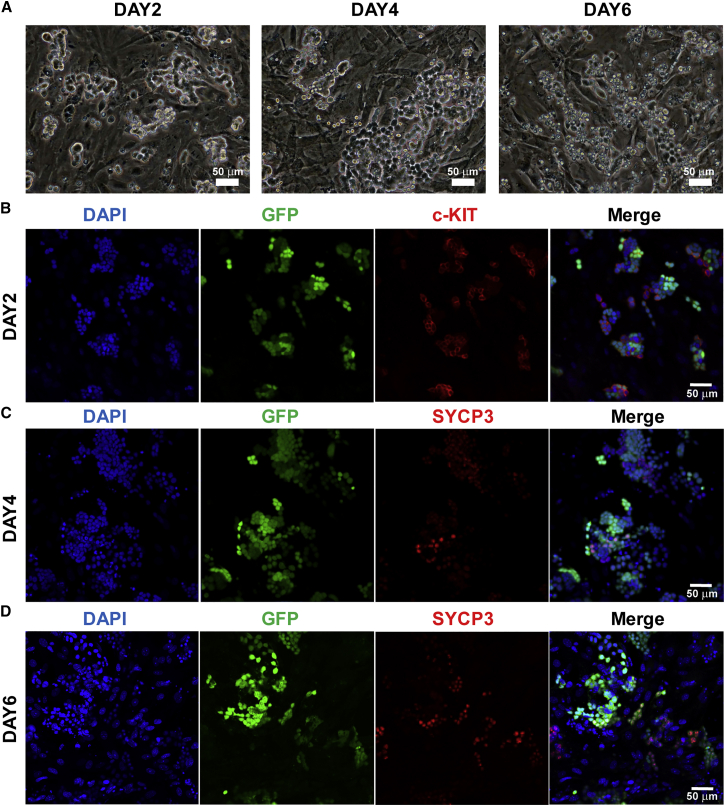
In the pup Sertoli cell co-cultures (Figure S5A), mSSCs underwent vigorous proliferation for at least 3 days and formed monolayer patches with clear cell boundaries when observed 4 days after plating (Figure 3A), indicating that these germ cells underwent differentiation. Thereafter most of the differentiated germ cells underwent apoptosis and detached from the feeder layer (Figure 3A). c-KIT+ cells were observed 1 day after plating, and these cells did not express SYCP3 based on immunostaining results (Figure S5B). The induced germ cells became the W cells on the third day of induction (Figures S5B and S5C), the S cells appeared on the fourth day (Figures 3C and S5C), and the proportion of S cells continued to increase by day 6 (Figures 3D and S5C).
Figure 3.
Induction of Spermatocytes from mSSCs Using Pup Sertoli Cell Co-cultures
(A) Bright-field images of germ cells from mSSCs, which were GFP-labeled by lentiviral infection (GFP-mSSCs), on mitomycin C-inactivated pup Sertoli cell co-cultures on different days after plating.
(B) Immunostaining of c-KIT in germ cells on day 2 of induction.
(C and D) Immunostaining of SYCP3 on day 4 (C) and day 6 (D) of induction.
See also Figures S4 and S5.
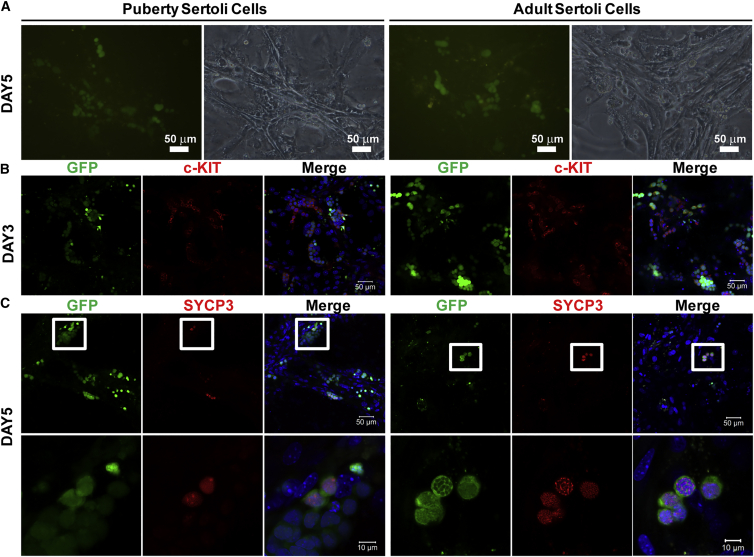
The puberty and adult Sertoli cells also support the differentiation of mSSCs (Figure 4A). The majority of the germ cells turned to c-KIT+ when observed on day 3 of induction (Figure 4B) while only a small number of S cells were detected on day 5 of induction (Figure 4C). Most of the germ cells were lost due to severe apoptosis on these two types of Sertoli cells (Figure 4A). These results indicated that the immature pup Sertoli cells supported both the proliferation and meiosis initiation of germ cells while the puberty and adult Sertoli cells supported the meiosis poorly, probably due to the compromised proliferation of germ cells on these cells.
Figure 4.
Induction of Spermatocytes from mSSCs Using Sertoli Cell Co-cultures from Puberty and Adult Mice
(A) Fluorescent and bright-field images of GFP-mSSCs on Sertoli cells on day 5 of induction.
(B) Immunofluorescence of c-KIT expression on day 3 of induction.
(C) Immunofluorescence of SYCP3 on day 5 of induction. Lower panel shows the enlarged images of the boxed areas in the corresponding images in the upper panel.
See also Figure S4.
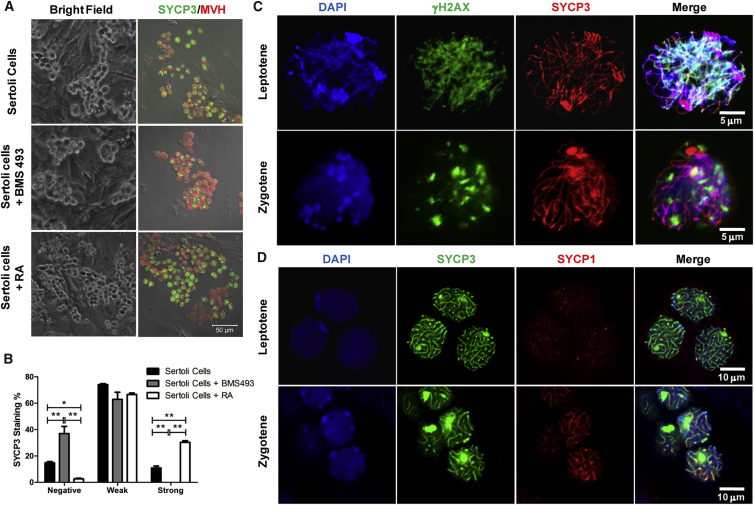
A More Efficient Meiosis-Inducing System Consists of Exogenous RA and Pup Sertoli Cells
We thought that the meiosis-inducing effect of the Sertoli cells was at least partially contributed by the endogenous RA signaling originating from these cells. To test this, we added 5 μM BMS493, a pan-RAR antagonist, to the mSSC and pup Sertoli cell co-cultures, whereby only W cells were observed, indicating a compromised meiosis (Figure 5A). We further hypothesized that addition of exogenous RA to the co-culture should improve the meiosis-inducing efficiency. Indeed, inclusion of 100 nM RA in the co-culture increased the percentages of SYCP3+ cells from 85.8% to 97.3% and the percentages of S cells from 10.6% to 28.3% (Figure 5B). Co-immunostaining of SYCP3 and γH2AX, or SYCP3 and SYCP1 showed that the S cells mainly contained the leptotene and zygotene spermatocytes (Figures 5C and 5D). Occasional patches of pachytene and diplotene spermatocytes were also detected using our induction system (Figure S6A).
Figure 5.
An Efficient Meiosis-Inducing In Vitro Model Using RA and Pup Sertoli Cell Co-cultures
(A) Bright-field and co-immunostaining images of germ cells on mitomycin C-treated pup Sertoli cells without or with additional treatment with either RA antagonist BMS 493 or RA.
(B) The percentages of SYCP3-negative, SYCP3-weak (W cells), and SYCP3-strong cells (S cells) in the MVH-positive germ cells corresponding to different treatments in (A). Error bars represent mean ± SD from three independent experiments. Asterisks denote statistically significant differences based on t test (∗p < 0.05, ∗∗p < 0.01).
(C) The identification of different spermatocytes by high-magnification imaging of the co-immunostaining of SYCP3 and γH2AX on chromosome spreads.
(D) High-magnification imaging of the co-immunostaining of SYCP1 and SYCP3.
See also Figure S6.
We also tested whether meiosis could be induced from mSSC cultures derived from adult mice using RA plus the pup Sertoli cell co-culture. After a 6-day induction, γH2AX and SYCP3 double-positive spermatocytes were also successfully observed (Figure S6B). Therefore, we have established an in vitro induction system that was suitable for meiosis induction of long-term SSC cultures from both pup and adult mice, and we named this system the RA/pup Sertoli cells (RA/pSC) meiosis model.
Characterization of Induced Spermatogenic Cells by Transcriptome Profiling
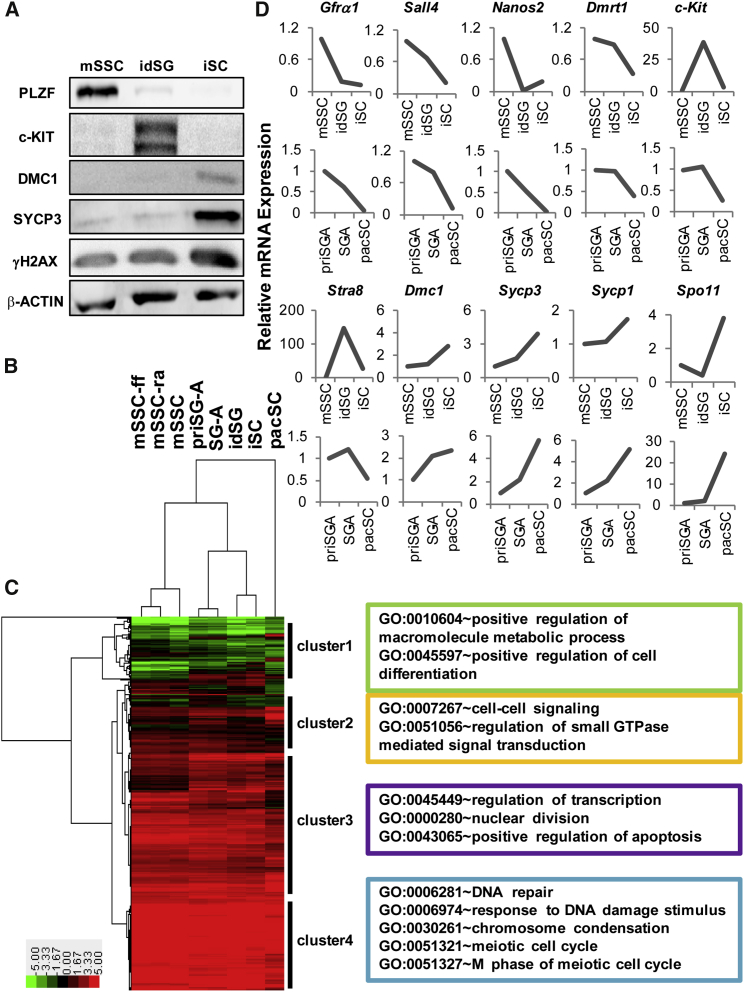
We next examined the gene-expression profiles of mSSCs and different induced spermatogenic cells and compared them with those of directly isolated spermatogenic cells. We used our RA/pSC model to derive c-KIT+ differentiating spermatogonia and spermatocytes from GFP-mSSCs. The GFP+c-KIThigh cells were harvested by fluorescence-activated cell sorting (FACS) using the c-Kit antibody on day 3 of induction, and therefore were regarded as the induced differentiating spermatogonia (idSG) (Figure S7A). The GFP+c-KITlow cells were collected by FACS on day 6 of induction and regarded as the induced spermatocytes (iSC) (Figure S7A). The purity of idSG was evaluated by measuring the immunofluorescent signal of c-Kit to be more than 95% (Figure S7C) while the purity of iSC was confirmed by SYCP3 immunostaining to be above 90% (Figure S7D). Moreover, we performed Western blotting of a panel of marker proteins to show that their expression patterns over the three types of spermatogenic cells were as expected (Figure 6A). For comparison, we also included RNA-seq data of the feeder-free mSSC (mSSC-ff) and RA-treated mSSCs (mSSC-ra) as well as isolated primitive spermatogonia (priSG-A), type A spermatogonia (SG-A), and pachytene spermatocytes (pacSC) reported in our previous study (Gan et al., 2013).
Figure 6.
Transcriptome Profiles of Induced Germ Cells and Isolated Spermatogenic Cells
(A) Characterization of three types of in vitro spermatogenic cells by Western blotting analysis of several marker proteins specific to different spermatogenic cells. idSG, induced differentiating spermatogonia; iSC, induced spermatocytes.
(B and C) Hierarchical clustering of in vitro- and in vivo-derived spermatogenic cells (B) and protein-coding genes (C) based on the RNA-seq data. mSSC-ff, feeder-free mSSCs; mSSC-ra, RA-treated mSSCs; priSG-A, primitive SG-A isolated from 6-dpp mice; SG-A, isolated type A spermatogonia; pacSC, isolated pachytene spermatocytes. Differentially expressed genes were identified using ANOVA and samples of mSSC-ff, mSSC-ra, mSSC, idSG, and iSC, each of which included two biological replicates. Four clusters of genes were labeled based on visual inspection of the hierarchical clustering of genes, and representative enriched gene ontology (GO) terms are listed on the right side of the heatmap.
(D) Comparison of the expression patterns of a set of genes involved in spermatogenesis in the in vitro- and in vivo-derived germ cells using the FPKM values (fragments per kilobase of transcript per million mapped reads) of the RNA-seq data.
See also Figure S7.
Hierarchical clustering was conducted using genes, whose expression values were significantly different among samples based on ANOVA test (Figures 6B and 6C). Three mSSC samples (mSSC-ff, mSSC-ra, mSSC) clustered together sequentially to form a bigger cluster; the two isolated spermatogonial samples (priSG-A, SG-A) formed a cluster, which then joined the cluster formed by the two induced cell types (idSG, iSC) to form another bigger cluster. These two bigger clusters joined together subsequently and finally with the isolated pacSC. Therefore, the two induced cell types (idSG and iSC) were more similar to spermatogonia than to mSSCs and pacSC. The gap in gene-expression profile between iSC and the isolated pacSC might explain why iSCs were arrested at the leptotene and zygotene stage of meiosis. We roughly identified four big clusters of genes, in which some particular gene ontology terms were enriched (Figure 6C and Table S4). We plotted the expression changes of some RA-regulated genes shown in Figure 2C along the in vitro and in vivo differentiation paths based on the RNA-seq data (Figure 6D). The trends of the in vitro and in vivo changes of each gene are similar although the change folds were different for some genes, suggesting that the in vitro differentiation path of mSSCs was a good model for the in vivo process.
Functional Studies of RA-Regulated Genes Using the RA/pSC In Vitro Meiosis-Inducing Model
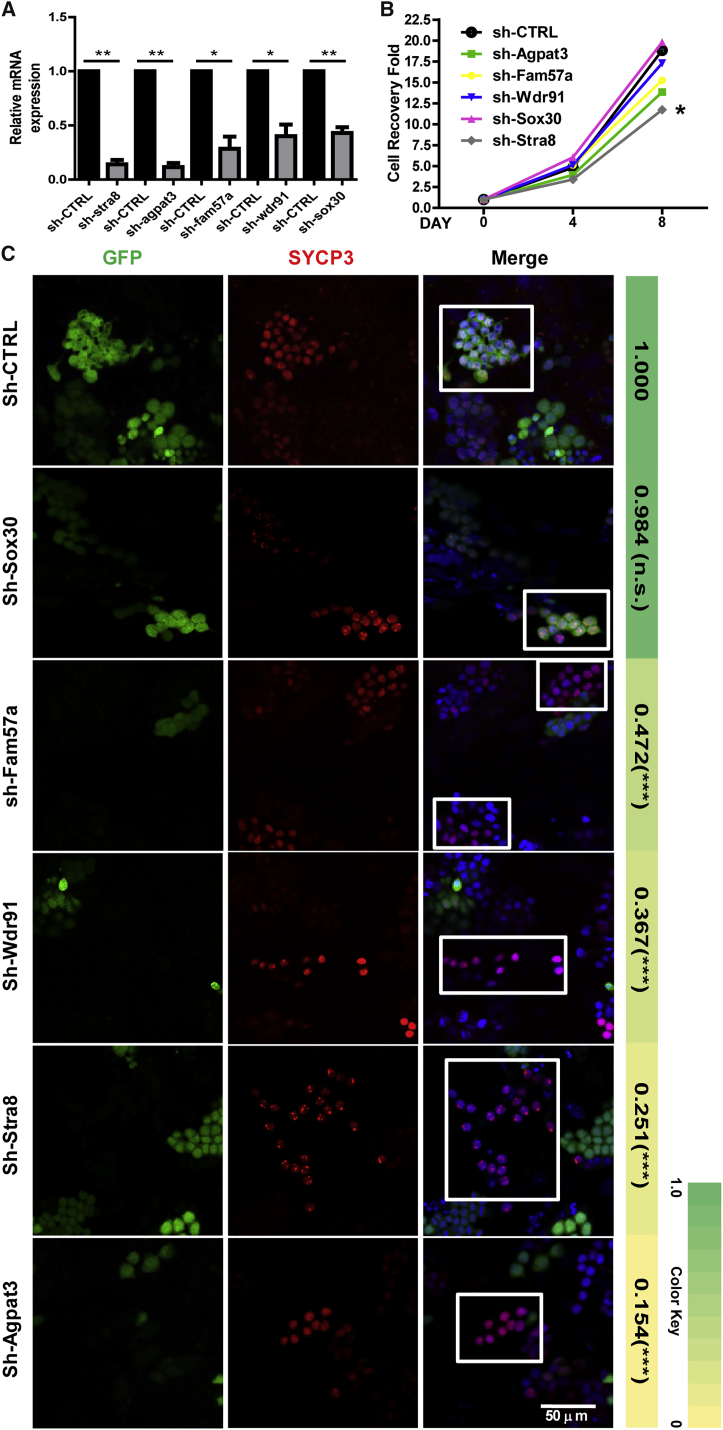
We next conducted proof-of-concept experiments to show that our RA/pSC model could be used to identify genes involved in meiosis. We selected Stra8 and four genes that are upregulated by RA based on our RNA-seq data, Sox30 (1.6-fold upregulation by RA), Wdr91 (2.42-fold upregulation), Fam57a (3.76-fold upregulation), and Agpat3 (16.6-fold upregulation). SOX30 is a transcription factor belonging to the Sox gene family, which contains more than 20 members in mammals, of which the sex-determining gene Sry is the prototypic member. The binding of RARG to the promoter of Sox30 gene was confirmed by ChIP-PCR (Figure 2D). Agpat3 (1-acylglycerol-3-phosphate O-acyltransferase 3) is an enzyme highly expressed in mouse spermatogenic cells and catalyzes the synthesis of highly polyunsaturated fatty acids, which is accumulated in the testis during mammalian puberty (Koeberle et al., 2012). Wdr91 (WD repeat domain 91) encodes a WD repeat domain-containing protein belonging to the WD40 superfamily, members of which are involved in many biological process, including signal transduction, regulation of cell cycle, and gene regulation (Gori et al., 2005). Fam57a (family with sequence similarity 57, member A) encodes a protein with conserved TRAM/Lag1/CLN8 (TLC) domain, and is involved in cancer cell growth and other cellular processes (Yamashita-Sugahara et al., 2013). The functions of Wdr91 and Fam57a in spermatogenesis are unknown. The short hairpin RNA (shRNA)-expressing constructs, from which GFP was also expressed, were introduced into mSSCs by lentiviral infection, and the knockdown of mRNAs of each gene was confirmed by qRT-PCR (Figure 7A). These cells were subjected to meiosis induction using our RA/pSC model, and the S-cell induction efficiency of each knockdown cell type was compared with that of the cells infected with the Sh-CTRL virus, i.e., the percentage of S cells among GFP+ cells from each sample was normalized by the percentage value of the Sh-CTRL sample. Knockdown of Stra8 reduced the proliferation of spermatogonia on MEF feeder cells significantly (Figure 7B) and also reduced the S-cell induction efficiency to 25.1% of the control induction efficiency (Figure 7C). Knockdown of Agpat3, Fam57a, and Wdr91 did not change the proliferation significantly but reduced the S-cell induction efficiency to 15.4%, 47.2%, and 36.7% of the control value, respectively, while the knockdown of Sox30 had no effect on either the proliferation or the meiosis of mSSCs (Figures 7B and 7C). These results show that our meiosis-inducing model can be used to identify genes that are involved in meiosis.
Figure 7.
Investigation of the Functions of RA-Regulated Genes Stra8, Sox30, Agpat3, Wdr91, and Fam57a in Spermatogenesis Using the RA/pSC Model
(A) Quantitative evaluation of mRNAs in feeder-free mSSCs, in which the shRNAs of different genes were expressed from the lentiviral GFP-expressing vector using qRT-PCRs. Error bars indicate mean ± SD from three independent experiments. Asterisks represent statistically significant differences (∗p < 0.05, ∗∗p < 0.01).
(B) The growth curves of gene-knockdown mSSC lines on MEF feeders. The data were from three independent experiments. Asterisk denotes statistically significant difference (∗p < 0.05).
(C) Effects of RNA knockdown on the induction of SYCP3-positive cells using the RA/pSC model. The images were taken on day 6 of induction. The white boxes frame areas where the co-localization of GFP and SYCP3 signals were or were not observed. The ratio of S cells to the total GFP+ cells was calculated based on the results from three independent assays (n = 3). The quantification data were normalized by the ratio of S cells in the Sh-CTRL sample, and the mean value for each gene is shown on the right of the immunostaining images. Asterisks denote statistically significant differences (∗∗∗p < 0.001; n.s., no significance).
Discussion
We report in the present study that RA alone is sufficient for inducing the initiation of meiosis of cultured mSSCs and the derivation of spermatocytes under feeder-free and serum-free conditions. A previous study showed that purified prenatal male PGCs isolated before but not after 14.5 days post coitum (dpc) can be induced by RA to initiate meiosis and to generate spermatocytes in the absence of any somatic cells (Ohta et al., 2010). However, the question of whether the lost co-culture-independent RA responsiveness of the male germ cells after 14.5 dpc can be resumed later in germ cell development remains open. Moreover, the number of isolated PGCs is low and they are refractory to mitotic proliferation even in the presence of 20% serum, thus having limited use in further applications. By taking advantage of cultured mSSCs that can be amplified infinitely while keeping their spermatogenic function, we have been able to address this important question and conduct more mechanistic studies.
Our finding is somewhat surprising given that in vivo spermatogenesis, which occurs in the testis containing multiple hormone and/or cytokine-producing cell types, seems to be regulated by diverse factors. That RA plays a key role in meiosis initiation in mammals and other vertebrates has been supported by multiple lines of evidence. However, whether RA is the only essential factor remains unknown, probably due to the lack of an in vitro culture system for germ cells. If RA is indeed the only meiosis-inducing signal in vivo, as suggested by our in vitro data presented here, we can argue that the major roles of other components in the gonads in terms of meiosis initiation are to regulate the production and degradation of this key molecule. This regulation can be complex for reasons that are poorly understood currently. For example, Raverdeau et al. (2012) reported that development of undifferentiated spermatogonia to differentiating A1 spermatogonia in the first spermatogenic cycle was dependent on the Sertoli cell-derived RA but the subsequent spermatogenic cycles were initiated by RA produced by spermatocytes.
Few genes regulated by RA in germ cells have been identified despite the important role of RA in meiosis. Stra8 is the best known direct target gene of RA (Oulad-Abdelghani et al., 1996). A recent study indicated that RA regulated two parallel pathways represented by Stra8 and Rec8 (Koubova et al., 2014). Our RNA-seq data of the RA-treated mSSCs not only confirmed these observations but also provided clues for dissecting the mechanisms of meiosis. First, a much larger number of genes are regulated by RA than previously known. Although we are still unable to discern how many of them are direct targets of RA, we think the number should be high based on the 1,041 upregulated and 1,768 downregulated genes in the intersecting sets of the RA-regulated and RA + CHX-regulated genes. More evidently, the binding of RARG to the predicted RAREs in a small panel of RA-regulated genes was mostly confirmed by ChIP-PCR results. Second, RA not only upregulate genes that favor the differentiation of spermatogonia (c-Kit) and the initiation (Stra8) and progression (Rec8, Dmc1, Prdm9, Sycp3) of meiosis, but also downregulate genes beneficial to the self-renewal of mSSCs (Oct4, Plzf, Lin28a, to name a few; see Figure 2 for more), the proliferation of progenitor spermatogonia (Ngn3, Sox3), and the genes safeguarding the premature meiosis (Dmrt1). Third, RA regulates the expression of a diverse family of genes encoding, for example, signaling proteins, metabolizing enzymes, transcription factors, and many others. The results expand far more our recognition of the molecular mechanisms underlying the action of RA as the key regulator of meiosis.
Although RA is sufficient to promote the development of spermatogenic cells from mSSCs to spermatocytes, the Sertoli cell co-culture increases further the induction efficiency. Severe apoptosis of the germ cells were observed under both the feeder-free and the co-culture conditions, indicating that the in vitro meiosis model needs to be optimized in the future. Sertoli cell co-culture alleviates the apoptosis to some degree, but the problem persists in being a barrier for the efficient derivation of haploid spermatids. Notably, spermatogonia in the first wave of spermatogenesis in vivo supported by the immature Sertoli cells undergo more severe apoptosis compared with the subsequent spermatogenic cycles, during which Sertoli cells mature (Mori et al., 1997). The mature Sertoli cells, and probably other somatic cell types in the adult testis, are more likely to slow down the initiation of meiosis so that the cells have enough time to go through the lengthy pre-meiotic development, during which genes that are needed to overcome the meiotic barrier are readily expressed. Unfortunately, we found that Sertoli cells from puberty and adult mice were even worse than pup Sertoli cells in supporting the induction of meiotic cells. We observed that pup Sertoli cells were better than the other two types in supporting the proliferation of co-cultured germ cells. Therefore, the net result of using the pup Sertoli cell co-cultures, which contribute to a better pro-proliferation and probably the same anti-apoptosis effects on the germ cells, is a higher chance of meiotic cell induction.
It is interesting that feeder-free mSSCs with and without RA treatment first cluster together despite a large number of genes changing their expression in response to RA. This suggests that the 24-hr RA treatment at the supra-physiological concentration (100 nM) does not change the stem cell character, although many marker genes of differentiating spermatogonia start to express. The mSSCs cultured on the MEF feeder layer then join in and form the stem cell cluster. The isolated priSG-A and SG-A together form another bottom-level cluster, which subsequently merges with the other bottom-level cluster of the two types of induced germ cells, idSG and iSC. These results suggest that our induced cells are different from mSSCs with or without a short-term RA treatment, but similar to differentiating germ cells. Moreover, the idSG and iSC used in the RNA profiling experiments are sorted by FACS on days 3 and 6 of induction based on EGFP and c-KIT, and therefore represent the population of differentiating/differentiated cells that are induced in a stepwise manner. This is apparent in the time-course dynamic changes of c-KIT as well as other spermatogonial and spermatocyte markers shown in Figure S5. Interestingly, the isolated pacSC are the most distant to the other cell types. This is not surprising, considering that pacSC are quite different from other types of spermatocytes due to the dramatic remodeling of chromatins during synapsis. In other words, it can be understood that our iSC are more different from pacSC than from other types of spermatogenic cells, as they are mostly leptotene/zygotene spermatocytes. The gap between iSC and pacSC in the gene-expression profile explained why it is difficult to induce pacSC in vitro. Above all, the clustering pattern of the in vitro- and in vivo-derived spermatogenic cells indicates that our in vitro model basically reflects the normal pathway of spermatogenesis. The effectiveness of this model is further supported by the consistency between the reported expression patterns of a selected set of key genes in spermatogenesis and their patterns in the induced germ cells (Figure 6D).
To prove that our model has application values in identifying genes involved meiosis, we selected five RA-regulated genes to study their function in spermatogenesis using our model system and the RNAi technique. Stra8 has been well known for its essential role in meiosis initiation and was chosen as a positive control. Meiosis of Stra8 knockout mice is not initiated on the C57BL/6 background, while in some cells on a mixed background it is initiated but fails to complete the prophase (Mark et al., 2008). Using our in vitro model, we not only confirmed the failed meiosis of the Stra8 knockdown cells but also found that the proliferation of the mSSCs and the idSGs was also reduced. This proliferation-promoting effect is either specific to the in vitro model or a neglected fact of the in vivo process reported by most of the previous studies. Recently, Endo et al. (2015) found that periodic RA-STRA8 signaling not only regulates meiotic initiation but also regulates spermatogonial differentiation. According to this study, a majority of the differentiating spermatogonia were missing in the Stra8 knockout mice, consistent with our observation that Stra8 also promotes the proliferation of mSSCs. Sox30 was chosen because it represents an important transcription factor family with multiple members of diverse functions. It turned out that its knockdown had no effect on either the proliferation or the meiosis initiation of the germ cells. This result can be explained by the redundancy of this protein family, and serves as a negative control of our in vitro model. Alternatively, SOX30 protein may be involved in post-meiotic development, although its mRNAs can be transcribed at an earlier stage. Indeed, we only recently generated the Sox30 knockout mice and found that the spermatogenesis was arrested at the early round spermatid stage (D.Z. and C.H., unpublished data). Agpat3, Wdr91, and Fam57a were chosen because they represent the large number of RA-induced genes whose function needs to be annotated. Interestingly, their knockdowns all blocked the initiation of meiosis. These results show that our in vitro model, once coupled with high-throughput screening, will contribute to identifying many more genes of different categories that are involved in processes before, during, and after meiosis. Our work has also established a solid foundation for the eventual establishment of gametogenesis in vitro.
Experimental Procedures
Animals
Mouse husbandry and surgical procedures followed the guidelines of the Animal Care and Use Committee of the Institute of Zoology, Chinese Academy of Sciences. Male F1 progenies of C57BL6 and DBA/2 mice were used to derive mSSC and Sertoli cell cultures.
Culture of Mouse Spermatogonial Stem Cells
Protocol for the establishment and maintenance of mSSC cultures basically followed the methods developed by the Shinohara and Brinster groups with minor modifications (Kanatsu-Shinohara et al., 2003, Kubota et al., 2004) and was detailed in our recently study (Wang et al., 2015, Zhang et al., 2012). In brief, seminiferous tubules from the testis of pup (5–7 dpp) or adult mice were digested with collagenase IV into small fragments and cultured in mouse embryonic fibroblast medium (MEFM) (DMEM plus 10% fetal bovine serum). 24 hr (pup) and 48 hr (adult) later, spermatogonia could be obtained and transferred to mitomycin C-inactivated MEF feeder cells for later culture.
Culture of Mouse Sertoli Cells
Three types of Sertoli cells from the F1 progenies of C57BL6 and DBA/2 mice were prepared for inducing the differentiation of mSSCs. The pup Sertoli cells were from 6- to 7-dpp mice, the puberty Sertoli cells were from 21-dpp mice, and the adult Sertoli cells were from 7- to 8-wpp mice. Segments of seminiferous tubules were prepared and cultured for 24 hr to prepare the pup Sertoli cells as described in the previous section. After the spermatogonia were removed from the P0 culture, the remaining cells, mostly Sertoli cells, were cultured in MEFM. When the cells were confluent, a 1:3 passage was performed. The cells were treated with mitomycin C when needed and stored in liquid nitrogen for future use. To prepare the puberty and adult Sertoli cells, we collected the segments of seminiferous tubules after the collagenase IV digestion at 37°C for 10 min. After culturing in MEFM for 72 hr, the Sertoli cells were treated with 20 mM Tris-HCl (pH 7.4) for 2.5 min to destroy contaminated germ cells. The remaining Sertoli cells were washed twice with Dulbecco’s PBS, supplied with fresh MEFM, and cultured in a normal incubator. When these two types of Sertoli cells were confluent, the cells were used for induction assays immediately or stored in liquid nitrogen for future use.
Induction of Meiotic Germ Cells
mSSCs were maintained on MEF feeder layer in serum-free medium, and then induced to produce meiotic germ cells using two methods. mSSC cultures from a well of a six-well plate were digested with Accutase for 6 min, resuspended in SSC medium (SSCM) (α-minimum essential medium (GIBCO, catalog no. 12561-056), 20 ng/ml glial cell-derived neurotrophic factor, 5 ng/ml basic fibroblast growth factor, 20 μg/ml insulin, and other components as described previously; Wang et al., 2015), and plated on a 6-cm dish. 30 min later, MEFs but not mSSCs attached to the dish bottom firmly. Floating mSSCs were collected and plated to either a feeder-free laminin-coated six-well plate or a plate in which mitomycin C or Tris-HCl-treated Sertoli cells had been prepared beforehand. The density of plated mSSCs was 106 cells per well of the six-well plate. 100 nM RA was added 24 hr later. Medium was changed once every 2 days. The induced germ cells were harvested for characterization on different days.
Statistics
All experiments were independently repeated at least three times. Data are presented as mean ± SD. Statistical analysis was determined using t test or ANOVA. All statistical tests were performed using SPSS 13.0 software.
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (2012CB966702, 2013CB945001, and 2015CB943002) and the National Natural Science Foundation of China (31271379, 31471349).
Published: June 23, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.05.013.
Accession Numbers
All of the RNA-seq data have been deposited in the Gene Expression Omnibus under the accession number GEO: GSE77051.
Supplemental Information
References
- Anderson E.L., Baltus A.E., Roepers-Gajadien H.L., Hassold T.J., de Rooij D.G., van Pelt A.M., Page D.C. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc. Natl. Acad. Sci. USA. 2008;105:14976–14980. doi: 10.1073/pnas.0807297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltus A.E., Menke D.B., Hu Y.C., Goodheart M.L., Carpenter A.E., de Rooij D.G., Page D.C. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat. Genet. 2006;38:1430–1434. doi: 10.1038/ng1919. [DOI] [PubMed] [Google Scholar]
- Bowles J., Knight D., Smith C., Wilhelm D., Richman J., Mamiya S., Yashiro K., Chawengsaksophak K., Wilson M.J., Rossant J. Retinoid signaling determines germ cell fate in mice. Science. 2006;312:596–600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- Bowles J., Feng C.W., Spiller C., Davidson T.L., Jackson A., Koopman P. FGF9 suppresses meiosis and promotes male germ cell fate in mice. Dev. Cell. 2010;19:440–449. doi: 10.1016/j.devcel.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Clark A.T., Bodnar M.S., Fox M., Rodriquez R.T., Abeyta M.J., Firpo M.T., Pera R.A. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum. Mol. Genet. 2004;13:727–739. doi: 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- Endo T., Romer K.A., Anderson E.L., Baltus A.E., de Rooij D.G., Page D.C. Periodic retinoic acid-STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc. Natl. Acad. Sci. USA. 2015;112:E2347–E2356. doi: 10.1073/pnas.1505683112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H., Wen L., Liao S., Lin X., Ma T., Liu J., Song C.X., Wang M., He C., Han C. Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat. Commun. 2013;4:1995. doi: 10.1038/ncomms2995. [DOI] [PubMed] [Google Scholar]
- Geijsen N., Horoschak M., Kim K., Gribnau J., Eggan K., Daley G.Q. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature. 2004;427:148–154. doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- Gori F., Friedman L., Demay M.B. Wdr5, a novel WD repeat protein, regulates osteoblast and chondrocyte differentiation in vivo. J. Musculoskelet. Neuronal Interact. 2005;5:338–339. [PubMed] [Google Scholar]
- Griswold M.D., Hogarth C.A., Bowles J., Koopman P. Initiating meiosis: the case for retinoic acid. Biol. Reprod. 2012;86:35. doi: 10.1095/biolreprod.111.096610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Surani M.A. Self-renewing epiblast stem cells exhibit continual delineation of germ cells with epigenetic reprogramming in vitro. Development. 2009;136:3549–3556. doi: 10.1242/dev.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ogushi S., Kurimoto K., Shimamoto S., Ohta H., Saitou M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science. 2012;338:971–975. doi: 10.1126/science.1226889. [DOI] [PubMed] [Google Scholar]
- Hubner K., Fuhrmann G., Christenson L.K., Kehler J., Reinbold R., De La Fuente R., Wood J., Strauss J.F., 3rd, Boiani M., Scholer H.R. Derivation of oocytes from mouse embryonic stem cells. Science. 2003;300:1251–1256. doi: 10.1126/science.1083452. [DOI] [PubMed] [Google Scholar]
- Irie N., Weinberger L., Tang W.W., Kobayashi T., Viukov S., Manor Y.S., Dietmann S., Hanna J.H., Surani M.A. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2014;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Ogonuki N., Inoue K., Miki H., Ogura A., Toyokuni S., Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol. Reprod. 2003;69:612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- Kee K., Angeles V.T., Flores M., Nguyen H.N., Reijo Pera R.A. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeberle A., Shindou H., Harayama T., Yuki K., Shimizu T. Polyunsaturated fatty acids are incorporated into maturating male mouse germ cells by lysophosphatidic acid acyltransferase 3. FASEB J. 2012;26:169–180. doi: 10.1096/fj.11-184879. [DOI] [PubMed] [Google Scholar]
- Koubova J., Menke D.B., Zhou Q., Capel B., Griswold M.D., Page D.C. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc. Natl. Acad. Sci. USA. 2006;103:2474–2479. doi: 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubova J., Hu Y.C., Bhattacharyya T., Soh Y.Q., Gill M.E., Goodheart M.L., Hogarth C.A., Griswold M.D., Page D.C. Retinoic acid activates two pathways required for meiosis in mice. Plos Genet. 2014;10:e1004541. doi: 10.1371/journal.pgen.1004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H., Avarbock M.R., Brinster R.L. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Gill M.E., Koubova J., Page D.C. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Science. 2008;322:1685–1687. doi: 10.1126/science.1166340. [DOI] [PubMed] [Google Scholar]
- MacLean G., Li H., Metzger D., Chambon P., Petkovich M. Apoptotic extinction of germ cells in testes of Cyp26b1 knockout mice. Endocrinology. 2007;148:4560–4567. doi: 10.1210/en.2007-0492. [DOI] [PubMed] [Google Scholar]
- Mark M., Jacobs H., Oulad-Abdelghani M., Dennefeld C., Feret B., Vernet N., Codreanu C.A., Chambon P., Ghyselinck N.B. STRA8-deficient spermatocytes initiate, but fail to complete, meiosis and undergo premature chromosome condensation. J. Cell Sci. 2008;121:3233–3242. doi: 10.1242/jcs.035071. [DOI] [PubMed] [Google Scholar]
- Mori C., Nakamura N., Dix D.J., Fujioka M., Nakagawa S., Shiota K., Eddy E.M. Morphological analysis of germ cell apoptosis during postnatal testis development in normal and Hsp 70-2 knockout mice. Dev. Dyn. 1997;208:125–136. doi: 10.1002/(SICI)1097-0177(199701)208:1<125::AID-AJA12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Ohta K., Lin Y.L., Hogg N., Yamamoto M., Yamazaki Y. Direct effects of retinoic acid on entry of fetal male germ cells into meiosis in mice. Biol. Reprod. 2010;83:1056–1063. doi: 10.1095/biolreprod.110.085787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulad-Abdelghani M., Bouillet P., Decimo D., Gansmuller A., Heyberger S., Dolle P., Bronner S., Lutz Y., Chambon P. Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J. Cell Biol. 1996;135:469–477. doi: 10.1083/jcb.135.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panula S., Medrano J.V., Kee K., Bergstrom R., Nguyen H.N., Byers B., Wilson K.D., Wu J.C., Simon C., Hovatta O. Human germ cell differentiation from fetal- and adult-derived induced pluripotent stem cells. Hum. Mol. Genet. 2010;20:752–762. doi: 10.1093/hmg/ddq520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raverdeau M., Gely-Pernot A., Feret B., Dennefeld C., Benoit G., Davidson I., Chambon P., Mark M., Ghyselinck N.B. Retinoic acid induces Sertoli cell paracrine signals for spermatogonia differentiation but cell autonomously drives spermatocyte meiosis. Proc. Natl. Acad. Sci. USA. 2012;109:16582–16587. doi: 10.1073/pnas.1214936109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa R., Neves R., Fernandes S., Alves C., Carvalho F., Silva J., Cremades N., Malheiro I., Barros A., Sousa M. Cytological and expression studies and quantitative analysis of the temporal and stage-specific effects of follicle-stimulating hormone and testosterone during cocultures of the normal human seminiferous epithelium. Biol. Reprod. 2008;79:962–975. doi: 10.1095/biolreprod.107.067546. [DOI] [PubMed] [Google Scholar]
- Saba R., Kato Y., Saga Y. NANOS2 promotes male germ cell development independent of meiosis suppression. Dev. Biol. 2013;385:32–40. doi: 10.1016/j.ydbio.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Sato T., Katagiri K., Gohbara A., Inoue K., Ogonuki N., Ogura A., Kubota Y., Ogawa T. In vitro production of functional sperm in cultured neonatal mouse testes. Nature. 2011;471:504–507. doi: 10.1038/nature09850. [DOI] [PubMed] [Google Scholar]
- Sato T., Katagiri K., Yokonishi T., Kubota Y., Inoue K., Ogonuki N., Matoba S., Ogura A., Ogawa T. In vitro production of fertile sperm from murine spermatogonial stem cell lines. Nat. Commun. 2011;2:472. doi: 10.1038/ncomms1478. [DOI] [PubMed] [Google Scholar]
- Song H.W., Wilkinson M.F. In vitro spermatogenesis: a long journey to get tails. Spermatogenesis. 2012;2:238–244. doi: 10.4161/spmg.22069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawa F., Arauzo-Bravo M.J., Yoon J., Kim K.P., Aramaki S., Wu G., Stehling M., Psathaki O.E., Hubner K., Scholer H.R. Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J. 2015;34:1009–1024. doi: 10.15252/embj.201488049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S., Kanatsu-Shinohara M., Tanaka T., Morimoto H., Inoue K., Ogonuki N., Jijiwa M., Takahashi M., Ogura A., Shinohara T. Functional differences between GDNF-dependent and FGF2-dependent mouse spermatogonial stem cell self-renewal. Stem Cell Rep. 2015;4:489–502. doi: 10.1016/j.stemcr.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y., Tsunekawa N., Akasu R., Noce T. Embryonic stem cells can form germ cells in vitro. Proc. Natl. Acad. Sci. USA. 2003;100:11457–11462. doi: 10.1073/pnas.1932826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tres L.L., Kierszenbaum A.L. Viability of rat spermatogenic cells in vitro is facilitated by their coculture with Sertoli cells in serum-free hormone-supplemented medium. Proc. Natl. Acad. Sci. USA. 1983;80:3377–3381. doi: 10.1073/pnas.80.11.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven F.J., Amon A. Regulation of entry into gametogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:3521–3531. doi: 10.1098/rstb.2011.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang X., Wu Y., Han C. IGF-1R signaling is essential for the proliferation of cultured mouse spermatogonial stem cells by promoting the G2/M progression of the cell cycle. Stem Cell Dev. 2015;24:471–483. doi: 10.1089/scd.2014.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West F.D., Mumaw J.L., Gallegos-Cardenas A., Young A., Stice S.L. Human haploid cells differentiated from meiotic competent clonal germ cell lines that originated from embryonic stem cells. Stem Cell Dev. 2010;20:1079–1088. doi: 10.1089/scd.2010.0255. [DOI] [PubMed] [Google Scholar]
- Yamashita-Sugahara Y., Tokuzawa Y., Nakachi Y., Kanesaki-Yatsuka Y., Matsumoto M., Mizuno Y., Okazaki Y. Fam57b (family with sequence similarity 57, member B), a novel peroxisome proliferator-activated receptor gamma target gene that regulates adipogenesis through ceramide synthesis. J. Biol. Chem. 2013;288:4522–4537. doi: 10.1074/jbc.M112.440792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang S., Wang X., Liao S., Wu Y., Han C. Endogenously produced FGF2 is essential for the survival and proliferation of cultured mouse spermatogonial stem cells. Cell Res. 2012;22:773–776. doi: 10.1038/cr.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.