Summary
The molecular mechanisms regulating neural progenitor (NP) proliferation are fundamental in establishing the cytoarchitecture of the mammalian neocortex. The rate of cell-cycle progression and a fine-tuned balance between cell-cycle re-entry and exit determine the numbers of both NPs and neurons as well as postmitotic neuronal laminar distribution in the cortical wall. Here, we demonstrate that the microRNA (miRNA) miR-210 is required for normal mouse NP cell-cycle progression. Overexpression of miR-210 promotes premature cell-cycle exit and terminal differentiation in NPs, resulting in an increase in early-born postmitotic neurons. Conversely, miR-210 knockdown promotes an increase in the radial glial cell population and delayed differentiation, resulting in an increase in late-born postmitotic neurons. Moreover, the cyclin-dependent kinase CDK7 is regulated by miR-210 and is necessary for normal NP cell-cycle progression. Our findings demonstrate that miRNAs are essential for normal NP proliferation and cell-cycle progress during neocortical development.
Keywords: CDK7, cell cycle, microRNA, miR-210, neocortex, neural progenitor
Graphical Abstract

Highlights
-
•
miR-210 level is essential for cell-cycle progression in cortical neural progenitors
-
•
Cdk7 and miR-210 control neural progenitor proliferation
-
•
miR-210 promotes premature cell-cycle exit and differentiation in neural progenitors
-
•
miR-210 expression induces a deep-layer neuronal fate in the neocortex
In this article, Sun, Tang, and colleagues demonstrate that miR-210 and its target gene cyclin-dependent kinase Cdk7 co-regulate neural progenitor proliferation in the developing mouse neocortex. They further demonstrate that miR-210 expression induces premature cell-cycle exit and differentiation in neural progenitors, thereby promoting a deep cortical layer fate in postmitotic neurons.
Introduction
The neocortex, also called the dorsal cortex, is an immensely complex mammalian brain structure that controls higher cognitive functions, such as problem solving and reasoning, sensory perception, emotional processing, and attention. The developing neocortex is populated by several major types of neural progenitors (NPs) that primarily give rise to the excitatory neurons that populate the cortical wall (Florio and Huttner, 2014, Gertz et al., 2014). The mouse neocortex consists of self-renewing radial glial cells (RGCs) that reside in the ventricular zone (VZ) and give rise to postmitotic neurons as well as intermediate progenitors (IPs), which populate the subventricular zone (SVZ). These NP populations generate most of the neurons in the cortical plate (CP) (Englund et al., 2005, Franco and Muller, 2013, Sessa et al., 2008). As progenitors exit the cell cycle and terminally differentiate, early-born neurons migrate away from the VZ/SVZ to form the deeper layers of the cortex, and late-born neurons migrate past these cells to populate the outer layers, thereby forming an inside-out six-layer structure characteristic of the mammalian cortex (Kowalczyk et al., 2009, Toma et al., 2014).
Regulation of proliferation in NPs must be tightly controlled, as the length of the cell cycle and the timing of cell-cycle exit determine the size of the NP and postmitotic neuron population, and the laminar positioning of mature neurons within the six layers of the cortical wall (Taverna et al., 2014). Regulators of cell-cycle progression play crucial roles in neocortical development and expansion (Chenn and Walsh, 2002, Chenn and Walsh, 2003, Pilaz et al., 2009). Consequently, investigating the genes and processes involved in neocortical development should provide insight into the development and remarkable evolutionary expansion of the cortex observed in higher mammals, as well as neurodevelopmental disorders that arise from abnormal NP proliferation and differentiation (Kriegstein et al., 2006, Rakic, 2009, Sun and Hevner, 2014).
MicroRNAs (miRNAs) are a class of small, noncoding RNAs that have recently emerged as important regulators of neural development (Fineberg et al., 2009, Shibata et al., 2011). Previous work has demonstrated that miRNAs are essential for normal neocortical development (Bian et al., 2013, Kawase-Koga et al., 2010, Nowakowski et al., 2013, Pollock et al., 2014). Mice lacking dorsal cortical expression of the miRNA-processing molecule Dicer, and thus, expression of most miRNAs, develop abnormally small cortices (Davis et al., 2008, Kawase-Koga et al., 2009). This evidence suggests that miRNAs are required for cortical development; however, the specific miRNAs that regulate neurogenesis in the neocortex are still being uncovered.
miRNA-210 (miR-210) has previously been described to regulate tissue development, tumorigenesis, and post-ischemia (Biswas et al., 2010, Huang et al., 2009). Overexpression of miR-210 in non-neural tissues impairs proliferation (He et al., 2013, Tsuchiya et al., 2011, Zhang et al., 2009). Further, gene ontology analysis of predicted and verified mRNA targets of miR-210 reveal that many are involved in regulating the cell cycle and, possibly, in neural development (Fasanaro et al., 2009). Taken together, these data suggests that miR-210 is a promising potential regulator of NP proliferation and differentiation.
Here, we demonstrate that miR-210 is expressed in NPs throughout neocortical development and is an important regulator of NP proliferation. We show that altering miR-210 expression levels disrupts cell-cycle progression, ultimately altering the number and final laminar position of NP-derived postmitotic neurons. We also verify cyclin-dependent kinase 7 (Cdk7) as a target of miR-210 and an important regulating factor in the NP cell cycle. Our results suggest that proper expression levels of CDK7 and miR-210 are required for normal neocortical development.
Results
miR-210 Regulates Proliferation in Radial Glial Cells and Intermediate Progenitors
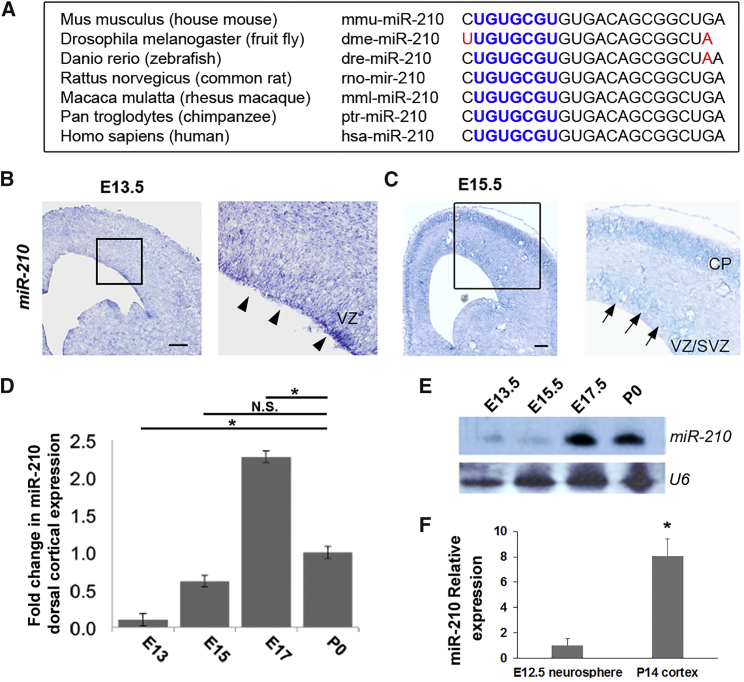
In our preliminary screen, we compared miRNA expression levels in the mouse neocortex at embryonic day 12.5 (E12.5) and postnatal day 0 (P0). We identified an miRNA, miR-210, which shows high expression at E12.5 and P0. We speculated that miR-210 plays a role in regulating NP proliferation. Based on analysis of its seed sequence and genomic context, miR-210 does not belong to a large family or cluster of miRNAs, and its seed sequence is conserved in invertebrates and vertebrates (Figure 1A). Consequently, we analyzed only the predominant mouse miR-210-3p strand, termed miR-210 for this study.
Figure 1.
miR-210 Is Expressed in the Developing Mouse Neocortex
(A) The mature miR-210 sequence is highly conserved across species. The conserved seed sequence of miR-210 is shown in blue and the variation in sequences among species is shown in red.
(B and C) Expression pattern of miR-210 in embryonic day 13.5 (E13.5) (B) and E15.5 (C) mouse cortices, as detected by in situ hybridization. miR-210 was expressed in neural progenitors in the ventricular zone (VZ, arrowheads) of the E13.5 cortex (B), and the VZ and subventricular zone (SVZ, arrows) at E15.5 (C). miR-210 was also expressed in the cortical plate (CP) of E15.5 neocortex (C).
(D) Expression of miR-210 in mouse dorsal cortices at different developmental stages, as detected by qRT-PCR. Expression was normalized to P0. The ubiquitously expressed Gapdh was used as an internal control.
(E) Expression of miR-210 in mouse dorsal cortices at different developmental stages, as detected by the northern blotting assay. The ubiquitously expressed small noncoding RNA U6 was used as a loading control.
(F) Expression of miR-210 in mouse neurospheres, cultured from E12.5 cortical neural stem cells, and in P14 cortex, detected by qRT-PCR. U6 was used as an internal control.
Scale bar, 20 μm. Data are presented as means ± SEM; n = 3 individual brain samples at each time point; p values in relation to control (∗p < 0.05). N.S., not significant.
We first examined miR-210 expression in the mouse neocortex during development using in situ hybridization. miR-210 was expressed in NPs in the VZ at E13.5, and in the VZ, SVZ, and early-born neurons in the CP at E15.5 (Figures 1B and 1C). Northern blotting assay and real-time quantitative RT-PCR (qRT-PCR) confirmed that miR-210 is expressed in the neocortex during early neurogenesis (E13.5). Its expression increased at the peak of neurogenesis (E15.5–E17.5) and slightly decreased at the postnatal stage (Figures 1D and 1E). Moreover, we cultured neural stem cells, collected from E12.5 neocortex, to form neurospheres and compared miR-210 expression in neurospheres and cortical tissues at P14. miR-210 expression was detected in neurospheres and its expression increased at P14 (Figure 1F).
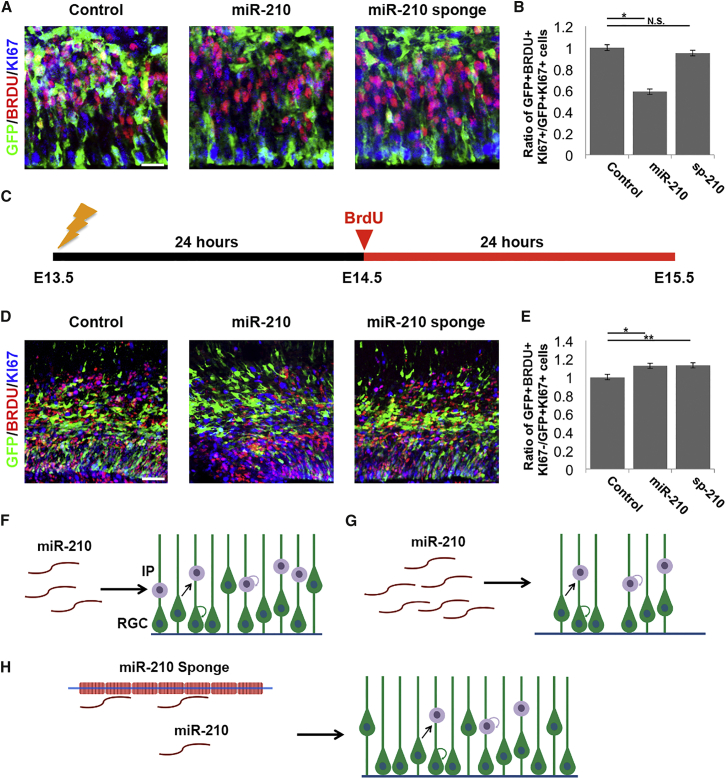
To determine the function of miR-210 in the regulation of the NP pool in the developing neocortex, we manipulated miR-210 expression in the ventricular NPs using in utero electroporation at E13.5 and collected the tissue on E14.5, 24 hr post-electroporation (Figure 2A). To overexpress miR-210, we designed a construct that contains the mouse miR-210 precursor sequence. Moreover, miR-210 knockdown was achieved through application of an miRNA sponge, specific to miR-210, here called miR-210 sponge (sp-210) (Otaegi et al., 2011). Overexpression of miR-210 resulted in a pronounced reduction in proliferation compared with vector controls, as measured by incorporation of the cell-cycle S-phase marker bromodeoxyuridine (BRDU) (Figures 2B and 2C). Interestingly, miR-210 did not appear to impair other mitotic phases, as KI67 labeling was unchanged in miR-210 overexpression cells (Figures 2D and 2E). However, sp-210 impaired S-phase entry compared with the control and resulted in a more pronounced cell-cycle impairment, and induced a reduction in KI67 expression, suggesting that inhibition of miR-210 function affects more than one mitotic phase (Figures 2B–2E).
Figure 2.
miR-210 Regulates Neural Progenitor Proliferation
(A) Illustration of in utero electroporation tissue collection. Electroporation was performed at E13.5, followed by BrdU incorporation at 23 hr, and tissue analyzed at E14.5.
(B–E) Overexpression (miR-210) and sponge-mediated knockdown (sp-210) of miR-210 decreased expression of proliferative markers BRDU (B and C) and KI67 (D and E) in GFP-positive cells.
(F–I) Overexpression of miR-210 decreased RGC marker PAX6 expression in GFP-positive cells, while sp-210 expression increased it (F and G). Both miR-210 and sp-210 expression decreased expression of the IP marker TBR2 (H and I).
Scale bar, 20 μm. Data are presented as means ± SEM; n = 6 individual brains for all constructs; p values in relation to control (∗p < 0.05, ∗∗∗p < 0.001). N.S., not significant.
Furthermore, because PAX6 and TBR2 have been shown to label RGCs and IPs, respectively, we examined whether the number of RGCs and IPs is affected by miR-210 (Englund et al., 2005). miR-210 overexpression resulted in a reduction in the size of both of the RGC (PAX6+ cells) and IP (TBR2+ cells) populations, which indicates that the miR-210-induced cell-cycle defect impairs proliferation in the entire NP pool (Figures 2F–2I). Noticeably, sp-210 overexpression induced a similar reduction in the size of the IP population compared with control. However, sp-210 promoted a striking increase in the size of the RGC pool (Figures 2F–2I). These results indicate that sp-210 negatively regulates cell-cycle progression in RGCs and reduces their transition to IPs, thereby decreasing the size of the IP pool.
To verify the specificity of sp-210, we generated an miR-210 sponge mutation (mut-sp-210), which is identical to sp-210 except for three mismatches in the target-binding site. Mut-sp-210 had no effect on proliferation in NP populations and RGCs and IPs, indicating that sp-210, not its mutation, can specifically knock down miR-210 in vivo (Figure S1). Our results suggest that precise levels of miR-210 expression are crucial for regulating proliferation in NPs and maintaining the size of distinct NP populations in the embryonic neocortex.
miR-210 Regulates Neural Progenitor Cell-Cycle Exit
We next investigated the function of miR-210 in regulating cell-cycle parameters, specifically, the labeling index and the cell-cycle-exit index. The labeling index, the proportion of cells entering the cell cycle, is measured by quantifying BrdU incorporation in all cycling cells (GFP+KI67+). Cell-cycle exit is measured by the number of cells that exit the cell cycle within 24 hr of BRDU incorporation (GFP+BRDU+KI67−) versus the total number of cycling cells (GFP+KI67+).
In brain tissue collected 24 hr post-electroporation, cells overexpressing miR-210, but not sp-210, displayed a decreased labeling index compared with controls, suggesting that miR-210 overexpression but not its knockdown reduces the fraction of cycling NPs reentering the cell cycle (Figures 3A and 3B). For cell-cycle exit analysis, brain tissue was collected 48 hr post-electroporation and 24 hr post-BrdU incorporation (Figure 3C). miR-210 overexpression resulted in an increased rate of NP exit from the cell cycle. Similarly, sp-210 expressing cells also exit the cell cycle at an elevated rate (Figures 3D and 3E). These results indicate that miR-210 regulates the S-G1 cell-cycle transition in NPs, where cells decide either to reenter or exit the cell cycle (Figure 3F). Elevated miR-210 levels cause decreased NP cell-cycle entry, increased cell-cycle exit, and a reduction in the entire progenitor pool (Figure 3G). miR-210 knockdown results in increased RGCs, decreased IP proliferation, and decreased population size, likely because the RGCs remain arrested in the cell cycle and fail to give rise to IPs (Figure 3H).
Figure 3.
miR-210 Regulates S-Phase Entry and Cell-Cycle Exit in Neural Progenitors
(A and B) miR-210 but not sp-210 overexpression in the E13.5 mouse brain, analyzed at E14.5, decreased the proportion of cells entering the S phase or the BrdU labeling index.
(C) Illustration of in utero electroporation timeline for tissue collection. Electroporation was performed at E13.5, followed by BrdU incorporation at E14.5, and tissue analyzed at E15.5.
(D and E) miR-210 and sp-210 increased the proportion of cells exiting the cell cycle.
(F) Model of miR-210 regulation of NP proliferation. Endogenous expression of miR-210 promotes normal proliferation in RGCs and IPs.
(G) Elevated miR-210 expression induces a decrease in proliferation and an increase in cell-cycle exit in both RGCs and IPs, thus reducing the NP pool.
(H) Attenuated miR-210 expression using miR-210 sponge increases the population size of the RGCs, which fail to transition to IPs, resulting in an increased RGC pool and decreased IP pool.
Scale bar, 20 μm. Data are presented as means ± SEM; n = 5 individual brains for all constructs; p values in relation to control (∗p < 0.05, ∗∗p < 0.01). N.S., not significant.
miR-210 Targets Cdk7, a Cell-Cycle Regulator and Cyclin Activating Kinase
miRNAs function through silencing target genes, as a few studies have shown a targeting effect of miR-210 (Friedman et al., 2009). Because we found that miR-210 regulates cell-cycle progression in the developing neocortex, we sought to identify miR-210 target genes that might play a role in controlling the cell cycle in NPs. Among predicted miR-210 targets, we found that the 3′UTR of Cdk7 contains a binding site for miR-210 (Figure 4A). CDK7, a cyclin-dependent kinase (CDK) activating kinase (CAK) that targets and activates numerous other CDKs, regulates several cell-cycle checkpoints and is required for S-phase entry (Ganuza et al., 2012, Ganuza and Santamaria, 2012, Rossi et al., 2001, Schachter et al., 2013).
Figure 4.
Cdk7 Is a Putative Target of miR-210 in the Regulation of Proliferation in Neural Progenitors
(A) The Cdk7 3′UTR contains a target-binding site for miR-210.
(B) Luciferase activity in a luciferase vector containing the Cdk7 3′UTR was reduced by miR-210 expression. Co-expression of sp-210, but not mutated sp-210 (mut-sp-210), partially rescued the luciferase activity. n = 3 biological replicates.
(C) Expression pattern of CDK7 in E13.5 and E15.5 mouse cortices, as detected by immunohistochemistry. CDK7 was co-expressed with the NP marker NESTIN in the ventricular zone (VZ) and subventricular zone (SVZ) (arrowheads).
(D and E) shRNA-mediated knockdown of Cdk7 (shCdk7) decreased BRDU and KI67 expression in GFP-positive cells.
(F and G) shCdk7 decreased PAX6 and TBR2 expression in GFP-positive cells.
Scale bar, 20 μm. Data are presented as means ± SEM; n = 5 individual brains for all constructs; p values in relation to control (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). N.S., not significant.
To verify Cdk7 as a target of miR-210, we used a luciferase assay by cloning the 3′UTR of Cdk7 into a luciferase vector, which was then co-transfected with miR-210. Luciferase activity in constructs containing the Cdk7 3′UTR was significantly reduced by miR-210, while the control miRNA miR-9 did not affect luciferase activity, suggesting that miR-210 specifically binds the 3′UTR of Cdk7 (Figure 4B). Moreover, co-transfection of sp-210 with miR-210 partially rescued the reduction in luciferase activity, while mut-sp-210 had no rescuing effect, demonstrating the effectiveness and specificity of sp-210 at reducing miR-210 target-binding activity (Figure 4B). These results indicate that Cdk7 is a putative target for miR-210.
Reducing Expression of the miR-210 Target Gene Cdk7 Impairs Neural Progenitor Proliferation
We next determined if CDK7 is expressed in NPs by co-staining the embryonic neocortex with CDK7 and the NP marker NESTIN. CDK7 was expressed in NPs in the VZ at E13.5 and E15.5, as well as in the CP, similar to the expression pattern of miR-210, suggesting an overlapping expression of miR-210 and its target CDK7 in the developing neocortex (Figure 4C).
We next investigated the role of Cdk7 in NP proliferation by manipulating expression levels of Cdk7 using in utero electroporation at E13.5. To thoroughly examine Cdk7 function, four shRNA constructs were designed and examined to knock down expression of Cdk7. Among them, shCdk7-1 and shCdk7-4 showed a stronger knockdown effect, with the former inducing a more pronounced reduction in Cdk7 expression, as detected by both qRT-PCR and western blotting assay (Figures S2A and S2B). Moreover, shCdk7-1 also reduced endogenous Cdk7 expression in the E17.5 cortex, electroporated at E13.5 (Figures S2C and S2D). Moreover, overexpression of Cdk7 resulted in a dramatic increase in cell death, such that electroporated cells did not survive longer than 24 hr for analysis (Figures S2E and S2F).
Furthermore, shCdk7-1 resulted in a reduction in the number of NPs and the RGC and IP populations (Figures 4D–4G). These results are similar to that observed in miR-210 overexpression (Figure 2). shCdk7-4 induced a similar but less pronounced reduction in NP proliferation compared with shCdk7-1, which further confirms the effect of Cdk7 knockdown (Figure S3). Moreover, while shCdk7-1 did not promote cell-cycle exit in NPs, it induced a decrease in the NP labeling index (Figure 5). These results suggest that miR-210 and CDK7 are both important regulators of the cell cycle in NPs, and their balanced expression maintains normal mitotic progression.
Figure 5.
Knockdown of Cdk7 Impairs Cell-Cycle Entry but Has No Effect on Cell-Cycle Exit
(A and B) shCdk7 expression in the E13.5 mouse brain, analyzed at E14.5, decreased the proportion of cells entering the S phase or the BrdU labeling index. To compare the effect of shCdk7, miR-210, and sp-210 on the BrdU labeling index, the same control shown in Figure 3A is displayed here.
(C and D) shCdk7 expression in the E13.5 mouse brain, analyzed at E15.5, had no effect on the proportion of cells exiting the cell cycle compared with the control.
Scale bar, 20 μm. Data are presented as means ± SEM; n = 5 individual brains for all constructs; p values in relation to control (∗p < 0.05). N.S., not significant.
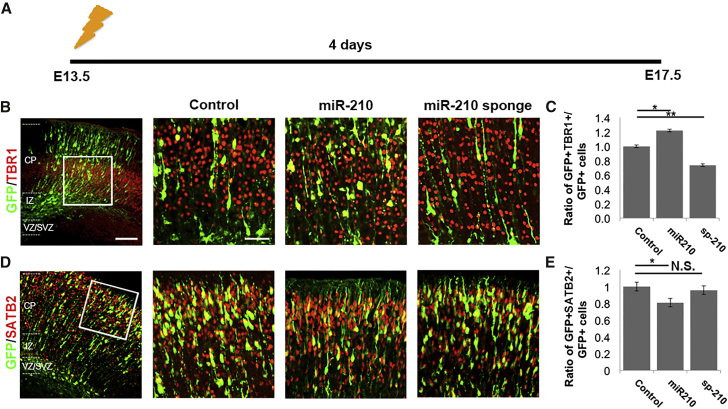
miR-210 Promotes Premature Differentiation of Neural Progenitors
Because altered cell-cycle regulation of NPs affects neurogenesis, we next investigated the function of miR-210 in NP terminal differentiation, using markers for early-born deep-layer neurons and late-born superficial-layer neurons, TBR1 and SATB2, respectively. To analyze NP differentiation, tissue was collected 4 days post-electroporation (Figure 6A). At E17.5, a higher proportion of miR-210 overexpressing postmitotic neurons co-expressed the deep-layer marker TBR1, while a lower proportion co-expressed the superficial-layer marker SATB2 compared with controls (Figures 6B–6E). Furthermore, cells overexpressing miR-210 populated deeper layers in the cortical wall compared with controls, suggesting that they were born earlier (Figure 7). Conversely, a decreased proportion of sp-210-expressing neurons co-expressed TBR1, with no significant change in SATB2-expressing cells (Figures 6B–6E), and an increased proportion populated the outermost layers of the cortex compared with controls, suggesting that they were born later (Figure 7).
Figure 6.
miR-210 Promotes Premature Differentiation in Neural Progenitors
(A) Illustration of in utero electroporation timeline for tissue collection. Electroporation was performed at E13.5 and tissue analyzed at E17.5.
(B–E) Overexpression of miR-210 increased the early-born neuronal marker TBR1 (B and C) and decreased the late-born neuronal marker SATB2 (D and E) in GFP-positive cells. Sp-210 decreased TBR1 expression in GFP-positive cells (C).
VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate.
Scale bar, 20 μm. Data are presented as means ± SEM; n = 5 individual brains for all constructs; p values in relation to control (∗p < 0.05, ∗∗p < 0.01). N.S., not significant.
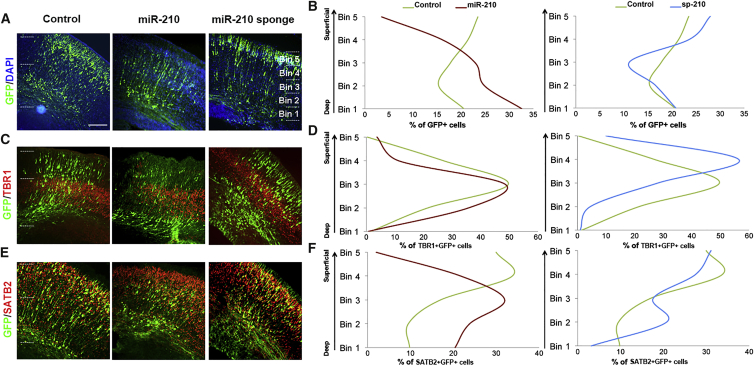
Figure 7.
miR-210 Promotes a Deep-Layer Location of Neurons in the Cortex
(A and B) Electroporation was performed at E13.5 and tissue analyzed at E17.5. Overexpression of miR-210 promoted a deep location of neurons in the cortical wall, while sp-210 induced a superficial location compared with empty vector controls (A). Relative numbers of GFP+ cells in each bin in the cortical wall are plotted in (B).
(C–F) Electroporation was performed at E13.5 and tissue analyzed at E17.5. Overexpression of miR-210 promoted deep localization within the TBR1 and SATB2 zones of expression. Sp-210 induced outer localization within the TBR1 and SATB2 zones of expression. Relative numbers of GFP+ cells in each bin in the cortical wall are plotted in (D and F). Control brains for GFP/TBR1 and GFP/SATB2 labeling, shown in Figures 6B and 6D, are displayed here for quantification of neuronal migration.
Scale bar, 20 μm. n = 5 individual brains for all constructs.
Furthermore, because migration of postmitotic neurons continues into the first postnatal week after the neurogenesis peak by E17.5 (Marin and Rubenstein, 2003), we then analyzed postmitotic cells on P5, 10 days post-electroporation (Figure S4A). We found that miR-210- and sp-210-expressing cells populate the deeper and more superficial layers of the cortex, respectively (Figures S4B–S4E). Moreover, sp-210-expressing cells were more likely to express the late-born neuronal marker SATB2 (Figures S4F and S4G), suggesting that the altered position of postmitotic neurons in the cortical wall is indicative of their birth date and is not transient, and that miR-210 is promoting deep lamination (Figure S5).
Discussion
Expansion of the NP population is thought to have contributed to the dramatic increase in the surface area of the neocortex (or neocorticalization) observed in higher mammals (Kriegstein et al., 2006, Rakic, 2009, Sun and Hevner, 2014). Thus, maintenance of the NP pool size and the rate of proliferation are crucial for normal cortical development and expansion. Here, we demonstrate that CDK7 and miR-210 play an important role in regulating cell-cycle progression as well as terminal differentiation in mouse NP populations, thereby helping to determine the birth date and laminar positioning of postmitotic neurons in the cortex.
In the mouse neocortex, generation of neurons peaks at approximately E15.5, at which time NP divisions have switched from asymmetric proliferative divisions to primarily symmetric neurogenic divisions (Miyata et al., 2004, Noctor et al., 2004). Lengthening of the NP cell cycle is thought to be one of the major factors regulating this switch (Dehay and Kennedy, 2007). Many cell-cycle regulators, both coding genes and miRNAs, are involved in regulating cell-cycle parameters in NPs (Buttitta and Edgar, 2007, Quinn et al., 2007). In this study, we show that overexpression and knockdown of miR-210 in NPs is sufficient to hinder the cell cycle, suggesting that optimal levels of miR-210 maintain proper cell-cycle progression. Overexpression of miR-210 consistently attenuates cell-cycle progression, across tissue types and environmental contexts, evincing that miR-210 plays a conserved role and directly or indirectly inhibits mitogenic function (He et al., 2013, Tsuchiya et al., 2011, Zhang et al., 2009).
Like most miRNAs, miR-210 functions by binding to and inhibiting its mRNA targets and modulates their expression levels (Bartel, 2004, Schratt, 2009). Here, we have found that miR-210 regulates the expression of the cell-cycle regulating molecule Cdk7, a kinase that is essential for all eukaryotic cell cycles. In non-neural cell types, ablation of Cdk7 expression induces cell-cycle arrest. We found that reduction of Cdk7 expression in NPs, much like overexertion of miR-210, yields a pronounced reduction in proliferation in both RGCs and IPs. This evidence indicates that Cdk7 and miR-210 may coordinate to promote NP cell-cycle progression. miR-210 may modulate Cdk7 expression levels to ensure that NPs enter and exit the cell cycle at an appropriate rate during cortical development.
Elevated miR-210 expression promotes cell-cycle exit, decreases the RGC and IP populations, and results in a reduction of the entire NP pool. miR-210 overexpression appears to force NPs out of the cell cycle prematurely, likely by inhibiting transition from G1 to S phase. As a consequence, later in development, at E17.5 and P5, cells overexpressing miR-210 populate the deeper layers of the cortex and become early-born neurons at higher proportions than controls. miR-210 knockdown also induces a proliferation impairment, selectively reducing the IP population and increasing the RGC population. This suggests that sp-210-expressing RGCs remain in the cell cycle and do not transition to IPs at the same rate as controls, possibly due to RGC cell-cycle arrest at G0 resulting from disrupted S-phase entry (Figure S5). At P5, sp-210-expressing cells are more likely to be late-born superficial neurons than controls (Figure S5). Our finding indicates that overexpression and knockdown of miR-210 have similar but distinct effects on cell-cycle progression in NPs. Together with CDK7, miR-210 fine-tunes proliferation and differentiation of cortical NPs and ensures establishment of proper lamination in the neocortex.
Because miR-210 is a conserved miRNA expressed in NPs at crucial points during neocortical development, it is likely that there are targets of miR-210 other than Cdk7, which also function in the regulation of cell-cycle progression in the cortex. Identifying targets and regulators of miR-210 that may contribute to its function of cell-cycle regulation in NPs will help provide a more complete picture of the role of miR-210 in the developing neocortex. In this study, our results suggest that miR-210 is an important regulator of cell-cycle progression, exit, and terminal differentiation in NPs of the developing neocortex, acting at least partially through regulation of Cdk7. Our work provides further insight into the vital role of miRNAs in cortical development.
Experimental Procedures
Tissue Preparation and Immunohistochemistry
Whole mouse brains were fixed overnight in 4% paraformaldehyde (PFA) in PBS, incubated in 30% sucrose in PBS overnight, embedded in optimal cutting temperature compound, and stored at −80°C until use. Brains were sectioned (12–16 μm) using a cryostat. For antigen retrieval, sections were immersed in heated (95–100°C) antigen recovery solution (1 mM EDTA, 5 mM Tris [pH 8.0]) for 15–20 min, and cooled for 20–30 min at 4°C. Sections were blocked in 10% normal goat serum in PBS with 0.1% Tween 20 (PBT) for 1 hr before adding antibodies. Sections were incubated with primary antibodies overnight at 4°C and visualized using goat anti-rabbit and/or anti-chick IgG-Alexa Fluor 488, goat anti-mouse and/or rabbit IgG-Alexa Fluor 546 (1:300; Molecular Probes), and goat anti-rabbit IgG-Cy5 (1:300; Invitrogen) for 1.5 hr at room temperature. Images were captured using a Leica digital camera under a fluorescent microscope (Leica DMI6000B) or a Zeiss confocal microscope. Primary antibodies against the following antigens were used: BRDU (1:50; DSHB, catalog no. G3G4, mouse), KI67 (1:500; Abcam, catalog no. ab15580, rabbit), PAX6 (1:200; Covance, catalog no. PRB-278P, rabbit), TBR1 (1:2500; Abcam, catalog no. ab31940, rabbit), TBR2 (1:500; Abcam, catalog no. ab23345, rabbit), SATB2 (1:1000; Abcam, catalog no. ab51502, rabbit), CDK7 (1:300; Santa Cruz, catalog no. sc-529, rabbit), NESTIN (1:10; DSHB, catalog no. Rat-401, mouse), GFP (1:1000; Abcam, catalog no. ab13970, chicken), and CASP3 (1:1000; R&D Systems, catalog no. AF835, rabbit).
Quantitative Real-Time RT-PCR
Total RNA was isolated from the dorsal cortex of E13.5, E15.5, E17.5, and P0 wild-type CD1 mice using the RNeasy Mini kit (QIAGEN) according to the manufacturer's instructions. All samples were treated with DNase to remove genomic DNA. Reverse transcription was performed using a Random Hexamer primer (Roche). qRT-PCR was performed using Power SYBR Green PCR Master Mix (Life Science) and Mx4000 Multiplex qPCR System (Stratagene) according to the manufacturer's instructions. RT primers for primary miR-210 (pri-miR-210) and internal control glyceraldehyde 3-phosphate-dehydrogenase (Gapdh) are listed as follows:
pri-miR-210 F: 5′-GGG GGC GGA GAG GAG GAC-3′, R: 5′-AGA TCA GCC GCT GTC ACA C-3′; Gapdh: F: 5′-ACT CCA CTC ACG GCA AAT TC-3′, R: 5′-CTA AGC AGT TGG TGG TGC AG-3′.
Statistics
For immunostaining, at least four sections from each brain and three brains were chosen for antibody labeling. For qRT-PCR, each sample was tested in triplicate. Statistical comparisons were made by an ANOVA (unpaired Student's t test).
Full details on the methods used are presented in the Supplemental Experimental Procedures.
Author Contributions
A.I.A., H.Z., W.T., and T.S. conceived the study and designed experiments, A.I.A. performed immunohistochemistry, analyzed data, and verified the target, H.Z. performed the in utero electroporation, western blotting assay, analyzed data, and verified the target, Y.N. performed neurosphere cultures and extracted RNA, and A.I.A. H.Z., and T.S. wrote the paper.
Acknowledgments
We thank Drs. Katherine Hajjar, M. Elizabeth Ross, and Barry Kosofsky for their insight and discussions of unpublished data. We are grateful to Dena Almeida for assistance with microscopy. This work was supported by the NIH Institutional T32 Training Grant (A.I.A.), the Hirschl/Weill-Caulier Trust (T.S.), and an R01-MH083680-08 grant from the NIH/NIMH (T.S.).
Published: July 12, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and five figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.06.005.
Contributor Information
Wei Tang, Email: tina_tangwei@163.com.
Tao Sun, Email: tas2009@med.cornell.edu.
Supplemental Information
References
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bian S., Hong J., Li Q., Schebelle L., Pollock A., Knauss J.L., Garg V., Sun T. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 2013;3:1398–1406. doi: 10.1016/j.celrep.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., Roy S., Banerjee J., Hussain S.R., Khanna S., Meenakshisundaram G., Kuppusamy P., Friedman A., Sen C.K. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc. Natl. Acad. Sci. USA. 2010;107:6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta L.A., Edgar B.A. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr. Opin. Cell Biol. 2007;19:697–704. doi: 10.1016/j.ceb.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A., Walsh C.A. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Chenn A., Walsh C.A. Increased neuronal production, enlarged forebrains and cytoarchitectural distortions in beta-catenin overexpressing transgenic mice. Cereb. Cortex. 2003;13:599–606. doi: 10.1093/cercor/13.6.599. [DOI] [PubMed] [Google Scholar]
- Davis T.H., Cuellar T.L., Koch S.M., Barker A.J., Harfe B.D., McManus M.T., Ullian E.M. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C., Kennedy H. Cell-cycle control and cortical development. Nat. Rev. Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- Englund C., Fink A., Lau C., Pham D., Daza R.A., Bulfone A., Kowalczyk T., Hevner R.F. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro P., Greco S., Lorenzi M., Pescatori M., Brioschi M., Kulshreshtha R., Banfi C., Stubbs A., Calin G.A., Ivan M. An integrated approach for experimental target identification of hypoxia-induced miR-210. J. Biol. Chem. 2009;284:35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineberg S.K., Kosik K.S., Davidson B.L. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Florio M., Huttner W.B. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- Franco S.J., Muller U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77:19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganuza M., Santamaria D. Cdk7: open questions beyond the prevailing model. Cell Cycle. 2012;11:3519–3520. doi: 10.4161/cc.21888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganuza M., Saiz-Ladera C., Canamero M., Gomez G., Schneider R., Blasco M.A., Pisano D., Paramio J.M., Santamaria D., Barbacid M. Genetic inactivation of Cdk7 leads to cell cycle arrest and induces premature aging due to adult stem cell exhaustion. EMBO J. 2012;31:2498–2510. doi: 10.1038/emboj.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz C.C., Lui J.H., LaMonica B.E., Wang X., Kriegstein A.R. Diverse behaviors of outer radial glia in developing ferret and human cortex. J. Neurosci. 2014;34:2559–2570. doi: 10.1523/JNEUROSCI.2645-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Wu J., Xu N., Xie W., Li M., Li J., Jiang Y., Yang B.B., Zhang Y. MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes. Nucleic Acids Res. 2013;41:498–508. doi: 10.1093/nar/gks995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ding L., Bennewith K.L., Tong R.T., Welford S.M., Ang K.K., Story M., Le Q.T., Giaccia A.J. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y., Otaegi G., Sun T. Different timings of dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev. Dyn. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y., Low R., Otaegi G., Pollock A., Deng H., Eisenhaber F., Maurer-Stroh S., Sun T. RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J. Cell Sci. 2010;123:586–594. doi: 10.1242/jcs.059659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk T., Pontious A., Englund C., Daza R.A., Bedogni F., Hodge R., Attardo A., Bell C., Huttner W.B., Hevner R.F. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb. Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A., Noctor S., Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Marin O., Rubenstein J.L. Cell migration in the forebrain. Annu. Rev. Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- Miyata T., Kawaguchi A., Saito K., Kawano M., Muto T., Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Noctor S.C., Martinez-Cerdeno V., Ivic L., Kriegstein A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Nowakowski T.J., Fotaki V., Pollock A., Sun T., Pratt T., Price D.J. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proc. Natl. Acad. Sci. USA. 2013;110:7056–7061. doi: 10.1073/pnas.1219385110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaegi G., Pollock A., Sun T. An optimized sponge for microRNA miR-9 affects spinal motor neuron development in vivo. Front Neurosci. 2011;5:146. doi: 10.3389/fnins.2011.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz L.J., Patti D., Marcy G., Ollier E., Pfister S., Douglas R.J., Betizeau M., Gautier E., Cortay V., Doerflinger N. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc. Natl. Acad. Sci. USA. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock A., Bian S., Zhang C., Chen Z., Sun T. Growth of the developing cerebral cortex is controlled by microRNA-7 through the p53 pathway. Cell Rep. 2014;7:1184–1196. doi: 10.1016/j.celrep.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.C., Molinek M., Martynoga B.S., Zaki P.A., Faedo A., Bulfone A., Hevner R.F., West J.D., Price D.J. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev. Biol. 2007;302:50–65. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D.J., Londesborough A., Korsisaari N., Pihlak A., Lehtonen E., Henkemeyer M., Makela T.P. Inability to enter S phase and defective RNA polymerase II CTD phosphorylation in mice lacking Mat1. EMBO J. 2001;20:2844–2856. doi: 10.1093/emboj/20.11.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter M.M., Merrick K.A., Larochelle S., Hirschi A., Zhang C., Shokat K.M., Rubin S.M., Fisher R.P. A Cdk7-Cdk4 T-loop phosphorylation cascade promotes G1 progression. Mol. Cell. 2013;50:250–260. doi: 10.1016/j.molcel.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. Fine-tuning neural gene expression with microRNAs. Curr. Opin. Neurobiol. 2009;19:213–219. doi: 10.1016/j.conb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Sessa A., Mao C.A., Hadjantonakis A.K., Klein W.H., Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M., Nakao H., Kiyonari H., Abe T., Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Hevner R.F. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat. Rev. Neurosci. 2014;15:217–232. doi: 10.1038/nrn3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E., Gotz M., Huttner W.B. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- Toma K., Kumamoto T., Hanashima C. The timing of upper-layer neurogenesis is conferred by sequential derepression and negative feedback from deep-layer neurons. J. Neurosci. 2014;34:13259–13276. doi: 10.1523/JNEUROSCI.2334-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S., Fujiwara T., Sato F., Shimada Y., Tanaka E., Sakai Y., Shimizu K., Tsujimoto G. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J. Biol. Chem. 2011;286:420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Sun H., Dai H., Walsh R.M., Imakura M., Schelter J., Burchard J., Dai X., Chang A.N., Diaz R.L. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.