Abstract
We describe a reliable and semi-automated method for Killer-cell Immunoglobulin-like Receptor (KIR) 3DL1/S1 genotyping using DNA recovered from frozen plasma. The primers and protocol were first validated using two independent genomic DNA reference panels. To confirm the approach using plasma-derived DNA, total nucleic acids were extracted from 69 paired frozen PBMC and plasma specimens representing all common KIR3DL1/S1 genotypes (3DS1/3DS1, 3DS1/3DL1 and 3DL1/3DL1, including rare allele 3DL1*054), and analyzed in a blinded fashion. The method involves independent nested PCR amplification of KIR3DL1/S1 Exon 4, and if required Exon 3, using universal sequence-specific primers, followed by bidirectional sequencing. The free basecalling software RECall is recommended for rapid, semi-automated chromatogram analysis. KIR3DL1/S1 type assignment is based on two key nucleotide polymorphisms in Exon 4 and, if required, up to two additional polymorphisms in exon 3. Assignment can be performed manually or using our web-based algorithm, KIR3D. Extractions from plasma yielded median [IQR] nucleic acid concentrations of 0.9 [below the limit of detection-2.45] ng/μl. PCR was successful for 100% of exon 4 (69/69) and exon 3 (29/29) plasma amplifications. Chromatogram quality was high and concordance between PBMC and plasma-derived types was 100%. The estimated lower limit of input DNA required for reliable typing is 0.01 ng/μl. This method provides reliable and accurate KIR3DL1/S1 typing when conventional sources of high-quality genomic DNA are unavailable or limiting.
Keywords: Killer-cell immunoglobulin receptor, KIR3DL1/S1, Plasma, Genotyping
1. INTRODUCTION
The Killer cell Immunoglobulin-like Receptor (KIR) gene family encodes receptors that regulate the function of Natural Killer (NK) cells (Dupont et al., 1997 and Bashirova et al., 2006). Approximately 150 kb long, the KIR region maps within the Leukocyte Receptor Complex (LRC) on human chromosome 19q13.4. This region, along with the Human Leukocyte Antigen (HLA) complex on chromosome 6p21.3, which encodes many known or presumed KIR ligands, ranks among the most polymorphic regions of the human genome (Campbell and Purdy, 2011). To date, 15 distinct KIR gene loci, plus two pseudogenes, have been defined (Marsh et al., 2003). KIR gene nomenclature reflects the structure of the receptor molecule encoded. The first digit following the KIR acronym indicates the number of Immunoglobulin (Ig)-like domains (D) while the subsequent letter indicates the length of the cytoplasmic tail: “L” (long), “S” (short) or “P” (pseudogene). In general, “L” and “S” genes encode inhibitory and activating KIR, respectively (Middleton and Gonzelez, 2010). The final digit corresponds to the specific protein characterized within the group (numbered in order of their characterization) (Marsh et al., 2003). Analogous to HLA nomenclature, the KIR gene name is followed by a “*” separator followed by the numerical allele designation (Marsh et al., 2003). KIR genes can vary from 4 to 16 Kb in length (genomic DNA) and encode four to nine exons encoding proteins 306–456 amino acids long (Selvakumar et al., 1996). For the KIR3DL1/S1 genes, exons 1–2 encode the leader peptide, exons 3–5 the D0, D1 and D2 extracellular domains, exon 6 the stem, exon 7 the transmembrane and exons 8–9 the cytoplasmic domains, respectively (Uhrberg et al., 1997).
KIR, alone and in combination with their specific ligands, modulate the susceptibility to, and pathogenesis of infectious (Gaudieri et al., 2005, Martin and Carrington, 2005, Boulet et al., 2008b, Bashirova et al., 2011, Dring et al., 2011, Guerini et al., 2011 and Korner and Altfeld, 2012) and autoimmune (Williams et al., 2005, Jiao et al., 2010 and Korner and Altfeld, 2012) diseases and influence transplantation outcomes (Nelson et al., 2004, Gagne et al., 2009 and Bao et al., 2010). Recently, interactions between highly polymorphic KIR3DL1/S1 molecules (of which 73 variants of the inhibitory KIR3DL1 and 16 variants of the activating KIR3DS1 molecules were characterized as of January 2013, http://www.ebi.ac.uk/ipd/kir/stats.html) and their putative HLA-B ligands, have been implicated in modulating Human Immunodeficiency Virus Type 1 (HIV-1) infection in some (Martin et al., 2002, Qi et al., 2006 and Martin et al., 2007) though not all (Gaudieri et al., 2005 and Barbour et al., 2007) HIV natural history studies. KIR3DL1 receptors are believed to interact with HLA-B molecules belonging to the Bw4 subfamily (determined by amino acids 77–83 of the HLA-B molecule), notably those harboring isoleucine at position 80 (Bw4-80I) (Cella et al., 1994), and to a lesser extent to those harboring threonine at this position (Bw4-80T) (Alter et al., 2007 and Eller et al., 2011). KIR3DS1 is suspected to interact directly with Bw4-80I, although this has not been confirmed (Korner and Altfeld, 2012). Protective effects of high-expressing, high inhibitory capacity KIR3DL1 alleles in combination with HLA-Bw4-80I on HIV acquisition have also been reported (Boulet et al., 2008a).
Molecular epidemiology investigations of the impact of immunogenetic variation on disease outcomes require genotyping of large cohorts. KIR genotyping can be performed using a variety of approaches including PCR using sequence-specific primers (PCR-SSP) (Kulkarni et al., 2010), allele-specific real-time PCR (Koehler et al., 2009 and Hong et al., 2011), single strand conformation polymorphism PCR (PCR-SSCP) (Witt et al., 2000) or sequence-based methods (Hou et al., 2012). These methods utilize genomic DNA extracted from peripheral blood mononuclear cells (PBMC) or other high-quality source as starting material, however methods utilizing lower-yield genomic DNA sources such as plasma or serum represent powerful research tools when PBMCs are unavailable (Martin et al., 1992, Fowke et al., 1995, Dixon et al., 1998 and Lin and Floros, 1998). Furthermore, though high-resolution allotyping is desirable for some applications, the ability to discriminate inhibitory (KIR3DL1) from activating (KIR3DS1) alleles is sufficient to inform many investigations (Martin et al., 2002, Martin et al., 2007, Boulet et al., 2008b and Guerini et al., 2011). To complement a protocol that our group previously developed for high-resolution HLA class I typing using genomic DNA extracted from plasma (Cotton et al., 2012), we describe a reliable, semiautomated sequence-based genotyping method for KIR3DL1/S1 genotyping using DNA recovered from this same material.
2. METHODS
2.1 Overview
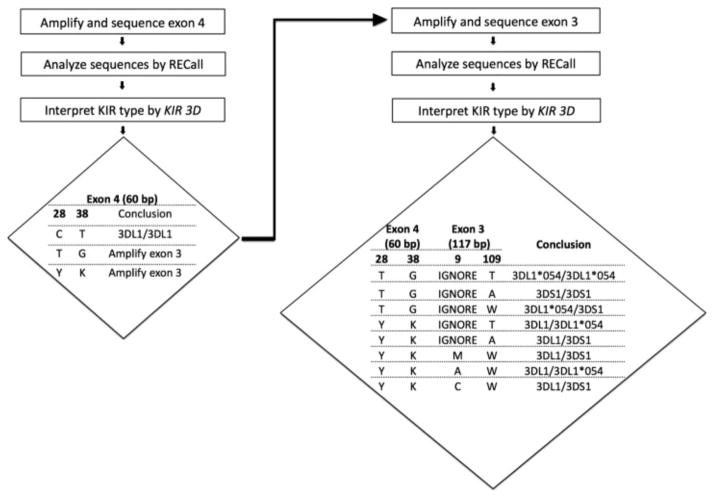
In developing our method, our goal was to achieve reliable typing by amplifying the smallest fragments and utilizing the minimum genetic information as input for type assignment. Examination of all known KIR3DL1/S1 allele sequences (available at http://www.ncbi.nlm.nih.gov/projects/gv/lrc/main.fcgi?cmd=init) revealed polymorphic sites in close proximity in KIR3DL1/S1 exons 3 and 4 that could discriminate 3DL1 from 3DS1 alleles. Our workflow and interpretation algorithm are presented in Fig. 1. Briefly, the first step involves nested PCR amplification, bulk sequencing and analysis of a 60 base pair region in exon 4 of the KIR3DL1/S1 gene, followed by interpretation of the respective KIR type based on nucleotide polymorphisms at positions 28 and 38 of the reference sequence. For approximately 60% of samples, this yields sufficient information to assign a type of 3DL1/3DL1. For the remaining ~ 40% of samples, nested PCR amplification, bulk sequencing and analysis of an 117-base pair region in exon 3 is required, followed by results interpretation based on nucleotide polymorphisms at position 109, and (if necessary) 9.
Figure 1.
Schematic workflow and interpretation algorithm for KIR3DL1/S1 typing. Interpretation can be performed manually, or using our web-based tool, KIR3D, developed for this purpose (see Methods section). Type assignment begins with inspection of nucleotide polymorphisms at positions 28 and 38 of the exon 4 reference sequence. Possible results are C–T, T–G or the mixed base calls Y–K. A type of C–T (observed in ~ 60% of cases) is sufficient to assign a type of KIR3DL1/3DL1. If T–G or Y–K combinations are observed, exon 3 sequencing is required. Exon 4 polymorphisms are then interpreted alongside polymorphisms at exon 3 nucleotide positions 109 and (if necessary) 9, yielding the eight possible KIR types shown. Types Y–K–M–W and Y–K–A–W incorporate a small amount of uncertainty in their assignments (see Methods section).
2.2. Samples and DNA reference panels
Validation of primers and protocol was first performed on the KIR Phase I genomic DNA Reference panel from the International Histocompatibility Working Group (IHWG) Cell and DNA bank (N = 48 samples; http://www.ihwg.org/reference/index.html), and secondly on a blinded reference panel provided by Dr. Nicole Bernard (McGill University, Montreal, QC, Canada; N = 23 samples originally typed using PCR-SSP (Boulet et al., 2008b)). Validation of the plasma-based typing protocol was performed using a panel of matched PBMC and plasma samples from 69 HIV-negative participants of the Vancouver Injection Drug Users Study I (VIDUS-I) (Strathdee et al., 1997) enrolled between March to July 1999. Blood was collected in BD Vacutainer® CPT™ Cell Preparation tubes containing Sodium Citrate (Becton, Dickinson, NJ, USA). Plasma and PBMCs were separated by centrifugation according to the manufacturer’s instructions and stored at −80 °C until use. The panel was selected to reflect a diversity of KIR3DL1/S1 types and contained 2, 26 and 41 individuals with 3DS1/3DS1, 3DS1/3DL1, 3DL1/3DL1 types, respectively, where the latter included one individual expressing rare allele 3DL1*054 (the only 3DL1 allele that possesses 3DS1-like polymorphisms at key exon 4 positions). All participants provided written informed consent.
2.3. DNA extraction and quantification
Total nucleic acids were extracted from ~ 2.5 million PBMC and 500 μl of plasma using a standard silica-based method, omitting the RNAase treatment step (NucliSENS® easyMAG®, BioMérieux, the Netherlands (Cotton et al., 2012)) and resuspended in 55 μl elution buffer (BioMérieux, the Netherlands). Extracts were assigned numeric ID codes and subsequent procedures were performed in a blinded manner. Total nucleic acid concentrations of extracts were measured on a NanoDrop™ 2000 spectrophotometer (Thermo Scientific Wilmington, DE, USA).
2.4. Amplification of KIR3DL1/S1 exon 4 and exon 3 (if required), from plasma-derived genomic DNA
Amplification of KIR3DL1/S1 exon 4, and if necessary exon 3, is performed via independent nested PCR using sequence-specific primers (Table 1). In addition to being more sensitive than a single round, nested PCR improves amplicon specificity and purity, yielding a cleaner DNA sequence, and is therefore recommended regardless of template DNA concentration. Primers were purified by standard desalting (Integrated DNA Technologies, Coralville, IA, USA). Reactions were prepared using the Roche Expand™ High Fidelity PCR system (Roche Applied Science, Laval, PQ, Canada).
Table 1.
Nested PCR and sequencing primers used for KIR3DL1/S1 typing.
| Exon | 1st round PCR | 2nd round PCR (and sequencing) |
|---|---|---|
| 4 | 3DL1_S1_356F: GAAACCACAGAAAACCTTCCCTC (3213–3235) | 3DL1_S1E4F: CTTCCCTCCTGGCCCACC (3228–3245) |
| 3DL1_S1_649R: CCACGATGTCCAGGGGATC (3488–3506) | 3DL1_S1E4R: GGATCACTGGGAGCTGACAACTG (3470–3492) | |
| 3 | 3DL1_S1_157F: GTCAGGACAAGCCCTTCCTG (1818–1837) | 3DL1_3DS1_Exon3F1: CCTTCCTGTCTGCCTGG(1830– 1846) |
| 3DL1_S1_258R: TGGGTGCCGACCACC (2058–2072) | 3DL1_3DS1_Exon3R1: ACCCAGTGGGGGAGTGT (2044–2060) |
Names, sequences and binding coordinates (in brackets) of primers used for KIR3DL1/S1 typing. All primers are in 5′–> 3′ orientation. Coordinates are numbered according to the KIR3DL1*0010101 genomic DNA reference sequence (EMBL-EBI, IPD KIR database access number: AC011501); alignments retrievable from (http://www.ebi.ac.uk/ipd/kir/align.html).
The first round PCR master mix for exon 4, yielding a 294 base pair amplicon, was prepared as follows for a final reaction volume of 50 μl per sample: 31.5 μl of molecular-grade H2O, 5 μl of 10 × Expand™ High Fidelity Buffer containing 15 mM MgCl2 (supplied by manufacturer), 2 μl of 25 mM MgCl2 (supplied by manufacturer), 0.5 μl of 25 mM dNTPs (Roche Applied Science, Laval, PQ, Canada), 1 μl each of 10 μM forward and reverse primers (Table 1) and 1 μl Expand™ High Fidelity Enzyme Mix (3.5 U/μl; supplied by manufacturer). 8 μl nucleic acid extract was added to each reaction. Negative (water) controls were included in each run. PCR amplification was performed using an ABI 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA), as follows: denaturation at 94 °C for 2 min, followed by 18 cycles of (denaturation at 94 °C for 15 s, annealing at 60 °C for 20 s and extension at 72 °C for 30 s), with a final extension at 72 °C for 5 min.
Second round PCR master mix for exon 4 was prepared as follows, with a final total reaction volume of 20 μl per sample: 11.75 μl of molecular-grade H2O, 2 μl 10 × Expand™ High Fidelity Buffer containing 15 mM MgCl2, 0.8 μl 25 mM MgCl2, 2 μl of 60% sucrose buffer containing 0.08% Cresol Red (note: this reagent is optional and can be substituted with molecular-grade H2O; its purpose is to serve as a pH indicator and to allow direct loading of amplicons onto to agarose gel without addition of loading dye), 0.16 μl of 25 mM dNTPs, 0.5 μl of 10 μM forward and reverse primers (Table 1) and 0.29 μl Expand™ High Fidelity Enzyme Mix. A total of 2.6 μl of first round PCR product was transferred to each reaction as template. Thermal cycling conditions were: denaturation at 94 °C for 2 min, followed by 30 cycles of (denaturation at 94 °C for 15 s, annealing at 55 °C for 20 s and extension at 72 °C for 30 s), with a final extension at 72 °C for 5 min.
If required (see 2.5 and 2.6), exon 3 was amplified using nested primers (Table 1) under conditions identical to those described for exon 4.
PCR master mix ingredients for plasma-based KIR3DL1/S1 typing, and a slightly modified protocol for typing from PBMC-derived genomic DNA, are summarized in Table 2. Thermal cycling conditions are identical for both protocols.
Table 2.
Per-reaction master mixes for KIR3DL1/S1 amplification from PBMC or plasma-derived genomic DNA.
| Specimen type | 1st round PCR | Volume (μl) | 2nd round PCR | Volume (μl) |
|---|---|---|---|---|
| PBMC | H2O | 20 | H2O | 6.5 |
| 10 × buffer w/15 mM MgCl2 a | 2.5 | 10 × buffer w/15 mM MgCl2 | 1 | |
| 25 mM dNTP | 0.25 | Sucrose buffer c | 1 | |
| 10 μM Forward primer | 0.5 | 25 mM dNTP | 0.1 | |
| 10 μM Reverse primer | 0.5 | 10 μM Forward primer | 0.25 | |
| Enzyme mix b | 0.25 | 10 μM Reverse primer | 0.25 | |
| Template DNA | 1 | Enzyme mix | 0.15 | |
| Total | 25 | 1st round PCR product | 1 | |
| Total | 10.25 | |||
| Plasma | H2O | 31.5 | H2O | 11.75 |
| 10 × buffer w/15 mM MgCl2 | 5 | 10× buffer w/15 mM MgCl2 | 2 | |
| 25 mM MgCl2 | 2 | Sucrose buffer | 0.8 | |
| dNTP | 0.5 | 25 mM MgCl2 | 2 | |
| 10 μM Forward primer | 1 | dNTP | 0.16 | |
| 10 μM Reverse primer | 1 | Forward primer | 0.5 | |
| Enzyme mix | 1 | Reverse primer | 0.5 | |
| Template DNA | 8 | Enzyme mix | 0.29 | |
| Total | 50 | 1st round PCR product | 2.6 | |
| Total | 20.6 |
We recommend a nested PCR approach even for higher DNA-concentration templates, as nested PCR improves specificity and amplicon purity, thereby yielding a higher-quality DNA sequence.
10 × Expand™ High Fidelity Buffer containing 15 mM MgCl2.
Expand™ High Fidelity Enzyme Mix (3.5 U/μl).
60% sucrose buffer containing 0.08% Cresol Red.
Due to the small amplicon size, successful amplification of products was visually confirmed on a 3% agarose gel prior to DNA sequencing.
2.5. DNA sequencing
Before bulk (“direct”) sequencing, second round amplicons were diluted approximately 15-fold with molecular-grade H2O. Dilution renders amplicon concentrations appropriate for sequencing, and removes the need for PCR purification using commercial kits. Amplicons were sequenced bidirectionally using the same primers used for 2nd round PCR (Table 1) in small-volume (6 μl) reactions consisting of 2.6 μl of 2 μM sequencing primer, 2.1 μl dilution buffer (175 mM Trizma HCl, 1.25 mM MgCl2, pH = 9.0) and 0.3 μl of ABI Prism® BigDye® Terminator v3.1 sequencing mix (Applied Biosciences by Life Technologies, Carlsbad, California), to which 1 μl diluted 2nd round amplicon was added as a template. Plates were capped, vortexed and briefly centrifuged to ensure reactions were positioned at the bottom of each well. Thermal cycling consisted of 25 cycles of (10 s at 96 °C, 5 s at 50 °C and 55 s at 60 °C). Following amplification, reactions were purified via ethanol precipitation by adding 4 μl of 1 × EDTA-Sodium Acetate solution (containing 375 mM Sodium Acetate and 62.5 mM EDTA) and 40 μl of chilled (−20 °C) 95% ethanol to each well. Plates were incubated at −20 °C for 30 min and then centrifuged for 20 min at 2000 g after which the wash was discarded. A second wash using 155 μl of 95% chilled ethanol was performed without centrifugation, and wells were allowed to air dry. DNA pellets were resuspended in 10 μl of Hi-Di™ Formamide (Applied Biosciences by Life Technologies, Carlsbad, California) and denatured at 90 °C for 2 min. Data were collected on an ABI 3130 × l automated DNA analyzer (Applied Biosciences by Life Technologies, Carlsbad, California).
2.6. Analysis and interpretation
Chromatograms were aligned to 60 base pair (exon 4) and 117 base pair (exon 3) reference standards spanning 3DL1*0010101 nucleotide CDS coordinates 523–582 (exon 4) and 115–231 (exon 3) (reference sequences supplied in FASTA format asSupplementary data). For chromatogram assembly and basecalling, we employed the free, web-based, semi-automated software RECall (http://pssm.cfenet.ubc.ca/) (Woods et al., 2012) with the threshold of nucleotide mixture detection set at 20% of the dominant peak area. Commercial basecalling softwares such as Sequencher® v4.9 (Gene Codes Corporation, Ann Arbor, MI, USA) can also be used; in this case a secondary peak height of 25% is recommended for nucleotide mixture detection. Analyzed sequences can be exported from RECall as concatenated (batch) FASTA files labeled with their individual IDs.
Assignment of KIR types can be performed manually according to the algorithm outlined in Fig. 1, or using our web-based algorithm KIR 3D, which accepts batch FASTA sequences as input. Access to this tool is available to interested researchers (please contact corresponding author for link and password). As outlined in Fig. 1, the tool assigns KIR types based on nucleotide polymorphisms at positions 28 and 38 in the exon 4 reference sequence. All known 3DL1 alleles except KIR3DL1*054 exhibit C-T polymorphisms at these positions. Therefore, if a C-T genotype is observed a type of KIR3DL1/3DL1 is assigned and typing is complete. Approximately 60% of samples fall into this category. If T-G or mixed base Y-K polymorphisms are observed, the KIR 3D tool will instruct the user to perform exon 3 amplification and sequencing. Upon subsequent input of both exon 4 and exon 3 sequences with matching IDs, the KIR 3D tool will additionally incorporate information from exon 3 nucleotide 109, and if necessary 9, to assign the type. Briefly, in exon 3, all 3DS1 alleles exhibit 109A and the vast majority of 3DL1 alleles (including 3DL1*054) exhibit 109T. Therefore, for all types that contain at least one single (non-mixed) base in exon 4 or 3, the combination of exon 4 and exon 3 base 109 is sufficient to assign type as KIR3DL1/3DL1 (where one or both alleles will be KIR3DL1*054), KIR3DL1/3DS1 or KIR3DS1/3DS1. For samples with mixed bases in exon 4, and exon 3 base 109, exon 3 base 9 is additionally required to assign the type. Importantly, note that the assignments for Y–K–M–W and Y–K–A–W incorporate a small amount of uncertainty. Briefly, all 3DS1 alleles except 3DS1*014 display a C at exon 3 position 9, while all 3DL1 alleles except 3DL1*009, 3DL1*042 and 3DL1*057 display A. However, since at time of submission these alleles had extremely low (or no) reported frequencies in most countries (http://www.allelefrequencies.net, (Gonzalez-Galarza et al., 2011)), we assign the most probable types of KIR3DL1/3DS1 and KIR3DL1/3DL1*054, respectively, in these cases.
Finally note that the KIR 3D algorithm features quality-control features. The tool will return errors if input sequences are the incorrect length, incorrect orientation, if exon 3/4 fragments from the same sample are labeled with non-matching IDs, or if nucleotide combinations other than those expected are observed.
3. RESULTS AND DISCUSSION
3.1. Justification for exon 4 and 3 fragments as basis for typing
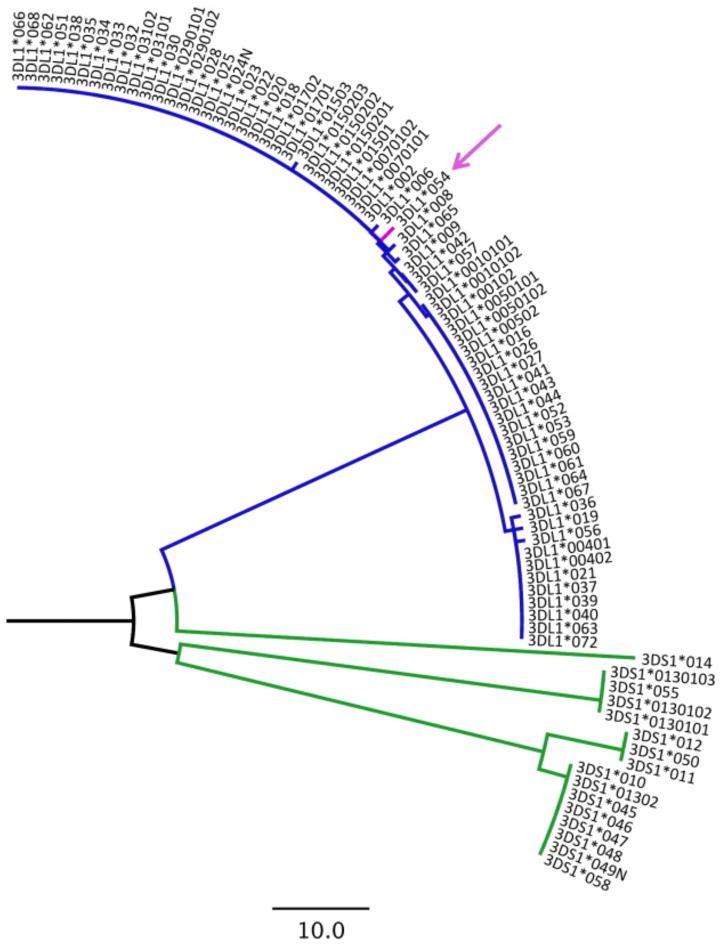
When amplifying genomic DNA from low-copy/low-quality sources such as plasma, amplicon lengths should be kept to a minimum to increase reaction success rates. In addition, interpretation algorithms should use the fewest number of polymorphic sites in the type assignment process while preserving accuracy. For this reason, we sought to design a minimalist method based on small amplicons, and an interpretation simple enough to be performed manually if desired. Our method is therefore based on the amplification, sequencing, analysis and interpretation of a 60 base pair region in KIR3DL1/S1 exon 4, and if necessary a 117 base pair region in exon 3, in which a minimum of two, and a maximum of four polymorphisms are required for interpretation. A maximum-likelihood phylogenetic tree constructed by Molecular Evolutionary Genetic Analysis (MEGA) package 5 (Tamura et al., 2011), using a concatenated alignment of these two small fragments for all known KIR3DL1/S1 alleles (http://www.ncbi.nlm.nih.gov/projects/gv/lrc/main.fcgi?cmd=init, accessed January 19, 2013) indicates that these regions contain sufficient genetic information to clearly segregate 3DL1 from 3DS1 alleles (Fig. 2).
Figure 2.
Maximum-likelihood phylogenetic tree constructed from a concatenated nucleotide alignment of exon 4 (60 base-pair) and 3 (117 base-pair) reference fragments for all known KIR3DL/S1 alleles. The tree indicates that these small fragments contain sufficient information to clearly discriminate KIR3DL1 (blue) from KIR3DS1 (green) alleles. Note that rare allele KIR3DL1*054 (pink arrow) differs from all other KIR3DL1 alleles in that it harbors KIR3DS1-like polymorphisms (T-G) at codons 28 and 38 of the exon 4 reference fragment, thus necessitating exon 3 sequencing.
3.2. Methods validation
A modified version of the plasma typing protocol (see Methods section and Table 2) was first validated in a blinded fashion using reference DNA panels obtained from commercial (IHWG KIR Phase I genomic DNA Reference panel; N = 48) and academic (Dr. Nicole Bernard, McGill University, Montreal, QC, N = 23 samples) sources, which comprised samples with 3DL1/3DL1, 3DL1/3DS1 and 3DS1/3DS1 types. We achieved 100% typing accuracy for all reference samples (not shown). Validation of the plasma-based typing protocol was performed using 69 frozen paired PBMC/plasma samples from HIV-negative participants of the Vancouver Injection Drug Users Study I (VIDUS-I) (Strathdee et al., 1997). This represented the first thawing and manipulation of these specimens, which had been stored at −80 °C for 13.5 years.
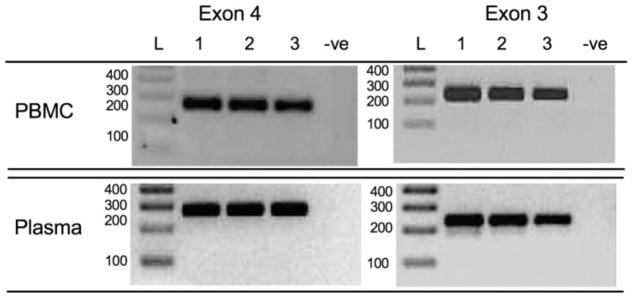
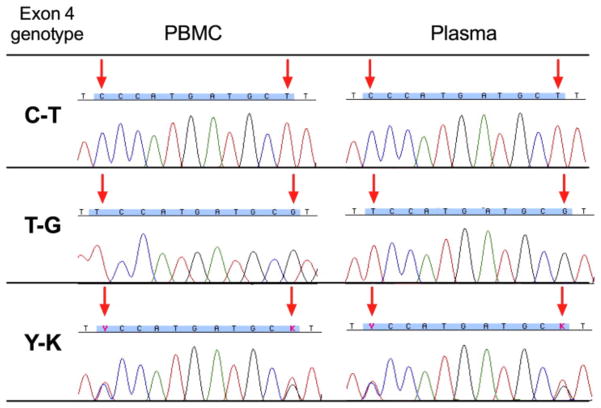
The median and interquartile ranges [IQR] of total nucleic acid yields from PBMC and plasma specimens were 11.5 [3.45–33.05] ng/μl and 0.9 [below the limit of detection-2.45] ng/μl, respectively. Since the extraction procedure does not include an RNAse treatment step ( Cotton et al., 2012), these concentrations include cellular RNA as well as any plasma viral RNA (HCV in particular is highly prevalent in the studied population (Patrick et al., 2001)); genomic DNA concentrations may therefore be lower. 8 μl of plasma-derived extract was added as template per 1st round PCR, representing a median [IQR] template input quantity of 7.2 [8–19.6] ng total nucleic acid per reaction. Nested PCR amplification of both exons 3 and 4 yielded high quality bands for PBMC and plasma-derived DNA in all cases (Fig. 3). DNA sequence chromatograms were of uniformly high quality with clearly distinguishable mixed bases regardless of DNA source (Representative exon 4 sequences shown in Fig. 4).
Figure 3.
PCR amplification of KIR3DL1/S1 exon 4 and 3 from PBMC vs. plasma-derived genomic DNA. Representative amplicons (and negative control) of KIR3DL1/S1 exon 4 and 3, respectively, amplified from PBMC (upper panels) and plasma (lower panels). Amplicons were resolved on 3% agarose gel and bands were visualized under UV light alongside DNA size standards (Invitrogen 1 kb + DNA ladder; Life Technologies, Burlington, ON, Canada) after staining with SYBR safe DNA Gel Stain (Invitrogen).
Figure 4.
Sequence chromatograms for KIR3DL1/S1 exon 4 sequences from PBMC vs. plasma-derived genomic DNA. Representative chromatograms from the exon 4 region spanning nucleotides 28–38 (blue highlighting) for matched PBMC (left panels) and plasma (right panels) specimens displaying the three possible nucleotide combinations at key positions 28 and 38 (arrows). Mixed bases are denoted using International Union of Pure and Applied Chemistry (IUPAC) ambiguity codes: Y (C/T) and K (G/T).
Concordance between PBMC and plasma-based KIR 3DL1/3DS1 typing results was 100%. Out of a total of 11,160 nucleotides analyzed for exons 3 and 4, no mismatches were observed between PBMC and plasma-derived sequences.
3.3. Estimating the lower limit of typing reliability
To estimate the lower limit of template nucleic acid required for reliable typing, we performed serial ten-fold dilutions of plasma-derived extracts from seven patients with 3DS1/3DL1 types and three patients with 3DL1/3DL1 types (mean concentration, 3.42 ng/μl). Extracts were serially diluted up to 1000-fold, yielding a panel of 30 diluted extracts with concentrations ranging from 0.0019 to 7.3 ng/μl. At a concentration of 0.01 ng/μl, amplification, sequencing and interpretation were successful for all diluted samples (not shown). Typing was possible at 0.001 ng/ul for some samples, but amplification was not reliable at this threshold. The lower limit of template nucleic acid concentration required for reliable typing is therefore estimated to be 0.01 ng/μl.
3.4. Method limitations and other recommendations
Some limitations of this new protocol merit mention. Firstly, although the short fragments sequenced contain sufficient genetic information to discriminate 3DL1 from 3DS1 alleles (and to some extent can further discriminate some 3DS1 alleles from one another; Fig. 2), there is insufficient information to discriminate 3DL1 and 3DS1 alleles at the individual level. Sequencing of additional regions would be necessary to perform high-resolution KIR3DL/S1 allotyping. Furthermore, two of our typing assignments (Y–K–M–W and Y–K–A–W) contain a very small (estimated < 1%) uncertainty due to polymorphisms found in very rare alleles (see Methods section).
In addition, our method is not designed to detect KIR3DL1/S1 copy number variation, nor was it validated on specimens from individuals with documented KIR3DL1/S1 locus deletions (present in an estimated 5% and 3.6% of individuals of European ancestry, respectively) (Pelak et al., 2011). Although we estimate a minimum of 0.01 ng/μl template nucleic acid is needed for reliable typing, template quality will also play a major role in determining this lower limit. Finally, although the RECall base calling software is designed to eliminate human subjectivity during chromatogram analysis, DNA sequence quality can be reduced when typing from low-copy number or low-quality templates. To avoid potential missed or erroneous calls, we recommend that users view their chromatograms, paying particular attention to key polymorphic positions listed in Fig. 1, prior to performing KIR3DL1/S1 type interpretation.
3.5. Simplified protocol based on exon 4 sequencing only: consider with caution
The rare allele 3DL1*054 differs from all other 3DL1 alleles in that it exhibits 3DS1-like “T–G” polymorphisms at key positions in exon 4, rather than the “C–T” motif expressed by all other 3DL1 alleles. KIR3DL1*054 is the reason that exon 3 sequencing is additionally required to discriminate it from 3DS1 alleles. If one were to accept the risk of erroneously mis-typing KIR3DL1*054 as 3DS1, one could simplify the protocol to comprise exon 4 genotyping only. In this case, C–T would designate 3DL1/3DL1, T–G would designate 3DS1/3DS1, and Y–K would designate 3DL1/3DS1. Moreover, if chromatogram quality is consistently excellent, throughput could be further enhanced and costs reduced by limiting exon 4 sequencing to the forward primer only.
If implemented correctly, the simplified protocol could yield time and cost savings of > 50% with only modest reductions in accuracy, which could be useful if very large numbers of specimens need to be typed rapidly. Note however that it should be considered with caution. Of the 581 patients typed thus far in our laboratory, 5 (0.86%) are 3DL1/3DL1*054 (not shown); therefore, we estimate that eliminating exon 3 sequencing will lead to erroneous types in a minimum of 1% of cases in North American populations. The error rate could be much higher in other populations, and could also be impacted by the future discovery of additional KIR alleles. Before the simplified protocol is considered, we strongly recommend that the original protocol be first implemented to confirm consistent chromatogram quality and to quantify KIR3DL1*054 prevalence in the studied population, so that informed decisions regarding method accuracy vs. cost-saving can be made.
3.6. Conclusion
We have developed a reliable and accurate sequence-based genotyping method to perform KIR3DL1/S1 typing using genomic DNA extracted from frozen plasma. A modified version of this method can also be used to type from genomic DNA extracted from PBMCs or other conventional source. Use of the free web-based sequence analysis software RECall (Woods et al., 2012) coupled with our web-based automated interpretation algorithm KIR3D renders analysis and KIR3DL1/S1 type interpretation virtually automated, although type assignment is simple enough to be performed manually if desired. The result is a method that is scalable for high-throughput typing. This method may be especially useful for retrospective molecular epidemiological studies where plasma represents the only archived specimen available, or where high quality genomic DNA extracts are inaccessible or limited in quantity.
Highlights.
Reliable, accurate method for KIR3DL1/S1 typing using genomic DNA from plasma.
Nested PCR/sequencing of short region within exon 4 (and if necessary exon 3).
Genotype interpretations can be performed manually or automatically (via web tool).
Estimated lower limit of input DNA required for reliable typing is 0.01 ng/μl.
Useful for studies where high quality genomic DNA is unavailable or limited.
Acknowledgments
Funding statement
This work was supported by an operating grant from the Canadian Institutes for Health Research (CIHR) (HOP-115700) to ZLB. The VIDUS and ACCESS projects were funded by the National Institute on Drug Abuse, NIH (RO1DA011591 and RO1DA021525). EM was supported by a Master’s Scholarship from the Canadian Association of HIV Research and Abbott Virology. MJM is the recipient of fellowships from the Michael Smith Foundation for Health Research and CIHR (Bisby Fellowship). ZLB is the recipient of a CIHR New Investigator Award and a Scholar Award from the Michael Smith Foundation for Health Research. The study sponsors were not involved in the study design, the collection, analysis or interpretation of the data, the writing of the report, or the decision to submit the article for publication.
The authors thank Nicole Bernard for providing a blinded genomic DNA reference panel for initial assay validation. The authors also thank Tsoarello Mabanga and Manal Abdur Rahman for technical assistance, and Mark Brockman for critical input on the manuscript.
Appendix A. Supplementary data
KIR3DL1 exon 4 and exon 3 reference standard sequences can be found online as supplementary data, at http://dx.doi.org/10.1016/j.jim.2013.03.005.
References
- Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao XJ, Hou LH, Sun AN, Qiu QC, Yuan XN, Chen MH, Chen ZX, He J. The impact of KIR2DS4 alleles and the expression of KIR in the development of acute GVHD after unrelated allogeneic hematopoietic SCT. Bone Marrow Transplant. 2010;45:1435. doi: 10.1038/bmt.2009.357. [DOI] [PubMed] [Google Scholar]
- Barbour JD, Sriram U, Caillier SJ, Levy JA, Hecht FM, Oksenberg JR. Synergy or independence? Deciphering the interaction of HLA Class I and NK cell KIR alleles in early HIV-1 disease progression. PLoS Pathog. 2007;3:e43. doi: 10.1371/journal.ppat.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- Bashirova AA, Thomas R, Carrington M. HLA/KIR restraint of HIV: surviving the fittest. Annu Rev Immunol. 2011;29:295. doi: 10.1146/annurev-immunol-031210-101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet S, Kleyman M, Kim JY, Kamya P, Sharafi S, Simic N, Bruneau J, Routy JP, Tsoukas CM, Bernard NF. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS. 2008a;22:1487. doi: 10.1097/QAD.0b013e3282ffde7e. [DOI] [PubMed] [Google Scholar]
- Boulet S, Sharafi S, Simic N, Bruneau J, Routy JP, Tsoukas CM, Bernard NF. Increased proportion of KIR3DS1 homozygotes in HIV- exposed uninfected individuals. AIDS. 2008b;22:595. doi: 10.1097/QAD.0b013e3282f56b23. [DOI] [PubMed] [Google Scholar]
- Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology. 2011;132:315. doi: 10.1111/j.1365-2567.2010.03398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3- specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton LA, Abdur Rahman M, Ng C, Le AQ, Milloy MJ, Mo T, Brumme ZL. HLA class I sequence-based typing using DNA recovered from frozen plasma. J Immunol Methods. 2012;382:40. doi: 10.1016/j.jim.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Dixon SC, Horti J, Guo Y, Reed E, Figg WD. Methods for extracting and amplifying genomic DNA isolated from frozen serum. Nat Biotechnol. 1998;16:91. doi: 10.1038/nbt0198-91. [DOI] [PubMed] [Google Scholar]
- Dring MM, Morrison MH, McSharry BP, Guinan KJ, Hagan R, O’Farrelly C, Gardiner CM. Innate immune genes synergize to predict increased risk of chronic disease in hepatitis C virus infection. Proc Natl Acad Sci USA. 2011;108:5736. doi: 10.1073/pnas.1016358108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont B, Selvakumar A, Steffens U. The killer cell inhibitory receptor genomic region on human chromosome 19q13.4. Tissue Antigens. 1997;49:557. doi: 10.1111/j.1399-0039.1997.tb02802.x. [DOI] [PubMed] [Google Scholar]
- Eller MA, Koehler RN, Kijak GH, Eller LA, Guwatudde D, Marovich MA, Michael NL, de Souza MS, Wabwire-Mangen F, Robb ML, Currier JR, Sandberg JK. Human immunodeficiency virus type 1 infection is associated with increased NK cell polyfunctionality and higher levels of KIR3DL1+ NK cells in ugandans carrying the HLA-B Bw4 motif. J Virol. 2011;85:4802. doi: 10.1128/JVI.00111-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowke KR, Plummer FA, Simonsen JN. Genetic analysis of human DNA recovered from minute amounts of serum or plasma. J Immunol Methods. 1995;180:45. doi: 10.1016/0022-1759(94)00297-a. [DOI] [PubMed] [Google Scholar]
- Gagne K, Busson M, Bignon JD, Balere-Appert ML, Loiseau P, Dormoy A, Dubois V, Perrier P, Jollet I, Bois M, Masson D, Moine A, Absi L, Blaise D, Charron D, Raffoux C. Donor KIR3DL1/3DS1 gene and recipient Bw4 KIR ligand as prognostic markers for outcome in unrelated hematopoietic stem cell transplantation. Biology Blood Marrow Trans- plant. 2009;15:1366. doi: 10.1016/j.bbmt.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Gaudieri S, DeSantis D, McKinnon E, Moore C, Nolan D, Witt CS, Mallal SA, Christiansen FT. Killer immunoglobulin-like receptors and HLA act both independently and synergistically to modify HIV disease progression. Genes Immun. 2005;6:683. doi: 10.1038/sj.gene.6364256. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39:D913. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerini FR, Lo Caputo S, Gori A, Bandera A, Mazzotta F, Uglietti A, Zanzottera M, Maserati R, Clerici M. Under representation of the inhibitory KIR3DL1 molecule and the KIR3DL1+/BW4+ complex in HIV exposed seronegative individuals. J Infect Dis. 2011;203:1235. doi: 10.1093/infdis/jir020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong HA, Loubser AS, de Assis Rosa D, Naranbhai V, Carr W, Paximadis M, Lewis DA, Tiemessen CT, Gray CM. Killer-cell immunoglobulin-like receptor genotyping and HLA killer-cell immunoglobulin-like receptor– ligand identification by real-time polymerase chain reaction. Tissue Antigens. 2011;78:185. doi: 10.1111/j.1399-0039.2011.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Chen M, Steiner N, Kariyawasam K, Ng J, Hurley CK. Killer cell immunoglobulin-like receptors (KIR) typing by DNA sequencing. Methods Mol Biol. 2012;882:431. doi: 10.1007/978-1-61779-842-9_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao YL, Zhang BC, You L, Li JF, Zhang J, Ma CY, Cui B, Wang LC, Chen ZJ, Zhao YR. Polymorphisms of KIR gene and HLA-C alleles: possible association with susceptibility to HLA-B27-positive patients with ankylosing spondylitis. J Clin Immunol. 2010;30:840. doi: 10.1007/s10875-010-9444-z. [DOI] [PubMed] [Google Scholar]
- Koehler RN, Walsh AM, Moqueet N, Currier JR, Eller MA, Eller LA, Wabwire-Mangen F, Michael NL, Robb ML, McCutchan FE, Kijak GH. High-throughput genotyping of KIR2DL2/L3, KIR3DL1/S1, and their HLA class I ligands using real-time PCR. Tissue Antigens. 2009;74:73. doi: 10.1111/j.1399-0039.2009.01265.x. [DOI] [PubMed] [Google Scholar]
- Korner C, Altfeld M. Role of KIR3DS1 in human diseases. Front Immunol. 2012;3:326. doi: 10.3389/fimmu.2012.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Martin MP, Carrington M. KIR genotyping by multiplex PCR-SSP. Methods Mol Biol. 2010;612:365. doi: 10.1007/978-1-60761-362-6_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Floros J. Genomic DNA extraction from small amounts of sera to be used for genotype analysis. Biotechniques. 1998;24:937. doi: 10.2144/98246bm07. [DOI] [PubMed] [Google Scholar]
- Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, Vilches C, Carrington M, Witt C, Guethlein LA, Shilling H, Garcia CA, Hsu KC, Wain H. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics. 2003;55:220. doi: 10.1007/s00251-003-0571-z. [DOI] [PubMed] [Google Scholar]
- Martin MP, Carrington M. Immunogenetics of viral infections. Curr Opin Immunol. 2005;17:510. doi: 10.1016/j.coi.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Martin M, Carrington M, Mann D. A method for using serum or plasma as a source of DNA for HLA typing. Hum Immunol. 1992;33:108. doi: 10.1016/0198-8859(92)90060-z. [DOI] [PubMed] [Google Scholar]
- Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O’Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O’Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D, Gonzelez F. The extensive polymorphism of KIR genes. Immunology. 2010;129:8. doi: 10.1111/j.1365-2567.2009.03208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson GW, Martin MP, Gladman D, Wade J, Trowsdale J, Carrington M. Cutting edge: heterozygote advantage in autoimmune disease: hierarchy of protection/susceptibility conferred by HLA and killer Ig-like receptor combinations in psoriatic arthritis. J Immunol. 2004;173:4273. doi: 10.4049/jimmunol.173.7.4273. [DOI] [PubMed] [Google Scholar]
- Patrick DM, Tyndall MW, Cornelisse PG, Li K, Sherlock CH, Rekart ML, Strathdee SA, Currie SL, Schechter MT, O’Shaughnessy MV. Incidence of hepatitis C virus infection among injection drug users during an outbreak of HIV infection. CMAJ. 2001;165:889. [PMC free article] [PubMed] [Google Scholar]
- Pelak K, Need AC, Fellay J, Shianna KV, Feng S, Urban TJ, Ge D, De Luca A, Martinez-Picado J, Wolinsky SM, Martinson JJ, Jamieson BD, Bream JH, Martin MP, Borrow P, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Carrington M, Goldstein DB, Alter G. Copy number variation of KIR genes influences HIV-1 control. PLoS Biol. 2011;9:e1001208. doi: 10.1371/journal.pbio.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Martin MP, Gao X, Jacobson L, Goedert JJ, Buchbinder S, Kirk GD, O’Brien SJ, Trowsdale J, Carrington M. KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PLoS Pathog. 2006;2:e79. doi: 10.1371/journal.ppat.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvakumar A, Steffens U, Dupont B. NK cell receptor gene of the KIR family with two IG domains but highest homology to KIR receptors with three IG domains. Tissue Antigens. 1996;48:285. doi: 10.1111/j.1399-0039.1996.tb02647.x. [DOI] [PubMed] [Google Scholar]
- Strathdee SA, Patrick DM, Currie SL, Cornelisse PG, Rekart ML, Montaner JS, Schechter MT, O’Shaughnessy MV. Needle exchange is not enough: lessons from the Vancouver injecting drug use study. AIDS. 1997;11:F59. doi: 10.1097/00002030-199708000-00001. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- Williams AP, Bateman AR, Khakoo SI. Hanging in the balance. KIR and their role in disease Mol Interv. 2005;5:226. doi: 10.1124/mi.5.4.6. [DOI] [PubMed] [Google Scholar]
- Witt CS, Martin A, Christiansen FT. Detection of KIR2DL4 alleles by sequencing and SSCP reveals a common allele with a shortened cytoplasmic tail. Tissue Antigens. 2000;56:248. doi: 10.1034/j.1399-0039.2000.560307.x. [DOI] [PubMed] [Google Scholar]
- Woods CK, Brumme CJ, Liu TF, Chui CK, Chu AL, Wynhoven B, Hall TA, Trevino C, Shafer RW, Harrigan PR. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol. 2012;50:1936. doi: 10.1128/JCM.06689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]