Abstract
IMPORTANCE
The important depletion of mitochondrial DNA (mtDNA) and the general depression of mitochondrial respiratory chain complex levels (including complex II) have been confirmed, implying an increasing paucity of mitochondria in the muscle from patients with types I, II, and III spinal muscular atrophy (SMA-I, -II, and -III, respectively).
OBJECTIVE
To investigate mitochondrial dysfunction in a large series of muscle biopsy samples from patients with SMA.
DESIGN, SETTING, AND PARTICIPANTS
We studied quadriceps muscle samples from 24 patients with genetically documented SMA and paraspinal muscle samples from 3 patients with SMA-II undergoing surgery for scoliosis correction. Postmortem muscle samples were obtained from 1 additional patient. Age-matched controls consisted of muscle biopsy specimens from healthy children aged 1 to 3 years who had undergone analysis for suspected myopathy. Analyses were performed at the Neuromuscular Unit, Istituto di Ricovero e Cura a Carattere Scientifico Foundation Ca’ Granda Ospedale Maggiore Policlinico-Milano, from April 2011 through January 2015.
EXPOSURES
We used histochemical, biochemical, and molecular techniques to examine the muscle samples.
MAIN OUTCOMES AND MEASURES
Respiratory chain activity and mitochondrial content.
RESULTS
Results of histochemical analysis revealed that cytochrome-c oxidase (COX) deficiency was more evident in muscle samples from patients with SMA-I and SMA-II. Residual activities for complexes I, II, and IV in muscles from patients with SMA-I were 41%, 27%, and 30%, respectively, compared with control samples (P < .005). Muscle mtDNA content and cytrate synthase activity were also reduced in all 3 SMA types (P < .05). We linked these alterations to downregulation of peroxisome proliferator–activated receptor coactivator 1α, the transcriptional activators nuclear respiratory factor 1 and nuclear respiratory factor 2, mitochondrial transcription factor A, and their downstream targets, implying depression of the entire mitochondrial biogenesis. Results of Western blot analysis confirmed the reduced levels of the respiratory chain subunits that included mitochondrially encoded COX1 (47.5%; P = .004), COX2 (32.4%; P < .001), COX4 (26.6%; P < .001), and succinate dehydrogenase complex subunit A (65.8%; P = .03) as well as the structural outer membrane mitochondrial porin (33.1%; P < .001). Conversely, the levels of expression of 3 myogenic regulatory factors—muscle-specificmyogenic factor 5, myoblast determination 1, and myogenin—were higher in muscles from patients with SMA compared with muscles from age-matched controls (P < .05).
CONCLUSIONS AND RELEVANCE
Our results strongly support the conclusion that an altered regulation of myogenesis and a downregulated mitochondrial biogenesis contribute to pathologic change in the muscle of patients with SMA. Therapeutic strategies should aim at counteracting these changes.
Spinal muscular atrophy (SMA) is characterized by degeneration of anterior horn cells of the spinal cord, which leads to weakness and muscle wasting.1–3 Types of SMA are divided into the following 4 main groups: severe (SMA-I), intermediate (SMA-II), mild (SMA-III), and adult onset (SMA-IV). Spinal muscular atrophy is caused by mutations in the SMN1 gene (NCBI Entrez Gene 6606) that drastically reduce the expression of the survival motor neuron (SMN) protein.4–6 The SMN protein is part of a macromolecular complex that directly regulates the assembly of specific RNA-protein complexes, the spliceosomal U small nuclear ribonucleoproteins.7,8 Moreover, SMN protein is implicated in pre–messenger RNA splicing of several genes.8–11
The human genome also harbors the SMN2 gene (NCBI Entrez Gene 6607), which slightly differs from SMN1. Deletion or gene conversion events render patients with SMA homozygous null for SMN1, whereas a variable number of SMN2 copies is retained. A higher number of SMN2 copies corresponds to higher amounts of full-length protein and a milder clinical phenotype.12 Spinal muscular atrophy usually manifests at postnatal stages; however, pathologic alterations are reported to start during prenatal development.13–15 Increasing evidence suggests that maturation of all parts of the neuromuscular system is delayed in patients with SMA,16 and delayed neuromuscular development also has been reported in mouse models of SMA.17 In addition, a few studies hypothesize a delay in myogenesis showing constitutive abnormalities in muscle cultures from patients with SMA and an altered expression of several muscle components, including developmental myosins, desmin, and vimentin.18
A possible pathogenic role of mitochondria also has been proposed. Severe depletion of mitochondrial DNA (mtDNA)was reported in muscle samples from patients with SMA but was considered a consequence of the neurogenic fiber atrophy.19 Mitochondrial dysfunction has been shown in a neural cell culture model of SMA.20 Phenocopies of SMA have been associated with primary mitochondrial cytochrome-c oxidase (COX) deficiency, including mutations in the COX assembly gene SCO221–23 (NCBI Entrez Gene 9997) and mutations in TK2 (NCBI Entrez Gene 7084).24,25 Murine Tk2 models show severe paralysis accompanied by a reduced mtDNA copy number, decreased steady-state levels of respiratory chain enzymes in the brain, and abnormal vacuolar changes in spinal cord neurons.26,27
These observations prompted us to perform a systematic review of oxidative metabolism in our collection of skeletal muscle samples from 24 patients with genetically defined SMA. Moreover, we studied the expression of the main regulatory factors related to mitochondrial biogenesis and myogenesis.
Methods
Study Subjects
We studied biopsy specimens of the quadriceps muscle from 24patientswith a confirmed molecular diagnosis of SMA. They had undergone muscle biopsy before the advent of genetic testing. In addition, paraspinal muscle samples were obtained from 3 patients with SMA-II during surgery for scoliosis correction. All muscle biopsies came from the Telethon Network of Genetic Biobanks at the Neuromuscular Unit, Istituto di Ricovero e Cura a Carattere Scientifico Foundation Ca’ Granda Ospedale Maggiore Policlinico-Milano. All muscle biopsies were taken after ethical committee approval in agreement with the Italian law and the Biobank rules (Telethon Network for Genetics Biobanks and Eurobiobank). Postmortem muscle samples (psoas, diaphragm, intercostal, and paraspinal) from 1 additional patient with SMA-I were obtained from Columbia University Medical Center.
For histologic studies, we selected muscle biopsy specimens from 6 pediatric patients with hereditary neuropathies, namely, 3 patients with mitofusin 2–related Charcot-Marie-Tooth disease, 2 with myelin protein zero–related Charcot- Marie-Tooth disease, and 1 with a severe congenital undiagnosed polyneuropathy as a positive control sample for denervation. We also chose muscle specimens from healthy children aged 1 to 3 years who had undergone muscle biopsy for suspected myopathy that was excluded after analysis. The study was performed from April 2011 through January 2015. Further details about controls and their use in the study are listed in eTable 1 in the Supplement.
Sample Analysis
Muscle specimens were processed according to standard procedures (eMethods in the Supplement).28,29 Mitochondrial respiratory chain enzyme and cytrate synthase activities were measured using spectrophotometric analysis as described.30 We extracted DNA from muscle by standard procedures. Loci for SMN1 and SMN2 were genotyped using a previously validated assay.31,32 We ruled out TK2 mutations by results of direct sequencing.32 We measured levels of mtDNA by quantitative polymerase chain reaction (PCR) analysis, using primers and probes for the mitochondrial CYTB gene (mtDNA) and the nuclear APP gene (nuclear DNA) as previously described.33
Gene Expression Analysis
Primary antibodies used for Western blot analysis are listed in the eMethods in the Supplement. In addition, we obtained RNA from muscle samples by a standard protocol and performed random hexamer-primed retrotranscription. The resulting complementary DNA was used as a template in quantitative reverse transcription–PCR analysis to assess gene expression profiles of selected transcripts (eMethods in the Supplement).
Statistical Analysis
Results of quantitative analysis from SMA specimens and relative control samples were analyzed using the unpaired 2-tailed t test. Unless otherwise indicated, data are expressed as mean (SD).
Results
Clinical, histologic, and molecular features of the 24 patients with SMA are presented in the Table. Molecular analysis at the genomic level disclosed homozygous deletion of SMN1 in 21 patients. Three individuals were compound heterozygous for an exon 7 deletion and a pathogenic single-nucleotide mutation. Results of screening for TK2 mutations were negative.
Table.
Clinical and Morphologic Data of 24 Patients With SMA
| Patient No./Sex |
Clinical Diagnosis |
Hypotonia Muscle Weakness |
Respiratory Impairment |
Age at Onset |
Age at Biopsy |
Genotype | Severity of COX Deficiency, % |
Residual mtDNA, % |
|
|---|---|---|---|---|---|---|---|---|---|
| SMN1 | SMN2a | ||||||||
| 1/M | SMA-I | Severe | Severe | Birth | 4 mo | Δ7,8 Hom | 2 | >76 | 14 |
| 2/M | SMA-I | Severe | Severe | Birth | 2 mo | Δ7,8 Hom | 2 | 51–75 | 32 |
| 3/F | SMA-I | Severe | Severe | Birth | 4 mo | Δ7 Het c.815A>G | 2 | >76 | 21 |
| 4/F | SMA-I | Severe | Severe | Birth | 6 mo | Δ7 Het c.469C>T | 2 | >76 | 16 |
| 5/F | SMA-I | Severe | Severe | Birth | 24 mo | Δ7,8 Hom | 2 | >76 | 12 |
| 6/F | SMA-I | Severe | Severe | Birth | 5 mo | Δ7,8 Hom | NE | 26–50 | NE |
| 7/F | SMA-I | Severe | Severe | Birth | 3 mo | Δ7,8 Hom | NE | <25 | NE |
| 8/F | SMA-I | Severe | Severe | Birth | 3 mo | Δ7,8 Hom | NE | <25 | 40 |
| 9/F | SMA-I | Severe | Severe | Birth | 12 mo | Δ7,8 Hom | NE | 51–75 | 40 |
| 10/F | SMA-II | Severe | Severe | 9 mo | 9 mo | Δ7,8 Hom | 3 | >76 | 27 |
| 11/F | SMA-II | Severe | Moderate | 12 mo | 16 mo | Δ7,8 Hom | 2 | 51–75 | 73 |
| 12/M | SMA-II | Severe | Severe | 9 mo | 9 mo | Δ7,8 Hom | 2 | >76 | 33 |
| 13/M | SMA-II | Moderate | Moderate | 16 mo | 24 mo | Δ7,8 Hom | 2 | 51–75 | 30 |
| 14/M | SMA-II | Moderate | Severe | 16 mo | 18 mo | Δ7,8 Hom | NE | >76 | NE |
| 15/F | SMA-II | Severe | Severe | 12 mo | 12 mo | Δ7,8 Hom | NE | 51–75 | 18 |
| 16/M | SMA-II | Moderate | Severe | 15 mo | 20 mo | Δ7,8 Hom | NE | <25 | 29 |
| 17/M | SMA-II | Moderate | Severe | 12 mo | 12 mo | Δ7,8 Hom | NE | 51–75 | NE |
| 18/F | SMA-III | Mild | None | 14 mo | 24 mo | Δ7,8 Hom | 3 | <25 | NE |
| 19/F | SMA-III | Moderate | None | 6 mo | 27 mo | Δ7 Hom | 3 | 51–75 | 25 |
| 20/F | SMA-III | Moderate | None | 3 y | 4 y | Δ7,8 Hom | 4 | 26–50 | 84 |
| 21/F | SMA-III | Mild | None | 16 mo | 18 mo | Δ7,8 Hom | 4 | <25 | 85 |
| 22/F | SMA-III | Moderate | None | 3 y | 3 y | Δ7,8 Hom | 3 | 51–75 | 32 |
| 23/M | SMA-III | Severe | Moderate | 8 y | 14 y | Δ7 Het c.389A>G | 1 | 51–75 | 50 |
| 24/F | SMA-III | Mild | None | 6 y | 14 y | Δ7,8 Hom | NE | None | 31 |
Abbreviations: COX, cytochrome-c oxidase; Δ, deletion; Het, heterozygous; Hom, homozygous; mtDNA, mitochondrial DNA; NE, not evaluated; SMA, spinal muscular atrophy.
Indicates the number of copies evaluated for the SMN2 gene.
Morphologic Studies
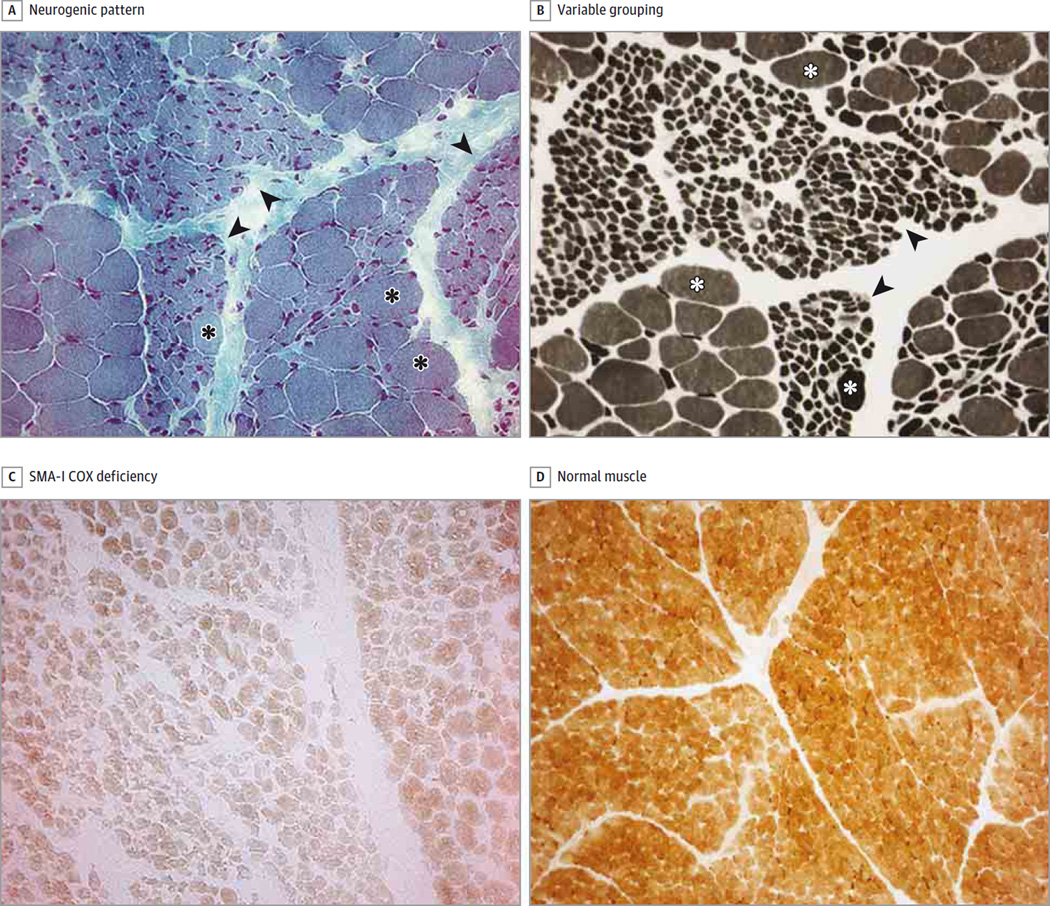
In all patients, results of muscle biopsies revealed a chronic neurogenic pattern, with large groups of atrophic fibers interspersed with hypertrophic ones (Figure 1A and B). Variable type grouping was also present (Figure 1B).
Figure 1. Severe Cytochrome-c Oxidase (COX) Deficiency in Patients With Spinal Muscular Atrophy Type I (SMA-I).
A, Typical histologic neurogenic pattern of SMA-I, with groups (often large) of atrophic fibers (arrowheads) interspersed with groups of hypertrophic fibers (asterisks) (Gomori trichrome stain). B, In addition to the neurogenic pattern (arrowheads), hypertrophic fibers (asterisks) and a variable-type grouping are seen (adenosine triphosphate stain; pH, 9). C, Severe COX deficiency in SMA-I, evident in atrophic and normal/hypertrophic fibers (COX stain). D, COX activity in normal human muscle (COX stain) (A through D, original magnification ×25).
We found several COX-negative fibers in muscle biopsy samples from all 24 patients. The deficiency was particularly severe in patients with SMA-I and SMA-II but was also evident, albeit less marked, in patients with SMA-III (Figure 1 and Figure 2). The enzyme defect was not restricted to atrophic fibers but was also found in normal and hypertrophic fibers of SMA-I (Figure 1C), SMA-II, (Figure 2C), and SMA-III (Figure 2A and B) samples. No histochemical defect was found in samples from patients with other chronic neurogenic disorders (Figure 2D and E). Activity of succinate dehydrogenase (complex II) was also reduced and, although less marked, paralleled the distribution pattern seen for complex IV.
Figure 2. Severe Cytochrome-c Oxidase (COX) Deficiency in Patients With Spinal Muscular Atrophy Types II and III (SMA-II and -III).
A and B, Severe COX deficiency in SMA-III. C, Severe COX deficiency in SMA-II. D, Normal COX activity in normal and atrophic fibers of patients with chronic neurogenic disorders (Charcot-Marie-Tooth disease). E, Positive control sample for denervation in a patient with severe congenital undiagnosed polyneuropathy (A through E, COX stain; original magnification ×25).
Biochemical Studies
We performed biochemical analysis of mitochondrial enzyme activities in muscle from 11 patients, including 5 with SMA-I (patients 4, 5, 6, 7, and 8), 3 with SMA-II (patients 14, 15, and 17), and 3 with SMA-III (patients 18, 21, and 24) and in 5 age-matched controls. Biochemical results are summarized in eTable 2 in the Supplement. The levels of cytrate synthase normalized to protein content were decreased in SMA-I (69.9%; P = .03) and SMA-II (71.0%; P = .055) samples. After normalization of cytrate synthase levels, residual activities of complexes I, II, II + III, and IV were reduced significantly in patients’ muscles, with more pronounced biochemical defects in SMA-I samples. Residual activities for complexes I, II, and IV were 41% (P = .001), 27% (P < .001), and 30% (P < .001), respectively (eTable 2 in the Supplement). Muscle specimens from patients with SMA-III also showed a combined deficiency of respiratory chain complexes compared with age-matched controls, although residual activities were higher than those in patients with severe SMA-III. Finally, we assayed respiratory chain complexes in paraspinal muscles obtained during surgery from3 patients with SMA-II, and we observed a reduction of complex II + III, IV, and cytrate synthase activities (eFigure 1 in the Supplement).
Evaluation of Mitochondrial Content
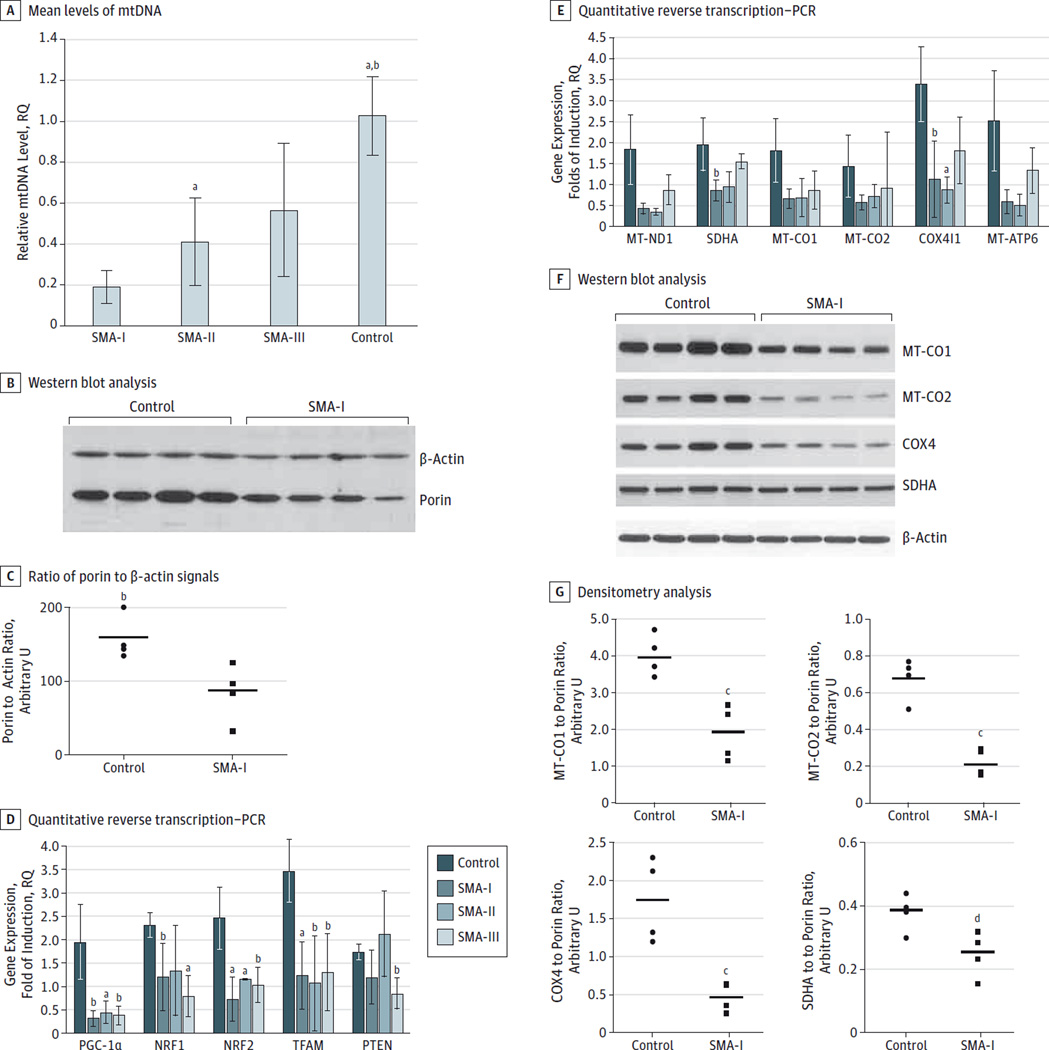
We assessed the muscle mtDNA amount in 19 affected patients and in a panel of age-matched healthy controls by results of quantitative PCR. The content of mtDNA in muscle from the patients with SMA-I (n = 7) was significantly reduced compared with that from age-matched controls (0–3 years) (n = 4) (mean relative quantification value, 0.25 [0.12]; P < .01). The degree of depletion was greater than 50% in the patients examined (Figure 3A and the Table).
Figure 3. Evaluation of Mitochondrial Content in Muscle Specimens With Spinal Muscular Atrophy (SMA) and Controls.
Mitochondrial DNA (mtDNA) copy number was determined by quantitative real-time polymerase chain reaction (PCR) in skeletal muscle samples obtained from 7 patients with SMA-I, 5 with SMA-II, and 4 with SMA-III and from 4 age-matched (12–36 months) healthy control samples. All determinations were performed in quadruplicate using 25 ng of total DNA as a template. The mtDNA levels were normalized to nuclear DNA and expressed as relative quantification (RQ) values; reference (RQ = 1.0), the amount of mtDNA detected in skeletal muscle from age-matched controls. A, Mean (SD) levels of mtDNA in each group after normalization to nuclear DNA content. B, Western blot analysis of protein extracts obtained from 4 SMA-I and 4 control biopsy samples using primary antibodies against porin and β-actin for normalization. C, Ratio of porin to β-actin signals expressed in arbitrary units for control and SMA-I samples. Horizontal bars indicate mean values. D and E, Quantitative reverse transcription–PCR studies to evaluate the levels of the indicated transcripts in skeletal muscle samples obtained from 4 patients with SMA-I, 3 with SMA-II, and 4 with SMA-III and 4 age-matched controls. 18S was used as the control housekeeping gene. All determinations were performed in replicates (n = 4). Results are presented as mean (SD) levels in each group. F, Western blot analysis of protein extracts obtained from 4SMA-I and 4 control biopsy specimens using primary antibodies against respiratory chain subunits encoded by mtDNA (MT-CO1 and MT-CO2) and nuclear DNA (COX4 and SDHA) compared with β-actin levels. G, Results of densitometry analysis of respiratory chain subunits after normalization to β-actin expressed in arbitrary units for controls and SMA samples. Horizontal bars indicate mean values. All P values were calculated using the unpaired 2-tailed t test. COX4I1 indicates cytochrome-c oxidase (COX) 4 isoform 1; MT-ATP6, mitochondrially encoded adenosine triphosphate 6;MT-CO1 and -CO2, mitochondrially encoded COX1 and COX2, respectively; MT-ND1, mitochondrially encoded nicotinamide adenine dinucleotide dehydrogenase; NRF, nuclear respiratory factor; PGC-1α, peroxisome proliferator–activated receptor coactivator 1α; PTEN, phosphatase and tensin homolog; SDHA, succinate dehydrogenase complex subunit A; and TFAM, mitochondrial transcription factor A.
a P < .01 compared with SMA-I.
b P < .05 compared with SMA-II.
c P < .05 compared with control.
d P < .01 compared with control.
The ratio of mtDNA to nuclear DNA was also reduced in patients with SMA-II (n = 6)(mean relative quantification value, 0.35 [0.19]; P = .04) and was especially pronounced in the muscle from patient 10 (27% residual amount). Results of molecular analysis performed in paraspinal muscles from 3 patients with SMA-II were consistent with these findings (P < .05; eFigure 1 in the Supplement).
Results from patients with SMA-III were heterogeneous. Residual levels of mtDNA were low in patients 19 and 22 (25% and 32%, respectively) but were 84% and 85% in patients 20 and 21, respectively. The patients with SMA-III who underwent biopsy in late childhood showed only moderate depletion compared with adult controls; residual levels of muscle mtDNA were 50% in patient 23 and 31% in patient 24.
Collectively, mtDNA content in patients with SMA-I was significantly reduced compared with SMA-II samples (P = .04). Also, mtDNA levels in SMA-III samples were higher when compared with patients with SMA-I and SMA-II but lower compared with healthy controls (P = .08). Residual levels of mtDNA were inversely related to disease severity.
Examination of postmortem specimens from an additional patient with SMA-I who died at 5 months of age showed less than 30%residual mtDNA levels in intercostal and psoas muscles but greater than 40% in paraspinal and diaphragm muscles (eFigure 2A in the Supplement). Similarly, results of histochemical studies showed the oxidative defect in the psoas muscle, whereas paraspinal and diaphragm muscles were relatively spared (eFigure2B–G in the Supplement). Cytochrome-c oxidase activity was also preserved in intrafusal muscle fibers from COX-deficient psoas (eFigure 2H in the Supplement).
To check whether biochemical defects and severe mtDNA depletion were hints of a general reduction of muscle mitochondrial content, we performed immunoblot analysis using an antibody against the outer mitochondrial membrane protein porin. We analyzed biopsy samples from patients with SMA-I and age-matched controls. In the samples with SMA-I, the porin signal was moderately reduced, which was more evident after normalization to β-actin (Figure 3B). Lane densitometry confirmed the ratio in the reduction of porin to β-actin signals in the SMA samples compared with controls (mean signals, 1.65 [0.39] vs 4.97 [0.33]; 33.1% compared with controls; P < .001) (Figure 3C).
Defective Mitochondrial Biogenesis in SMA Muscles
To verify whether mitochondrial dysfunction was caused by reduced mitochondrial content, we performed gene expression analysis of factors involved in the regulation of mitochondrial content and mtDNA expression. Quantitative reverse transcription–PCR experiments showed a significant (P < .05) downregulation of the main coactivator of mitochondrial biogenesis, peroxisome proliferator–activatedreceptor coactivator 1α (PGC-1α), in specimens from patients with SMA-I (n = 4), SMA-II (n = 3), and SMA-III (n = 4), without major differences among the 3 phenotypes. Expression of nuclear respiratory factors 1 (NRF1) and 2 (NRF2) was also reduced in all 3 groups, although a comparison between SMA-II samples and controls did not reach significance for NRF1 (P = .21). Levels of mitochondrial transcription factor A (TFAM) transcript, which encodes the key activator of mitochondrial transcription, were also severely decreased in SMA tissues (P < .05). Conversely, we found no difference in the expression of the phosphatase and tensin homolog, a tumor suppressor that regulates the mitogenic PI3 kinase-AKT signaling pathway, in patients with SMA-I and SMA-II compared with controls (P = .16 and P = .84, respectively) (Figure 3D). We determined the expression levels of genes encoding for respiratory chain subunits, which are downstream targets of NRF1 and NRF2. Nuclear (succinate dehydrogenase complex subunit A and COX4 isoform 1 [COX4I1]) and mitochondrial (mitochondrially encoded nicotinamide adenine dinucleotide dehydrogenase 1, COX1, COX2, and adenosine triphosphate 6) transcripts were found to be downregulated in all SMA subtypes. This tendency was more prominent in SMA-I and SMA-II specimens, although statistical significance was only observed for COX4I1 (P < .05, Figure 3E). These results were confirmed by results of immunoblot analysis of the corresponding proteins in available samples from patients with SMA-I (Figure 3F). The difference between the SMA and control samples was significant after β-actin normalization (mitochondrially encoded COX1, 47.5% [P = .004]; mitochondrially encoded COX2, 32.4% [P < .001]; COX4, 26.6% [P < .001]; and succinate dehydrogenase complex subunit A, 65.8% [P = .03]) (Figure 3G).
Impaired Maturation in SMA Muscles
Observations by different authors suggested that the myogenic program is deregulated in SMA mice models,34 human myoblasts,35 and postnatal muscle samples18 from patients with SMA. We quantified expression levels of 3 myogenic regulatory factors, namely, myoblast determination 1 (MYOD), myogenin (MYOG), and the muscle-specific myogenic factor 5 (MYF5) in SMA-I samples (n = 3) and age-matched controls (n = 4). Levels of expression were found to be significantly higher in muscles from patients with SMA (P < .05) (Figure 4A). Samples from patients with SMA also showed the upregulation of DES, encoding for desmin, a muscle protein that is abundant in immature and developing fibers and downregulated in mature muscles. Conversely, levels of CAV3, which encodes the muscle caveolin isoform and is expressed in terminally differentiated myocytes, were significantly reduced in SMA-I samples compared with controls. Western blot studies followed by densitometry analysis confirmed the difference of desmin content between samples with SMA and controls (P < .01) (Figure 4B and C). Gene expression levels were also evaluated on a subset of patients with SMA-III (n = 3). Notably, their differentiation also halted compared with those of controls, although statistical significance was only reached for MYOG and MYOD (P < .05) (Figure 4A). The results of quantitative reverse transcription–PCR analysis were significantly different between groups with SMA-I and SMA-III for all the tested assays (P < .05).
Figure 4. Levels of Expression of Myogenic Regulatory Factors and Muscle Transcripts.
A, Relative quantification (RQ) levels of selected transcripts in 3 muscle samples of types I and III spinal muscle atrophy (SMA-I and SMA-III, respectively) each compared with 3 age-matched control samples. 18S was used as the control housekeeping gene. All determinations have been performed in replicates (n = 4). Results are presented as mean (SD) levels (RQ) in each group. CAV3 encodes the muscle caveolin isoform; DES, desmin. MYF5 indicates muscle-specific myogenic factor 5; MYOD, myoblast determination 1; MYOG, myogenin. B, Immunoblot analysis of desmin, confirming the quantitative reverse transcription–polymerase chain reaction results. C, Ratio of desmin to β-actin signals expressed in arbitrary units for control and SMA samples. Horizontal bars represent mean values of the 2 groups. All P values were calculated using the unpaired 2-tailed t test.
a P < .01 compared with control.
b P < .05 compared with control.
Discussion
Spinal muscular atrophy is caused by mutations in the gene encoding the SMN protein.1,5,6,29 The mechanisms underlying the pathophysiological features of the disease are not clear.8–11
Expression of SMN is ubiquitous, and its complete absence is incompatible with life, from yeast to humans.36,37 Analysis on animal models and model systems of cellular differentiation demonstrated a tissue-specific temporal regulation of SMN,38–40 which is high during embryonic and early postnatal development of the central nervous system and decreases dramatically at the onset of myelination, reaching a low steady level maintained throughout life.38 Recent investigations have demonstrated that SMN has a role in myogenesis and that normal muscle differentiation requires adequate levels of SMN34,35; these considerations support the hypothesis that a delay in muscle maturation is one of the primary pathologic components of SMA.18
To investigate a possible altered regulation of myogenesis in our patients with SMA, we evaluated the expression of 3 myogenic regulatory factors—MYF5, MYOD, and MYOG— and found higher levels in samples from patients with SMA compared with samples from age-matched controls. These findings suggest that the developmental programs of myoblast activation (MYF5) and myogenic determination (MYOD and MYOG) are still active in muscle from patients with SMA. The delay in postnatal development of muscle impairs the maturation of functional and competent myofibers, as demonstrated by the residual expression of immature proteins and the relatively low levels of a marker of terminal differentiation. These findings support the conclusions of previous studies that reported altered myogenesis in human SMA myoblasts41 and satellite cells42 and in muscles from transgenic murine models.34
Remels et al43 demonstrated the association between normal myogenesis and increase doxidative metabolism. This finding agrees with those of previous studies and illustrates development of the oxidative phenotype during myogenesis.44,45 Expression of PGC-1α increases during myogenesis and governs mitochondrial biogenesis through control over other regulatory proteins, including NRF1 and the master mitochondrial regulator TFAM.46 The impaired differentiation of the C2C12 cell model owing to experimental PGC-1α downregulation was recently observed.47
In our patients, biomolecular analyses demonstrated significant reduction of mitochondrial biogenesis regulatory factors, including PGC-1α; the transcription factors NRF1, NRF2, and TFAM; and several immediate targets encoding for respiratory chain subunits. This defect can depend on the anomalous process of myogenesis and correlates with the reduction in mitochondrial expression, which is also documented by the decreased amount of the mitochondrial structural protein porin and by the reduced activity of complex II, the only entirely nuclear DNA–encoded respiratory chain complex.
Deregulated myogenesis and impaired mitochondrial biogenesis seem to be inversely proportional to SMN availability, being more prominent in muscle from patients with SMA-I than in muscle from patients with SMA-III. These events could reflect the severity of muscle dysfunction observed in patients, but speculation about the existence of 1 or more factors ruling muscle maturation and metabolism in which expression or editing are directly dependent on SMN is tempting. No obvious candidate so far has emerged from studies addressing global gene expression of SMA muscles.
Further analyses on muscle biopsy samples from our patients compared with age-matched controls provided evidence of mitochondrial dysfunction in skeletal muscle. Using histochemical and biochemical techniques, we have documented decreased activity of COX in limb muscles from all patients. Also, we have shown that the activities of all respiratory chain enzymes (including succinate dehydrogenase) were defective regardless of their genetic control (nuclear DNA or mtDNA). By means of quantitative PCR analysis, we have additionally observed a concomitant reduction in mtDNA content that directly correlated with disease severity (Table).
Berger and coauthors19 previously reported the impairment of the respiratory chain, mtDNA depletion, and reduction of mitochondrial protein content in pooled muscle specimens from all types of SMA. Our data do not support their proposed hypothesis that COX deficiency is a consequence of muscle atrophy and denervation.19 We have documented COX deficiency in normal-sized and hypertrophic muscle fibers in patients with SMA, whereas we failed to observe COX deficiency in skeletal muscle from individuals with other forms of neurogenic atrophy. Control samples of muscles from children with polyneuropathies were quadriceps muscles and thus presumably were not so severely affected as samples from patients with SMA-I. However, the control samples displayed neurogenic alterations that paralleled those seen in samples from patients with SMA-II and SMA-III, and, more importantly, they all showed several COX-positive atrophic fibers.
Moreover, biochemical and molecular studies on a muscle sample displaying severe neurogenic features gave results undistinguishable from those of age-matched reference controls but significantly different from those of patients with SMA. These findings are coherent with previous studies on isolated SMA myoblasts35 or comparing muscles from SMA mice models and controls that underwent nerve axotomy.34
Mitochondrial dysfunction was not restricted to limb muscles but was also observed in other specimens, including paraspinal muscles from patients with SMA-II, which is relevant because limb and paraspinal muscles are different in terms of embryonic origin, differentiation process, fiber-type proportion, and physiologic features. Paraspinal muscles show type I fiber predominance and perform isometric rather than dynamic contractions, which makes them more sensitive to oxidative damage.48
Also, we examined different skeletal muscles from1 postmortem case with SMA-I. Histochemical data from psoas muscle confirm the COX deficiency, whereas results in paraspinal and, more specifically, diaphragm muscles do not show any clear defect, probably because, as reported, some muscles are spared in the early stages of SMA and other disease models.49–51 Indeed, paraspinal and diaphragm muscles already show reduction of mtDNA levels compared with age-matched controls (eFigure 2A in the Supplement). In addition, paraspinal muscles in cases of SMA-II, which have undergone biopsy in a later disease stage, show mitochondrial dysfunction (eFigure 1 in the Supplement). The preservation of COX activity in intrafusal muscle fibers from COX-deficient psoas has been reported and is supposedly related to the presence of a different COX isoenzyme.52–54 We finally considered the effect of the SMN2 copy number in our patients, but we did not find a clear relationship between the SMN1/SMN2 genotype and COX deficiency (Table).
The best-known explanation for the neuromuscular phenotype in SMA is to assume that insufficient SMN protein causes motor neuron dysfunction or death and that muscle atrophy is a secondary consequence of denervation. Herein, we documented that a delayed myogenic program likely results in an altered maturation and oxidative metabolism in SMA muscles. These findings support the hypothesis of an autonomous pathogenic mechanism acting in muscle as a consequence of reduced SMN protein levels. Because innervation directly stimulates the late phase of muscle differentiation, motor neuron abnormalities might further contribute to halt the proper maturation of striated muscle. Recent observations of immature protein composition of neuromuscular junctions in SMA models and patients support this notion.55,56 Maturation of muscle fibers and activation of mitochondrial biogenesis are closely related. The reduced mitochondrial content makes SMA muscle unable to sustain muscle fiber maturation and contraction properly, contributing to patient weakness and hypotonia. Therefore, muscle mitochondrial dysfunction should be more accurately regarded as a pathogenic event underlying muscle abnormalities in SMA rather than as a mere consequence of muscle atrophy. Recent clinical observations reporting an incomplete response to aerobic exercise in patients with SMA also sustain this scenario.57
These findings open the possibility of novel therapeutic interventions for SMA and target mitochondria and myogenic regulatory pathways. Different pharmacological and genetic strategies aimed at improving mitochondrial biogenesis have been tried in several neurologic disorders.58–61 Notably, increasing PGC-1α activity in the muscle of a mouse model of amyotrophic lateral sclerosis delayed muscle atrophy and significantly improved muscle endurance, even at late disease stages.62 Recently, resveratrol, a natural polyphenol known to stimulate mitochondrial biogenesis, was found to promote the myogenic program in murine C2C12 cells.63
Conclusions
Our study strengthens the relevance of these muscle-specific mechanisms for SMA pathogenesis and the development of strategies aimed at improving residual muscle function and delaying disease progression. We demonstrated that the altered regulation of myogenesis and a downregulated mitochondrial biogenesis most likely contribute to pathologic change in the muscle of patients with SMA. Our study strengthens the relevance of these muscle-specific mechanisms for SMA pathogenesis and the development of strategies aimed at improving residual muscle function and delaying disease progression.
Supplementary Material
Acknowledgments
Funding/Support: This study was supported by grant P01HD032062-20 from the National Institutes of Health (Dr DiMauro) and by grant 2012-0513 from the Foundation Cariplo (Dr Corti). The Italian Association of Myology; Associazione Amici del Centro Dino Ferrari, University of Milan; Eurobiobank; and Telethon network of Genetic biobanks (grant number GTB12001) provided human biological samples.
Role of the Funder/Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Drs Ripolone and Ronchi contributed equally to the present work. Both had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Ripolone, Ronchi, Berardinelli, Mora, Sciacco, Comi, Moggio.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Ripolone, Ronchi, Sciacco, DiMauro, Comi, Moggio.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Ronchi, Colombo, Villa.
Administrative, technical, or material support: Violano, Vallejo, Fagiolari, Barca, Lucchini, Bordoni, Fortunato, Parisi.
Study supervision: Sciacco, DiMauro, Comi, Moggio.
Additional Contributions: The postmortem biopsy samples were provided by Wendy K. Chung, MD, PhD, Columbia University Medical Center.
Conflict of Interest Disclosures: None reported.
Supplemental content at jamaneurology.com
REFERENCES
- 1.Lunn MR, Wang CH. Spinal muscular atrophy. Lancet. 2008;371(9630):2120–2133. doi: 10.1016/S0140-6736(08)60921-6. [DOI] [PubMed] [Google Scholar]
- 2.Sossi V, Giuli A, Vitali T, et al. Premature termination mutations in exon 3 of the SMN1 gene are associated with exon skipping and a relatively mild SMA phenotype. Eur J Hum Genet. 2001;9(2):113–120. doi: 10.1038/sj.ejhg.5200599. [DOI] [PubMed] [Google Scholar]
- 3.Engel AG, Armstrong-Franzini C. Myology. 2nd. New York, NY: McGraw-Hill; 1994. [Google Scholar]
- 4.Brzustowicz LM, Lehner T, Castilla LH, et al. Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2-13.3. Nature. 1990;344(6266):540–541. doi: 10.1038/344540a0. [DOI] [PubMed] [Google Scholar]
- 5.Lefebvre S, Bürglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy–determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 6.Coovert DD, Le TT, McAndrew PE, et al. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6(8):1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 7.Terns MP, Terns RM. Macromolecular complexes: SMN—the master assembler. Curr Biol. 2001;11(21):R862–R864. doi: 10.1016/s0960-9822(01)00517-6. [DOI] [PubMed] [Google Scholar]
- 8.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90(6):1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 9.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90(6):1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 10.Meister G, Hannus S, Plöttner O, et al. SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J. 2001;20(9):2304–2314. doi: 10.1093/emboj/20.9.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298(5599):1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 12.Gavrilov DK, Shi X, Das K, Gilliam TC, Wang CH. Differential SMN2 expression associated with SMA severity. Nat Genet. 1998;20(3):230–231. doi: 10.1038/3030. [DOI] [PubMed] [Google Scholar]
- 13.Burlet P, Huber C, Bertrandy S, et al. The distribution of SMN protein complex in human fetal tissues and its alteration in spinal muscular atrophy. Hum Mol Genet. 1998;7(12):1927–1933. doi: 10.1093/hmg/7.12.1927. [DOI] [PubMed] [Google Scholar]
- 14.Ito Y, Shibata N, Saito K, Kobayashi M, Osawa M. New insights into the pathogenesis of spinal muscular atrophy. Brain Dev. 2011;33(4):321–331. doi: 10.1016/j.braindev.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-Hernández R, Soler-Botija C, Also E, et al. The developmental pattern of myotubes in spinal muscular atrophy indicates prenatal delay of muscle maturation. J Neuropathol Exp Neurol. 2009;68(5):474–481. doi: 10.1097/NEN.0b013e3181a10ea1. [DOI] [PubMed] [Google Scholar]
- 16.Hausmanowa-Petrusewicz I, Vrbová G. Spinal muscular atrophy: a delayed development hypothesis. Neuroreport. 2005;16(7):657–661. doi: 10.1097/00001756-200505120-00001. [DOI] [PubMed] [Google Scholar]
- 17.Lee YI, Mikesh M, Smith I, Rimer M, Thompson W. Muscles in a mouse model of spinal muscular atrophy show profound defects in neuromuscular development even in the absence of failure in neuromuscular transmission or loss of motor neurons. Dev Biol. 2011;356(2):432–444. doi: 10.1016/j.ydbio.2011.05.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Hernández R, Bernal S, Alias L, Tizzano EF. Abnormalities in early markers of muscle involvement support a delay in myogenesis in spinal muscular atrophy. J Neuropathol Exp Neurol. 2014;73(6):559–567. doi: 10.1097/NEN.0000000000000078. [DOI] [PubMed] [Google Scholar]
- 19.Berger A, Mayr JA, Meierhofer D, et al. Severe depletion of mitochondrial DNA in spinal muscular atrophy. Acta Neuropathol. 2003;105(3):245–251. doi: 10.1007/s00401-002-0638-1. [DOI] [PubMed] [Google Scholar]
- 20.Acsadi G, Lee I, Li X, et al. Mitochondrial dysfunction in a neural cell model of spinal muscular atrophy. J Neurosci Res. 2009;87(12):2748–2756. doi: 10.1002/jnr.22106. [DOI] [PubMed] [Google Scholar]
- 21.Tarnopolsky MA, Bourgeois JM, Fu MH, et al. Novel SCO2 mutation (G1521A) presenting as a spinal muscular atrophy type I phenotype. Am J Med Genet A. 2004;125A(3):310–314. doi: 10.1002/ajmg.a.20466. [DOI] [PubMed] [Google Scholar]
- 22.Pronicki M, Kowalski P, Piekutowska-Abramczuk D, et al. A homozygous mutation in the SCO2 gene causes a spinal muscular atrophy like presentation with stridor and respiratory insufficiency. Eur J Paediatr Neurol. 2010;14(3):253–260. doi: 10.1016/j.ejpn.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Salviati L, Sacconi S, Rasalan MM, et al. Cytochrome-c oxidase deficiency due to a novel SCO2 mutation mimics Werdnig-Hoffmann disease. Arch Neurol. 2002;59(5):862–865. doi: 10.1001/archneur.59.5.862. [DOI] [PubMed] [Google Scholar]
- 24.Mancuso M, Salviati L, Sacconi S, et al. Mitochondrial DNA depletion: mutations in thymidine kinase gene with myopathy and SMA. Neurology. 2002;59(8):1197–1202. doi: 10.1212/01.wnl.0000028689.93049.9a. [DOI] [PubMed] [Google Scholar]
- 25.Götz A, Isohanni P, Pihko H, et al. Thymidine kinase 2 defects can cause multi-tissue mtDNA depletion syndrome. Brain. 2008;131(pt 11):2841–2850. doi: 10.1093/brain/awn236. [DOI] [PubMed] [Google Scholar]
- 26.Akman HO, Dorado B, López LC, et al. Thymidine kinase 2 (H126N) knockin mice show the essential role of balanced deoxynucleotide pools for mitochondrial DNA maintenance. Hum Mol Genet. 2008;17(16):2433–2440. doi: 10.1093/hmg/ddn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartesaghi S, Betts-Henderson J, Cain K, et al. Loss of thymidine kinase 2 alters neuronal bioenergetics and leads to neurodegeneration. Hum Mol Genet. 2010;19(9):1669–1677. doi: 10.1093/hmg/ddq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sciacco M, Fagiolari G, Lamperti C, et al. Lack of apoptosis in mitochondrial encephalomyopathies. Neurology. 2001;56(8):1070–1074. doi: 10.1212/wnl.56.8.1070. [DOI] [PubMed] [Google Scholar]
- 29.Bürglen L, Lefebvre S, Clermont O, et al. Structure and organization of the human survival motor neurone (SMN) gene. Genomics. 1996;32(3):479–482. doi: 10.1006/geno.1996.0147. [DOI] [PubMed] [Google Scholar]
- 30.Bresolin N, Moggio M, Bet L, et al. Progressive cytochrome C oxidase deficiency in a case of Kearns-Sayre syndrome: morphological, immunological, and biochemical studies in muscle biopsies and autopsy tissues. Ann Neurol. 1987;21(6):564–572. doi: 10.1002/ana.410210607. [DOI] [PubMed] [Google Scholar]
- 31.Gómez-Curet I, Robinson KG, Funanage VL, Crawford TO, Scavina M, Wang W. Robust quantification of the SMN gene copy number by real-time TaqMan PCR. Neurogenetics. 2007;8(4):271–278. doi: 10.1007/s10048-007-0093-1. [DOI] [PubMed] [Google Scholar]
- 32.Galbiati S, Bordoni A, Papadimitriou D, et al. New mutations in TK2 gene associated with mitochondrial DNA depletion. Pediatr Neurol. 2006;34(3):177–185. doi: 10.1016/j.pediatrneurol.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Spinazzola A, Viscomi C, Fernandez-Vizarra E, et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet. 2006;38(5):570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- 34.Boyer JG, Deguise MO, Murray LM, et al. Myogenic program dysregulation is contributory to disease pathogenesis in spinal muscular atrophy. Hum Mol Genet. 2014;23(16):4249–4259. doi: 10.1093/hmg/ddu142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bricceno KV, Martinez T, Leikina E, et al. Survival motor neuron protein deficiency impairs myotube formation by altering myogenic gene expression and focal adhesion dynamics. Hum Mol Genet. 2014;23(18):4745–4757. doi: 10.1093/hmg/ddu189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2007;2(9):e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paushkin S, Gubitz AK, Massenet S, Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr Opin Cell Biol. 2002;14(3):305–312. doi: 10.1016/s0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- 38.Gabanella F, Carissimi C, Usiello A, Pellizzoni L. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum Mol Genet. 2005;14(23):3629–3642. doi: 10.1093/hmg/ddi390. [DOI] [PubMed] [Google Scholar]
- 39.La Bella V, Cisterni C, Salaün D, Pettmann B. Survival motor neuron (SMN) protein in rat is expressed as different molecular forms and is developmentally regulated. Eur J Neurosci. 1998;10(9):2913–2923. doi: 10.1111/j.1460-9568.1998.00298.x. [DOI] [PubMed] [Google Scholar]
- 40.Germain-Desprez D, Brun T, Rochette C, Semionov A, Rouget R, Simard LR. The SMN genes are subject to transcriptional regulation during cellular differentiation. Gene. 2001;279(2):109–117. doi: 10.1016/s0378-1119(01)00758-2. [DOI] [PubMed] [Google Scholar]
- 41.Guettier-Sigrist S, Hugel B, Coupin G, Freyssinet JM, Poindron P, Warter JM. Possible pathogenic role of muscle cell dysfunction in motor neuron death in spinal muscular atrophy. Muscle Nerve. 2002;25(5):700–708. doi: 10.1002/mus.10081. [DOI] [PubMed] [Google Scholar]
- 42.Hayhurst M, Wagner AK, Cerletti M, Wagers AJ, Rubin LL. A cell-autonomous defect in skeletal muscle satellite cells expressing low levels of survival of motor neuron protein. Dev Biol. 2012;368(2):323–334. doi: 10.1016/j.ydbio.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Remels AH, Langen RC, Schrauwen P, Schaart G, Schols AM, Gosker HR. Regulation of mitochondrial biogenesis during myogenesis. Mol Cell Endocrinol. 2010;315(1–2):113–120. doi: 10.1016/j.mce.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 44.Duguez S, Féasson L, Denis C, Freyssenet D. Mitochondrial biogenesis during skeletal muscle regeneration. Am J Physiol Endocrinol Metab. 2002;282(4):802–809. doi: 10.1152/ajpendo.00343.2001. [DOI] [PubMed] [Google Scholar]
- 45.Leary SC, Battersby BJ, Hansford RG, Moyes CD. Interactions between bioenergetics and mitochondrial biogenesis. Biochim Biophys Acta. 1998;1365(3):522–530. doi: 10.1016/s0005-2728(98)00105-4. [DOI] [PubMed] [Google Scholar]
- 46.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25(4):1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baldelli S, Aquilano K, Ciriolo MR. PGC-1α buffers ROS-mediated removal of mitochondria during myogenesis. Cell Death Dis. 2014;5:e1515. doi: 10.1038/cddis.2014.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell GR, Reeve A, Ziabreva I, et al. Mitochondrial DNA deletions and depletion within paraspinal muscles. Neuropathol Appl Neurobiol. 2013;39(4):377–389. doi: 10.1111/j.1365-2990.2012.01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubowitz V. Very severe spinal muscular atrophy (SMA type 0): an expanding clinical phenotype. Eur J Paediatr Neurol. 1999;3(2):49–51. doi: 10.1053/ejpn.1999.0181. [DOI] [PubMed] [Google Scholar]
- 50.Simonds AK. Respiratory complications of the muscular dystrophies. Semin Respir Crit Care Med. 2002;23(3):231–238. doi: 10.1055/s-2002-33031. [DOI] [PubMed] [Google Scholar]
- 51.Di Meo I, Fagiolari G, Prelle A, Viscomi C, Zeviani M, Tiranti V. Chronic exposure to sulfide causes accelerated degradation of cytochrome C oxidase in ethylmalonic encephalopathy. Antioxid Redox Signal. 2011;15(2):353–362. doi: 10.1089/ars.2010.3520. [DOI] [PubMed] [Google Scholar]
- 52.Zeviani M, Nonaka I, Bonilla E, et al. Fatal infantile mitochondrial myopathy and renal dysfunction caused by cytochrome C oxidase deficiency: immunological studies in a new patient. Ann Neurol. 1985;17(4):414–417. doi: 10.1002/ana.410170422. [DOI] [PubMed] [Google Scholar]
- 53.Müller-Höcker J, Droste M, Kadenbach B, Pongratz D, Hübner G. Fatal mitochondrial myopathy with cytochrome-c-oxidase deficiency and subunit-restricted reduction of enzyme protein in two siblings: an autopsy-immunocytochemical study. Hum Pathol. 1989;20(7):666–672. doi: 10.1016/0046-8177(89)90154-8. [DOI] [PubMed] [Google Scholar]
- 54.Oldfors A, Sommerland H, Holme E, Tulinius M, Kristiansson B. Cytochrome c oxidase deficiency in infancy. Acta Neuropathol. 1989;77(3):267–275. doi: 10.1007/BF00687578. [DOI] [PubMed] [Google Scholar]
- 55.Kong L, Wang X, Choe DW, et al. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci. 2009;29(3):842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wadman RI, Vrancken AF, van den Berg LH, van der Pol WL. Dysfunction of the neuromuscular junction in spinal muscular atrophy types 2 and 3. Neurology. 2012;79(20):2050–2055. doi: 10.1212/WNL.0b013e3182749eca. [DOI] [PubMed] [Google Scholar]
- 57.Montes J, Garber CE, Kramer SS, et al. A randomized, controlled clinical trial of exercise in patients with spinal muscular atrophy: methods and baseline characteristics. J Neuromusc Dis. 2014;1(2):151–161. [PubMed] [Google Scholar]
- 58.Viscomi C, Bottani E, Civiletto G, et al. In vivo correction of COX deficiency by activation of the AMPK/PGC-1α axis. Cell Metab. 2011;14(1):80–90. doi: 10.1016/j.cmet.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srivastava S, Barrett JN, Moraes CT. PGC-1α/β upregulation is associated with improved oxidative phosphorylation in cells harboring nonsense mtDNA mutations. Hum Mol Genet. 2007;16(8):993–1005. doi: 10.1093/hmg/ddm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng B, Liao Z, Locascio JJ, et al. Global PD Gene Expression (GPEX) Consortium. PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med. 2010;2(52):52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsunemi T, Ashe TD, Morrison BE, et al. PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4(142):142ra97. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Da Cruz S, Parone PA, Lopes VS, et al. Elevated PGC-1α activity sustains mitochondrial biogenesis and muscle function without extending survival in a mouse model of inherited ALS. Cell Metab. 2012;15(5):778–786. doi: 10.1016/j.cmet.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Montesano A, Luzi L, Senesi P, Mazzocchi N, Terruzzi I. Resveratrol promotes myogenesis and hypertrophy in murine myoblasts. J Transl Med. 2013;11:310. doi: 10.1186/1479-5876-11-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.