Summary
The BRCT-domain protein, Rad4TopBP1, facilitates activation of the DNA damage checkpoint in S. pombe by physically coupling the Rad9-Rad1-Hus1 clamp, the Rad3ATR-Rad26ATRIP kinase complex and Crb253BP1 mediator. We have now determined crystal structures of the BRCT repeats of Rad4TopBP1, revealing a distinctive domain architecture, and have characterized their phosphorylation-dependent interactions with Rad9 and Crb253BP1. We identify a cluster of phosphorylation sites in the N-terminal region of Crb253BP1 that mediate interaction with Rad4TopBP1, and reveal a hierarchical phosphorylation mechanism in which phosphorylation of Thr215 and Thr235 promotes phosphorylation of the non-canonical Thr187 site by scaffolding CDK recruitment. Finally we show that simultaneous interaction of a single Rad4TopBP1 molecule with both Thr187 phosphorylation sites in a Crb253BP1 dimer, is essential for establishing the DNA damage checkpoint.
Introduction
Survival of acute DNA damage requires the coordinated recruitment and assembly of a set of multi-protein signaling complexes, localised to the substantial segments of ssDNA that result from double strand break resection or replication fork collapse (Smits et al., 2010). Rad4TopBP1, in the fission yeast Schizosaccharomyces pombe, plays a central role in assembling this DNA damage checkpoint apparatus. This multi-BRCT-domain protein interacts simultaneously with the Rad9-Rad1-Hus1 clamp (Furuya et al., 2004) loaded at the 5’-recessed margin of the ssDNA by the Rad17-RFC clamp loader, and with the PI3K-like kinase Rad3ATR that is recruited via its Rad26ATRIP subunit, to RPA-coated ssDNA.
As well as coupling the two halves of the checkpoint apparatus, Rad4TopBP1 also interacts with the mediator protein, Crb253BP1. This in turn couples the checkpoint apparatus to chromatin via interaction of its C-terminal Tudor and BRCT domains with epigenetic modifications on histones H4 and H2A respectively in the G1/S phase of the cell cycle (Lin et al., 2012), and facilitates recruitment of the checkpoint kinase, Chk1 (Qu et al., 2012). In a parallel exclusive role in replication, Rad4TopBP1 also facilitates assembly of the CMG replicative DNA helicase via interaction with Sld3 (Fukuura et al., 2011; Taylor et al., 2011), a function that is conserved in the interaction of TopBP1 with treslin in mammalian systems (Boos et al., 2011; Kumagai et al., 2011).
We have now determined the crystal structures of the BRCT1,2 and BRCT3,4 segments of Rad4TopBP1, and have structurally and biochemically characterized their phospho-specific interactions with Rad9 and Crb253BP1. Unexpectedly we find that the Rad4TopBP1 interaction with Crb253BP1, both in vitro and in vivo, is mediated principally by phosphorylation of Crb2-Thr187, which is essential for establishing the DNA damage checkpoint. Phosphorylation of Thr187 is dependent on phosphorylation of two canonical CDK phosphorylation sites (Thr215, Thr235), which also mediate interaction with Rad4TopBP1. Our studies reveal a hierarchical phosphorylation system that facilitates assembly of a core complex within the DNA damage checkpoint apparatus in which a single Rad4TopBP1 molecule connects the 9-1-1 checkpoint clamp to a Crb253BP1 dimer.
Results
Structure of Rad4TopBP1 BRCT-repeats
Crystals of the separate BRCT1,2 and BRCT3,4 segments of S. pombe Rad4 were obtained using protein expressed in, then purified from E. coli (FIGURE 1A). Their respective structures were solved by molecular replacement (FIGURE1B,C; see also EXPERIMENTAL PROCEDURES).
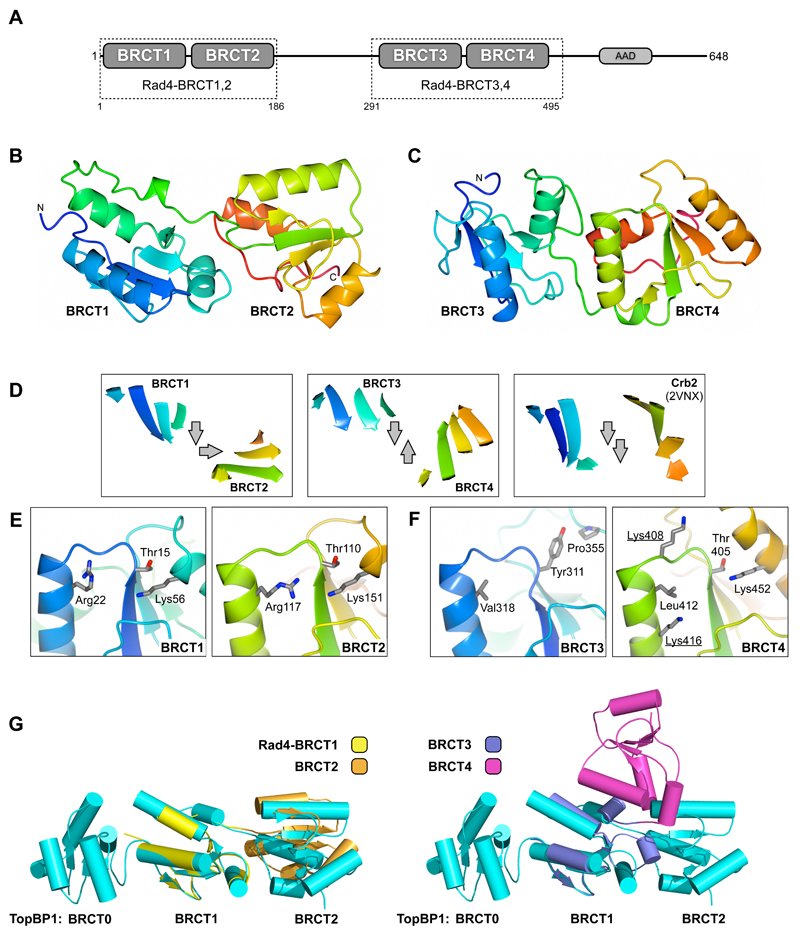
Figure 1. Structure of Rad4 BRCT repeats.
A) Schematic of S.pombe Rad4TopBP1. The protein consists of two BRCT tandem repeats, separated by ~100 residues that are predicted to be highly flexible. The C-terminus contains an ATR-activating domain (labelled AAD) conserved in mammalian TopBP1 (Kumagai et al., 2006)
B) Secondary structure cartoon of the crystal structure of Rad4-BRCT1,2 rainbow coloured blue:N-terminus → red:C-terminus
C) As B) but for Rad4-BRCT3,4
D) Comparison of the orientation of the core β-sheets between the first and second BRCT domain in the Rad4-BRCT1,2 and BRCT3,4 tandem repeats (left and middle), with the BRCA1/MDC1-family tandem BRCT repeat from Crb2. The orthogonal arrangement of Rad4-BRCT1,2 is distinct from the anti-parallel arrangement seen in Rad4-BRCT3,4, and both are different to the ‘canonical’ parallel arrangement found in Crb2 and related proteins.
E) BRCT domains 1, 2 and 4 of Rad4 possess characteristic clusters of polar and charged residues that are implicated in mediating interaction with phosphorylated peptides in other proteins. By contrast the equivalent site in Rad4-BRCT3 is predominantly hydrophobic and unlikely to be involved in phospho-mediated interactions.
F) The arrangement of Rad4-BRCT1,2 is identical to that previously observed between BRCT domains 1 and 2 of the mammalian Rad4 orthologue TopBP1 (left). Rad4-BRCT3,4 has a completely different domain arrangement to Rad4-BRCT1,2 and TopBP1-BRCT1,2, and is so far unique.
Rad4-BRCT1,2 has the same architecture as previously described for human TopBP1-BRCT0,1,2 (Rappas et al., 2011), with the central β-sheets in the two BRCT domains arranged perpendicularly to each other, rather than parallel as in canonical phosphopeptide-binding tandem BRCT2 structures - such as those from BRCA1, MDC1 and Crb2 (Shiozaki et al., 2004; Williams et al., 2004; Lee et al., 2005; Stucki et al., 2005; Kilkenny et al., 2008) (FIGURE 1D). The additional N-terminal BRCT domain 0 found in human TopBP1 is not present in the yeast protein. Like the mammalian TopBP1 equivalents, Rad4-BRCT1 and BRCT2 each possess a cluster of residues that are implicated in mediating specific interactions with the phospho-group on phosphorylated serine or threonine residues (Kilkenny et al., 2008; Rappas et al., 2011) (FIGURE 1E), suggesting that the tandem BRCT1,2 structure is capable of interacting with two phosphorylated residues simultaneously.
The arrangement of the BRCT domains in Rad4-BRCT3,4 is distinct from both the BRCA1/MDC1/Crb2 and Rad4-BRCT1,2 classes, with the central β-sheets of the two domains having an anti-parallel orientation (FIGURE 1D). Unlike the canonical class, where the N-terminal domain provides a phospho-binding site, or the Rad4-BRCT1,2 class in which both domains do, only BRCT4 possesses a suitable configuration of residues to mediate interaction with a phosphorylated residue (FIGURE 1F).
Phospho-dependent interaction of Rad9 with Rad4TopBP1
Previous studies identified a phosphorylation-dependent interaction, near the end of the C-terminal tail of Xenopus Rad9, in the 9-1-1 clamp with the BRCT1,2 segment of TopBP1, essential to establishment of a DNA damage checkpoint (Lee et al., 2007). Further studies identified the essential Rad9 phosphorylation in humans as a constitutive modification of Ser387 by CK2 (Takeishi et al., 2010; Rappas et al., 2011), which promotes interaction with TopBP1-BRCT1 (Rappas et al., 2011). In the S. pombe system the C-terminal tail of Rad9 is phosphorylated in S-phase and after DNA damage at SQ/TQ sites Thr412 and Ser423, and its interaction with Rad4 is mediated by the C-terminal BRCT3,4 domains rather than the N-terminal domains as in vertebrate TopBP1 (Furuya et al., 2004; Garcia et al., 2005).
To define the specificity of the S.pombe Rad9 / Rad4 interaction biochemically, we used a fluorescence polarization (FP) assay and measured the binding of fluorescein-labelled phosphopeptides derived from the Rad9 tail to both BRCT1,2 and BRCT3,4 constructs (FIGURE 2A). Consistent with previous mapping results, we found that a Rad9 peptide phosphorylated on Thr412 and Ser423 showed little, if any specific binding to the Rad4-BRCT1,2 construct in vitro, whereas it bound with low micromolar affinity to the BRCT3,4 construct. Peptides phosphorylated on either Thr412 or Ser423 alone, both bound to BRCT3,4 (FIGURE 2B), but with the pThr412 peptide binding nearly five times tighter (1.9 μM) than a pSer423 peptide (8.6 μM), whose interaction was slightly weaker than the bis-phosphorylated peptide (6.4 μM). This behavior is fully consistent with BRCT3,4 interacting with either pThr412 or pSer423 but not both simultaneously, and with some internal binding competition between the two phosphorylated residues in the bis-phosphorylated peptide. We also measured the interaction of BRCT3,4 with a Rad9-pThr412 / pSer423 peptide that had been pretreated with λ-phosphatase (see EXPERIMENTAL PROCEDURES), and observed no binding, confirming that the interaction is driven by the phosphorylation of the Rad9 sequence (FIGURE 2C). To test whether the putative phospho-binding site identified in the BRCT3,4 crystal structure is involved in the interaction, we expressed a mutant containing a charge reversal mutation (K452E) in a residue expected to be essential for interaction with a phosphate group, and measured its interaction with the tight-binding pThr412 peptide. Unlike wild-type, the K452E mutant showed no detectable binding, confirming BRCT4 as the binding site for the phosphorylated C-terminal tail of Rad9. Additionally, maps from crystals of Rad4-BRCT3,4 grown in buffers containing LiSO4 displayed strong electron density consistent with a sulphate ion (iso-structural with phosphate) bound to the putative phospho-binding site in BRCT4 (FIGURE 2C inset), thereby confirming the importance of the Lys452 / phosphate interaction.
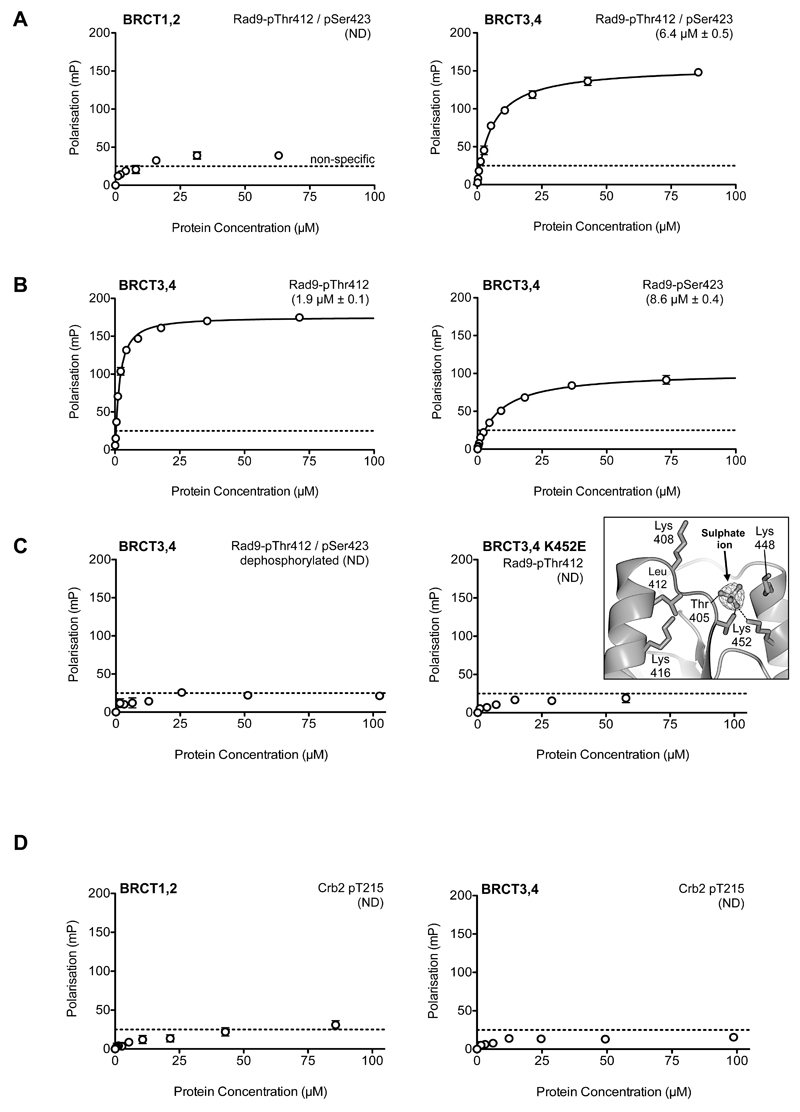
Figure 2. Phosphopeptide binding to Rad4 BRCT repeats.
A) Fluorescence polarization analysis of interaction of the C-terminus of S. pombe Rad9 phosphorylated at the DNA damage dependent (S/T-Q) sites Thr412 and Ser423, with Rad4 BRCT repeats. No significant interaction is seen with BRCT1,2 (left), but the bis-phosphorylated peptide binds with micromolar affinity to BRCT3,4 (right). Data points are averages of three measurements; error bars indicate one standard deviation.
B) A Rad9 C-terminal peptide binds Rad4-BRCT3,4 >4-fold tighter when mono-phosphorylated at Thr412 than when mono-phosphorylated at Ser423, however both bind with sufficient affinity that they are likely to be mutually redundant in vivo. The lower affinity of the bis-phosphorylated peptide results from internal competition between the two phosphorylated residues for the single binding-site, as previously described (Rappas et al., 2011). Data points are averages of three measurements; error bars indicate one standard deviation.
C) (left) Dephosphorylation of a Rad9 C-terminal peptide abolishes interaction with Rad4-BRCT3,4. (right) Charge reversal mutation of Lys452 in BRCT4 abolishes interaction with phosphorylated Rad9 C-terminal peptide. (right inset) – Lys452 interacts with a sulphate ion in a Rad4-BRCT3,4 crystal structure implicating it as a likely phosphate-binding residue. Data points are averages of three measurements; error bars indicate one standard deviation.
D) Contrary to expectation, a Crb2-derived phosphopeptide incorporating the CDK phosphorylation site pThr215, previously shown to be important for a yeast two-hybrid interaction between Rad4 and Crb2, showed no interaction with either Rad4 BRCT tandem repeat. Data points are averages of three measurements; error bars indicate one standard deviation.
Phospho-dependent interactions of Crb253BP1 with Rad4TopBP1
Crb2 was identified in a yeast two-hybrid screen with Rad4 as bait (Saka et al., 1997), and is essential for establishment of the DNA damage checkpoint in S. pombe (Saka et al., 1997; Willson et al., 1997). Previous studies have also identified a CDK-dependant phosphorylation of Crb2 at Thr215 (Esashi and Yanagida, 1999) that when mutated to alanine weakened the interaction between Rad4 and Crb2 and compromised recruitment of Crb2 to persistent DSBs (Du et al., 2006).
To test the suggestion that pThr215 mediates interaction with Rad4, we made a fluorescently labeled Crb2-pThr215 phosphopeptide and analysed its interaction with the Rad4-BRCT1,2 and BRCT3,4 constructs. We observed essentially no interaction with Rad4-BRCT3,4, and contrary to expectation, only a weak non-specific level of binding to BRCT1,2 (FIGURE 2D). Thus, while phosphorylation of Crb2-Thr215 may play a role in the DNA damage checkpoint, our in vitro data would suggest that it alone does not mediate the interaction with Rad4.
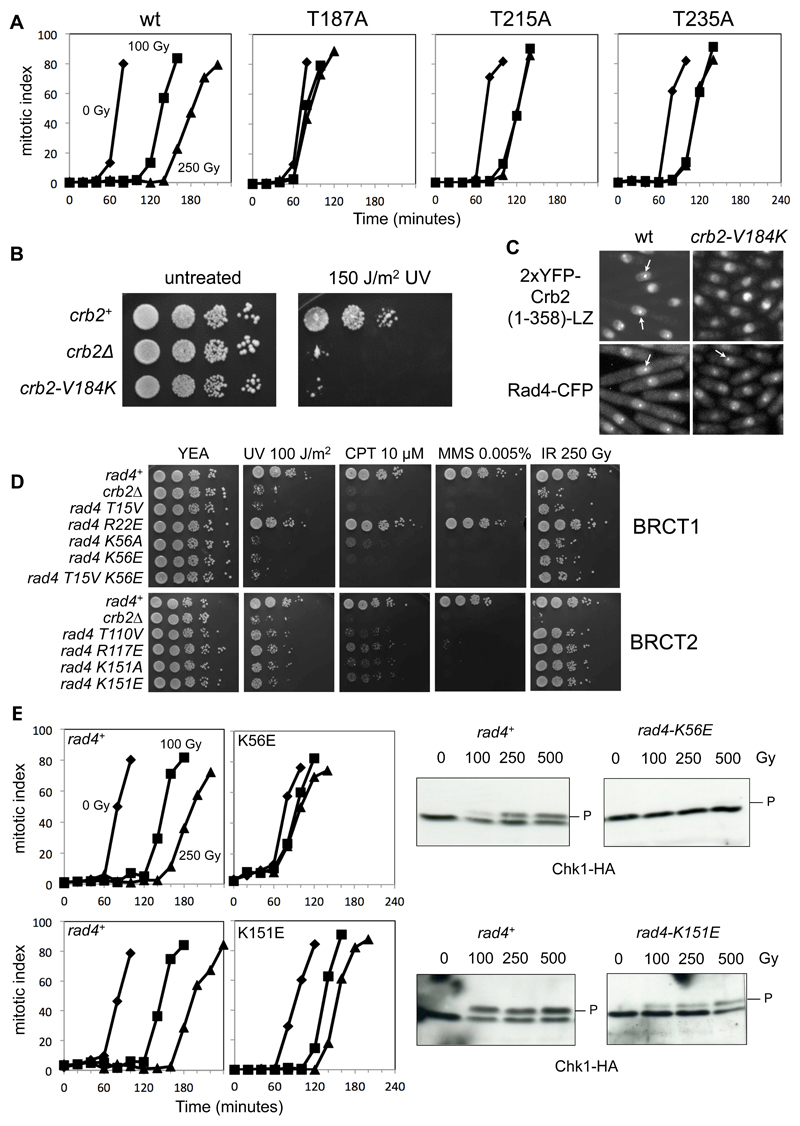
The region of Crb2 essential for interaction with Rad4 had previously been localized to between amino acid residues 141 and 245 (Du et al., 2006). To identify other potential CDK phosphorylation sites in Crb2 that might mediate interaction with Rad4, we performed a multiple sequence alignment of this region from a range of fission yeast species, and identified a conserved TP motif at Thr235 (FIGURE 3A). Consistent with a role for Thr235 in mediating a CDK phosphorylation-dependent interaction with Rad4, a Crb2-T235A mutation abolished formation of radiation-induced foci by a Crb2 N-terminal fragment fused to a leucine zipper (Du et al., 2006) (FIGURE 3B), and cells arrested in M-phase showed strong reactivity to a pThr235-phospho-specific antibody (FIGURE S1A). Mutation of the only other possible phosphorylation site (Ser234) within this region of Crb2, had no affect on focus formation.
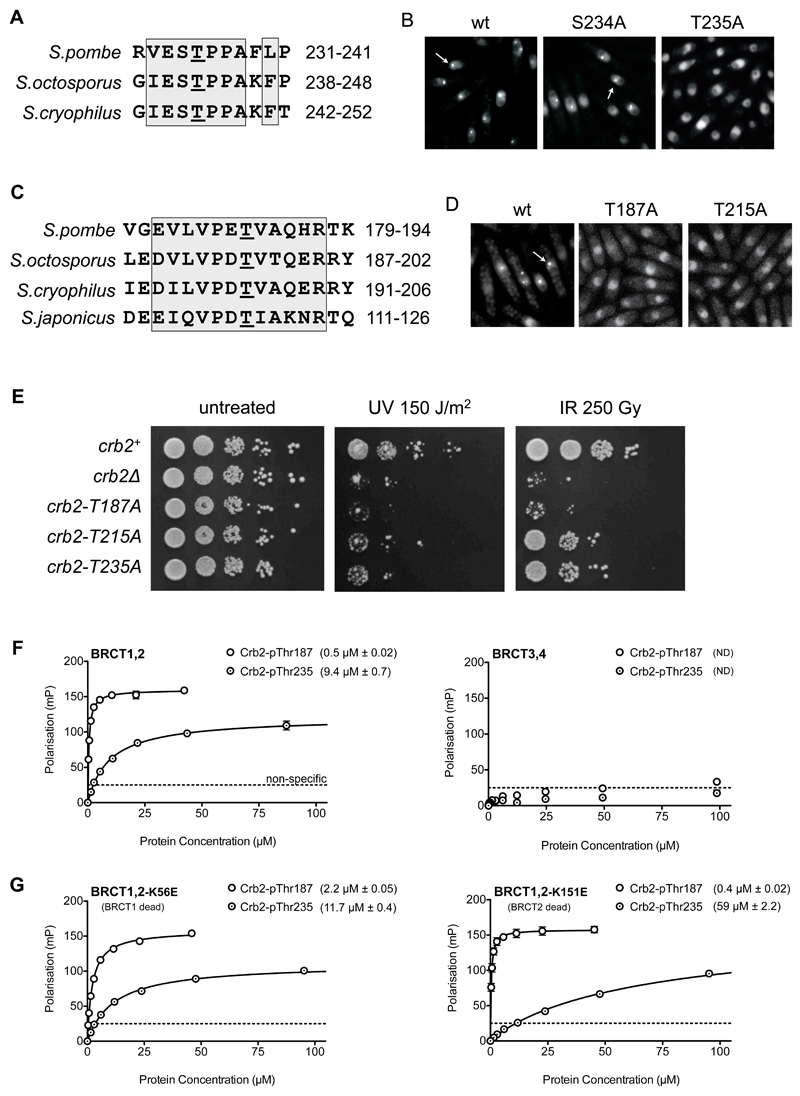
Figure 3. Identification of Crb2 phosphorylation sites mediating interaction with Rad4.
A) Thr235 is a consensus CDK phosphorylation site conserved across Schizosaccharomyces species.
B) Compared to wild-type (left) or an S234A mutation (middle), T235A mutation abolished recruitment of a YFP-tagged Crb2(67-358)-LZ reporter to DNA double-strand breaks (see METHODS). Punctate fluorescent foci are indicated by arrows. In all focus formation figures, cells were exposed to 80 Gray of ionising radiation.
C) Truncation analysis (SUPPLEMENTARY FIGURE 1) identifies a Crb2 segment, from amino acids 180 to 200, as essential for DNA damage focus formation by the fluorescent Crb2 reporter, with Thr187 being the only conserved phosphorylatable residue in the vicinity.
D) Mutation of Thr187 or Thr215 also abolished recruitment of the fluorescent Crb2 reporter to DNA damage foci.
E) Alanine mutation of Thr187, Thr215 or Thr235 all impaired survival of exposure to UV or ionizing radiation, compared to unmutated Crb2 (see METHODS). T215A and T235A displayed some residual resistance whereas T187A phenocopied the Crb2 null strain.
F) A Crb2-derived peptide incorporating pThr187 bound with sub-micromolar affinity to Rad4-BRCT1,2, but not BRCT3,4. A pThr235 peptide bound with modest micromolar affinity to Rad4-BRCT1,2, but not BRCT3,4. Data points are averages of three measurements; error bars indicate one standard deviation.
G) Charge-reversal mutation of the phosphate-binding site in either BRCT1 (left) or BRCT2 (right) did not significantly affect the tight binding of the Crb2-pThr187 peptide, suggesting that it is able to bind to either site. By contrast, the modest affinity of the Crb2-pThr235 peptide was decreased 5-fold by a BRCT2 mutation, identifying this as its preferred binding site. Data points are averages of three measurements; error bars indicate one standard deviation.
In parallel we constructed a series of deletions within the N-terminal region of Crb2, and identified the region between residues 180 and 200 as important for DSB focus formation by a Crb2 N-terminal fragment (FIGURE S1B). Within this region, there is a stretch of 12 amino acids (181-192) that is strongly conserved amongst Crb2 homologs in other fission yeast species (FIGURE 3C). Mutation of the only conserved phosphorylatable residue in this stretch, Thr187, resulted in a complete loss of focus formation by a Crb2-leucine zipper fusion, like the phenotype caused by either T215A or T235A mutation (FIGURE 3D). Furthermore, similar to the previously reported phenotype of the T215A mutation, a T187A mutation completely abolished, and a T235A substantially diminished the ability of Crb2 to restore the IR and UV hypersensitivity of crb2Δ cells (FIGURE 3E, FIGURE S1C). As with Thr235, cells arrested in M-phase showed strong reactivity to a pThr187-phospho-specific antibody, but only weak reactivity was observed with a T187A mutant Crb2 (FIGURE S1D), strongly implicating Thr187 in mediating a phosphorylation-dependent interaction with Rad4. Unlike Thr215 and Thr235, Thr187 is not present in a TP motif that would identify it as an obvious CDK target, and the sequence context resembles no known protein kinase consensus.
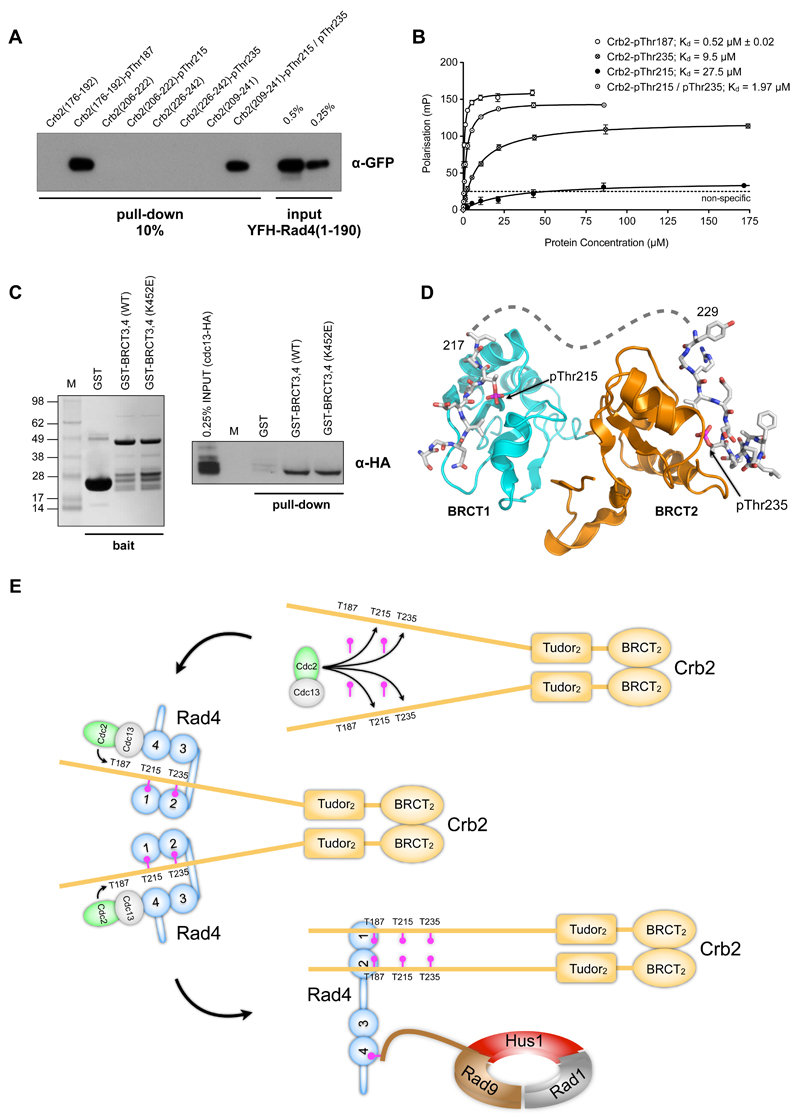
To better understand the role these previously unreported phosphorylation sites might play in mediating Crb2 – Rad4 interactions, we synthesized fluorescently-labelled Crb2 phosphopeptides and analysed their interaction with Rad4 in vitro, as above. Neither the pThr187 or pThr235 peptides bound to the BRCT3,4 construct (FIGURE 3F). However, a strong interaction of pThr187 (0.5 µM), and a weaker interaction of pThr235 (9.4 µM) were both observed with the BRCT1,2 construct. Consistent with this, only a Crb2-pThr187 phosphopeptide was effective in co-precipitating BRCT1,2 in a pull-down assay (FIGURE S2A).
In order to fully dissect, and understand, the specific interactions of the Crb2-phosphopeptides with the Rad4-BRCT1,2 construct, we used our previous strategy of making defined charge-reversal mutants in either BRCT1 (K56E) or BRCT2 (K151E) of the BRCT1,2 construct, specifically designed to abrogate any phospho-interaction with that particular domain (Boos et al., 2011; Rappas et al., 2011). Unexpectedly, the FP experiments showed that the pThr187 peptide was able to bind to both BRCTs, albeit with higher affinity to BRCT1 over BRCT2 – 0.4 vs 2.2 μM respectively (FIGURE 3G). The pThr235 peptide, however, showed much weaker binding, plus a clear preference for binding to BRCT2 (11.7 μM) over BRCT1 (59 μM). All phosphopeptide binding was lost with a K56E/K151E double mutant (FIGURE S2B).
Crystal structure of Rad4TopBP1-BRCT1,2 bound to Crb253BP1-pThr187
Co-crystallisation of Rad4-BRCT1,2 with a phosphorylated peptide derived from residues 179-194 of Crb2, gave a crystal structure with peptide molecules bound to both BRCT domains (FIGURE 4A). The core Crb2 sequence 184 – Val Pro Glu pThr Val – 188 bound with an essentially identical conformation to each BRCT domain, with the phosphate groups attached to Crb2-Thr187 forming a network of hydrogen bonding interactions with a set of conserved phosphate-binding residues in each BRCT domain - (FIGURE 4B,C). While the phosphate interactions are similar to those observed in complexes with canonical phosphopeptide-binding tandem BRCT domains (Williams et al., 2004; Stucki et al., 2005; Kilkenny et al., 2008; Gong et al., 2010), the trajectory of the phosphopeptide across the binding surface of the domain is radically different to these, with the bound phosphopeptide lying in a shallow groove on the outer face of each BRCT domain. In both cases Crb2-Val184 three residues upstream of the phosphothreonine 187 packs into a hydrophobic pocket; formed by Rad4 residues Phe40, Pro55, Lys56 and Phe59 in BRCT1 and by Leu 135, Arg150, Lys151, Tyr154 and Trp158 in BRCT2. The pocket in BRCT1 is more extended and also accommodates the side chain of Crb2-Leu183 packed between the side chains of Phe59 and Lys42, whereas this residue is directed away from Rad4 in the BRCT2 interaction. This additional interaction may underlie the higher affinity of the BRCT1 site.
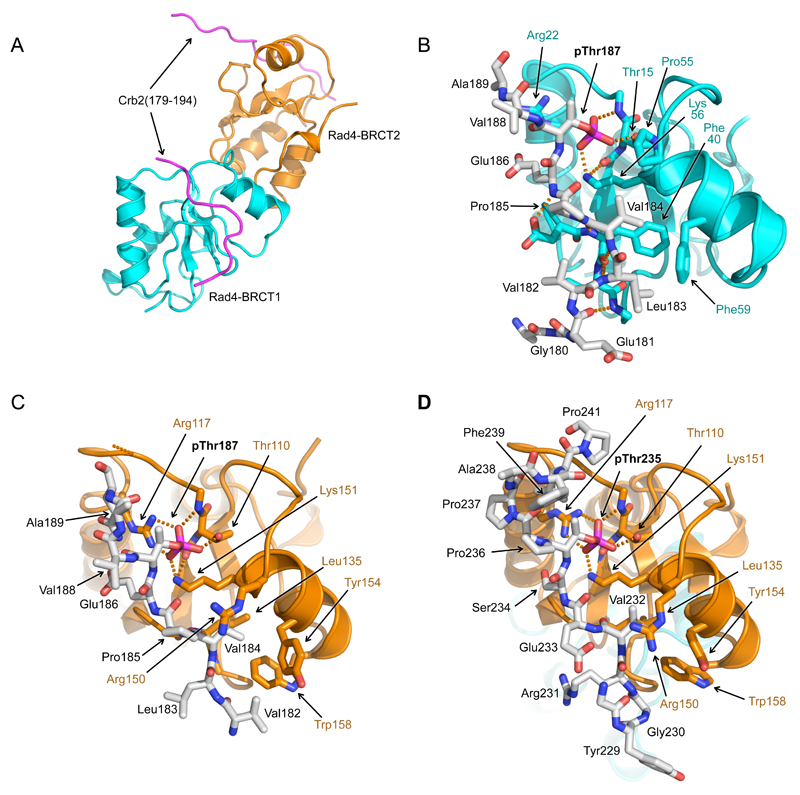
Figure 4. Structure of Rad4 – Crb2 phosphopeptide complexes.
A) Overall structure of Rad4-BRCT1,2 bound to two copies of a Crb2-derived peptide incorporating pThr187. BRCT1 is coloured cyan, BRCT2 orange, and Crb2 peptides in magenta.
B) Detail of interaction of Crb2-pThr187 peptide with Rad4-BRCT1. The phosphate group of pThr187 interacts with the side-chain of Lys56 and the main and side chains of Thr15. Upstream of pThr187, Val184 binds in a hydrophobic pocket on the BRCT domain. This orientation of peptide binding to a BRCT domain has not previously been observed.
C) As B) but for Rad4-BRCT2. As well as Lys151 and Thr110, the phosphate of Crb2-pThr187 also interacts with Arg117 of Rad4. The peptide binds in the same orientation as to BRCT1, with Val184 again interacting with a hydrophobic pocket.
D) As C) but for a Crb2-derived peptide incorporating pThr235. Consistent with the measured affinities the pThr235 peptide was only found bound to BRCT2 in the crystal structure. The peptide binds in the same orientation as the pThr187 peptide, with the phosphate group and the upstream valine residue (Val232) making the same interactions as in the pThr187 peptide complex.
Crystal structure of Rad4TopBP1-BRCT1,2 bound to Crb253BP1-pThr235
We were also able to define conditions for the co-crystallisation of a Crb2 phosphopeptide (residues 229-241) encompassing the weaker binding pThr235 phosphorylation site with Rad4-BRCT1,2. Consistent with the much lower affinity we observed for the Rad4-BRCT1 phospho-binding site, only the BRCT2 site was occupied by the phosphopeptide in the crystals (FIGURE 4D). The core sequence 232 – Val Glu Ser pThr Pro – 236 binds with a very similar conformation to the Crb2-pThr187 peptide, with the phosphate groups engaging the conserved phosphate-binding triad of Thr110, Arg117 and Lys151. As with the pThr187 peptide, the valine (Val232) three residues upstream of the phosphothreonine binds into the hydrophobic pocket formed by residues Leu 135, Arg150, Lys151, Tyr154 and Trp158, strengthening the notion that a small hydrophobic residue at this position is an important specificity determinant for phosphopeptide binding to this BRCT domain. The C-terminal part of the Crb2-pThr235 phosphopeptide, 236 – Pro Pro Ala Phe Leu Pro – 241, forms a rigid 310-helix that buries the Cγ of the threonine side-chain. While this structured segment is accommodated in the Rad4-BRCT2 site, it generates some steric clashes when modeled into the BRCT1 site, consistent with its much lower affinity for binding at that site.
In vivo function of Crb253BP1-Rad4TopBP1 – phospho-dependent interactions
Previous studies have highlighted the importance of the overall Crb2-Rad4 interaction in establishing the DNA damage checkpoint in fission yeast (Du et al., 2006). To define the individual contributions of the phosphorylation sites in Crb2 and their binding partners in Rad4, we established a series of yeast strains in which the components of the overall interaction could be mutated on either side, and the effects on cell viability, DNA damage resistance and checkpoint determined.
Yeast strains with the null allele crb2Δ are fully viable in the absence of DNA damage, but show severe sensitivity to a range of DNA damaging agents (Saka et al., 1997; Willson et al., 1997). The crb2-T215A and crb2-T235A mutants are only partially sensitive to damaging agents, while the crb2-T187A mutant was as defective as the null allele (see FIGURE 3E). Consistent with their DNA damage sensitivity phenotypes, all three crb2 mutants showed G2/M checkpoint defects, with crb2-T187A again being the most severe (FIGURE 5A). These data confirm the importance of all these phosphorylation sites in Crb2 function, but implicate Thr187 as having a primary role. The null mutant-like phenotype of crb2-T187A shows that the role of the Crb2-Rad4 interaction it mediates is more important in establishing the DNA damage checkpoint than was previously inferred from analysis of the crb2-T215A mutant (Du et al., 2006). That the main function of these phosphorylations is to mediate Rad4-Crb2 interaction can be seen from the behavior of a Rad4-Crb2 fusion protein, which is able to rescue the DNA damage sensitivity of a Crb2 null allele regardless of T187A, T215A or T235A mutations (FIGURE S2C)
Figure 5. Rad4-Crb2 phospho-dependent interactions are essential for a DNA-damage checkpoint response.
A) Cell cycle arrest due to DNA damage, in wild-type cells and those with mutations in Thr187, Thr215 and Thr235. While wild-type cells experience a delay of 60 minutes in the time for 50% of the cells to go through mitosis, this is greatly reduced in the phosphorylation site mutants. As with survival of DNA damage, the Thr187 mutant is the most severely affected.
B) Val184 binds to the hydrophobic pockets of both Rad4-BRCT1 and BRCT2 in the crystal structure of the complex with the Crb2-pThr187 peptide. Mutation to a residue incompatible with that interaction (V184K), disrupts survival of DNA damage to a similar degree as the Crb2 null strain.
C) Crb2-V184K mutation disrupts co-localisation of Rad4 and Crb2 at DNA damage induced foci.
D) Mutation of residues in the phosphate-binding sites of either BRCT1 or BRCT2 domains of Rad4 significantly impair survival of a range of genotoxic insults. Unlike the topologically equivalent Arg117 in BRCT2, Arg22 in BRCT1 does not interact directly with the phosphate group in the bound phosphopeptide, and its mutation has little effect on sensitivity to DNA damage.
E) Phosphate-binding site mutations in either BRCT1 or BRCT2 domains of Rad4 significantly reduce DNA damage-dependent cell cycle arrest (left), and associated Chk1 phosphorylation and activation by Rad3ATR (middle and right).
We also mutated Val184, which the co-crystal structures identify as likely to be a key specificity determinant in phosphopeptide binding. A crb2-V184K mutation designed to prevent accommodation of the residue in the hydrophobic pocket in Rad4-BRCT1 and BRCT2, showed comparable defects in DNA damage resistance to a crb2-null allele (FIGURE 5B) and abolished Rad4-dependent focus formation by the fluorescent Crb2(1-358)-LZ reporter, confirming the role of this residue in mediating Crb2-Rad4 interactions (FIGURE 5C).
Unlike Crb2, Rad4 is essential for yeast viability under all circumstances (Fenech et al., 1991). Strains with Rad4 mutations aimed at disrupting phosphopeptide binding to either the BRCT1 or BRCT2 domains showed normal growth and were fully viable, with no temperature sensitivity (FIGURE S3A,B). However, simultaneous mutation of both BRCT1 and BRCT2 was lethal, suggesting that BRCT1 and BRCT2 are redundant for the essential Rad4 functions. Phosphopeptide-binding mutants in either BRCT1 or BRCT2 were sensitive to a range of DNA damaging agents (FIGURE 5D), showed defects in cell cycle arrest and Chk1 phosphorylation on DNA damage (FIGURE 5E) and did not recruit a fluorescent Crb2 reporter to sites of DNA damage (FIGURE S4). Interestingly, mutants proposed to abolish phospho-binding to BRCT1 were as defective for DNA damage phenotypes as the crb2 null mutant, whereas those proposed to abolish phospho-binding to BRCT2 retained residual resistance and Chk1 phosphorylation. When combined with a crb2Δ background, the Rad4 BRCT1,2 phospho-binding mutations showed enhanced sensitivity to camptothecin (FIGURE S4C), consistent with the additional role of Rad4/TopBP1 in mediating interaction with Treslin/Sld3, which may be required for replication re-start following fork collapse (Kumagai et al., 2010).
Thr187 phosphorylation and Crb253BP1 dimerisation
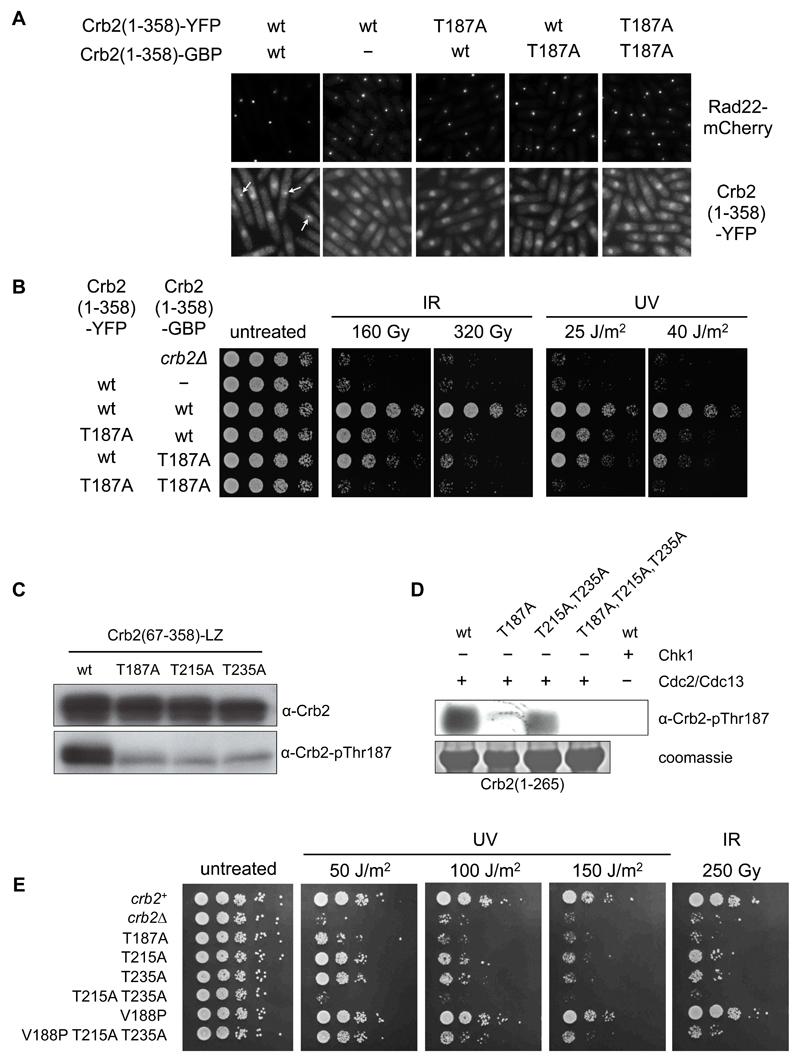
Previous studies have shown that dimerization, mediated by the C-terminal tandem BRCT domains, is essential for Crb2 function in the DNA damage checkpoint (Du et al., 2004; Kilkenny et al., 2008). We were therefore concerned to discover whether the essential phosphorylation of Thr187 we have demonstrated here, is required on one or both of the Crb2 molecules present in a functionally active dimer. To do this we made an inherently monomeric Crb2 construct (1-358) lacking the C-terminal dimerization domains but fused to Yellow Fluorescent Protein (YFP), and similarly a monomeric Crb2(1-358) construct fused to GFP-binding protein (GBP), a single-chain anti-GFP antibody that binds strongly to GFP and its variants, such as YFP (Rothbauer et al., 2008).
Expressed alone, Crb2(1-358)-YFP was unable to form DSB-associated foci in crb2Δ cells following DNA damage with ionizing radiation (FIGURE 6A). However when Crb2(1-358)-YFP and Crb2(1-358)-GBP were simultaneously expressed in crb2Δ cells, the interaction between YFP and GBP facilitates formation of a functional heterodimer between the two Crb2(1-358) fragments, and allows formation of fluorescent foci at DSBs. Consistent with this observation, the DNA damage sensitivities of crb2Δ cells were largely restored by co-expression of Crb2(1-358)-YFP and Crb2(1-358)-GBP (FIGURE 6B). This functional Crb2 heterodimer, allowed us to separately mutate the essential Thr187 phosphorylation site on each of the individual subunits. We found that a T187A mutation on either subunit abolished DNA damage induced focus formation and severely compromised the ability of the dimer to confer DNA damage resistance, indicating that both Thr187 residues in a Crb2 dimer must be phosphorylated for a functional DNA damage checkpoint response.
Figure 6. Phosphorylation of both Thr187 residues in a Crb2 dimer is required for the DNA damage checkpoint.
A) Heterodimerisation of a wild-type N-terminal Crb2 construct via C-terminal YFP and GFP-binding protein (GBP) fusions, promotes co-localistion of fluorescently-tagged Crb2 with Rad22 at DNA damage-induced foci. Omission of the GBP construct or mutation of Thr187 on either Crb2 construct prevents co-localisation.
B) As in A) heterodimerisation of Crb2-YFP and Crb2-GBP fusion constructs only rescues resistance to UV or ionizing radiation when Thr187 in both Crb2 molecules is available for phosphorylation. Crb2-YFP in isolation is unable to homodimerise and does not rescue damage resistance regardless of a wild-type Thr187.
C) Thr187 in a Crb2 reporter construct is phosphorylated in cells arrested in mitosis by the cold-sensitive nda3-KM311 mutation. The Crb2-pThr187 reactive signal is greatly reduced in a T187A mutant, but also in T215A and T235A mutants, indicating a dependence of Thr187 phosphorylation on the ability of the other two sites to also be phosphorylated.
D) In vitro, an N-terminal Crb2 construct can be phosphorylated on Thr187 by the CDK complex Cdc2/Cdc13, but not by Chk1. As in C) Thr187 phosphorylation is reduced by mutation of Thr215 and Thr235, although the effect is less pronounced than in vivo.
E) Conversion of the Thr187 phosphorylation site to a CDK consensus by a V188P mutation makes DNA damage survival independent of phosphorylation of Thr215 and Thr235.
See also FIGURE S5
Hierarchical phosphorylation of Crb253BP1 sites
Our data implicate three phosphorylation sites in Crb2, pThr187, pThr215 and pThr235, as important in mounting a robust DNA damage checkpoint response, with pThr187 playing a critical role. Thr215 and Thr235 are in consensus CDK sites and are phosphorylated in M-phase arrested cells most likely by Cdc2. Thr187 was also phosphorylated in M-phase arrested cells, but this appears to be dependent on Thr215 and Thr235 also being available for phosphorylation (FIGURE 6C). In contrast, phosphorylation of Thr235 still occurred in M-phase arrested cells with a Crb2-T187A mutation (FIGURE S5A) indicating that the dependence is unidirectional. Surprisingly, although not within a consensus CDK site, we found that Thr187 within a recombinant Crb2(1-265) construct could be phosphorylated in vitro by recombinant Cdc2/Cdc13 (FIGURE 6D). This site was not phosphorylated in vitro by purified Chk1 and Crb2(1-358)-YFP still formed foci in a chk1Δ background (FIGURE S5B), eliminating Chk1 as an alternative kinase for this site. Consistent with the in vivo phosphorylation, we found that in vitro Thr187 phosphorylation was reduced when Thr215 and Thr235 were mutated to alanine.
Taken together these data suggest a hierarchical system in which phosphorylation of Thr215 and Thr235 is required to prime the non-canonical Thr187 site for CDK phosphorylation. To test this hypothesis, we mutated Val188 to proline, thereby creating a consensus CDK TP motif. This crb2-V188P mutation did not affect the ability of Crb2 to confer DNA damage resistance (FIGURE 6E), nor did it affect the affinity of a Crb2 phosphopeptide for Rad4-BRCT1,2 in vitro (FIGURE S5C) confirming that Val188 is not critical for the Crb2-Rad4 interaction, consistent with the lack of direct interaction of Val188 with Rad4-BRCT1,2 in the co-crystal structures (FIGURE 4B,C). Remarkably, while mutation of both Thr215 and Thr235 to alanine in the wild-type background confers sensitivity to DNA damaging agents equivalent to loss of Thr187, these mutations had a significantly less profound effect on sensitivity in the V188P background (FIGURE 6E) and substantially restored DNA damage checkpoint function (FIGURE S5D). Thus, artificially converting Thr187 to a CDK consensus site largely dispenses with the requirement for phosphorylation of the two bona fide CDK sites, confirming a hierarchical phosphorylation mechanism for Thr187.
We have shown that the high affinity interaction mediated by the phosphorylated Crb2-Thr187 peptide is sufficient for mediating the functional Crb2-Rad4 interaction in vivo, and is dependent on phosphorylation of Thr215 and Thr235, but the biochemical mechanism by which these sites function together to prime Thr187 for phosphorylation is not defined. While a Crb2 peptide phosphorylated on Thr215 shows little affinity for Rad4, Crb2-pT235 binds Rad4 with a modest affinity sufficient to allow co-crystallisation (see above), suggesting that the pT235 interaction might have a biological role. To explore this further we synthesized a Crb2 peptide phosphorylated on both Thr215 and Thr235 and found that unlike peptide phosphorylated on only one site, this was effective in co-precipitating Rad4-BRCT1,2 (FIGURE 7A). Consistent with this, the affinity of the pThr215 / pThr235 peptide for BRCT1,2 as determined by FP assay (see above) was 1.9 micromolar, ~ 5 fold-tighter than the affinity of the pThr235 site in isolation (FIGURE 7B).
Figure 7. Hierarchical phosphorylation and checkpoint complex assembly.
A) A GFP-tagged Rad4 construct is efficiently co-precipitated by Crb2-derived peptides incorporating either pThr187, or bis-phosphorylated on pThr215 and pThr235, but not by unphosphorylated peptides or peptides with singly phosphorylated Thr215 or Thr235.
B) Phosphorylation of both Thr215 and Thr235 substantially increases the affinity of Crb2 peptides for Rad4-BRCT1,2 compared with those phosphorylated in isolation. The measured affinity (1.97 micromolar) is comparable to that for peptides incorporating pThr187. Data points are averages of three measurements; error bars indicate one standard deviation.
C) HA-tagged cyclin Cdc13 can be co-precipitated from S.pombe lysates by GST-Rad4-BRCT3,4, and GST-Rad4-BRCT3,4(K452E) containing a charge-reversal mutation in the phosphate-binding site of BRCT4, but not by GST alone.
D) Model of bi-dentate interaction of Crb2 segment phosphorylated on both Thr215 and Thr235. The perpendicular orientation of the BRCT domains allows pThr215 to bind to BRCT1 and pThr235 to BRCT2 with the intervening 12 amino acids connecting comfortably between the two.
E) Mechanism of hierarchical phosphorylation – (top) Cdc2-Cdc13 phosphorylates the canonical CDK target sites Thr215 and Thr235 in Crb2 in a cell-cycle dependent manner; (middle) Phosphorylation on Thr215 and Thr235 enables binding of Rad4 to Crb2, which in turn recruits Cdc2-Cdc13 via a phosphorylation-independent interaction with BRCT3,4 of Rad4. The ‘scaffold’ effect provided by simultaneous interaction of Rad4 with the CDK complex and its substrate, facilitates phosphorylation of the non-canonical site Thr187; (bottom) A single Rad4 is able to bind simultaneously to both pThr187 sites in a Crb2 dimer via BRCT domains 1 and 2, bridging Crb2 to the Rad9-Rad1-Hus1 complex via interaction of the phosphorylated C-terminus of Rad9 with BRCT4 of Rad4.
The observation that Crb2 phosphorylated on both Thr215 and Thr235 binds Rad4 with low micromolar affinity, together with the dependence of Thr187 phosphorylation on the prior phosphorylation of Thr215 and Thr235, suggests that a Crb2-pThr215 / pThr235 interaction with Rad4 might play a role in Thr187 phosphorylation, possibly by facilitating recruitment of the Cdc2/Cdc13 kinase to the N-terminal region of Crb2. This notion is supported by our previous identification of Cdc2-Cdc13 as binding partners of Rad4 by mass spectrometry (data not shown) and the observation that Cdc13 co-precipitated from yeast lysates with Rad4 (Lin et al., 2012).
To test this possibility, we repeated the pull-down experiment, and found that Cdc13 could be co-precipitated specifically from yeast lysates by a GST-fusion of Rad4-BRCT3,4. The charge-reversal mutation (K452E), which abolished phospho-dependent binding of Rad9 to Rad4, had no effect on the ability of GST-Rad4-BRCT3,4 to co-precipitate Cdc13 (FIGURE 7C). Thus, while recruitment of Cdc13 (and thereby Cdc2) to Rad4 is dependent on interaction with one or more of the BRCT domains, it does not appear to require phosphorylation of Cdc13.
Discussion
Mechanism of hierarchical phosphorylation
We show here that assembly of the Rad4TopBP1-Crb253BP1 core of the DNA damage checkpoint apparatus is driven by a complex pattern of phosphorylation at three sites in the N-terminal region of Crb253BP1. DNA damage-dependent phosphorylation of Thr187 in both molecules of a Crb253BP1 dimer is both necessary and sufficient for mediating the key interaction with Rad4TopBP1, but only occurs when Thr215 and/or Thr235 have previously been phosphorylated. All three sites share a V-x-x-pT motif, but bind Rad4TopBP1 with different affinities and BRCT domain selectivity. Thus, while the pThr187 site in isolation binds tightly to both BRCT1 and BRCT2, and pThr235 in isolation binds weakly to BRCT2, the extended segment including Thr215 and Thr235 only binds Rad4TopBP1-BRCT1,2 with high affinity when both threonines are phosphorylated. Although we have not determined the structure of that complex directly, modeling based on the experimentally determined structures suggests that this involves cooperative binding of the pThr215 site and the pThr235 site to BRCT1 and BRCT2 respectively (FIGURE 7D). The valine -3 relative to the phosphorylated residue plays an important role in the specific interaction, and mutations of this residue severely affect Crb2 – Rad4 interactions in vitro and in vivo (FIGURE S5C, FIGURE 5B,C).
Thr187 lacks the consensus target sequence for CDK phosphorylation, being followed by valine rather than proline. Despite this, it is phosphorylated in vivo so long as Thr215 and Thr235 are both phosphorylated and thereby able to bind Rad4TopBP1 via BRCT1 and 2. Rad4TopBP1 is also able to bind the active CDK complex Cdc2CDK1/Cdc13CyclinA,B via a phosphorylation-independent interaction of the CDK with BRCT3,4. This suggests a mechanism for Thr187 phosphorylation in which binding of Rad4TopBP1 to the phosphorylated consensus CDK sites Crb2-pThr215 / pThr235 scaffolds the interaction of CDK with the non-canonical site Crb2-Thr187, facilitating its phosphorylation (FIGURE 7E - top and middle). Significantly, mutation of the Thr187 site to a CDK consensus, abolishes the dependence of Thr187 phosphorylation on prior phosphorylation of Thr215 and Thr235.
In both S phase and in G2, the Rad3ATR pathway is activated by the same biochemical entity, RPA-coated ssDNA, but the downstream signal this generates must be tailored to the specific phase of the cell cycle in which activation occurs. In S phase, activated Rad3ATR signals through Cds1Chk2 via the Mrc1Claspin mediator, resulting in cell cycle arrest and stabilization of stalled replication forks (Alcasabas et al., 2001; Tanaka and Russell, 2001). However in G2, Rad3ATR signals through Chk1, which is recruited to the ssDNA bound complex via the Crb253BP1 mediator (Qu et al., 2012), to both arrest cell cycle progression and regulate enzymes involved in DNA repair. In that context, the likely role of the CDK-dependent Crb253BP1 recruitment to Rad4TopBP1 we describe here, is to contribute to this cell cycle-dependent choice of downstream kinase following Rad3ATR activation by sensing the progressive increase in CDK activity as the cell cycle progresses through S into G2. We suggest that the hierarchical phosphorylation mechanism identified here provides a means of integrating incremental changes in CDK activity to provide a step-change in the affinity of Crb2 for Rad4TopBP1 that facilitates recruitment and Rad3ATR activation of Chk1 in G2, rather than Cds1Chk2 in S phase.
The role of Crb253BP1 dimerisation in checkpoint complex assembly
We have previously shown that dimerization of Crb253BP1 via its C-terminal tandem BRCT2 domain is essential for its function in the DNA damage checkpoint (Du et al., 2004; Kilkenny et al., 2008). The biochemical and structural analyses presented here show that Rad4TopBP1-BRCT1,2 can bind simultaneously, with high affinity, and in a phosphorylation-dependent manner, to two copies of the region of Crb253BP1 that encompasses pThr187. Our in vivo data show that the functional interaction between Crb253BP1 and Rad4TopBP1 is dependent on phosphorylation of Thr187, and that mutation of either phosphopeptide-binding site in Rad4TopBP1-BRCT1,2 or of either Thr187 in a Crb253BP1 dimer abolishes an effective DNA damage response. Taken together these data strongly imply a model for assembly of a key DNA damage checkpoint complex in which BRCT1,2 of a single Rad4TopBP1 molecule bridges the pThr187 residues of both molecules in a functional Crb253BP1 dimer, ensuring the proximity of the N-terminal regions of the two Crb253BP1 molecules which are themselves implicated in recruitment of the downstream checkpoint kinase Chk1 (Qu et al., 2012). We speculate that the N-terminal proximity facilitated by the bidentate interaction of Rad4TopBP1 with the Crb253BP1 dimer plays an important role in Chk1 recruitment and/or activation, but whether phosphorylation of all or some of the sites we have characterized is sufficient for Crb2 recruitment and subsequent phosphorylation by Rad3 remains to be determined. Finally, Rad4TopBP1 couples the Crb253BP1 dimer and its associated ligand proteins to a single 9-1-1 complex via interaction of BRCT4 with DNA damage-dependent phosphorylation sites in the C-terminal tail of Rad9 (FIGURE 7E - bottom).
All the components of this system have clear orthologues throughout the eukarya, and all the key interactions appear to be conserved at the level of the molecule, but with some subtle variations in the underlying structural biochemistry. Thus, while both yeast Rad4TopBP1 and metazoan TopBP1 bind the phosphorylated tail of Rad9 in the 9-1-1 clamp complex, in yeast this is a DNA damage-dependent phosphorylation site binding specifically to the second BRCT tandem pair of Rad4TopBP1-BRCT3,4, while in metazoa it is a constitutive CK2 site that binds specifically to the first phosphopeptide-binding BRCT tandem pair of TopBP1-BRCT1,2.
Like Rad4TopBP1 and Crb253BP1 in yeast, mammalian TopBP1 and 53BP1 also interact in a DNA damage-dependent manner, but via the BRCT4,5 of TopBP1 rather than BRCT1,2 as in the yeast system (Cescutti et al., 2010). Sequence and structure analysis (Rappas et al., 2011) identify Rad4TopBP1-BRCT3,4 as the equivalent of TopBP1-BRCT4,5, with the key phosphate-binding residues in BRCT4 of Rad4TopBP1 conserved in BRCT5 of TopBP1. This strongly suggests that the interaction of TopBP1 with 53BP1 is also likely to be mediated by phosphorylation, however the phosphorylation site (or sites) involved have not yet been determined, and whether they are also regulated by a hierarchical phosphorylation mechanism coupled to cell cycle progression, comparable to that described here, is currently unknown.
Experimental Procedures
Full details of experimental procedures are provided in the Supplemental Information.
Protein production, Structure Determination and Peptide Binding Experiments
Rad4 constructs used in the biochemical and structural experiments were expressed in E.coli and and purified by affinity tags and conventional column chromatography. Crystals of proteins and complexes were grown by vapour diffusion in hanging drops, and data for each structure collected from single crystals either on a home source or at the Diamond Light Source, Didcot, UK. Structures were phased by molecular replacement and refined. Full crystallographic statistics are given in TABLE 1. Peptide affinities were determined by fluorescence polarization using Fluorescein labeled peptides purchased from Peptide Protein Research Ltd, Fareham, UK.
Table 1. Crystallographic data collection and refinement statistics.
| Rad4-BRCT1,2 | Rad4-BRCT1,2 / Crb2-pThr187 | Rad4-BRCT1,2 /Crb2-pThr235 | Rad4-BRCT3,4 | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 1.00 | 0.9795 | 1.5419 | 0.97130 |
| Space group | P21 | P21212 | P21212 | P3121 |
| Cell dimensions | ||||
| a, b, c (Å) | 36.7, 39.3, 72.1 | 84.82, 63.72, 53.56 | 76.84 92.14 65.44 | 93.0, 93.0, 75.4 |
| αβγ (°) | 90, 104, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) | 39.27 -1.98 (2.09-1.98)* |
50.92-1.5 (1.58-1.5) |
59.01-2.1 (2.18-2.10) |
55.05-2.5 (2.64-2.5) |
| Rsym | 0.119 (0.473) | 0.129 (0.78) | ||
| Rmerge | 0.062 (0.251) | 0.108 (0.358) | ||
| Mn I / σI | 7.0 (2.5) | 14.2 (4.4) | 7.9 (1.8) | 10.3 (2.8) |
| Completeness (%) | 97.1 (93.0) | 99.8 (100) | 99.2 (93.7) | 98.2 (99.0) |
| Multiplicity | 3.3 (3.2) | 3.9 (3.6) | 4.61 (2.39) | 4.8 (XXX) |
| Refinement | ||||
| Total reflections | 44611 | 184733 | 127133 | 58608 |
| Unique reflections | 13673 | 47028 | 27555 | 13113 |
| Rwork / Rfree | 0.19 / 0.24 | 0.18 / 0.20 | 0.23/ 0.27 | 0.18/ 0.22 |
| Number of atoms | 1724 | 2138 | 3605 | 1752 |
| Macromolecules | 1490 | 1756 | 3124 | 1595 |
| Ligand | 7 | 6 | 140 | 13 |
| Water | 227 | 376 | 341 | 144 |
| Wilson B-factor | 18.78 | 12.47 | 29.27 | 41.27 |
| Average B-factor | 22.50 | 20.80 | 35.00 | 44.50 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.003 | 0.014 | 0.013 | 0.002 |
| Bond angles (°) | 0.73 | 1.7 | 1.6 | 0.67 |
| Ramachandran favoured (%) | 98 | 100 | 98 | 97 |
| Ramachandran outliers | 0 | 0 | 0 | 0 |
| Molprobity clashscore | 4.7 | 2.3 | 5.9 | 5.9 |
Values in parentheses are for highest resolution shell.
Yeast Experiments
Strains used in this study are listed in SUPPLEMENTARY TABLES 1 and 2.
Focus formation was determined microscopically using a DeltaVision system equipped with a CFP/YFP/mCherry filter set (Chroma Technology Corp, Vermont, USA) and a Photometrics CoolSNAP HQ2 camera. Specific phosphorylation of Crb2 in vivo was determined by western blotting of YFP-immunoprecipitated constructs, detected with either an anti-Crb2 antibody (Du et al., 2003), an anti-pThr187 antibody (custom generated by Beijing B&M Biotech, China), or an anti-pThr235 antibody (custom generated by Beijing B&M Biotech). In vitro phosphorylation of Crb2 was performed with Chk1 or Cdc13 immunoprecipitated from yeast cell lysate. Detection of Cdc13 binding to Rad4 was achieved by co-precipitation with GST-Rad4 constructs from cell lysate of yeast expressing HA-tagged Cdc13, and detected by western blotting using an anti-HA antibody.
Supplementary Material
Highlights.
Rad4-BRCT domains mediate novel interactions with Crb2 phosphorylation sites
Rad4 facilitates hierarchical phosphorylation of Crb2 sites by CDK
Rad4 scaffolds CDK phosphorylation of a non-canonical site
Rad4 couples Crb2 dimer to single 9-1-1 checkpoint clamp
Acknowledgements
We thank Mark Roe for assistance with X-ray data collection and Mairi Kilkenny and Wei Yang for contributions at an early stage of the project. We are grateful to the Diamond Light Source Ltd., Didcot, UK, for access to synchrotron radiation and to the Wellcome Trust for support for X-ray diffraction facilities at the University of Sussex. This work was supported by: Cancer Research UK Project Grant C5514/A10722 (CPW, AMC), Cancer Research UK Programme Grant C302/A14532 (LHP and AWO), MRC Programme Grant G1100074 (AMC), and grants from the Chinese Ministry of Science and Technology and the Beijing Municipal Government (LLD). LHP and AMC acknowledge funding for the Genome Damage and Stability Centre from the Medical Research Council.
Footnotes
Accession Codes
Coordinates and structure factors have been deposited with the Protein Data Bank under accession codes: 4bmc, 4bmd, 4bu0 and 4bu1 for Rad4-BRCT1,2; Rad4-BRCT3,4; Rad4-BRCT1,2 / Crb2-pThr187 and Rad4-BRCT1,2 / Crb2-pThr235 respectively.
References
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nature cell biology. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- Boos D, Sanchez-Pulido L, Rappas M, Pearl LH, Oliver AW, Ponting CP, Diffley JF. Regulation of DNA Replication through Sld3-Dpb11 Interaction Is Conserved from Yeast to Humans. Current biology : CB. 2011;21:1152–1157. doi: 10.1016/j.cub.2011.05.057. [DOI] [PubMed] [Google Scholar]
- Cescutti R, Negrini S, Kohzaki M, Halazonetis TD. TopBP1 functions with 53BP1 in the G1 DNA damage checkpoint. The EMBO journal. 2010;29:3723–3732. doi: 10.1038/emboj.2010.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LL, Moser BA, Russell P. Homo-oligomerization is the essential function of the tandem BRCT domains in the checkpoint protein Crb2. J Biol Chem. 2004;279:38409–38414. doi: 10.1074/jbc.M403326200. [DOI] [PubMed] [Google Scholar]
- Du LL, Nakamura TM, Russell P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes & development. 2006;20:1583–1596. doi: 10.1101/gad.1422606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi F, Yanagida M. Cdc2 phosphorylation of Crb2 is required for reestablishing cell cycle progression after the damage checkpoint. Molecular cell. 1999;4:167–174. doi: 10.1016/s1097-2765(00)80364-0. [DOI] [PubMed] [Google Scholar]
- Fenech M, Carr AM, Murray J, Watts FZ, Lehmann AR. Cloning and characterization of the rad4 gene of Schizosaccharomyces pombe; a gene showing short regions of sequence similarity to the human XRCC1 gene. Nucleic Acids Res. 1991;19:6737–6741. doi: 10.1093/nar/19.24.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuura M, Nagao K, Obuse C, Takahashi TS, Nakagawa T, Masukata H. CDK promotes interactions of Sld3 and Drc1 with Cut5 for initiation of DNA replication in fission yeast. Molecular biology of the cell. 2011;22:2620–2633. doi: 10.1091/mbc.E10-12-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes & development. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA repair. 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Molecular cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny ML, Dore AS, Roe SM, Nestoras K, Ho JC, Watts FZ, Pearl LH. Structural and functional analysis of the Crb2-BRCT2 domain reveals distinct roles in checkpoint signaling and DNA damage repair. Genes & development. 2008;22:2034–2047. doi: 10.1101/gad.472808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Dunphy WG. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J Cell Biol. 2011;193:995–1007. doi: 10.1083/jcb.201102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Lee MS, Edwards RA, Thede GL, Glover JN. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the gamma-H2AX histone tail. J Biol Chem. 2005;280:32053–32056. doi: 10.1074/jbc.C500273200. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Wardlaw CP, Morishita T, Miyabe I, Chahwan C, Caspari T, Schmidt U, Carr AM, Garcia V. The Rad4(TopBP1) ATR-activation domain functions in G1/S phase in a chromatin-dependent manner. PLoS Genet. 2012;8:e1002801. doi: 10.1371/journal.pgen.1002801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu M, Yang B, Tao L, Yates JR, 3rd, Russell P, Dong MQ, Du LL. Phosphorylation-dependent interactions between Crb2 and Chk1 are essential for DNA damage checkpoint. PLoS Genet. 2012;8:e1002817. doi: 10.1371/journal.pgen.1002817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappas M, Oliver AW, Pearl LH. Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res. 2011;39:313–324. doi: 10.1093/nar/gkq743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes & development. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Molecular cell. 2004;14:405–412. doi: 10.1016/s1097-2765(04)00238-2. [DOI] [PubMed] [Google Scholar]
- Smits VA, Warmerdam DO, Martin Y, Freire R. Mechanisms of ATR-mediated checkpoint signalling. Front Biosci. 2010;15:840–853. doi: 10.2741/3649. [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Takeishi Y, Ohashi E, Ogawa K, Masai H, Obuse C, Tsurimoto T. Casein kinase 2-dependent phosphorylation of human Rad9 mediates the interaction between human Rad9-Hus1-Rad1 complex and TopBP1. Genes Cells. 2010;15:761–771. doi: 10.1111/j.1365-2443.2010.01418.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Russell P. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nature cell biology. 2001;3:966–972. doi: 10.1038/ncb1101-966. [DOI] [PubMed] [Google Scholar]
- Taylor M, Moore K, Murray J, Aves SJ, Price C. Mcm10 interacts with Rad4/Cut5(TopBP1) and its association with origins of DNA replication is dependent on Rad4/Cut5(TopBP1) DNA repair. 2011;10:1154–1163. doi: 10.1016/j.dnarep.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- Willson J, Wilson S, Warr N, Watts FZ. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Res. 1997;25:2138–2146. doi: 10.1093/nar/25.11.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.