Abstract
An expanded GGGGCC repeat in C9orf72 is the most common genetic cause of frontotemporal dementia and amyotrophic lateral sclerosis. A fundamental question is whether toxicity is driven by the repeat RNA itself and/or by dipeptide repeat proteins generated by repeat-associated, non-ATG translation. To address this question we developed in vitro and in vivo models to dissect repeat RNA and dipeptide repeat protein toxicity. Expression of pure repeats in Drosophila caused adult-onset neurodegeneration attributable to poly-(glycine-arginine) proteins. Thus expanded repeats promoted neurodegeneration through neurotoxic proteins. Expression of individual dipeptide repeat proteins with a non-GGGGCC RNA sequence showed both poly-(glycine-arginine) and poly-(proline-arginine) proteins caused neurodegeneration. These findings are consistent with a dual toxicity mechanism, whereby both arginine-rich proteins and repeat RNA contribute to C9orf72-mediated neurodegeneration.
Frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) are adult-onset, neurodegenerative diseases associated with personality change, language dysfunction and progressive muscle weakness. These syndromes overlap genetically and pathologically, and can also co-occur in individuals, and within families (1). An intronic GGGGCC hexanucleotide repeat expansion in C9orf72 is the most common genetic cause of both FTD and ALS (C9FTD/ALS) (2–4), and can be found in patients diagnosed with all common neurodegenerative diseases (5). Healthy individuals carry fewer than 33 hexanucleotide repeats, with 2 repeats being the most common, but C9FTD/ALS cases carry between 400 and 4400 repeats (2, 5, 6).
The repeat expansion could cause disease by three possible mechanisms: i) toxic sense and/or antisense repeat RNA species that sequester key RNA-binding proteins, ii) toxic dipeptide repeat (DPR) proteins, generated by repeat-associated, non-ATG (RAN) translation, or iii) reduced expression of C9orf72. The absence of a severe phenotype in a homozygous C9orf72 mutation case (7), and the lack of C9orf72 coding mutations (8) argue against loss-of-function as a primary mechanism. Neuronal aggregates of RNA, termed RNA foci, generated from both sense and antisense repeat transcripts are frequent in C9FTD/ALS patient brain (9–13). The GGGGCC repeat can be translated in all sense and antisense frames, two of which encode the same DPR, resulting in five DPR proteins, all of which form inclusions in widespread brain regions (10, 12, 14–18). It is therefore of fundamental importance to understand the contributions of repeat RNA and DPR proteins to C9orf72-mediated neurodegeneration.
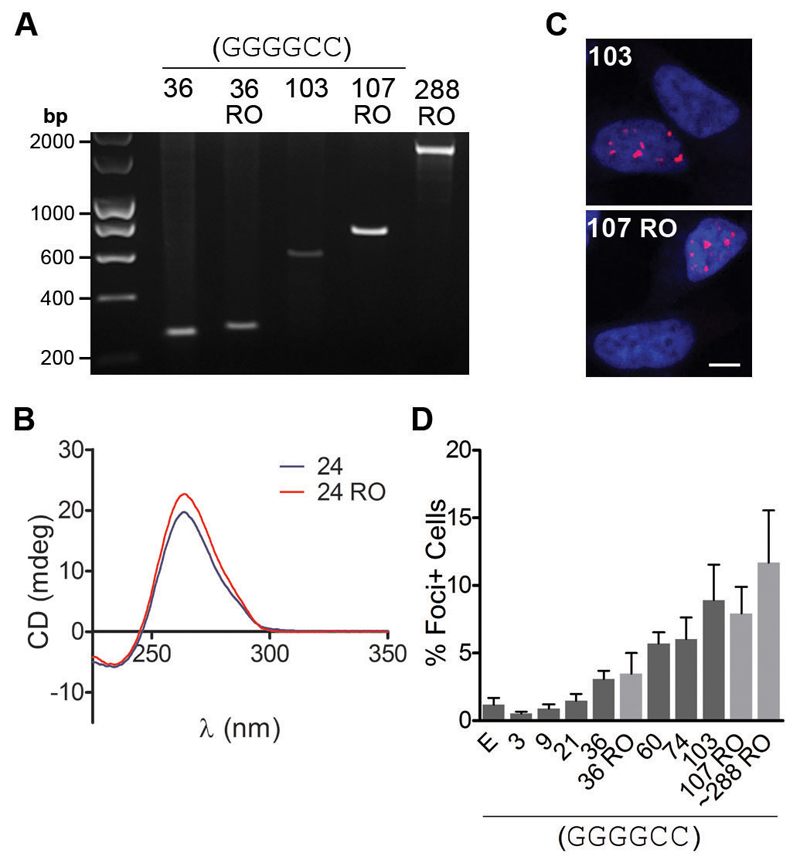
A major obstacle in the investigation of large expanded repeats is that they are inherently unstable. We used recombination-deficient E. coli and a cloning strategy termed recursive directional ligation (19) to sequentially build seamless pure repeats from small GGGGCC repeat units (fig. S1). This allowed generation of a stable range of pure repeats from 3 to a maximum of 103 (Fig. 1A). To dissect repeat RNA and DPR protein toxicity, we generated “RNA-only” repeats, using our cloning strategy to insert interruptions containing stop codons in all sense and antisense frames. In models of other non-coding repeat expansion disorders, interruptions comprising 4 to 11 percent of the total repeat sequence confer stability while maintaining pathogenicity in vitro and in vivo (20–22). One of three 6-base-pair interruptions, each containing one stop codon in the sense and one in the antisense direction, were inserted every 12 GGGGCC repeats, resulting in a stop codon for all 6 (sense and antisense) frames, and interruptions that comprised 8% of the total sequence (fig. S2). We generated stop codon-interrupted RNA-only repeats equivalent in length to our pure repeats and longer RNA-only repeats up to ~288 (Fig. 1A). GGGGCC repeat RNA forms a stable tertiary structure termed a G-quadruplex (23). Circular dichroism showed that the RNA-only repeats formed RNA G-quadruplexes similarly to pure repeat RNA (Fig. 1B), showing that the interruptions did not affect the tertiary structure of the RNA. To investigate the formation of RNA foci, constructs were expressed in human neuroblastoma cells. RNA fluorescence in situ hybridisation (FISH) showed that formation of RNA foci was length-dependent for both pure and RNA-only repeats which, at equivalent length, had the same propensity to form foci (Fig 1C, D).
Fig. 1. Generation and characterisation of expanded pure and RNA-only GGGGCC repeats.
(A) Agarose gel showing pure GGGGCC repeats and stop codon-interrupted RNA-only (RO) repeats. (B) Circular dichroism (CD) spectra of both 24 pure and 24 RO repeats showed characteristic RNA G-quadruplex structure with minima and maxima at 237 and 262 nm respectively (23). (C) Confocal microscope images of nuclei (blue) in RNA FISH-labelled SH-SY5Y cells showed that 103 pure and 107 RO repeats both produced nuclear RNA foci (red). Scale bar represents 5 µm. (D) Quantification of the number of SH-SY5Y cells containing RNA foci after transfection with pure and RO repeats of different lengths, and empty vector (E). No difference was observed between equivalent length pure and RNA-only repeats (36 vs. 36 RO, 103 vs. 107 RO, 1 way ANOVA with Bonferroni test (selected pairs), n > 3, error bars represent SEM).
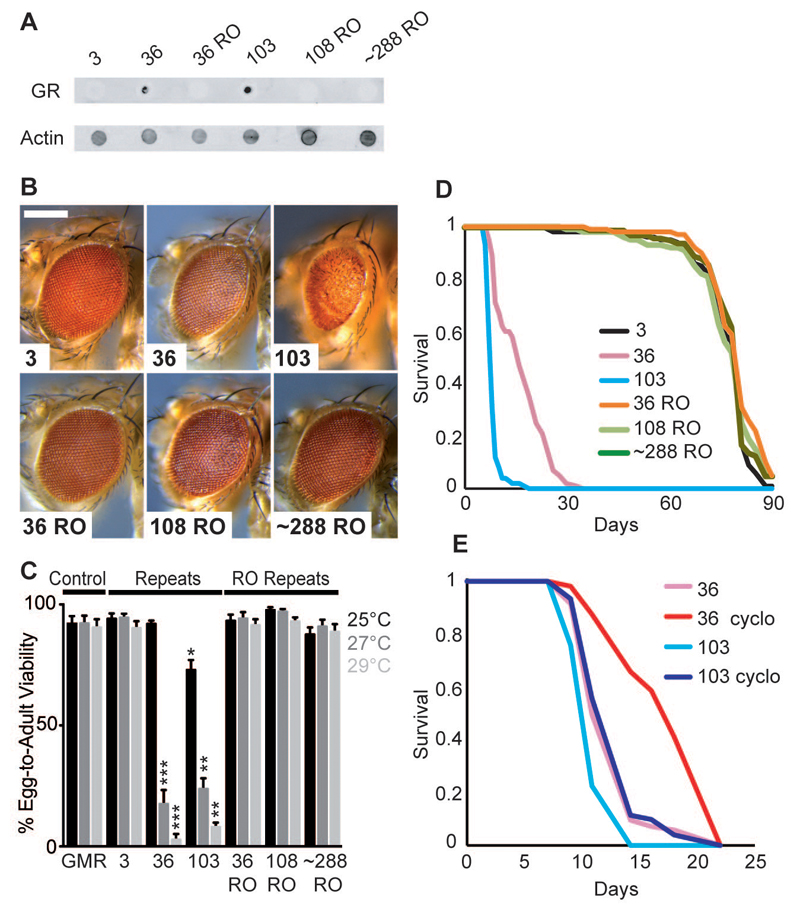
In order to differentiate between repeat RNA and DPR protein toxicity in vivo, we generated lines of the fruit fly Drosophila melanogaster carrying a range of our pure and RNA-only repeats under the UAS promoter, integrated into the same genomic location to ensure equivalent expression levels. When expressed specifically in the adult fly, the different repeats expressed sense transcripts at comparable levels and of the expected sizes, but no antisense transcripts (fig. S3A). RNA FISH showed that pure and RNA-only repeats were both able to generate RNA foci in Drosophila (fig. S4). Immunoblotting using an anti-poly-(GR) antibody (Fig. 2A) or an anti-poly-(GP) antibody (fig. S5B) showed that, as expected, the pure repeats generated DPR proteins and the RNA-only repeats did not. Expression of both 36 and 103 pure repeats in the eye caused eye degeneration, whereas 36, 108 and ~288 RNA-only repeats had no effect under the same conditions (Fig. 2B). The toxicity of the pure repeats was thus attributable to the presence of DPR proteins. Increasing expression levels of the pure repeats, by increasing the temperature (24), led to lethality from both 36 and 103 repeats (Fig. 2C, fig S6) but for the RNA-only repeats had no effect, again demonstrating that the pure repeats caused lethality through the production of DPR proteins.
Fig. 2. Pure GGGGCC repeats caused toxicity via DPR proteins.
(A) Dot blot showing that 36 and 103 pure repeats generated poly-(GR) proteins while 3 pure repeats, and 36, 108 and ~288 RNA-only (RO) repeats did not. Genotypes were: w; UAS-3/hsGal4, w; UAS-36/hsGal4, w; UAS-103/ hsGal4, w; UAS-36 RO/hsGal4, w; UAS-108 RO/ hsGal4, w; UAS-288 RO/hsGal4. (B) Stereomicroscopy images of representative Drosophila eyes expressing pure or RO repeats using the GMR-GAL4 driver. 36 pure repeats were mildly toxic. 103 pure repeats showed more overt toxicity. 3 repeats and 36 and 108 RO repeats had no effect. Genotypes were: w; GMR-Gal4/+, w; GMR-Gal4/UAS-3, w; GMR-Gal4/UAS-36, w; GMR-Gal4/UAS-103, w; GMR-Gal4/UAS-36RO, w; GMR-Gal4/UAS-108RO, w; GMR-Gal4/UAS-288RO. Scale bar represents 200 µm. (C) Quantification of egg-to-adult viability showed that 36 and 103 pure repeats were lethal at higher temperatures, whereas RO repeats had no effect (Kruskal Wallis test with Dunn's multiple comparison (selected pairs), ***p<0.001, **p<0.01, *p<0.05, error bars represent SEM). Genotypes were as in (B). (D) Survival of female flies expressing repeats in adult neurons using the elav-GeneSwitch (elavGS) driver. 36 and 103 pure repeats substantially decreased survival, while 36, 108 and 288 RO repeats had no effect (p<0.0001, log-rank test). Genotypes were: w; UAS-3/+; elavGS/+, w; UAS-36/+; elavGS/+, w; UAS-103/+; elavGS/+, w; UAS-36 RO/+; elavGS/+, w; UAS-108 RO/+; elavGS/+, w; UAS-288 RO/+; elavGS/+. (E) Flies expressing 36 and 103 pure repeats survived longer in the presence of cycloheximide than in its absence (p<0.001, log-rank test). Genotypes were: w; UAS-36/+; elavGS/+, w; UAS-103/+; elavGS/+.
The GMR-Gal4 driver is expressed throughout development. However, ALS and FTD are adult-onset diseases. To circumvent developmental effects, we confined expression of the repeat constructs to adult neurons, using the inducible elav-GeneSwitch driver. Expression of 36 and 103 repeats killed all flies by 30 days post-eclosion. 36, 108 and ~288 RNA-only repeats had no effect, showing that the neurotoxicity of the pure repeats was attributable to DPR protein production (Fig. 2D). To confirm this, we reduced protein synthesis in the 36 and 103 pure repeat-expressing flies with a sub-lethal dose of cycloheximide, which ameliorated the reduction in lifespan caused by the pure repeats (Fig. 2E), again showing that toxicity was attributable to DPR proteins.
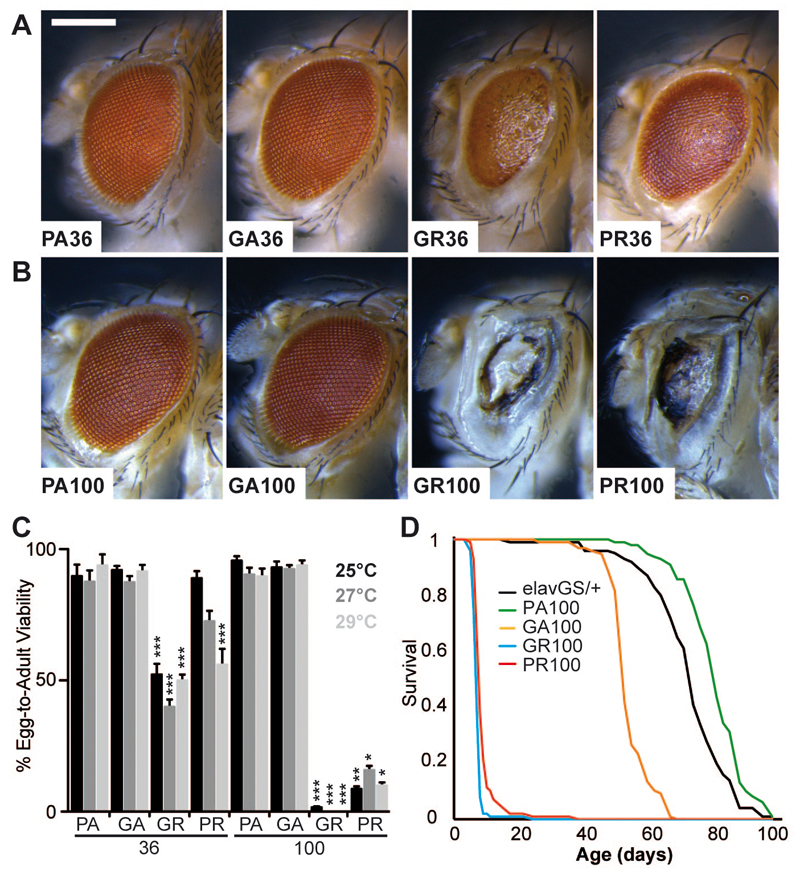
To assess whether DPR protein expression alone was sufficient for toxicity, “protein-only” constructs were generated, by using alternative codons to those found within the GGGGCC repeat. We compared the two arginine-containing DPR proteins, glycine-arginine (GR) and proline-arginine (PR), with two neutral DPR proteins, proline-alanine (PA) and glycine-alanine (GA). When constructs containing 36 dipeptide repeats (equivalent to 36 pure GGGGCC repeats) were expressed in the fly eye, the arginine-containing DPR proteins GR and PR caused eye degeneration and lethality, while GA and PA DPR proteins had no effect (Fig. 3A, C). Thus, the arginine-containing DPR proteins induced toxicity. We next generated longer protein-only sequences, of equivalent length to 103 pure repeats. Expression of (PR)100 or (GR)100 caused eye degeneration and increased lethality, while (PA)100 and (GA)100 had no effect (Fig. 3B, C). Expression of (PR)100 and (GR)100 in adult neurons caused a substantial decrease in survival (Fig. 3D); a late-onset reduction in survival was also observed in (GA)100-expressing flies, while (PA)100 had no effect. Expression levels varied among the individual protein-only constructs, but did not correlate with toxicity (fig. S3C), which was therefore attributable to the arginine-rich sequences. Thus, the highly basic arginine-containing DPR proteins drove C9orf72 GGGGCC repeat toxicity in Drosophila neurons.
Fig. 3. DPR toxicity was caused by poly-GR and poly-PR proteins.
“Protein-only” constructs for individual DPR proteins were expressed in the Drosophila eye (A-C), and the adult nervous system (D). (A) (GR)36 and (PR)36 caused eye degeneration, while (GA)36 and (PA)36 had no effect. Genotypes were: w; UAS-PA36/GMR-Gal4, w; UAS-GA36/GMR-Gal4, w; UAS-GR36/GMR-Gal4, w; UAS-PR36/GMR-Gal4. Scale bar represents 200 µm. (B) (GR)100 and (PR)100 caused extensive eye degeneration, while (GA)100 and (PA)100 had no effect. Genotypes were: w; UAS-PA100/GMR-Gal4, w; UAS-GA100/GMR-Gal4, w; UAS-GR100/GMR-Gal4, w; UAS-PR100/GMR-Gal4. (C) Quantification of egg-to-adult viability showed (GR)100 and (PR)100 caused a substantial reduction in survival, whereas (GA)100 and (PA)100 had no effect (Kruskal Wallis test with Dunn's multiple comparison, selected pairs, ***p<0.001, **p<0.01, error bars represent SEM). Genotypes were as in (A) and (B). (D) Expression of (GR)100 and (PR)100 in adult neurons using the elav-GeneSwitch (elavGS) driver caused a substantial decrease in viability (p<0.001, log-rank test); (GA)100 caused a late-onset decrease in survival, and (PA)100 or elavGS driver alone had no effect. Genotypes were: w; elavGS/+, w; UAS-PA100/+; elavGS/+, w; UAS-GA100/+; elavGS/+, w; UAS-GR100/+; elavGS/+, w; UAS-PR100/+; elavGS/+.
Our data identified GR and PR DPR proteins as the predominant toxic protein species, although all five DPR proteins form inclusions in affected brain regions. Similarly, the distribution of poly-(GA) inclusions does not correlate well with neurodegeneration (25). The presence of arginine in both of the highly toxic DPR species suggests a common pathological mechanism, perhaps attributable to their basic nature or a common structural motif. Restricted expression of C9orf72 to specific neuronal populations (26), or a deficit in the affected neurons’ ability to clear these particular proteins, may explain why these highly toxic proteins cause selective neurodegeneration. In patients all five DPR proteins may be produced in a single neuron. While our findings indicate that toxicity is driven by the arginine-rich DPR proteins it remains possible that high focal levels of the other DPR proteins could contribute to cytotoxicity.
We have been able to separate RNA and DPR toxicity associated with C9orf72 GGGGCC repeats and, surprisingly, our data suggests that the major toxic species are the DPR proteins. However, the DPR protein toxicity that we observed from over-expression of pure repeats does not rule out an additional contribution of RNA toxicity. Several lines of evidence suggest a toxic role of repeat RNA. In C9FTD patient brain, RNA foci are most abundant in the frontal cortex, which has the greatest degree of neuronal loss, and frontal cortex RNA foci burden correlates with age at onset in C9FTD cases (9). GGGGCC repeats also sequester several RNA-binding proteins, which could lead to toxicity (13, 27–31). However, modelling RNA toxicity may require longer repeats that are closer to the pathological range seen in disease, possibly because a toxic threshold of repeat number must be crossed. A continuing conundrum is why the same expanded repeat can cause either pure FTD or pure ALS. Our data raise the possibility that the different patient phenotypes could be caused by differences in the relative contributions of RNA or protein-mediated toxicity within distinct neuronal subtypes. A further prediction from this hypothesis is that genetic variants that affect RAN translation or DPR protein levels may also contribute to disease penetrance.
Supplementary Materials
Materials and Methods
Figs. S1-S7
References (32-35)
Acknowledgments
Funding was provided by Alzheimer’s Research UK (AMI), The Motor Neurone Disease Association (AMI, EF, PF), the MHMS General Charitable Trust (AMI), NIHR-UCLH-BRC (PF), NIHR ACF (IW), the UK Medical Research Council (AMI, EF, PF), the Wellcome Trust (LP), and the Max Plank Society (LP). We thank Dr J Wadsworth and Dr N Alic for helpful discussion. LP dedicates this work to the memory of Noreen Murray.
References and Notes
- 1.Lillo P, Hodges JR. Frontotemporal dementia and motor neurone disease: overlapping clinic-pathological disorders. J Clin Neurosci. 2009;16:1131–1135. doi: 10.1016/j.jocn.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Dejesus-Hernandez M, et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renton AE, et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majounie E, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck J, et al. Large C9orf72 Hexanucleotide Repeat Expansions Are Seen in Multiple Neurodegenerative Syndromes and Are More Frequent Than Expected in the UK Population. Am J Hum Genet. 2013;92:345–353. doi: 10.1016/j.ajhg.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Blitterswijk M, et al. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol. 2013;12:978–988. doi: 10.1016/S1474-4422(13)70210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fratta P, et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 2013;126:401–409. doi: 10.1007/s00401-013-1147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harms MB, et al. Lack of C9ORF72 coding mutations supports a gain of function for repeat expansions in amyotrophic lateral sclerosis. Neurobiol Aging. 2013;34:2234–2239. doi: 10.1016/j.neurobiolaging.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizielinska S, et al. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 2013;126:845–857. doi: 10.1007/s00401-013-1200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gendron TF, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagier-Tourenne C, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zu T, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YB, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ash PE, et al. Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lashley T, Hardy J, Isaacs AM. RANTing about C9orf72. Neuron. 2013;77:597–598. doi: 10.1016/j.neuron.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Mori K, et al. The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 17.Mori K, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 18.Zu T, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer DE, Chilkoti A. Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: examples from the elastin-like polypeptide system. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- 20.de Haro M, et al. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum Mol Genet. 2006;15:2138–2145. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- 21.Orengo JP, et al. Expanded CTG repeats within the DMPK 3' UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc Natl Acad Sci U S A. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto N, et al. GGA*TCC-interrupted triplets in long GAA*TTC repeats inhibit the formation of triplex and sticky DNA structures, alleviate transcription inhibition, and reduce genetic instabilities. J Biol Chem. 2001;276:27178–27187. doi: 10.1074/jbc.M101852200. [DOI] [PubMed] [Google Scholar]
- 23.Fratta P, et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 25.Mackenzie IR, et al. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 2013;126:859–879. doi: 10.1007/s00401-013-1181-y. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki N, et al. The mouse C9ORF72 ortholog is enriched in neurons known to degenerate in ALS and FTD. Nat Neurosci. 2013;16:1725–1727. doi: 10.1038/nn.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almeida S, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donnelly CJ, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori K, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125:413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- 30.Sareen D, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.