Abstract
Plant-associated fungi are considered a vast source for biotechnological processes whose potential has been poorly explored. The interactions and diversity of sugarcane, one of the most important crops in Brazil, have been rarely studied, mainly concerning fungal communities and their interactions with transgenic plants. Taking this into consideration, the purpose of this study was, based on culture dependent strategy, to determine the structure and diversity of the fungal community (root endophytes and rhizosphere) associated with two varieties of sugarcane, a non-genetically modified (SP80-1842) variety and its genetically modified counterpart (IMI-1, expressing imazapyr herbicide resistance). For this, the sugarcane varieties were evaluated in three sampling times (3, 10 and 17 months after planting) under two crop management (weeding and herbicide treatments). In addition, a strain of Trichoderma virens, an endophyte isolated from sugarcane with great potential as a biological control, growth promotion and enzyme production agent, was selected for the fungal-plant interaction assays. The results of the isolation, characterization and evaluation of fungal community changes showed that the sugarcane fungal community is composed of at least 35 different genera, mostly in the phylum Ascomycota. Many genera are observed at very low frequencies among a few most abundant genera, some of which were isolated from specific plant sites (e.g., the roots or the rhizosphere). An assessment of the possible effects upon the fungal community showed that the plant growth stage was the only factor that significantly affected the community’s structure. Moreover, if transgenic effects are present, they may be minor compared to other natural sources of variation. The results of interaction studies using the Green fluorescent protein (GFP)-expressing T. virens strain T.v.223 revealed that this fungus did not promote any phenotypic changes in the host plant and was found mostly in the roots where it formed a dense mycelial cover and was able to penetrate the intercellular spaces of the root epidermis upper layers. The ability of T. virens to colonize plant roots suggests a potential for protecting plant health, inhibiting pathogens or inducing systemic resistance.
Introduction
Sugarcane (Saccharum sp.) is an important crop in Brazil, mainly for the production of ethanol, a biofuel, as a renewable energy source that is already an alternative to petrol in Brazilian cars. To increase productivity and decrease costs, researchers are investing in new sugarcane varieties developed by classical breeding as well as genetically modified plants, searching for characteristics such as insect resistance, herbicide resistance, a higher sugar content, etc [1, 2]. Therefore, studies on biosecurity and environmental impact should be performed before releasing the genetically modified plants for commercial production
The microbial community of the host plant can have a positive, negative or neutral effect on the plant, caused mainly by those microorganisms that live in intimate contact with the plant tissues, such as endophytes or rhizoplane microbes [3]. Studies reported the effects of the genetically modified plant on the microbial community associated [4–8], but these effects were minor in comparison with the effect caused by cultivar [5,9], soil type [10,11], crop location [12,13] and plant growth stage [5, 14, 15, 16, 17].
Microbial diversity studies can help elucidate the role of native microorganisms in the ecosystem and the association of biological changes with microbial diversity [18,19]. A few studies have been performed with transgenic sugarcane, mainly involving beneficial fungi and plant interactions [20,21]. Fungi play a key role in several ecological processes essential for ecosystem maintenance [22]. Endophytic fungi colonize plant tissue internally without causing damage to the plant host [23], establishing a balanced and mutualistic interaction [24]. The fungus receives nutrients while the plant benefit from the inhibition of pathogens and stress resistance as well as increased growth [25].
On the other hand, the rhizosphere is influenced by root exudates and is intensely colonized by microbial communities [26], which have direct effects on plant growth and nutrient availability or protecting against pathogens [27, 28]. Several rhizosphere fungi degrade toxic compounds, such as xenobiotic and aromatic molecules, and are essential for the survival of plants in contaminated soils [29]. Other fungi, such as Fusarium oxysporum, a nonpathogenic strain, can suppress pathogenic strains [30]. Many fungi, such as members of the Trichoderma genus, can inhibit phytopathogens and act as a biological control [31]. Among these, Trichoderma virens has been reported to be aggressive mycoparasite [32, 33], able to parasitize not only hyphae but also fungal resistance structures [32, 33, 34]. In addition to its mycoparasitic activity, it can also produce extracellular chitinase [35] and several antibiotics and can induce the production of plant fitoalexins [36]. T. virens has been considered a versatile and effective biological control agent. Moreover, T. virens has been reported to be a plant endophyte [37], able to asymptomatically colonize the host plant and occupy the same niche as phytopathogens. In addition, Trichoderma spp. strains have been used to promote plant growth [31,38,39] and show a great potential for agricultural use.
In the present study we considered the hypothesis that if the genetically modified plant could have an impact on the microbial communities associated, the accumulation of these effects over the time could be detected after a long period of the plant in the field. Therefore, we investigated the effect of genetically modified sugarcane, sampling time (plant growth stage) and crop management (weeding and herbicide application) on the structure and diversity of the cultivable fungus communities associated with sugarcane plants. Rhizosphere and root endophytic fungi were isolated from two sugarcane varieties, a non-transgenic (SP80-1842) and a genetically modified (IMI-1, expressing imazapyr herbicide resistance) variety, treated and untreated with the herbicide imazapyr, at three different growth stages over a two-year period. Trichoderma, a genus abundant in sugarcane, which has potential agricultural use, was selected for these plant interaction studies.
Material and Methods
The sugarcane orchard was not in a protected area. Sugarcane plants (Saccharum officinarum) were grown in the experimental area of the Centro de Tecnologia Canavieira S.A. (CTC), a private company that present the CQB (Certificate for Biosecurity Quality) for the cultivation area, in Piracicaba, São Paulo, Brazil (22° 41’ S e 47° 33’ O), under supervision of Dr. Eugenio Cesar Ulian a Director of the CTC company. Therefore, the only one permission that we need for this experiment permission was gave by the experimental Company Supervisor, Dr. Eugenio Cesar Ulian."
Sugarcane experiments
Sugarcane plants (Saccharum officinarum) were grown in the experimental area of the Centro de Tecnologia Canavieira S.A. (CTC), Piracicaba, São Paulo, Brazil (22° 41’ S e 47° 33’ O) under supervision of Dr. Eugenio Cesar Ulian. We used the transgenic sugarcane IMI-1, which expresses resistance to the herbicide imazapyr, and the non-transgenic isoline cultivar SP80-1842. Three treatments were used in a randomized experimental design with four replicates: i) the non-transgenic sugarcane SP80-1842 + weeding (NW), ii) the transgenic sugarcane IMI-1 + weeding (TW) and iii) the transgenic sugarcane IMI-1 with imazapyr herbicide applied two months after planting (TH). In this experimental design the TW x TH comparison allowed evaluate the crop management (weeding x herbicide application) and NW x TW comparison allowed evaluate the plant genotype (non-transgenic X genetically modified variety). Samples were collected at 3, 10 and 17 months after planting (sampling times), and each sample was comprised by four replicates (with 3 plants each).
Isolation of sugarcane endophytic and rhizosphere fungi
Endophytic and rhizosphere fungi were isolated from sugarcane according to Stuart et al. [20]. For endophytic fungi isolation, roots were surface sterilized and for each three treatments (NW, TW and TH), the roots of three plants (from each replicate) were cut in 21 fragments (5 mm x 5mm each) and transferred onto Potato Dextrose Agar (PDA, Merck, Kenilworth, NJ, USA) with tetracycline (50 μg mL-1) to inhibit bacterial growth. For rhizosphere fungi isolation, 5 g of rhizosphere soil was diluted in 50 mL of PBS buffer (NaCl 140 mM, KCl 3 mM, Na2HPO4 10 mM and KH2PO4 2 mM, pH = 7.4) and shaken with glass beads for 1 h at 150 rpm. The suspension was diluted and the 10−3 and 10−1 dilutions were plated on PDA with tetracycline as used in previous study (Stuart et al., 2010). The plates were incubated at 28°C for 2 to 14 days. After fungal growth, the colonies were counted and grouped by morphological characteristics. Filamentous fungal isolates were purified by serial dilution and stored. The frequency of the colony grown per fragment and per soil gram was calculated and converted to the number of colony forming units per gram (CFU g-1).
DNA extraction and molecular identification of the endophytic and rhizosphere fungi
DNA extraction of the isolated fungi was performed according to Raeder & Broda [40] with modifications. The fungi were grown in 50 mL of PDB (Potato Dextrose Broth) for 5 days at 28°C. The fungal suspension was filtered, and the mycelia were ground in liquid nitrogen. The grounded mycelia (200 mg) were transferred to 1 mL of extraction buffer (SDS 1%, EDTA 25 mM, NaCl 250 mM and Tris-HCl 200 mM [pH 8,0]), incubated at 65°C for 20 min and centrifuged at 16,000 X g for 10 min at 4°C. The DNA was purified using phenol/chloroform and precipitated with isopropanol. After the DNA was extracted, the DNA was quantified and its integrity was assessed by electrophoresis in a 0.8% of agarose gel; the DNA was stored at -20°C.
The ITS1-5.8S-ITS2 region of rDNA was amplified using ITS-1 and ITS-4 primers [41]. After electrophoresis, the fragments were purified and sequenced at the Centro de Estudos do Genoma Humano (University of São Paulo, São Paulo, Brazil). To identify the fungal isolates, the sequences were compared to the GenBank database via BLASTn (http://www.ncbi.nlm.nih.gov/BLAST). Partial DNA sequences of the ITS1-5.8S-ITS2 region were deposited in the GenBank database under the accession numbers GQ495269 and GU973612-GU973859.
Sequence analysis and phylogeny
All the chromatograms were first trimmed for high quality bases (80% of bases with quality > 20) by means of Phred software, and the trimmed sequences were used for comparison in the GenBank database (nr/nt). The best hits of well-characterized strains were retrieved from the databases and subsequently used for alignment and phylogeny analysis with MEGA 4.0 version software [42]. The evolutionary history was inferred through the Neighbor-Joining method [43] and evolutionary distances were computed using the Jukes-Cantor method.
Determination of endophytic and rhizosphere fungal community structures
The diversity and richness of the fungal community in the three treatments (NW, TW and TH) for three isolation periods (3, 10 and 17 months) were quantified using the Shannon-Wienner (H’) and Chao1 index, respectively. These indices were calculated using the DOTUR program [44], which also generated rarefaction data used to infer coverage considering the different similarity levels of the ITS1-5.8S-ITS2 sequence of fungus rDNA. The similarity matrix was generated using the DOTUR program, aligning the sequence and calculating the distance matrix using the Phylip DNADist program (http://bioweb.pasteur.fr/seqanal/interfaces/dnadist-simple.html). The taxonomic levels were calculated with the following cutoffs: 100% (strains), 99% (species) and 97%-95% (genera). The similarity coefficient of Jaccard (J) was calculated, considering the generic level.
AMOVA was performed to estimate the genetic variation among the different fungal communities and to determinate how much of the genetic variation could be attributed to the differences among the sugarcane genotypes (conventional vs. transgenic), crop management (weeding vs. imazapyr), the sampling period (growth stages) and the rhizosphere and endophytic fungal community. All analyses of the DNA sequences from the fungal isolates were performed using the Arlequin program version 3.1 [45], considering 16.000 permutations.
A Principal Component Analysis (PCA) was used to verify whether a correlation existed between the fungal communities found in the rhizosphere and those found in the endosphere. The PCA analysis was performed using Canoco version 4.5 [46], using the frequency of each genus in each repetition of endophytic and rhizosphere samples as the input variable.
Agrobacterium-mediated transformation
The isolate T. virens strain T.v.223 (GenBank access number GQ495269) obtained from inner root tissues was selected to further studies. For this, transformation was carried out according to a previously described method [47] with modifications [21, 48]. Briefly, A. tumefaciens EHA105 transformed with the plasmid pFAT-gfp [49] was grown at 28°C for 16–20 h. The culture suspension was diluted to a λ660 = 0.15 in an induction medium broth (10 mM K2HPO4; 10 mM KH2PO4; 2.5 mM NaCl; 2 mM MgSO4; 0.7 mM CaCl2; 9 mM FeSO4; 4 mM NH4SO4; 10 mM glucose; 0.5% glycerol; 40 mM 2-morpholinoethanesulfonic acid, pH 5.3) supplemented with acetosyringone (200 mM) and incubated until a λ660 = 0.6 was reached. The cell suspension was mixed 1:1 with a conidial suspension (106 or 107 conidia. mL-1) from T. virens strain T.v.223. Aliquots were spread on different types of membranes (cellophane, filter paper, nitrocellulose, or nylon) on induction medium agar (5 mM glucose instead of 10 mM) supplemented with 200 mM or 400 mM of acetosyringone. After co-cultivation at 25°C for 24 h or 48 h, the membranes were transferred onto PDA or M-100 [50] media amended with hygromycin B (200 μg mL–1; to select transformants) and cefotaxime (200 μg mL–1; to eliminate the bacteria).
Individual transformants were subsequently transferred to PDA supplemented with 200 μg mL-1 hygromycin. The mitotic stability of the integrated T-DNA was tested by sub culturing five-fold randomly selected transformants on PDA supplemented with 200 μg mL-1 of hygromycin incubated at 28°C for 5 days.
Molecular and morphological characterization of transformants
The genomic DNA of several selected transformants was extracted according to Raeder and Broda [40], and the presence of the gfp gene that express the green fluorescent protein was verified by PCR. We used the primers glGFP5 (5’-GCCGGAATTCATGAGCAAGGGCGAGGAACTGTTC-3’) and glGFP3 (5’-GCCGAGCTCAGATCTCACTTGTACAGCTCGTCCATGCC-3’) that amplified 700 pb of gfp gene [49].
To check the number of inserts in the genome, four transformants (T2, T7, T10 and T20) and the wild type strain (used as a negative control) were analyzed by Southern blotting. The DNA (15 μg) of each strain was cleaved with 10 U μL-1 EcoRI (Invitrogen, Life Technologies) incubated at 37°C for 12 h [49]. The digested DNA was electrophoresed in a 1% agarose gel. The probe used in the Southern blot analysis, the 700 pb sequence of gfp gene, was labeled using a Gene Images™ AlkPhos Direct™ Labelling and Detection System (GE Healthcare) kit according to the manufacturer’s protocol.
The gel was washed with a 0.25 M HCl solution for 10 min, followed by distilled water, a denaturing solution (NaOH 0.5 M, NaCl 1.5 M) for 30 minutes, distilled water, and two washes with a neutralization solution (Tris-HCl 0.5 M; NaCl 1.5 M; EDTA 0.001 M [pH = 7.2]). The DNA in the gel was transferred to a nylon membrane (Hybond N+, Amersham) for 12 h in transference solution SSC 20X (NaCl 3 M; sodium citrate 0.3 M [pH = 7.0]). The membrane was dried for 2 h at 80°C and stored at 4°C.
Morphological characterization was performed to compare the wild type and mutant strains on the basis of growth rate and morphology (sporulation rate and color). The strains were grown for 5 days in PDA at 28°C, and the colony diameter was measured after 24, 48, 72 and 96 h. All tests were performed in triplicate.
Trichoderma virens–host plant interaction
Sugarcane plant reisolation of Trichoderma virens mutant T20
The mutant T20, which expressed the gfp gene and hygromycin resistance, was grown in PDA (Merck) at 28°C for 7 days to obtain conidia. In vitro sugarcane plants (variety SP80-1842) from Centro de Tecnologia Canavieira (CTC, Piracicaba, SP, Brazil), were transferred to glass flasks containing 20 mL of MS liquid broth [51] with 106 conidia mL-1 of mutant T20. The plants were incubated at 28°C under a 16 h light and 8 h dark cycle. After 48 h, the plants were transferred to pots containing a commercial substrate PlantMax (Eucatex) and placed in a greenhouse to acclimatize. Non-inoculated sugarcane plants (variety SP80-1842) were similarly treated as a control. After 20, 40 and 60 days in the greenhouse, the sugarcane plants were sampled and leaves, stem and roots were surface-disinfected according to Stuart et al. [20]. After processing, the leaf and stem tissues were cut in fragments of approximately 100 mm2 (10 mm x 10 mm) and roots were cut in 5 mm fragments. A total of 1512 plant fragments (504 fragment per sampling time) were transferred to PDA (Merck) with tetracycline (100 μg mL-1) to inhibit bacterial growth. Hygromycin B (200 μg mL-1) was also added to the culture medium for the T20 mutant. The isolation plates were incubated at 28°C and periodically evaluated up to 14 days. After growth, the total fungal colonies and Trichoderma colonies were counted, and T. virens strain T20 were confirmed by gfp fluorescence and hygromycin B resistance.
Microscopic analysis of Trichoderma virens and sugarcane interaction
Axenic sugarcane plants (variety SP80-1842) were inoculated with T. virens strain T20 conidia (with the gfp gene) and incubated at 28°C under a 16 h light and 8 h dark cycle. After 24, 48, 72 and 168 h, the roots were processed according to Ferreira et al. [52] and observed under an optical microscope (Zeiss Axiophot-2) using two filters, FITC (459–490 nm) and Rhodamine (546 nm), to differentiate sugarcane fluorescence from GFP fungus fluorescence. Images were captured separately with each filter and then overlapped using the ISIS software (Meta Systems, Germany).
The same roots were collected after 12, 24, 48 and 72 h, and fixed with Karnowsky solution and processed according to Rossetto et al. [53]. In addition to the colonization of the root surface, the inner root tissues were visualized via a freeze-fracture technique by fixing the sample with Karnowsky solution and washing with 0.05 M cacodylate buffer. The roots were treated with 30% glycerol for 2 h, wrapped in tissue paper and frozen in liquid nitrogen. The roots were fractured and dehydrated in ethanol, subjected to critical point drying and coated with metal. The samples were observed in a scanning electron microscope (LEO-Zeiss) at the Núcleo de Apoio à Pesquisa em Microscopia Eletrônica (NAP/MEPA), University of São Paulo.
Trichoderma virens–plant interaction assessment
The effect of the T. virens wild type strain (T.v.223) and a strain containing gfp (T20) on sugarcane (variety SP80-1842) growth were measured. The plants were inoculated with 106 conidia mL-1 of the wild type strain (T.v.223) or the gfp-tagged strain (T20) in a commercial PlantMax substrate (Eucatex). Non-inoculated plants were used as a control and were maintained in a greenhouse for two months, after which the plant fresh and dry mass were determined. A total of 23 plants per treatment were used in a completely randomized experimental design. To confirm fungal colonization, T. virens was reisolated from the inoculated plants as previously described.
Statistical analysis
The solation data were statistically analyzed using the fungus frequency (Ff = number of fungus colony/number of root fragments) for the endophytic fungal community and the fungus density (Df = number of forming unit colony/gram of soil) for the rhizosphere fungal community. An analysis of variance (ANOVA) was performed with SAS (Copyright (c) 1989–1996 by SAS Institute Inc., Cary, NC, USA) for a completely randomized experimental design.
Results
Isolation of fungi associated with sugarcane
To analyze the potential effects of genetically modified sugarcane on fungal frequency (Ef) (in root endophyte) and density (Df) (in rhizosphere), we isolated fungi from the rhizosphere and endorhizosphere of genetically modified plants that expressed an imazapyr resistance gene (IMI-1) and the conventional isoline (SP80-1842) for three different treatments (NW, TW and TH) at three growth stages of sugarcane plants (3, 10 and 17 months).
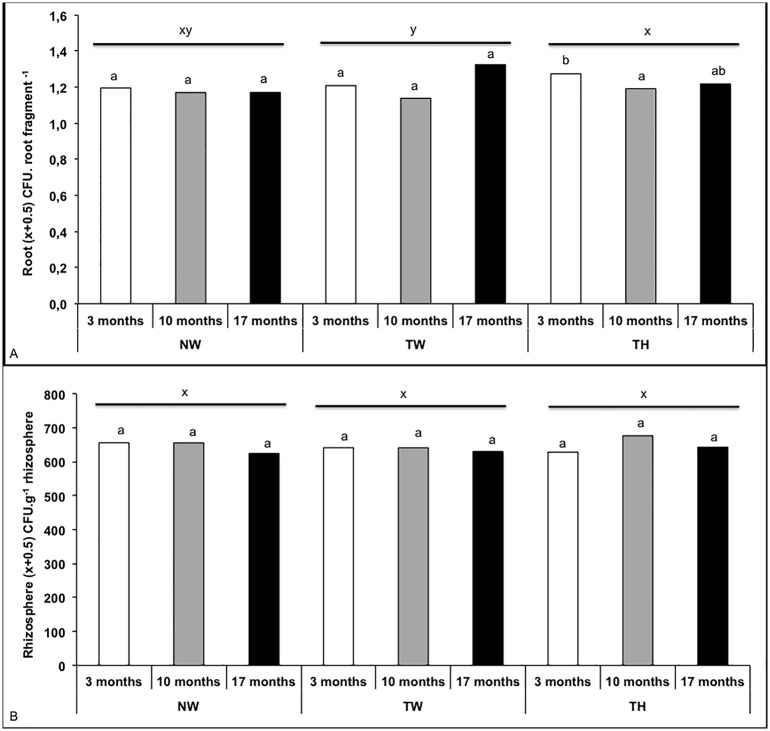
The experiment evaluated the effects of plant genotype, crop management and growth stage on the fungal community associated with sugarcane. Sugarcane roots had a high number of endophytic fungi, and from 756 root fragments (252 fragment per sampling time), 2,236 fungal colonies we isolated, being 781 from 3month-old plants, 659 from 10 month old plants and 796 from 17-month-old plants (Fig 1). Similarly, the plant rhizosphere was also found to be highly colonized by fungi, being isolated 1,224 fungi colonies from this plant site. The number of rhizosphere fungi ranged from 4.25 105 CFU g-1 of soil at 17 month-old plants to 8.20 105 CFU g-1 of soil at 10 month-old plants (Fig 1). The ANOVA for the different treatments (NW, TW and TH) and sampling time (3, 10 and 17 months) was performed on the fungus frequency (endophytic communities) and density (rhizosphere communities) data (S1 Table).
Fig 1. Filamentous fungal isolation of root entophytes frequency (a) and rhizosphere density (b).
The bars indicate the average of each effect in each variation factor: treatments (NW- non-transgenic plants with weeding, TW- genetically modified plants with weeding and TH- genetically modified plants treated with herbicide); sampling time (3, 10 and 17 months after planting). Different letters represent difference at 5% probability in the Tukey test. Comparison among sampling time (3, 10 and 17 months) we used letter a and b and comparison of treatments (NW, TW, TH) we used letters x and y.
The ANOVA results showed that regardless the fungal community evaluated (root endophyte or rhizosphere), the genetically modified sugarcane did not induce a significant change (P>0.05) in the fungal frequency or density (S1 Table), suggesting that neither the evaluated plant genotype nor crop management had a significant effect on the sugarcane fungal community. The frequency of root endophytic fungi was significantly (P = 0.0256) affected by sampling time (S1 Table), being the frequency 17 month-old > 10-month-old > 3-month-old sugarcane plants (Fig 1). Curiously, such growth effects were not observed in the density of the rhizosphere community (P>0.05) (S1 Table). Additionally, the ANOVA revealed the presence of an interaction (P = 0.0305) between treatment and sampling time for endophytic fungi, so the effect of the different treatments on the fungal frequency could depended on the plant growth stage.
Composition and distribution of the fungal community associated with sugarcane
Due to the large number of isolates obtained, the fungi were grouped according to morphological characteristics (type of mycelium, color, presence of spores), and representative samples were selected for subsequent analysis. The morphological diversity of fungus associated with sugarcane displayed a great variation in mycelial color, including white, beige, yellow, green, pink, lilac, brown, gray and black (S1 Fig). In addition to the variety of colors, the fungus also differed in mycelia growth habits (cottony, submerged aerial hyphae), the presence of reproductive structures (spores), the growth rate and the secretion of compounds that changed the color of the growth medium.
Therefore, a total of 249 fungi comprising 16 morphotypes (S1 Fig) from all three treatments were selected for molecular identification by sequencing of the ITS1-5.8S-ITS2 region of rDNA. The sequences obtained were used to evaluate the effects of genotype, management and growth stage using a diversity and richness index. The fungal isolates were taxonomically identified at the genus level via BLASTn by comparison with the ITS1-5.8S-ITS2 rDNA sequences available in GenBank and confirmed by morphological characteristics.
Analysis of the ITS1-5.8S-ITS2 sequences showed that the fungal community associated with sugarcane comprised at least 35 different genera (S2 Table). Overall, the phylum Ascomycote predominated among the fungi identified (96.0%), whereas the phyla Zygomycota/Mucoromycotina and Basidiomycota represented only 2.4% and 1.6% of the community, respectively. In the phylum Ascomycote, Eurotiomycetes (43.0%), the classes Sordariomycetes (38.5%) and Dothydeomycetes (7.6%) were predominant, and the most frequent genera were Penicillium (33.3%), Fusarium (16.9%), Aspergillus (7.2%) and Trichoderma (4.4%) (S2 Table).
Other genera were reported at a low frequency (<2.8%), and 16 (Acremonium, Alternaria, Cladophialophora, Colletotrichum, Curvularia, Exophiala, Mariannaea, Myrmecridium, Myrothecium, Paecilomyces, Paraphaeosphaeria, Phoma, Phomopsis, Pyricularia, Saccharicola and Sagenomella) were represented by only one isolate.
The genus Penicillium (teleomorphs Eupenicillium or Talaromyces), represented 33.3% of the fungi isolated from both the roots and the rhizosphere. A phylogenetic analysis indicated the similarity of 17 different species (P. restrictum, E. katangense, P. janthinellum, T. flavus, E. javanicum, P. ochrochloron, E. reticulisporum, E. brefeldianum, E. levitum, P. vinaceum, P. radicum, P. minioluteum, T. udagawae, T. trachyspermus, P. pinophilum, T. indigoticus and P. marneffei), and fourteen isolates were classified as T. trachyspermus; three isolates were classified as P. pinophilum and two isolates were classified as E. javanicum (S2A Fig).
The genus Fusarium (teleomorph Gibberella) was the second most frequent genus and included 16.9% of the isolates, and five different species were highly similar (F. oxysporum, F. acutatum, F. solani, F. dlaminii and G. moniliformis). In this group, 19 isolates were classified as F. oxysporum, and the remaining 23 isolates were classified only at the genus level (S2B Fig).
The third most frequent genus, with 7.2% of the isolates, was Aspergillus, which had 7 different species that were highly similar (A. brasiliensis, A. flavus, A. oryzae, A. versicolor, A. niger, A. terreus and A. fumigatus). The phylogenetic analysis showed that six isolates were classified as A. brasiliensis; three isolates were identified as A. terreus and one isolate was identified as A. fumigatus (S2C Fig).
The genera Trichoderma and Epicoccum occurred less frequently than other genera. Trichoderma (teleomorph Hypocrea), the fourth most frequent genus, was represented by 11 isolates corresponding to 4.4% of fungus community. The phylogenetic analysis allowed the classification of two isolates as H. virens, three isolates as T. asperellum and six isolates as Trichoderma sp. (S2D Fig). The genus Epicoccum was the fifth most abundant genus and represented 3.6% of the total isolates.
In addition to the most frequent genera (Penicillium, Fusarium, Aspergillus, Trichoderma and Epicoccum), the fungal community associated with sugarcane included 28 other genera in the phylum Ascomycota, the genus Resinicium (with 3 isolates) in the phylum Basidiomycota and the genus Cunninghamella (with 3 isolates) in the phylum Zigomycota/Mucoromycotina.
In the phylum Ascomycota, the class Sordariomycetes comprised a large number of the isolates (96), with at least 17 genera identified (Bionectria, Chaetomium, Chaetosphaeria, Colletotrichum, Diaporthe, Fusarium, Mariannaea, Microdochium, Myrmecridium, Myrothecium, Nigrospora, Paecilomyces, Phomopsis, Pyricularia, Thielavia, Thozetella and Trichoderma). Moreover, nine isolates were not similar to any reported genus, so they were classified as not identified (N.I.) (S2 Table).
In the phylum Ascomycota, besides the class Sordariomycetes, the fungal community associated with sugarcane was characterized by isolates from the classes Dothideomycetes, Eurotiomycetes, Leotiomycetos and mitosporic fungi. The class Dothideomycetes comprised 19 isolates, classified in six different genera (Alternaria, Cladosporium, Curvularia, Epicoccum, Paraphaeosphaeria and Saccharicola). The Eurotiomycetos class comprised 107 isolates; however, 94.4% were classified in the genera Aspergillus and Penicillium, and the other 6 isolates were identified as Cladophialophora, Exophiala, and Sagenomella. The class Leotiomycetes had only two representatives, both in the genus Acephala.
Sugarcane fungal community structure
Rarefaction curves were constructed to verify whether the sampling of the present work was adequate to assess the population diversity. Overall, the rarefaction curves of different populations using similarity levels of 95% and 97% were sufficient to encompass the diversity of fungal population (S3 Fig). Therefore, richness and diversity comparisons based on 95% and 97% similarities were performed.
The diversity (Shannon-Wienner) and richness (Chao1) index for the fungal population associated with sugarcane are presented in Table 1. The results show that the fungal community associated with sugarcane has great richness and diversity; the total diversity and richness indices (H’(95%) = 3.53; H’(97%) = 3.64; Chao1(95%) = 98; Chao1(97%) = 134) are considered high for the similarity index used. Moreover, rhizosphere seems to have a higher fungal diversity than root endophytes, although the values were not significantly different (Table 1).
Table 1. Diversity and Richness index of all sequence groups at 97% and 95% similarity.
| Index | Sequence group | Similarity | |
|---|---|---|---|
| 97% | 95% | ||
| Diversity (Shannon-Wiener) | 3 months | 3.17(±0.24) | 3.10(±0.23) |
| 10 months | 2.75(±0.28) | 2.61(±0.28) | |
| 17 months | 3.48(±0.19) | 3.39(±0.19) | |
| NW | 3.28(±0.25) | 3.18(±0.24) | |
| TW | 3.16(±0.22) | 3.09(±0.22) | |
| TH | 3.34(±0.25) | 3.28(±0.26) | |
| Root endophyte | 3.16 (±0.21) | 3.12 (0.20) | |
| Rhizosphere | 3.51 (±0.21) | 3.45 (±0.20) | |
| TOTAL | 3.64 (±0.17) | 3.53 (±0.17) | |
| Richness (Chao1) | 3 months | 68(46–130) | 47(36–80) |
| 10 months | 37(30–63) | 29(25–46) | |
| 17 months | 97(56–147) | 57(47–94) | |
| NW | 88(59–166) | 69(48–126) | |
| TW | 59(42–109) | 49(37–86) | |
| TH | 133(74–295) | 101(61–211) | |
| Root endophyte | 69 (50–123) | 52 (42–85) | |
| Rhizosphere | 145 (88–296) | 121 (76–244) | |
| TOTAL | 134 (100–210) | 98 (80–141) | |
Moreover, the richness and diversity of the fungal community associated with sugarcane changed according to the sampling time; 17-month-old sugarcane plants had the highest diversity and richness indices. Plants 17 and 10 months old had significantly different diversity and richness indices (Table 1). In addition, impact of plant genotype (NW x TW) and crop management (TW x TH) on fungal richness and diversity was not observed.
The AMOVA allowed an assessment with a defined statistical probability of how much of the variation in the fungal community associated with sugarcane is due to genetically modified sugarcane planting (genotype effects), weed management (management effects), sampling time or isolation site (root endophyte or rhizosphere). Analysis of the results showed that plant genotype, crop management and the fungal community did not have significant effects on fungal community (P>0.05). However, the plant growth stage caused significant variation, contributing to 3.3% of the total variation (P = 0.0035) (Table 2). This variation was mainly related to the root endophytic community (3.21%, P = 0.0079); other groups did not contribute significant variation (P>0.05) (Table 2). These data suggest that plant age affects the endophytic root community but not the rhizosphere community.
Table 2. Analysis of molecular variance (AMOVA) for different fungus populations according to the evaluated effect.
| Effect | Fungus Group | % variation 1 | P value2 |
|---|---|---|---|
| Plant Genotype (Non-transgenic vs. transgenic) | Total sample | -0.61 | 0.7019 n.s. |
| Plants with 3 months | 0.00 | 0.6673 n.s. | |
| Plants with 10 months | -4.76 | 1.0000 n.s. | |
| Plants with 17 months | -0.71 | 1.0000 n.s. | |
| Root endophyte | -1.16 | 0.7972 n.s. | |
| Rhizosphere fungus | -1.68 | 0.6976 n.s. | |
| Crop management (weeded vs. herbicide) | Total sample | 2.88 | 0.3041 n.s. |
| Plants with 3 months | 8.65 | 0.3394 n.s. | |
| Plants with 10 months | 6.78 | 0.3301 n.s. | |
| Plants with 17 months | 1.08 | 0.6706 n.s. | |
| Root endophyte | 0.56 | 0.2052 n.s. | |
| Rhizosphere fungus | -2.85 | 1.0000 n.s. | |
| Plant growth stage (3 months vs. 10 months vs. 17 months) | Total sample | 3.30 | 0.0035 |
| Non transgenic plants (NW) | 3.40 | 0.1323 n.s. | |
| Transgenic plants with weeded (TW) | 7.80 | 0.1299 n.s. | |
| Transgenic plants with herbicide (TH) | -0.05 | 0.5971 n.s. | |
| Root endophyte | 3.21 | 0.0079 | |
| Rhizosphere fungus | 2.71 | 0.0731 n.s. | |
| Fungus community (root endophyte vs. Rhizosphere fungus) | Total sample | 1.76 | 0.0971 n.s. |
| Non transgenic plants (NW) | -0.04 | 0.3971 n.s. | |
| Transgenic plants with weeded (TW) | 0.60 | 0.3023 n.s. | |
| Transgenic plants with herbicide (TH) | 0.68 | 0.2988 n.s. | |
| Plants with 3 months | 1.24 | 0.2033 n.s. | |
| Plants with 10 months | 2.46 | 0.2943 n.s. | |
| Plants with 17 months | 1.36 | 0.2002 n.s. |
1 Percentage of variation between analyses groups;
2 P–probability of a variation component higher than the observed for the fixation index;
n.s.–non significantly values at the level of 5% probability.
Correlation between genera and plant niche (root vs. rhizosphere)
The taxonomic similarities between fungi isolated from the roots (endophytes) and the rhizosphere were assessed by the Jaccard coefficient of similarity (J), considering the genera that occurred in each community. The value obtained (J = 0.343) indicated that these communities should have a greater number of genera that occurred infrequently than common genera.
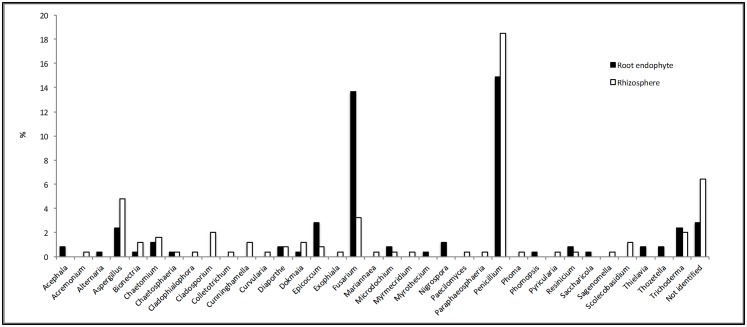
The proportion of rhizosphere and root endophytes in the community profile can be observed in Fig 2. The figure shows, for example, that 8 genera (Acephala, Alternaria, Myrothecium, Nigrospora, Phomopsis, Saccharicola, Thielavia and Thozetella) were observed only in roots and that 15 genera (Acremonium, Cladophialophora, Cladosporium, Colletotrichum, Cunninghamella, Curvularia, Exophiala, Mariannaea, Myrmecridium, Paecilomyces, Paraphaeosphaeria, Phoma, Pyricularia, Sagenomella and Scolecobasidium) were observed only in the rhizosphere. Moreover, some of the most frequent genera (such as Aspergillus, Penicillium, Fusarium and Trichoderma) were predominant in the roots or the rhizosphere. The genus Aspergillus was observed at a frequency two-fold higher in the rhizosphere than in the roots; on the other hand, 80% of the isolates identified as Fusarium were root endophytes (Fig 2), although this genus was also isolated from rhizosphere.
Fig 2. Percentage of the occurrence of each genus observed in the sugarcane fungal community and their relative proportions according to isolation location (rhizosphere or roots).
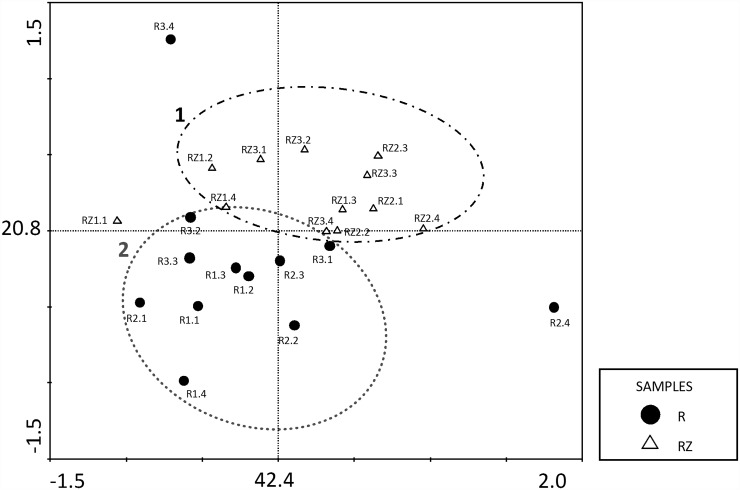
A principal component analysis (PCA) confirmed that the fungal communities in the roots and the rhizosphere were different (Fig 3). Although some fungi genera were shared by both inner roots and rhizosphere, the two principal components, which explain 63.2% of the variance, clustered the samples in two groups, being the first composed by rhizosphere community (RZ); and the second by endophytic community (R). These results indicated that the isolation site determined the variation of the frequency and the occurrence of such genera in the isolated fungal community.
Fig 3. Principal Component Analysis (PCA) of the occurrence of the different fungus genera in the root endophytic community (● R) and the fungal rhizosphere community (Δ RZ).
The values on both axes indicate the percentage of variance explained by sample distribution on each respective axis.
Genetic transformation of T. virens mediated by A. tumefaciens
The number of transformants that grew on selective medium was counted every 5 days for 30 days. The tests indicated that the optimal conditions for the transformation of T. virens fungus with Agrobacterium were the use of filter paper, inoculated with 107 conidia mL-1, in PDA medium and co-cultivated for 48 h. Other conditions, such as two selection conditions were tested (normal–no addition of culture media to the membrane after the transfer to selective medium and overlay–the addition of culture media to the membrane) as well as two concentrations (200 and 400 μM) of the bacterial virulence inductor acetosyringone (AS).
The results showed that transformants were obtained under all tested conditions with similar transformation efficiencies. Overall, the average number of transformants obtained was 12 transformants per 107 conidia. mL-1, and we obtained T. virens transformants with the chosen transformation conditions.
Analysis of transformants
Mitotic stability
The mitotic stability of 20 random transformants was evaluated after 5 successive generations in non-selective medium. Of the transformants tested, 70% maintained hygromycin B resistance and green fluorescence emission (observed under FITC filter– 459 to 490 nm). Moreover, we did not observe sector emission in the evaluated transformants.
Molecular analysis
Eight transformants (T2, T4, T7, T9, T10, T15, T20, T22) resistant to hygromycin B were selected to confirm the presence of gfp gene by PCR using the glGFP5 and glGFP3 primers. All transformants tested and the plasmid pFAT-gfp (positive control) led to the amplification of a 700 pb fragment. The wild type strain T.v.223 was used as a negative control and did not allow amplification of this fragment.
Four transformants (T2, T7, T10 and T20) were randomly picked for Southern blot analysis from the eight transformants obtained to confirm the integration of the T-DNA genome and determine the number of copies inserted using a probe containing the gfp sequence (700 pb) (S4 Fig). The results confirmed that T-DNA was integrated into the genomes of all T. virens transformants and indicated the random occurrence of this event because different band sizes indicated different integration events. Transformants T20 and T2 had only one copy inserted, whereas the remaining transformants had more than one copy of T-DNA integrated into the genome. The wild type strain T.v.223 used as a negative control and the pFAT-gfp vector used as a positive control showed no bands and a 15 kb band, respectively.
Comparison of the transformants and the wild type T. virens
The mitotically stable T20 transformant that had a single copy inserted was selected for further analysis. The morphological and physiological similarities with the wild type strain T.v.223 were evaluated. After 5 days of growth in PDA at 28°C, the morphological characteristics of both isolates were similar. Both showed weak and diffuse growth, with cottony mycelia and green conidia. The growth rates were similar; both grew approximately 2.7 cm per day in a Petri dish with PDA medium. Moreover, the T.v. T20 transformant and the wild type T.v.223 had similar conidial production (1.31 x 107 conidia cm-2 and 1.71 x 107 cm-2, respectively).
Interaction of T. virens with the host plant
Reisolation of T. virens strain T20 from sugarcane plants
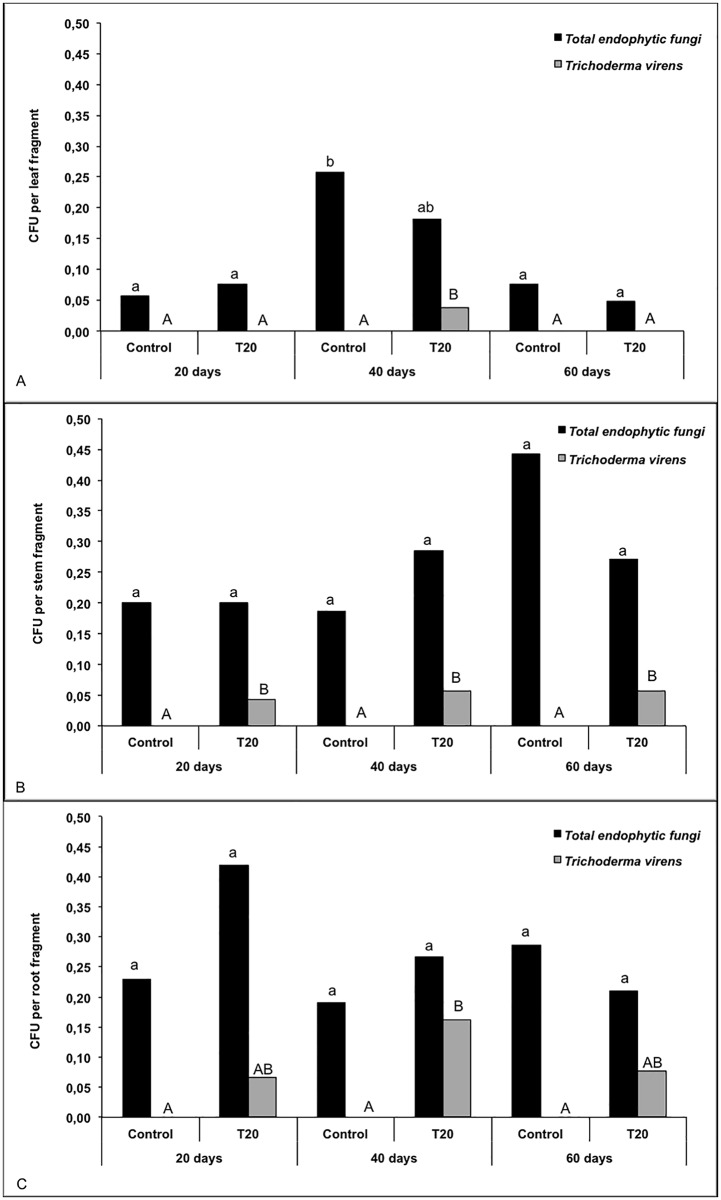
The results show that strain T20 endophytically colonized plant roots and stems but scarcely colonized plant aerial tissues. Root and stem colonization by T20 occurred at a low frequency (Fig 4) and did not lead to phenotypic changes in the plants. Moreover, the presence of T20 did not significantly affect the total frequency of endophytic fungi on the various plant tissues (Fig 4). Although this fungus is naturally found in sugarcane, in the present experiment T. virens could not be isolated from control (uninoculated) plants under any of the conditions tested (Fig 4); it was found only in inoculated plants.
Fig 4. Isolation frequency of total endophytic fungi and T. virens strain T20 from leaves (a), stems (b) roots (c) of sugarcane plants at 20, 40 and 60 days.
The same letter indicates that treatments are not significantly different according to Tukey’s test at 5% significance.
The statistical analysis of the fungal reisolation frequency from sugarcane leaves showed that “total endophytic fungi” did not differ significantly between the inoculated and non-inoculated plants (S3 Table). However, a significant difference (P = 0.0418) was observed for the isolation of endophytic fungi in sugarcane plants at 20, 40 and 60 days; after forty days, plants showed a higher frequency of endophytic fungi.
A statistically significant difference for “Total Trichoderma virens” isolation was observed between the inoculated and uninoculated plants (P = 0.0415) and among the different plant ages (P = 0.0198), and their interaction was also significant (P = 0.0198). These differences are shown in Fig 4A, which shows that T20 was presented in inoculated plants only after 40 days. Similar results were observed in the stems and the roots. For both roots and shoots the “Total endophytic fungus” were not significantly different for either inoculated or non-inoculated plants and different isolation data. The variable “Total Trichoderma virens” showed significant differences between inoculated and non-inoculated plants, with a probability of P = 0.0222 for stems and P = 0.0005 for roots, indicating that that the frequency of T. virens reisolated from the inoculated plants was significantly higher than for the control plants because T. virens could not be isolated from the control plants (Fig 4B and 4C). Moreover, the reisolation frequency was not significantly different for different plant ages.
Monitoring of fungal colonization by fluorescence microscopy (FM) and scanning electronic microscopy (SEM)
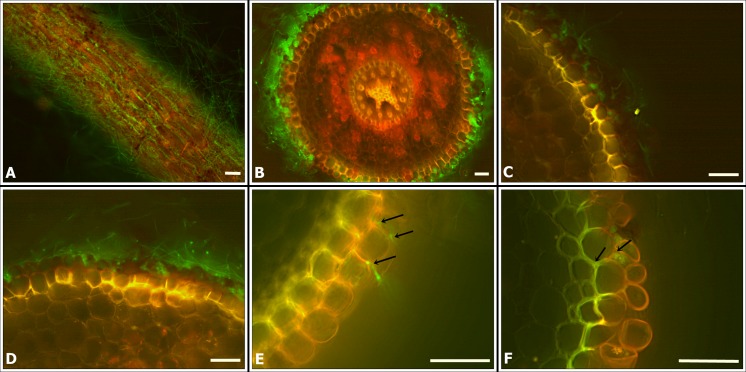
The superficial and internal colonization of sugarcane root tissue by T. virens fungus were observed by optical fluorescence microscopy (FM) and scanning electronic microscopy (SEM). The transformant T20 was selected for the microscopic analysis because it had a similar morphology, growth rate and sporulation as the wild type. Root tissue had an intense auto fluorescence, so two different filters were used: FITC (459–490 nm) and Rhodamine (546 nm). The fungus fluoresces only under the FITC filter, so root tissue could be distinguished from the fungus in an overlaid image. The conidia and hyphae of T20 transformant emit an intense green fluorescence (Fig 5), whereas the mycelium of the wild type did not. We were able to visualize dense mycelia in sugarcane roots inoculated with T.v. T20 (Fig 5A, 5B, 5C, 5D and 5F).
Fig 5. Optical fluorescence microscopy (OFM) with a FITC filter (459–490 nm), Rhodamine filter (546 nm) and an overlapped image with both filters, showing sugarcane root colonization (variety SP80-1842) by T20.
(A) Sugarcane root with T20; (B) Cross section of sugarcane root with T20 (C, D) A longitudinal section of sugarcane colonized by T20 (E, F) Cross section of sugarcane root with T20; the arrows indicate possible fungal penetration in the outer layers of the root epidermis. Scale bars = 50 μm.
Conidial production was also observed in samples collected after 4 days. In addition to root surface colonization, in some cross sections, we were able to observe intercellular colonization of the outer root layers (Fig 5).
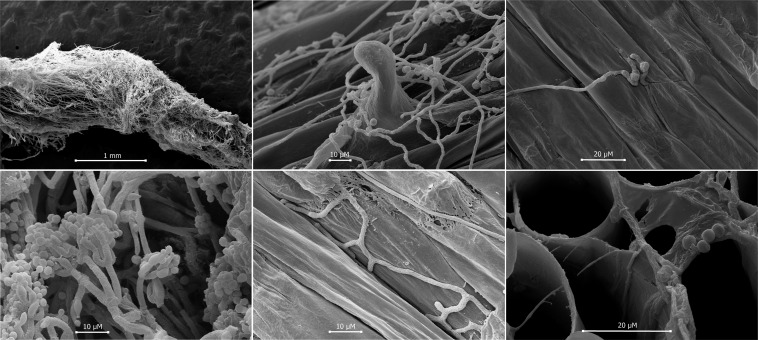
Sugarcane root tissue infected with T. virens were also analyzed by SEM, which revealed interesting aspects of fungal behavior during plant colonization (Fig 6). First, we were able to observe the conidial germination on the root tissue surface and the elongation of hyphae adhered to the cuticle (Fig 6C). Moreover, the hyphae showed preferential growth toward the cellular junctions (Fig 6C and 6E). We could not see any appressorium or alteration of the plant surface in the presence of the fungus. Seven days after conidial inoculation, we were able to visualize dense colonization on the root surface, hyphae ramification, conidiospore formation and conidial production (Fig 6A, 6D and 6E). Observation of roots fractured with liquid nitrogen revealed the presence of fungal hyphae in the root tissue, in the intercellular space (Fig 6F).
Fig 6. Scanning electronic microscopy images (SEM) showing sugarcane root colonization (variety SP80-1842) by T. virens strain T20.
(A, B) Root surface colonization by T20; (C) T. virens conidia germination adhered to the plant host cuticle; (D) Asexual reproduction structure (conidiophore and conidia) of T. virens fungi; (E) Mycelia growth toward the cellular junctions and (F) Inner root segment showing the presence of fungal hyphae in the intercellular space. Scale bars are indicated in the figure.
The effect of T. virens on sugarcane growth
The results show that fungal inoculation did not result in any detectable phenotypical alteration and that all plants were healthy and vigorous with growth similar to the control. In fact, when the fresh and dry masses were compared, no significant differences were observed in shoots, roots or the entire plant in any treatment (Table 3). These results show that neither wild type nor transformant T. virens did not affect the growth of sugarcane under the conditions tested. Moreover, genetic alterations present in strain T20 apparently did not interfere with its interaction with sugarcane compared to the wild type strain.
Table 3. Effect of the inoculation of strains T.v.223 and T.v T20 on the fresh and dry weight accumulation on root and shoot of sugarcane plants (variety SP80-1842) cultivated for 60 days in greenhouse.
| Treatment | Fresh mass (g) 1 | Dry mass (g) 1 | ||||
|---|---|---|---|---|---|---|
| Root | Shoot | Total | Root | Shoot | Total | |
| SP80-1842 (control) | 5.12 a | 2.90 a | 8.02 a | 0.51 a | 0.83 a | 1.34 a |
| SP80-1842 + T.v. strain 223 | 4.75 a | 2.53 a | 7.27 a | 0.43 a | 0.68 a | 1.12 a |
| SP80-1842 + T.v. strain T20 | 4.66 a | 2.66 a | 7.33 a | 0.47 a | 0.69 a | 1.16 a |
1 Values are the average of 23 repetitions;
Treatments with the same letters do not statistically differ in Tukey test, at 1% of significance.
To confirm that the fungal inoculation of the soil was sufficient to promote sugarcane endophytic colonization and that the effects observed occurred in the presence of T. virens, some plants were sampled and endophytic fungi were isolated from sugarcane. Reisolation showed a low frequency of endophytic Trichoderma in the leaves and stems, similar to the previous reisolation results. T. virens was isolated only from the roots of inoculated plants, indicating that inoculation of the soil leads to the colonization of plant roots by T. virens but does not translocate to the shoot nor affect the plant biomass.
Discussion
The indirect effect of genetically modified plants on ecosystem communities and functions are still being discussed [4, 54]. There is great concern about potential effects on non-target organisms such as mycorrhizae, rhizobium and other microorganisms that protect plants, as well as on phosphate solubilizing, decomposition and growth promotion.
The transgenic sugarcane plants in this study that are resistant to imazapyr (IMI-1) carry a plasmid containing a mutated wheat AHAS gene, which confers imazapyr resistance, that is under the control of a constitutive sugarcane promoter. The herbicide imazapyr inhibits the enzyme AHAS, which is responsible for the synthesis of some essential amino acids. This enzyme occurs in bacteria, fungi and plants, and has a similar amino acid sequence in these organisms, which suggests a common ancestor [55] and the potential that imazapyr could affect organisms other than weeds. Some studies have reported that nitrogen fertilizers affect the microbial community [56], as do imidazolinone-derived herbicides [20, 57], and transgenesis [20, 58, 59]; however, the results of this study indicated that neither the plant genotype (transgenic or non-transgenic) nor crop management (weeding or herbicide application) significantly changed the fungal community of cultivated sugarcane.
Interestingly, Stuart et al. [20] used the same field experiments as in the present study but evaluated only the first two growth stages (3 and 10 months) and observed an effect on the endophytic fungal community that colonized the leaves of the transgenic variety IMI-1 managed with imazapyr. They reported that the herbicide led to a rapid and transient alteration of the fungal community, whereas the transgenic variety displayed a slow and persistent change in the leaf fungal community [20]. Therefore, the effects of management and plant genotype seem to depend on the fungal community and probably on the plant growth stage.
However, similar to the present study, the lack of significant herbicide and/or genetically modified plants effects on the fungal community has been previously reported [5, 16, 60–62]. Indeed, the effect of genetic modification is low compared to that of other factors, such as the differences between plant varieties, which can be greater than those between genetically modified plants and their non-transgenic counterparts [63–65]. Therefore, it is likely that these studies did not detect any effect of plant genotype because other factors, such as the plant growth stage or crop management also significantly affected the microbial community, not mentioning other environmental factors, masking the lesser changes resulting from genetically modified plants.
Differently from genotype and crop management effects, the plant sampling time (3, 10 and 17 months) significantly affected the sugarcane endophytic fungal community, resulting in changes in the fungal frequency and the diversity and richness indices. This effect was also observed on endophyte in leaves by Stuart et al. [20]. Actually, the plant growth stage was previously reported to determine the microbial community structure, overcoming even the effects of genetically modified plants [11, 15, 66, 67]. Curiously though, the fungal community of the rhizosphere did not exhibit obvious changes in the parameters evaluated (fungal density, diversity and richness indices and molecular variance). Although culture dependent method could mask the presence of some important fungi, if the plant genotype or crop management had significant effect on these parameters, shifts in the fungi community would be observed.
The identification of the fungal community that colonizes the sugarcane roots and rhizosphere allowed us to calculate the diversity and richness indices. The results revealed that root endophytic fungi are less diverse than rhizosphere fungi, which we expected in view of the enormous fungal density in soil and the rhizosphere effect. The rhizosphere effect was demonstrated by Gomes et al. [68], who compared the DGGE profiles of the rhizosphere and soil fungal communities of maize and showed a significant increase in the relative abundance of fungi in the rhizosphere. Although the endophytic fungal diversity was lower than in the rhizosphere, the diversity index obtained (H’ = 3,12) is similar to those reported in studies that evaluated endophytes from other host plants, such as Guarea guidonia [69].
The fungal community are structured by the isolation site [70–72], and it is likely that the community differs due to the different micro-environments in each plant tissue, soil type and rhizosphere. In overall, it was observed that the cultivable fungal community associated with sugarcane comprised at least 35 different genera, distributed in the phyla Ascomycota (96.0%), Zygomycota/Mucoromycotina (2.4%) and Basidiomycota (1.6%). The predominance of cultivable Ascomycota fungi associated with plants has been previously reported [73–77], whereas the lack of basidiomycetes is probably because methods that use plant fragments plated in growth broth do not favor their isolation [78]. In fact, several fungi cannot be retrieved with cultivable isolation methods, in this way the characterization of fungal communities by either dependent or independent cultivation methods can generate different taxonomic community profiles [58, 79], but both method can support conclusion about the impact assessment.
In Ascomycota population, two classes (Eurotiomycetes, Sordariomycetes) represented approximately 80% of the fungal community, and among these, 65.4% of the isolates belongs to Penicillium, Fusarium, Aspergillus, Trichoderma and Epicoccum genera, corroborating previous studies which showed that the fungal community associated to the host plant is composed by a great number of species, but are dominated by few species [80–82]. The genus Epicoccum, the fifth most frequent associated with sugarcane, was previously reported as an endophyte in banana [83] and apple trees [84], in the wheat rhizosphere [85] and in sugarcane leaves [86]. The genera Penicillium, Fusarium, Aspergillus and Trichoderma have been isolated both as endophytes [87–89] and from the rhizosphere of different host plants [90,91] and are usually the most frequent genera.
The data collected in this study together with the literature on the sugarcane fungal community revealed the presence of some ubiquitous fungal genera associated with the culture of this plant. For example, common genera in the rhizosphere fungal community were Acremonium, Aspergillus, Cladosporium, Chaetomium, Cunninghamella, Curvularia, Fusarium, Penicillium, Trichoderma and Paecilomyces [92], whereas the cosmopolitan endophytic fungi were Aspergillus, Epicoccum, Fusarium, Penicillium and Trichoderma [20]. The finding that 9.2% of the sugarcane fungal community could not be assigned using ITS sequencing to any known fungal genera opens the possibility of the discovery of novel fungal species because the great diversity of higher plants has long been considered the greatest reservoir of new fungi [93].
The genera Penicillium (teleomorph Talaromyces or Eupenicillium) and Aspergillus are ascomycota of Trichocomaceae family. Several species of this family have medical, industrial and biotechnological importance [94]. These genera can contain plant pathogens, generally not of economic importance but also harbour endophytes in several host plants [72, 87, 88, 89]. In this study, at least three different Aspergillus (A. brasiliensis, A. fumigatus and A. terreus) and Penicillium species (P. pinophilum, E. javanicum and T. trachyspermus) were observed. Fusarium genera (teleomorfe Gibberella) have also been reported to be plant pathogens and to cause great economic losses. In sugarcane, F. verticillioides, F. moniforme and F. subglutinans species have been reported as the casual agents of Pokkah-boeng and Fusarium rot, which remain latent until stress conditions trigger the plant disease [95, 96]. This genus has also been reported to contain endophytes, for example, F. mangiferae and F. sterilihyphosum in mango [97], F. solani in palm trees [98], F. cf. avenaceum, F. decemcellulare and F. solani in Manilkara bidentata [99] and F. graminearum in wheat [89]. Moreover, some endophytic strains of F. oxysporum have great potential as biological control agents, for example, in cucumber against the pathogen Pythium ultimum [100]. In this study, 42 isolates were classified as Fusarium, and F. oxysporum was the predominant species.
The genus Trichoderma (teleomorph Hypocrea) is known for rapid growth, its ability to use different substrates and resistance to toxic compounds [101] showing a great plasticity which features could increase the fitness during plant interaction. These fungi colonize soil, roots and leaves, and have a ubiquitous distribution, predominating in soils in various climates [102, 103]. They have also been reported as endophytes in cacao [104, 105], Azadirachta indica [88], banana [106] and fig trees [107]. Some species are of great economic importance due to their ability to produce enzymes and antibiotics, their potential as biological control agents and the capacity to induce plant growth [108,109]. Specifically, in sugarcane, cellulase production was tested in T. viride to deconstruct the lignocellulose biomass from Saccharum spontaneum by fermentation [110] and the production of ethanol directly from sugarcane bagasse by a recombinant T. reesei [111]. This species has been reported to be an aggressive mycoparasite, able to synthesize antibiotics, induce phytoalexin production and plant host resistance [36], as well as to produce endochitinases used for biocontrol [112] and, in some cases, to promote plant growth [113–114]. Because of these features, T. virens, one of the most abundant species that is not described as a toxigenic fungus, was chosen to be used in the studies to understand the interaction of this sugarcane plants and the fungi communities, allowing not only to study the impact of plant genotype, sampling time and crop management, but also how these fungi may interact with the host plant.
The first step to study the Trichoderma-sugarcane interaction involved the generation of hygromycin-resistant fungal mutants expressing GFP by AMT transformation which has been used for transformation of several fungi [21, 47, 48]. A high mitotic stability was observed in the mutants we generated, similar to reports of transformations using gfp gene with the Agrobacterium system in other fungal species [21, 49, 115].
The ability of T. virens to colonize sugarcane tissues endophytically was confirmed by reisolation of the T20 mutant strain, using the hygromycin resistance and the GFP expression to confirm the identity of the reisolated strain. The results showed that T. virens could colonize sugarcane endophytically and was found mainly in the plant roots, since it was in low frequency in leaves T. virens was only detected 40 days after inoculation, indicating that this fungus may not translocate to the shoot. This result was in agreement with previous observations in which Trichoderma were not isolated from sugarcane leaves [20] but from sugarcane roots (this study). Likewise, preferential root colonization has been reported for other Trichoderma species [116, 117]. In fact, different plant tissue preferences by an endophytic fungus have been shown to be related to specific conditions present in each plant organ [21, 24, 71]. Overall, sugarcane colonization by the T. virens T20 mutant occurred at a low frequency and did not lead to plant phenotypic alterations or disease signs and did not affect the total endophytic fungal frequency. Given that endophytic colonization can be affected by the inoculum type and by the plant growth substrate [104], perhaps the low frequency of the T20 mutant may have resulted from the conditions used for the interaction study. Interestingly, in this pot interaction study, the fungal frequency varied according to plant age as previously observed in the field study, corroborating the fact that plant growth stage may significantly affect the fungal community. In addition, it was observed that the inserted genes (hph and gfp) were maintained in the T20 mutant even after colonizing plant tissues in the pot interaction experiment, confirming the high genetic stability of this strain.
Specific aspects of the colonization of T. virens in sugarcane roots were explored by microscopy, in which OFM and SEM were used to monitor fungal colonization of superficial and inner tissues during plant growth. The results showed that this fungus colonized the root surface, forming a dense mycelial mass. In addition, the observation of root transverse sections revealed that T. virens also entered the host plant tissues and colonized the intercellular space of the outer layers of the root epidermis. Some Trichoderma strains have been reported to be able to colonize only specific parts of the roots [118], although rhizosphere strains are known to be able to colonize the entire root surfaces for several weeks [119] or months [120].
Generally, the penetration of Trichoderma spp. into plant root tissue is limited to the outer cell layers [118, 121] and does not lead to disease signs [116]. The first step of an endophytic colonization may occur when the fungus enters and forms specialized structures such as an apressorium, as observed in Discula umbrinella on the leaf surface [122] and in Piriformospora indica on the root surface [123]. Curiously, in the present work, we did not observe apressorium formation or the presence of any other specialized structures, but growth proceeded toward the cell junction, suggesting that T. virens may penetrate plant root tissues by other mechanisms. Endophytic fungi, for instance, can enter the host plant through natural openings such as stomata or between epidermal cells through the cuticle via the action of lytic enzymes, as has been reported for the fungus Beauveria bassiana [124] and E. nigrum [21], and which could also occur with T. virens.
In addition to being largely known as biological control agents, Trichoderma species are also known for other indirect effects in plants [116], including plant growth promotion and raising productivity [120, 125]. In this regard, the effect of the T. virens fungus in the host plant has been broadly studied in cotton [36], but to date, no such results have been reported for sugarcane. This study also evaluated the effects of T. virens on sugarcane growth and the dry mass of the shoots and the roots. The results showed that the presence of this fungus on the roots did not affect the phenotypical characteristics, plant growth or dry mass accumulation. In addition, T. virens T20 imposed similar effects on sugarcane plants compared to the wild type strain. Transgenic strains do not always maintain similar characteristics in their interactions with the plant host, and changes of the fungal life style (from mutualistic to parasitic) during the interaction with the plant have been reported for the endophytic fungus Epichloë festucae due to a mutation in one gene (noxA) [126].
The exploration of the intimate aspects of fungal-plant interactions allowed the confirmation of an endophytic relationship between T. virens and sugarcane, the determination of the colonization pattern and its phenotypic effects. T. virens could possibly be a biological control agent in the sugarcane fungal community, inhibiting phytophatogens by competition or parasitism, but this should be further investigated. Finally, this is the first study that reports the interaction between genetically modified sugarcane and the root endophytic and rhizospheric fungi communities. In addition, a large number of fungal species was isolated, generating a rich strains collection, which could be used in future studies that seeking to explore fungus-plant interactions and for the evaluation of potential fungal isolates that have biotechnological and agronomic potential.
Supporting Information
Fungal colonies grown in PDA broth at 28°C for 5–10 days. (a) Epicoccum sp., (b) Penicillium sp., (c) Chaetomium sp., (d) Fusarium sp., (e) Trichoderma virens, (f) Not identified root endophyte fungi, (g) Eupenicillium javanicum, (h) Acremonium sp., (i) Talaromyces trachyspermus, (j) Aspergillus niger, (k) Penicillium sp., (l) Epicoccum nigrum, (m) Mariannaea sp., (n) Fusarium sp., (o) Bionectria sp., (p) Myrmecridium schulzeri.
(DOCX)
Reference sequences of GenBank were used and the sequences obtained in the present work were signaled with the following symbol ♦. Bootstrap values (n = 1000) lower than 50 are represented in nods. a) 70 fungi of Penicillium genera, using the ascomycota Buergenerula spartinae as outgroup. b) 39 fungi of Fusarium genera, using the ascomycota Bionectria ochroleuca as outgroup. c) 17 fungi of Aspergillus genera, using the ascomycota Penicillium pinophilum as outgroup. d) 11 fungi of Trichoderma genera, using the ascomycota Hypomyces aurantius as outgroup.
(DOCX)
(a) Comparison between the expected number of phylotype for endophytic and root fungal population; (b) Number of phylotype expected for all fungal community associated to sugarcane (root endophyte + rhizosphere fungi).
(DOCX)
Genomic DNA of the isolates were digested with restriction enzyme EcoRI, which cuts T-DNA twice and do not cut gfp gene sequence, a electrophoresis were performed in 0.8% agarose gel, transferring to nylon membrane and hybridized with the 700 pb fragment (gfp gene) labeled with digoxigenine. Column 1 positive control (pFAT-gfp plasmid); column 2 is the negative control (Wild strain T.v.223); columns 3–6 are the transformed strains (T20, T10, T7 and T2). Visualized bands are indicated with arrow.
(DOCX)
(DOCX)
(DOCX)
Experimental design totally randomized. Analysis were performed separately for each part of the plant (leave, stem and root).
(DOCX)
Acknowledgments
The Centro de Tecnologia Canavieira (CTC), for hosting the crop experiment performed by Dr. Eugênio César Ulian. Dra. Sabrina M. Chabregas (Centro de Tecnologia Canavieira—CTC, Piracicaba, SP, Brazil) for the courtesy of the in vitro sugarcane plants (variety SP80-1842). We thank Dr. K. M. Plummer (La Trobe University, Australia) for providing the pFAT-gfp plasmid to Dra. Léia C. L. Fávaro. Núcleo de Apoio à Pesquisa em Microscopia Eletrônica (NAP/MEPA) and Laboratório de Citogenética Molecular de Plantas, coordinated by Prof. Margarida L.R.A. Perecin, both at ESALQ-USP (Piracicicaba, SP, Brazil) for the SEM and fluorescence optical microscopy images, respectively.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by FAPESP (http://www.fapesp.br/), grant number: 2002/14143-3 and 2003/13834-5.
References
- 1.Dal-Bianco M, Carneiro MS, Hotta CT, Chapola RG, Hoffmann HP, Garcia AA, et al. Sugarcane improvement: how far can we go? Curr Opin Biotechnol. 2012;23(2):265–70. 10.1016/j.copbio.2011.09.002 [DOI] [PubMed] [Google Scholar]
- 2.Wu H, Altpeter F. Sugarcane (Saccharum spp. hybrids). Methods Mol Biol. 2015;1224: 307–316. 10.1007/978-1-4939-1658-0_24 [DOI] [PubMed] [Google Scholar]
- 3.Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, et al. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol Mol Biol Rev. 2015; 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh AK, Singh M, Dubey SK. Changes in Actinomycetes community structure under the influence of Bt transgenic brinjal crop in a tropical agroecosystem. BMC Microbiol. 2013: 29;13:122 10.1186/1471-2180-13-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh AK, Singh M, Dubey SK. Rhizospheric fungal community structure of a Bt brinjal and a near isogenic variety. J Appl Microbiol. 2014:117(3),750–765. 10.1111/jam.12549 [DOI] [PubMed] [Google Scholar]
- 6.Danielsen L, Thürmer A, Meinicke P, Buée M, Morin E, Martin F, et al. Fungal soil communities in a young transgenic poplar plantation form a rich reservoir for fungal root communities. Ecol Evol. 2012;2(8):1935–1948. 10.1002/ece3.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andow DA, Zwahlen C. Assessing environmental risks of transgenic plants. Ecol Lett. 2006;9: 196–214. [DOI] [PubMed] [Google Scholar]
- 8.Widmer F. Assessing effects of transgenic crops on soil microbial communities. Adv Biochem Eng Biot. 2007;107: 207–234. [DOI] [PubMed] [Google Scholar]
- 9.Dunfield KE, Germida JJ. Diversity of bacterial communities in the rhizosphere and root interior of field-grown genetically modified Brassica napus. FEMS Microbiol Ecol. 2001;38: 1–9. [Google Scholar]
- 10.Rasche F, Hodl V, Poll C, Kandeler E, Gerzabek MH, Van Elsas JD, et al. Rhizosphere bacteria affected by transgenic potatoes with antibacterial activities compared with the effects of soil, wild-type potatoes, vegetation stage and pathogen exposure. FEMS Microbiol Ecol. 2006;56: 219–235. [DOI] [PubMed] [Google Scholar]
- 11.Rasche F, Velvis H, Zachow C, Berg G, Van Elsas JD, Sessitsch A. Impact of transgenic potatoes expressing anti-bacterial agents on bacterial endophytes is comparable with the effects of plant genotype, soil type and pathogen infection. J Appl Ecol. 2006;43: 555–566. [Google Scholar]
- 12.Heuer H, Kroppenstedt RM, Lottmann J, Berg G, Smalla K. Effects of T4 lysozyme release from transgenic potato roots on bacterial rhizosphere relative to communities are negligible natural factors. Appl Environ Microbiol. 2002;68: 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamarche J, Hamelin RC. No evidence of an impact on the rhizosphere diazotroph community by the expression of Bacillus thuringiensis Cry1Ab toxin by Bt white spruce. Appl Environ Microbiol. 2007;73: 6577–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, Lu HH, Wu W, Wei QK, Chen YX, Thies JE. Transgenic Bt rice does not affect enzyme activities and microbial composition in the rhizosphere during crop development. Soil Biol Biochem. 2008;40: 475–486. [Google Scholar]
- 15.Van Overbeek L, Van Elsas JD. Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiol Ecol, 2008;64: 283–296. 10.1111/j.1574-6941.2008.00469.x [DOI] [PubMed] [Google Scholar]
- 16.Hannula SE, de Boer W, van Veen J. A 3-year study reveals that plant growth stage, season and field site affect soil fungal communities while cultivar and GM-trait have minor effects. PLoS One. 2012;7(4):e33819 10.1371/journal.pone.0033819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SH, Kim CG, Kang H. Temporal dynamics of bacterial and fungal communities in a genetically modified (GM) rice ecosystem. Microb Ecol. 2011;61(3): 646–659. 10.1007/s00248-010-9776-5 [DOI] [PubMed] [Google Scholar]
- 18.Atlas RM, Horowitz A, Krichevsky M, Bej AK. Response of microbial populations to environmental disturbance. Microbial Ecol. 1991;22: 249–256. [DOI] [PubMed] [Google Scholar]
- 19.Turco RP, Kennedy AC, Jawson MD. Microbial indicators of soil quality In: DORAN J. W.; COLEMAN D. C.; BEZDICEK D. F.; STEWART B. A. (Ed.). Defining soil quality for a sustainable environment. Madison: American Society of Agronomy Special Publication, 1994. p. 73–90. [Google Scholar]
- 20.Stuart RM, Romão AS, Pizzirani-Kleiner AA, Azevedo JL, Araújo WL. Culturable endophytic filamentous fungi from leaves of transgenic imidazolinone-tolerant sugarcane and its non-transgenic isolines. Arch Microbiol. 2010;192(4):307–313. 10.1007/s00203-010-0557-9 [DOI] [PubMed] [Google Scholar]
- 21.Fávaro LCL, Sebastianes FLS, Araújo WL. Epicoccum nigrum P16, a sugarcane endophyte, produces antifungal compounds and induces root growth. PLoS One. 2012;7(6):e36826 10.1371/journal.pone.0036826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson IC, Campbell CD, Prosser JI. Potential bias of fungal 18S rDNA and internal transcribed spacer polymerase chain reaction primers for estimating fungal biodiversity in soil. Environ Microbiol. 2003;5: 36–47. [DOI] [PubMed] [Google Scholar]
- 23.Azevedo JL, Araújo WL. Diversity and applications of endophytic fungi isolated from tropical plants In: GANGULI B. N.; DESMHMUKH S. K. (Ed.). Fungi: multifaceted microbes. Boca Raton: CRC Press; chap. 12, 2007; p. 191–209. [Google Scholar]
- 24.Schulz B, Boyle C. The endophytic continuum. Mycol Res. 2005;109: 661–686. [DOI] [PubMed] [Google Scholar]
- 25.Saikkonen K, Faeth SH, Helander M, Sullivan TJ. Fungal Eendophytes: a continuum of interactions with host plants. Annu Rev Ecol Syst. 1998;29: 319–343. [Google Scholar]
- 26.Sorensen J. The rhizosphere as a habitat for soil microorganisms In: VAN ELSAS J. D.; TREVORS J. T.; WELLINGTON E. M. H. (Ed.). Modern soil microbiology. New York: Marcel Dekker, 1997;p. 21–45. [Google Scholar]
- 27.Atkinson D, Watson CA (2000) The beneficial rhizosphere: a dynamic entity. Appl Soil Ecol. 15: 99–104. [Google Scholar]
- 28.Sylvia DM, Chellemi DO. Interactions among root inhabiting fungi and their implications for biological control of root pathogens. Adv Agron. 2001;73: 1–33. [Google Scholar]
- 29.Harvey PJ, Campanella BF, Castro PM, Harm H, Lichtfouse E, Schaffner AR, et al. Phytoremediation of polyaromatic hydrocarbons, anilines and phenols. Environ Sci Pollut Res Int. 2002;9: 29–47. [DOI] [PubMed] [Google Scholar]
- 30.Fravel D, Olivain C, Alabouvette C. Fusarium oxysporum and its biocontrol. New Phytologist. 2003;157: 493–502. [DOI] [PubMed] [Google Scholar]
- 31.Whipps JM, Lumsden RD. Commercial use of fungi as plant disease biological control agents: status and prospects In: Butt T, Jackson C, Magan N. (Ed.). Fungal Biocontrol Agents: progress, problems and potential. Wallingford: CABI Publishing, 2001;p. 9–22. [Google Scholar]
- 32.Howell CR. Effect of Gliocladium virens on Pythium ultimum, Rhizoctonia solani, and damping-off of cotton seedlings. Phytopathology. 1982;72: 496–498. [Google Scholar]
- 33.Tu JC. Gliocladium virens, a destructive mycoparasite of Sclerotinia sclerotiorum. Phytopathology. 1980;70: 670–674. [Google Scholar]
- 34.Aluko MO, Herring TF. The mechanisms associated with the antagonistic relationship between Corticium solani and Gliocladium virens. T Brit Mycol Soc. 1970;55: 173–179. [Google Scholar]
- 35.Baek J-M, Howell CR, Kenerley CM. The role of an extracellular chitinase from Trichoderma virens Gv29-8 in the biocontrol of Rhizoctonia solani. Curr Genet. 1999;35: 41–50. [DOI] [PubMed] [Google Scholar]
- 36.Howell CR. Understanding the mechanisms employed by Trichoderma virens to effect biological control of cotton diseases. Phytopathology. 2006;96: 178–180. 10.1094/PHYTO-96-0178 [DOI] [PubMed] [Google Scholar]
- 37.Tsavkelova EA, Aleksandrova AV, Cherdyntsera TA, Kolomeitseva GL, Netrusov AI. Fungi associated with the Vietnamian tropical orchids. Mikol Fitopatol. 2005;39: 46–52. [Google Scholar]
- 38.Colla G, Rouphael Y, Di Mattia E, El-Nakhel C, Cardarelli M.Co-inoculation of Glomus intraradices and Trichoderma atroviride acts as a biostimulant to promote growth, yield and nutrient uptake of vegetable crops. J Sci Food Agric. 2015;95(8):1706–1715. 10.1002/jsfa.6875 [DOI] [PubMed] [Google Scholar]
- 39.Martínez FD, Santos M, Carretero F, Marín F. Trichoderma saturnisporum, a new biological control agent. J Sci Food Agric. 2015. June 9 10.1002/jsfa.7301 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 40.Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1: 17–20. [Google Scholar]
- 41.White T, Burns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (Ed.). PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990; p. 315–322. [Google Scholar]
- 42.Tamura K, Dudley J, Nei M, Kumar S MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. [DOI] [PubMed] [Google Scholar]
- 44.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005; 71: 1501–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005; 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 46.Ter Braak CJF, Šmilauer P. CANOCO Reference Manual and CanoDraw for Windows User's Guide: Software for Canonical Community Ordination (version 4.5). Ithaca: Microcomputer Power; 2002; 500p. [Google Scholar]
- 47.De Groot MJA, Bundock P, Hooykaas PJJ, Beijersbergen AGM. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nature Biotechnol. 1998;16: 839–842. [DOI] [PubMed] [Google Scholar]
- 48.Araújo FDS, Fávaro LCL, Araújo WL, Oliveira FL, Aparicio R, Marsaioli AJ. Epicolactone—Natural product isolated from the sugarcane endophytic fungus Epicoccum nigrum. Eur J Org Chem. 2012;27: 5225–230. [Google Scholar]
- 49.Fitzgerald AM, Mudge AM, Gleave AP, Plummer KM. Agrobacterium and PEG-mediated transformation of the phytopathogen Venturia inaequalis. Mycol Res. 2003;107: 803–810. [DOI] [PubMed] [Google Scholar]
- 50.Stevens RB. Mycology Guidebook. Seatle: University of Washington Press, 1974; 703p. [Google Scholar]
- 51.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plantarum. 1962;15: 473–497. [Google Scholar]
- 52.Ferreira A, Quecine MC, Lacava PT, Oda S, Azevedo JL, Araújo WL. Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol Lett. 2008;287: 8–14. 10.1111/j.1574-6968.2008.01258.x [DOI] [PubMed] [Google Scholar]
- 53.Rossetto PB, Dourado MN, Quecine MC, Andreote FD, Araújo WL, Azevedo JL, et al. Specific plant induced biofilm formation in Methylobacterium species. Braz J Microbiol 2011;42:878–883. 10.1590/S1517-83822011000300006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, Yu M, Xu J, Du J, Ji F, Dong F, et al. Impact of transgenic wheat with wheat yellow mosaic virus resistance on microbial community diversity and enzyme activity in rhizosphere soil. PLoS One. 2014; 9(6):e98394 10.1371/journal.pone.0098394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pang SS, Duggleby RG. Expression, purification, characterization, and reconstitution of the large and small subunits of yeast acetohydroxyacid synthase. Biochemistry. 1999; 38: 5222–5231. [DOI] [PubMed] [Google Scholar]
- 56.Paungfoo-Lonhienne C, Yeoh YK, Kasinadhuni NR, Lonhienne TG, Robinson N, Hugenholtz P, et al. Nitrogen fertilizer dose alters fungal communities in sugarcane soil and rhizosphere. Sci Rep. 2015;5:8678 10.1038/srep08678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wyss GS, Charudattan R, Rosskopf EN, Littell RC. Effects of selected pesticides and adjuvants on germination and vegetative growth of Phomopsis amaranthicola, a biocontrol agent for Amaranthus spp. Weed Res. 2004;44: 469–482. [Google Scholar]
- 58.Hur M, Kim Y, Song HR, Kim JM, Choi YI, Yi H. Effect of genetically modified poplars on soil microbial communities during the phytoremediation of waste mine tailings. Appl Environ Microbiol. 2011: 77(21):7611–76119. 10.1128/AEM.06102-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Götz M, Nirenberg H, Krause S, Draeger HS, Buchner A, Lottmann J, et al. Fungal endophytes in potato roots studied by traditional isolation and cultivation-independent DNA-based methods. FEMS Microbiol Ecol. 2006;58: 404–413. [DOI] [PubMed] [Google Scholar]
- 60.Girlanda M, Bianciotto V, Cappellazzo GA, Casieri L, Bergero R, Martino E, et al. Interactions between engineered tomato plants expressing antifungal enzymes and nontarget fungi in the rhizosphere and phyllosphere. FEMS Microbiol Lett. 2008;288: 9–18. 10.1111/j.1574-6968.2008.01306.x [DOI] [PubMed] [Google Scholar]
- 61.De Azevedo LC, Stürmer SL, Lambais MR. Early changes in arbuscular mycorrhiza development in sugarcane under two harvest management systems. Braz J Microbiol. 2014;45(3):995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hart MM, Powell JR, Gulden RH, Dunfield KE, Pauls KP, Swanton CJ, et al. Separating the effect of crop from herbicide on soil microbial communities in glyphosate-resistant corn. Pedobiologia. 2009;52: 253–262. [Google Scholar]
- 63.Pasonen H-L, Lu J, Niskanen A-M, Seppänen S-K, Rytkönen A, Raunio J, et al. Effects of sugar beet chitinase IV on root-associated fungal community of transgenic silver birch in a field trial. Planta. 2009;230: 973–983. 10.1007/s00425-009-1005-4 [DOI] [PubMed] [Google Scholar]
- 64.Powell JR, Gulden RH, Hart MM, Campbell RG, Levy-Booth DJ, Dunfield KE, et al. Mycorrhizal and rhizobial colonization of genetically modified and conventional soybeans. Appl Environ Microbiol, 2007;73: 4365–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weinert N, Meincke R, Gottwald C, Heuer H, Gomes NCM, Schloter M, et al. Rhizosphere communities of genetically modified zeaxanthin-accumulating potato plants and their parent cultivar differ less than those of different potato cultivars. Appl Environ Microbiol. 2009;75: 3859–3865. 10.1128/AEM.00414-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sessitsch A, Gyamfi S, Tscerko D, Gerzabek MH, Kandeler E. Activity of microorganisms in the rhizosphere of herbicide treated and untreated transgenic glufosinate-tolerant and wild type oilseed rape grown in containment. Plant Soil. 2004;266: 105–116. [Google Scholar]
- 67.Tan F, Wang J, Feng Y, Chi G, Kong H, Qiu H, Wei S. Bt corn plants and their straw have no apparent impact on soil microbial communities. Plant Soil. 2009;329: 349–364. [Google Scholar]
- 68.Gomes NCM, Fagbola O, Costa R, Rumjanek NG, Buchner A, Mendona-Hagler L, Smalla K. Dynamics of fungal communities in bulk and maize rhizosphere soil in the tropics. Appl Environ Microbiol. 2003;69: 3758–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gamboa MA, Bayman P. Communities of endophytic fungi in leaves of tropical timber tree (Guarea guidonia: Meliaceae). Biotropica. 2001;33: 352–360. [Google Scholar]
- 70.Fröhlich J, Hyde KD, Petrini O. Endophytic fungi associated with palms. Mycol Res. 2000;104: 1202–1212. [Google Scholar]
- 71.Kumar DSS, Hyde KD. Biodiversity and tissue-recurrence of endophytic fungi in Tripterygium wilfordii. Fungal Divers. 2004;17: 69–90. [Google Scholar]
- 72.Santos RMG, Rodrigues-Fo E, Rocha WC, Teixeira MFS. Endophytic fungi from Melia azedarach. World J Microbiol Biotechnol. 2003;19: 767–770. [Google Scholar]
- 73.Bills GF. Isolation and analysis of endophytic fungal communities from woody plants In: REDLIN S. C, CARRIS L. M. (Ed.). Endophytic fungi in grasses and woody plants: systematics, ecology and evolution. St Paul: APS Press; 1996; p. 31–65. [Google Scholar]
- 74.Cui JL, Guo TT, Ren ZX, Zhang NS, Wang ML. Diversity and antioxidant activity of culturable endophytic fungi from alpine plants of Rhodiola crenulata, R. angusta, and R. sachalinensis. PLoS One. 2015;10(3):e0118204 10.1371/journal.pone.0118204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sridhar KR, Raviraja NS. Endophytes—a crucial issue. Curr Sci. 1995;69: 570–571. [Google Scholar]
- 76.Wang Y, Guo LD, Hyde KD. Taxonomic placement of sterile morphotypes of endophytic fungi from Pinus tabulaeformis (Pinaceae) in northeast China based on rDNA sequences. Fungal Divers. 2005;20: 235–260. [Google Scholar]
- 77.Zijlstra JD, Van’t Hof P, Baar J, Verkley GJM, Summerbell RC, Paradi I, et al. Diversity of symbiotic root endophytes of the Helotiales in ericaceous plants and the grass, Deschampsia flexuosa. Stud Mycol. 2005;53: 147–162. [Google Scholar]
- 78.Crozier J, Thomas SE, Aime MC, Evans HC, Holmes KA. Molecular characterization of fungal endophytic morphospecies isolated from stems and pods of Theobroma cacao. Plant Pathol. 2006;55: 783–791. [Google Scholar]
- 79.Zuccaro A, Schulz B, Mitchell J. Molecular detection of ascomycetes associated with Fucus serratus. Mycol Res. 2003;107: 1451–1466. [DOI] [PubMed] [Google Scholar]
- 80.Dunham SM, Larsson K-H, Spatafora JW. Species richness and community composition of mat-forming ectomycorrhizal fungi in old- and second-growth Douglas-fir forests of the HJ Andrews Experimental Forest, Oregon, USA. Mycorrhiza. 2007;17: 633–645. [DOI] [PubMed] [Google Scholar]
- 81.Maciá-Vicente JG, Jansson H-B, Abdullah SK, Descals E, Salinas J, Lopez-Llorca L V. Fungal root endophytes from natural vegetation in Mediterranean environments with special reference to Fusarium spp. FEMS Microbiol Ecol. 2008;64: 90–105. 10.1111/j.1574-6941.2007.00443.x [DOI] [PubMed] [Google Scholar]
- 82.Petrini O, Sieber TN, Toti L, Viret O. Ecology metabolite production, and substrate utilization in endophytic fungi. Natural Toxins. 1992;1: 185–196. [DOI] [PubMed] [Google Scholar]
- 83.Pereira JO, Carneiro-Vieira ML, Azevedo JL. Endophytic fungi from Musa acuminata and their reintroduction into axenic plants. World J Microbiol Biotechnol. 1999;15: 37–40. [Google Scholar]
- 84.Camatti-Sartori V, Azevedo JL, Sanhueza RMV, Ribeiro RTS, Echeverrigaray S. Endophytic yeasts and filamentous fungi associated with southern Brazilian apple (Malus domestica) orchards subjected to conventional, integrated or organic cultivation. J Basic Microbiol. 2005; 45: 397–402. [DOI] [PubMed] [Google Scholar]
- 85.Dal Bello GM, Sisterna MN, Monaco CI. Antagonistic effect of soil rhizosphere microorganisms on Bipolaris sorokiniana, the causal agent of wheat seedling blight. Int J Pest Manage. 2003;49: 313–317. [Google Scholar]
- 86.Fávaro LCL, Melo FL, Aguilar-Vildoso CI, Araújo WL. Polyphasic analysis of intraspecific diversity in Epicoccum nigrum warrants reclassification into separate species. PLoS One. 2011; 6: e14828 10.1371/journal.pone.0014828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cao LX, You JL, Zhou SN. Endophytic fungi from Musa acuminata leaves and roots in South China. World J Microbiol Biotechnol. 2002;18: 169–171. [Google Scholar]
- 88.Verma M, Brar SK, Tyagi RD, Surampalli RY, Valéro JR. Antagonistic fungi, Trichoderma spp.: Panoply of biological control. Biochem Eng J. 2007;37: 1–20. [Google Scholar]
- 89.Larran S, Perelló A, Simón MR, Moreno V. The endophytic fungi from wheat (Triticum aestivum L.). World J Microbiol Biotechnol. 2007;23: 565–572. [Google Scholar]
- 90.Vujanovic V, Hamelin RC, Bernier L, Vujanovic G, St-Arnaud M. Fungal diversity, dominance, and community structure in the rhizosphere of clonal Picea mariana plants throughout nursery production chronosequences. Microbial Ecol. 2007;54: 672–684. [DOI] [PubMed] [Google Scholar]
- 91.Orazova MK, Polyanskaya LM, Zvyagintsev DG. The structure of the microbial community in the barley root zone. Mikrobiologiya. 1999;68: 127–133. [Google Scholar]
- 92.El-Amin A-N, Saadabi AMA. Contribution to the knowledge of soil fungi in Sudan rhizosphere mycoflora of sugarcane at Kenana sugar Estate. International Journal of Botany. 2007; 3: 97–102. [Google Scholar]
- 93.Hawksworth DL. Fungal diversity and its implications for genetic resource collections. Stud Mycol. 2004;50: 9–18. [Google Scholar]
- 94.Seifert KA, Lévesque CA. Phylogeny and molecular diagnosis of mycotoxigenic fungi. Eur J Plant Pathol. 2004;110: 449–471. [Google Scholar]
- 95.Mendes R, Pizzirani-Kleiner AA, Araujo WL, Raaijmakers JM. Diversity of cultivated endophytic bacteria from sugarcane: genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl Environ Microbiol. 2007;73(22):7259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ricaud C., Egan B.T., Gillaspie A.G., Hughes C.G. Diseases of Sugarcane: Major Diseases. Elsevier, 2012;412p. [Google Scholar]
- 97.Britz H, Steenkamp ET, Coutinho TA, Wingfield BD, Marasas WF, Wingfield MJ. Two new species of Fusarium section Liseola associated with mango malformation. Mycologia. 2002;94: 722–730. [DOI] [PubMed] [Google Scholar]
- 98.Taylor JE, Hyde KD, Jones EBG. Endophytic fungi associated with the temperate palm, Trachycarpus fortunei, within and outside its natural geographic range. New Phytologist. 1999;142: 335–346. [Google Scholar]
- 99.Lodge DJ, Fisher PJ, Sutton BC. Endophytic fungi of Manikara bidentata leaves in Puerto Rico. Mycologia. 1996;88: 733–738. [Google Scholar]
- 100.Benhamou N, Garand C, Goulet A. Ability of nonpathogenic Fusarium oxysporum Strain Fo47 to induce resistance against Pythium ultimum infection in cucumber. Appl Environ Microbiol. 2002;68: 4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kubicek CP, Bissett J, Druzhinina I, Kullnig-Gradinger C, Szakacs G. Genetic and metabolic diversity of Trichoderma: a case study on South-East Asian isolates. Fungal Genet Biol. 2003;38: 310–319. [DOI] [PubMed] [Google Scholar]
- 102.Klein D, Eveleigh DE. Ecology of Trichoderma In: KUBICEK C. P, HARMAN G. E. (Ed.). Trichoderma and Gliocladium: basic biology, taxonomy and genetics. London: Taylor & Francis; chap. 3, 1998; 57–74. [Google Scholar]
- 103.Wardle DA, Parkinson D, Waller JE. Interspecific competitive interactions between pairs of fungal species in natural substrates. Oecologia. 1993;94: 165–172. [DOI] [PubMed] [Google Scholar]
- 104.Bailey BA, Bae H, Strem MD, Crozier J, Thomas SE, Samuels GJ, et al. Antibiosis, mycoparasitism, and colonization success for endophytic Trichoderma isolates with biological control potential in Theobroma cacao. Biol Control. 2008;46: 24–35. [Google Scholar]
- 105.De Souza JT, Bailey BA, Pomella AWV, Erbe EF, Murphy CA, Bae H, et al. Colonization of cacao seedlings by Trichoderma stromaticum, a mycoparasite of the witches’ broom pathogen, and its influence on plant growth and resistance. Biol Control, 2008;46: 36–45. [Google Scholar]
- 106.Photita W, Lumyong S, Lumyong P, Hyde KD. Endophytic fungi of wild banana (Musa acuminata) at Doi Suthep Pui National Park, Thailand. Mycol Res. 2001;105: 1508–1513. [Google Scholar]
- 107.Suryanarayanam TS, Vijaykrishna D. Fungal endophytes of aerial roots of Ficus benghalensis. Fungal Divers. 2001;8: 155–161. [Google Scholar]
- 108.Harman GE. Overview of mechanisms and uses of Trichoderma spp. Phytopathology. 2006;96: 190–194. 10.1094/PHYTO-96-0190 [DOI] [PubMed] [Google Scholar]
- 109.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species—opportunistic, avirulent plant symbionts. Nature Rev Microbiol. 2004;2: 43–56. [DOI] [PubMed] [Google Scholar]
- 110.Iqtedar M, Nadeem M, Naeem H, Abdullah R, Naz S, Syed QU, et al. Bioconversion potential of Trichoderma viride HN1 cellulase for a lignocellulosic biomass Saccharum spontaneum. Nat Prod Res. 2015; 29(11):1012–109. 10.1080/14786419.2014.971320 [DOI] [PubMed] [Google Scholar]
- 111.Huang J, Chen D, Wei Y, Wang Q, Li Z, Chen Y, et al. Direct ethanol production from lignocellulosic sugars and sugarcane bagasse by a recombinant Trichoderma reesei strain HJ48. ScientificWorldJournal. 2014;2014:798683 10.1155/2014/798683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Romão-Dumaresq AS, Araújo WL, Tabolt NJ, Thornton CR. RNA Interference of endochitinases in the sugarcane endophyte Trichoderma virens 223 Reduces its fitness as a biocontrol agent of pineapple disease. Plos One. 2012; 7(10): e47888 10.1371/journal.pone.0047888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rudresh DL, Shivaprakash MK, Prasad RD. Effect of combined application of Rhizobium, phosphate solubilizing bacterium and Trichoderma spp. on growth, nutrient uptake and yield of chickpea (Cicer aritenium L.). Appl Soil Ecol. 2005; 28: 139–146. [Google Scholar]
- 114.Doni F, Isahak A, Che Mohd Zain CR, Mohd Ariffin S, Wan Mohamad WN, Wan Yusoff WM. Formulation of Trichoderma sp. SL2 inoculants using different carriers for soil treatment in rice seedling growth. Springerplus. 2014;3:532 10.1186/2193-1801-3-532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martino E, Murat C, Vallino M, Bena A, Perotto S, Spanu P. Imaging mycorrhizal fungal transformants that express eGFP during ericoid endosymbiosis. Curr Genet. 2007;52: 65–75. [DOI] [PubMed] [Google Scholar]
- 116.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species—opportunistic, avirulent plant symbionts. Nature Rev Microbiol. 2004; 2: 43–56. [DOI] [PubMed] [Google Scholar]
- 117.Lo C-T, Nelson EB, Hayes CK, Harman GE. Ecological studies of transformed Trichoderma harzianum strain 1295–22 in the rhizosphere and on the phylloplane of creeping bentgrass. Phytopathology. 1998;88: 129–136. 10.1094/PHYTO.1998.88.2.129 [DOI] [PubMed] [Google Scholar]
- 118.Metcalf DA, Wilson CRT. The process of antagonism of Sclerotium cepivorum in white rot affected onion roots by Trichoderma koningii. Plant Pathology. 2001;50: 249–257. [Google Scholar]
- 119.Thrane C, Tronsmo A, Jensen DF. Endo-1,3-β-glucanase and cellulase from Trichoderma harzianum: purification and partial characterization, induction of and biological activity against plant pathogenic Pythium spp. Eur J Plant Pathol. 1997;103: 331–344. [Google Scholar]
- 120.Harman GE. Myths and dogmas of biocontrol changes in perceptions derived from research on Trichoderma harzianum T-22. Plant Dis. 2000;84: 377–393. [DOI] [PubMed] [Google Scholar]
- 121.Yedidia I, Benhamou N, Kapulnik Y, Chet I. Induction and accumulation of PR proteins activity during early stages of root colonization by the mycoparasite Trichoderma harzianum strain T-203. Plant Physiol Biochem. 2000;38: 863–873. [Google Scholar]
- 122.Viret O, Scheidegger C, Petrini O. Infection of beech leaves (Fagus sylvatica) by the endophyte Discula umbrinella (teleomorph: Apiognomoni aerrabunda): low temperature scanning electron microscopy studies. Can J Bot. 1993;71: 1520–1527. [Google Scholar]
- 123.Varma A, Verma S, Sahay N, Butehorn B, Franken P. Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl Environ Microbiol. 1999;65: 2741–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wagner BL, Lewis LC. Colonization of corn, Zea mays, by the entomopathogenic fungus Beauveria bassiana. Appl Environ Microbiol. 2000;66: 3468–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yedidia I, Srivastva AK, Kapulnik Y, Chet I. Effect of Trichoderma harzianum on microelement concentrations and increased growth of cucumber plants. Plant Soil. 2001;235: 235–242. [Google Scholar]
- 126.Tanaka A, Christensen MJ, Takemoto D, Park P, Scott B. Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell. 2006;18: 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fungal colonies grown in PDA broth at 28°C for 5–10 days. (a) Epicoccum sp., (b) Penicillium sp., (c) Chaetomium sp., (d) Fusarium sp., (e) Trichoderma virens, (f) Not identified root endophyte fungi, (g) Eupenicillium javanicum, (h) Acremonium sp., (i) Talaromyces trachyspermus, (j) Aspergillus niger, (k) Penicillium sp., (l) Epicoccum nigrum, (m) Mariannaea sp., (n) Fusarium sp., (o) Bionectria sp., (p) Myrmecridium schulzeri.
(DOCX)
Reference sequences of GenBank were used and the sequences obtained in the present work were signaled with the following symbol ♦. Bootstrap values (n = 1000) lower than 50 are represented in nods. a) 70 fungi of Penicillium genera, using the ascomycota Buergenerula spartinae as outgroup. b) 39 fungi of Fusarium genera, using the ascomycota Bionectria ochroleuca as outgroup. c) 17 fungi of Aspergillus genera, using the ascomycota Penicillium pinophilum as outgroup. d) 11 fungi of Trichoderma genera, using the ascomycota Hypomyces aurantius as outgroup.
(DOCX)
(a) Comparison between the expected number of phylotype for endophytic and root fungal population; (b) Number of phylotype expected for all fungal community associated to sugarcane (root endophyte + rhizosphere fungi).
(DOCX)
Genomic DNA of the isolates were digested with restriction enzyme EcoRI, which cuts T-DNA twice and do not cut gfp gene sequence, a electrophoresis were performed in 0.8% agarose gel, transferring to nylon membrane and hybridized with the 700 pb fragment (gfp gene) labeled with digoxigenine. Column 1 positive control (pFAT-gfp plasmid); column 2 is the negative control (Wild strain T.v.223); columns 3–6 are the transformed strains (T20, T10, T7 and T2). Visualized bands are indicated with arrow.
(DOCX)
(DOCX)
(DOCX)
Experimental design totally randomized. Analysis were performed separately for each part of the plant (leave, stem and root).
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.