Abstract
PIWI interacting RNAs (piRNAs), a member of non-coding RNA, originate from intergenic repetitive regions of the genome. piRNA expressions increase in various cancers and it is thought that this increase could be caused by hormones. We aimed to determine the effects of hormones on piRNA expression in breast and prostate cancer. High viability and a decrease in adhesion were observed at the concentrations of the highest proliferation. Furthermore, an increase in adhesion was also observed in MDA-MB-231 cells. After hormone treatment, while piR-651 expression had increased both breast and prostate cancer cell lines, piR-823 expressions increased in prostate cancer cell lines and only in the breast cancer cell line which was malignant. Thus, it was determined that piR-823 might show different expressions in different type of cancers.
Introduction
Gender dependent steroid hormones play an important role in the development and mechanism of cancer of the reproductive system, particularly in prostate cancer in males and uterus and breast cancer in females [1,2]. Androgen, a steroid hormone, plays an important role in the development of prostate cancer [3]. Prostate cancer develops in two ways, being either androgen-dependent or androgen-independent. Androgen-dependent prostate cancer cells require, in the early stages of the development of prostate cancer, the 5α-dihydrotestosterone to be converted from testosterone by the 5α-reductase enzyme system. Androgen-independent prostate cancer cells, however, are seen in the advanced stages of cancer development and do not need androgen in order to grow after these stages. The inefficacy of androgen in these types of cancer cells is associated with the changes, such as mutation, amplification or deletion, in the androgen receptor [2,4,5]. Breast cancer, the most common type of cancer after lung cancer, originates from cells in the tissues producing or carrying human breast milk, 80% of which are the epithelial layers of the lactiferous ducts [6] which contain estrogen receptors, and approximately 50 to 85% of breast tumors contain estrogen receptors and are seen in the cytosol [7].
The importance of non-coding RNAs in the development and prognosis of most diseases, particularly in cancer, has been increasing more and more, and the studies which have been carried out mark their importance as epigenetic regulators [8,9]. piRNAs and PIWI proteins are still being studied in order to obtain knowledge about their role in pathogenic mechanisms, such as tumorigenesis [10,11]. piRNAs maintain genome integrity by epigenetically silencing the transposons through DNA methylation, especially in gamete stem cells. piRNAs were identified in male germline cells during DNA methylation-mediated transposon silencing by affecting the expression of DNMT3, which is a DNA methyltransferase [10,12–14]. It is known that they are highly expressed at a length of 26–31nt and in the testes. In mammals, piRNAs were first identified in 2006 in the cloning of small RNAs associated with PIWI proteins in the mammalian system [15–17]. piRNAs act as a sequence-specific guide for PIWI proteins, and the PIWI protein-piRNA pathway in gamete stem cells is thought to be important for DNA methylation-mediated transposon silencing [12,13,18,19]. Genetic studies in mice, Drosophila, and Zebrafish reported that piRNAs are expressed more than any other short non-coding RNAs during germinal development and in the testes [20–22]. Though piRNAs are mostly thought to be specific to germline cells, recent studies have shown that they also play important roles in non-gonadal cells [10,23–25], such as in long-term memory functions, DNA methylation, epigenetic regulations, the silencing of transposable and non-transposable elements, and the stability of primary cells composing the gamete cells [26].
Our study investigates the piRNAs in hormone-dependent and hormone-independent cancer cells, and its aim is to determine whether these piRNAs play effective roles only in hormone-dependent cancers or the same is true for hormone-independent cancers as well. For this purpose, we treated androgen-dependent and androgen-independent prostate cancer cells (LNCaP and PC-3) with an androgen hormone, and estrogen-dependent and estrogen-independent breast cancer cells (MCF-7 and MDA-MB-231) with an estrogen hormone to investigate cell proliferation, viability, adhesion and hormone expression. Moreover, the present study identifies the expression of piRNA-651 (piR-651) and piRNA-823 (piR-823), which are among the piRNAs thought to be effective in gender-dependent cancers.
Materials and Methods
Cell Culture and Concentration of Androgen and Estrogen Hormones
Prostate cancer cell lines (LNCaP and PC-3) and breast cancer cell lines (MCF-7 and MDA-MB-231) were purchased from the American Type Culture Collection (ATCC, Washington D.C., USA), and the cells were incubated at 37°C with 5% CO2 in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, United Kingdom) containing 1% penicillin/streptomycin (Gibco, UK) and 10% foetal bovine serum (FBS) (Gibco, UK). Before hormone treatment, the breast and prostate cancer cells were incubated in 6-well plates (Greiner, Germany) to make sure the cells grew to 80% confluence. Estrogen (E2257) and androgen (A0887) were obtained from Sigma (St. Louis, USA). To observe the appropriate concentration, hormones with certain concentration ranges were used to treat cells according to the instructions of the manufacturer. Using the XTT method (Biological Industries, Israel), the most proliferative concentration and hours were determined to be the 24th, 48th and 72nd hours. By deciding on the optimal concentration of hormones and optimal time, viability, proliferation, adhesion and piRNA expression, assays were applied to these specified conditions.
Viability, Proliferation and Adhesion
The cell viability rate was determined using the trypan blue method. Hormone treated cells were exposed to trypan blue (Gibco, UK) after being trypsinized from a flask. The live and dead cells were counted using the Thoma Slide (Marienfeld, Germany).
The XTT Kit (Biological Industries, Israel) was used to determine the impacts of hormones on the proliferation of breast and prostate cancer cell lines. In brief, the cells were plated into 96-well plates (Greiner, Germany) and, after optimal hormone treatment, XTT was pipetted to all wells and cultured at 37°C with 5% CO2 for 2 hours. The microplate spectrophotometer (Lab Systems, Finland)) was used to determine the absorbance at 450nm.
The XTT Kit (Biological Industries, Israel) was again used, this time to determine the impacts of hormones on the adhesion of breast and prostate cancer cell lines, and, as before, the cells were plated into 96-well plates (Greiner, Germany). After optimal hormone treatment, the wells were flushed three times with phosphate-buffered saline (PBS; Sigma, Saint Louis, USA). XTT was then pipetted to all wells and cultured at 37°C with 5% CO2 for 2 hours, and the microplate spectrophotometer (Lab Systems, Finland)) was used to determine the absorbance at 450nm.
Real Time Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated from cells using the Paris Total RNA Isolation Kit (Ambion, Carlsbad, USA) in accordance with the instructions of the manufacturer. The RNAs isolated from the hormone treated cells were converted to cDNA through a reverse transcription (Bioneer, Alameda, USA). Primer sets for the amplification of piR–651, piR–823 and Glyseraldehide-3-phosphate dehydrogenase (GAPDH) were designed and supplied by Alpha DNA, Montreal, Quebec. RT-PCR was carried out inside a Strategene MxPro3000 (Strategene, UK). GAPDH (Alpha DNA, Montreal, Quebec) was used as an internal control, and the expression of piR–651 and piR–823 was normalized in line with the expression of GAPDH. Gene expression changes were quantified using the delta-delta CT method. The forward and reverse primer sequences used in RT-PCR are shown in Table 1 [10,25,27].
Table 1. The forward and reverse primer sequences used in RT-PCR.
| Primer | Forward Primer | Reverse Primer |
|---|---|---|
| piR-651 | 5′-AGAGAGGGGCCCGTGCCTTG-‘3 | 3’-CCAGTCTCAGGGTCCGAGGTATTC-‘5 |
| piR-823 | 5′-AGCGTTGGTGGTATAGTGGT-‘3 | 3’-CCAGTCTCAGGGTCCGAGGTATTC-‘5 |
| GAPDH | 5’-CGAGGGGGGAGCCAAAAGGG-‘3 | 3’-GAAACTGCGACCCCGACCGT-‘5 |
Statistical Analysis
A normal distribution of the continuous variables was enabled using the Kolmogorov-Smirnov suitability test. Comparisons between groups of normally distributed variables were evaluated using One-Way variance analysis (ANOVA). The Tukey HSD test was used for multiple comparisons. Comparisons between groups of variables which were not normally distributed were evaluated using the Kruskal-Wallis test. Multiple comparisons of these groups were evaluated using the Dunn test, while normally distributed piRNA values were compared using the Student t test. All analyses were carried out using the IBM SPSS Statistics 21.0 software package (S1–S10 Files). The obtained data were indicated as mean ± standard deviation (sd). In the figures, only mean values have been shown.
Results
Determining the Optimal Hormone Concentrations on Prostate and Breast Cancer Cell Lines
LNCaP androgen-dependent prostate cancer cells were treated with androgen at concentrations of 200nM, 100nM, 50nM, 25nM, 10nM, 1nM, and 0.1nM. Ethanol was used to dissolve the androgen and estrogen hormones in the medium, an additional ethanol group being formed for this reason. In the measurement carried out 24 hours after treatment with the hormone, statistically significant increases were observed in the 10nM androgen group (847229±19949) (P < 0.05; data not shown) and the 1nM androgen group (915657±25069) (P < 0.01; data not shown) when compared with the control group (156729±22278). The PC-3 cells were treated with 10nM, 1nM and 0.1nM androgen, and, when compared with the control group (41961±907), there was a statistically significant decrease in the 1nM androgen group (25990±5334) (P < 0.05; data not shown). The MCF–7 cells were treated with 10nM, 1nM and 0.1nM estrogen, but no statistically significant difference was found between the groups (P >0.05; data not shown). The MDA-MB–231 cells were treated with 10nM, 1nM and 0.1nM estrogen, but no statistically significant difference was found between the groups (P >0.05; data not shown).
The Effects of Optimal Hormone Concentrations on Proliferation, Viability, and Adhesion of Prostate and Breast Cancer Cells
When compared with the control group (156728.57±58943.07), a significant increase in the number of cells in the 1nM androgen group (915657.14±66326.66) was detected in the LNCaP cells (P < 0.001). While cells were detected in the control group (41961.43±2398.41) and in the 10nM androgen group (40561.43±5547.29) of PC-3 cells, but no statistically significant difference was found between the groups (P >0.05). In the MCF-7 cells, an increase was detected in the proliferation of the 10nM estrogen group (97704.71±7623.37) when compared with the ethanol group (82252.29±14261.99) (P < 0.05). In the MDA-MB-231 cells, white cells were detected in the control group (20816.71±7496.41) and 1nM estrogen group (32489.29±10916.76) (Table 2) (P <0.05).
Table 2. The Proliferation Values of the LNCaP, PC-3, MCF-7 and MDA-MB-231 cells.
| PROLIFERATION | ||||
|---|---|---|---|---|
| Group | Mean ±sd | p value | Multiple comparisons and p value | |
| LNCaP | CONTROL(1) | 156728.57±58943.07 | 0.001† | 1–3 p = 0.003†† |
| ETHANOL(2) | 166157.14±68929.74 | 2–3 p = 0.008†† | ||
| 1 nM ANDROGEN(3) | 915657.14±66326.66 | |||
| PC-3 | CONTROL(1) | 41961.43±2398.41 | 0.066* | - |
| ETHANOL(2) | 34061.43±9023.99 | |||
| 10 nM ANDROGEN(3) | 40561.43±5547.29 | |||
| MCF-7 | CONTROL(1) | 83514.29±9812.17 | 0.029* | 2–3 p = 0.041** |
| ETHANOL(2) | 82252.29±14261.99 | |||
| 10 nM ESTROGEN(3) | 97704.71±7623.37 | |||
| MDA-MB-231 | CONTROL(1) | 20816.71±7496.41 | 0.048† | 1–3 p = 0.018†† |
| ETHANOL(2) | 27525±7442.84 | |||
| 1 nM ESTROGEN(3) | 32489.29±10916.76 | |||
*: One-Way ANOVA,
**: Tukey HSD Test,
†:Kruskal-Wallis Test,
††:Dunn Test.
All obtained data were compared with the control group (n = 7 for each cell line)
In the LNCaP cells, a statistically significant increase was detected in the viable cell count percentage (86.75±6.84) of the 1nM androgen group when compared with the control (80.88±9.92) and ethanol groups (68±9.21) (P < 0.001). In the PC-3 cells, while the viable cell count percentage was detected to have increased in the 10nM androgen group (78.75±20.83) when compared to the ethanol group (73.88±19.48), no statistically significant difference was found between the groups (P >0.05). It was observed that the treatment of the MCF-7 cells with estrogen increased the viable cell count percentage of the 10nM estrogen group (86.75±9.54) when compared with the control (82.13±12.41) and ethanol (71.88±8.27) groups (P < 0.05). In the MDA-MB-231 cells, while the viable cell count % was detected to have increased in the 1nM estrogen group (86.13±15.41) when compared with the control (72.88±29.53) and ethanol (77±21.58) groups, no statistically significant difference was found between the groups (Table 3) (P >0.05).
Table 3. The Cell Viability Rate of LNCaP, PC-3, MCF-7 and MDA-MB-231.
| VIABILITY | ||||
|---|---|---|---|---|
| Group | Mean±sd | p value | Multiple comparisons and p value | |
| LNCaP | CONTROL(1) | 80.88±9.92 | 0.001* | 1–2 p = 0.020** |
| ETHANOL(2) | 68±9.21 | 1–3 p = 0.001** | ||
| 1 nM ANDROGEN(3) | 86.75±6.84 | |||
| PC-3 | CONTROL(1) | 86.13±15.41 | 0.406† | - |
| ETHANOL(2) | 73.88±19.48 | |||
| 10 nM ANDROGEN(3) | 78,75±20.83 | |||
| MCF-7 | CONTROL(1) | 82.13±12.41 | 0.025* | 2–3 p = 0.022** |
| ETHANOL(2) | 71.88±8.27 | |||
| 10 nM ESTROGEN(3) | 86.75±9.54 | |||
| MDA-MB-231 | CONTROL(1) | 72.88±29.53 | 0.588† | - |
| ETHANOL(2) | 77±21.58 | |||
| 1 nM ESTROGEN(3) | 86.13±15.41 | |||
*: One-way ANOVA,
**: Tukey HSD test,
†: Kruskal-Wallis test.
All obtained data were compared with the control group (n = 8 for each cell line)
In the LNCaP cells, it was observed that adhesion decreased in the 1nM androgen group (0.79±0.12) when compared with the control group (0.93±0.09) (P < 0.05), while in the PC-3 cells, a statistically significant increase was detected in the adhesion in ethanol (0.86±0.06) and the 10nM androgen group (0.84±0.06) when compared with the control group (0.74±0.05) (P < 0.01). In the MCF-7 cells, while it was observed that adhesion increased in the 10nM estrogen group (1.18±0.58) when compared to the control (0.69±0.17) and ethanol (0.66±0.12) groups, no statistically significant difference was found between the groups (P >0.05). In the MDA-MB–231 cells, it was observed that adhesion decreased in the 1nM estrogen group (0.69±0.05) when compared to the control (0.74±0.06) and ethanol (0.70±0.03) groups, but no statistically significant difference was found between the groups (Table 4) (P >0.05). Based on these results, it was determined that adhesion decreased in the LNCaP cells treated with androgen while it increased in PC-3 cells.
Table 4. The Adhesion Values of LNCaP, PC-3, MCF-7 and MDA-MB-231 cells.
| ADHESION | ||||
|---|---|---|---|---|
| Group | Mean±sd | p value | Multiple comparisons and p value | |
| LNCaP | CONTROL(1) | 0.93±0.09 | 0.018† | 1–3 p = 0.048†† |
| ETHANOL(2) | 0.90±0.07 | 2–3 p = 0.037†† | ||
| 1 nM ANDROGEN(3) | 0.79±0.12 | |||
| PC-3 | CONTROL(1) | 0.74±0.05 | 0.001* | 1–2 p = 0.002** |
| ETHANOL(2) | 0.86±0.06 | 1–3 p = 0.008** | ||
| 10 nM ANDROGEN(3) | 0.84±0.06 | |||
| MCF-7 | CONTROL(1) | 0.69±0.17 | 0.077† | - |
| ETHANOL(2) | 0.66±0.12 | |||
| 10 nM ESTROGEN(3) | 1.18±0.58 | |||
| MDA-MB-231 | CONTROL(1) | 0.74±0.06 | 0.2* | - |
| ETHANOL(2) | 0.70±0.03 | |||
| 1 nM ESTROGEN(3) | 0.69±0.05 | |||
*: One-way ANOVA,
**: Tukey HSD test,
†: Kruskal-Wallis test,
††: Dunn test.
All obtained data were compared with the control group (n = 7 for each cell line)
Determining Expressions of piR-651 and piR-823 in Optimal Hormone Concentrations for Prostate and Breast Cancer Cells
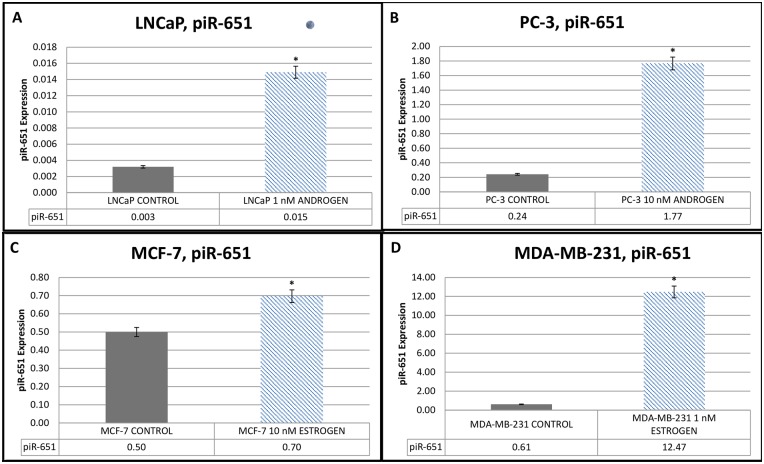
Statistically significant increases in the expression levels of piR-651 were observed in the 1nM androgen group (0.015±0.0002) when compared with the control group (0.003±0.0002) for the LNCaP cells (Fig 1A), in the 10nM androgen group (1.77±0.0002) when compared with the control group (0.24±0.0002) for the PC-3 cells (Fig 1B), in the 10nM estrogen group (0.7±0.0002) when compared with the control group (0.5±0.0002) for the MCF–7 cells (Fig 1C), and in the 1nM estrogen group when compared with the control group (0.61±0.0002) for the MDA-MB–231 cells (Fig 1D) (P < 0.001).
Fig 1. piR-651 Expressions of androgen dependent and independent prostate cancer cell lines and estrogen-dependent and estrogen-independent breast cancer cell lines.
(A) piR-651 Expression of androgen-dependent LNCaP cells before and after 1nM androgen hormone treatment. (B) piR-651 Expression of androgen-independent PC-3 cells before and after 10nM androgen hormone treatment. (C) piR-651 Expression of estrogen-dependent MCF-7 cells before and after 10nM estrogen hormone treatment. (D) piR-651 Expression of estrogen-independent MDA-MB-231 cells before and after 1nM estrogen hormone treatment. All obtained data were compared with the control group *P < 0.001. (n = 7 for each cell line).
Statistically significant increases in the expression levels of piR-823 were observed in the 1nM androgen group (0.018±0.0002) when compared with the control group (0.005±0.0002) for the LNCaP cells (Fig 2A), in the 10nM androgen group (1.24±0.0002) when compared with the control group (0.56±0.0002) for the PC-3 cells (Fig 2B), and in the 1nM estrogen group (9.51±0.0002) when compared with the control group (2.04±0.0002) for the MDA-MB–231 cells (Fig 2D) (P < 0.001). Contrary to the other cell lines, a statistically significant decrease was observed in the level of piR-823 expression in the 10nM estrogen group (0.27±0.0002) when compared with the control group (0.54±0.0002) for the MCF–7 cells (Fig 2C) (P < 0.001).
Fig 2. piR-823 Expression of androgen-dependent and androgen-independent prostate cancer cell lines and estrogen-dependent and estrogen-independent breast cancer cell lines.
(A) piR-823 Expression of androgen-dependent LNCaP cells before and after 1nM androgen hormone treatment. (B) piR-823 Expression of androgen-independent PC-3 cells before and after 10nM androgen hormone treatment. (C) piR-823 Expression of estrogen-dependent MCF-7 cells before and after 10nM estrogen hormone treatment. (D) piR-823 Expression of estrogen-independent MDA-MB-231 cells before and after 1nM estrogen hormone treatment. All obtained data were compared with the control group *P < 0.001. (n = 7 for each cell line).
Discussion
In recent times, studies investigating the relationship between piRNAs and cancer have been increasing rapidly. In our study, the effects of hormones in a medium containing cancer cells on piRNAs were determined. The literature review which was carried out to determine the hormone concentrations with which the cancer cells would be treated yielded varied information. In previous studies on an androgen-dependent prostate cancer cell line LNCaP, the cells were treated with androgen at concentrations ranging from 1nM to 10nM. [7,28–31]. In our study, we detected that the concentration at which androgen increased cell proliferation the greatest amount was 1nM at the 24th hour for the androgen-dependent prostate cancer cell line LNCaP. Considering the possibility of interaction with hormones, we did not use serum in our study. The absence of serum causes a shortage in the cells and induces other mechanisms [32–38]. Moreover, several previous studies reported that ethanol in cancer cells induced proliferation by increasing the cyclic AMP (cAMP) level after a certain period of time [39–43]. For this reason, the duration of a serum-free trial was discovered to be 24 hours. While we were planning the study, we wanted to compare the piRNAs which have known expression profiles in other cancers. Therefore, we gave priority to the comparison of piR-651 and piR-823 expressions in breast and prostate cancer when androgen and estrogen hormones were both present and absent. On the other hand, we also wanted to observe the effect of androgen and estrogen on the viability, proliferation and adhesion of breast and prostate cancer cell lines. These assays concerning cell behaviour can be carried out in further studies.
Androgen-independent cancer cells were treated with an androgen hormone at concentrations ranging from 10−13 to 10-7nM [44–46]. However, since PC-3 prostate cancer cells are androgen-independent cells, they did not present significant responses to the androgen used in the studies. Based on the data we obtained, we determined that the concentration at which the highest proliferation for PC-3 cells treated with androgen was observed to be 10nM at the 24th hour.
In the studies investigating the effects of estrogen on MCF–7 and MDA-MB–231 breast cancer cells, the cells were treated with androgen hormones at various concentrations and the effects of this treatment on cell proliferation was investigated. The studies showed that the optimum concentration of estrogen was between 0.01 and 10nM for MCF-7 cells [38,47–51], and the conclusion was that cell proliferation was equal at each one of a variety of concentrations for MDA-MB–231 cells and that estrogen did not have any effects on these cells [35,49,50]. Based on the data we obtained, we determined that the concentration at which the highest proliferation for MCF-7 cells treated with estrogen was observed to be 10nM at the 24th hour. For the estrogen-independent cancer cell line MDA-MB–231, 1nM at the 24th hour was determined to be the concentration of the highest proliferation.
In order to determine the effects of the substances and/or chemicals with which the cells are treated on the cell count, cell proliferation assays are carried out in the studies on the diagnosis or treatment of most diseases. According to previous studies, the androgen hormone increases the survival and proliferation of androgen-dependent prostate cancer cells [28,46,52,53]. The studies investigating the prostate cancer cell line LNCaP argue that tumor development is correlated with androgen levels, and report that cell proliferation depends on the androgen level [7,28,46]. In our study, increases were observed in the proliferation and viable cell count of LNCaP cells treated with 1nM androgen. The studies determined that androgen is not effective in the proliferation and viability of PC-3 cells [44,46,54]. As with these studies, we also determined that the androgen with which the cells were treated was not effective for PC-3 cells in terms of proliferation and viability.
The proliferation, development, transcriptional and translational mechanisms of breast cancer cells are regulated by many signal molecules and cell cycles, among which hormones also play important roles. Various studies have reported that estrogen at various concentrations caused an increase in the proliferation and viability of the estrogen-dependent breast cancer cell line MCF-7 [33,34,37,38,48,51,55]. Our study shows that 10nM estrogen may be effective in the proliferation and viability of MCF–7 cells. MDA-MB–231 cells are estrogen-independent cells and studies have revealed that estrogen at various concentration had effects on the proliferation and viability of the cells [36,56,57]. The data obtained in our study shows that estrogen does not affect the proliferation and viability of MDA-MB–231 cells.
Adhesion is important both pathologically and physiologically in cancer and inflammatory diseases. The effectiveness of anti-cancer drugs administered to tumor tissue is affected by cell adhesion and the density of the tumor cells [3]. The application of androgen into the medium increased metastatic characteristics and reduced adhesion (LNCaP, LNCaP TβRII and PC-3) [52]. Contrary to these studies, it was determined that 10nM androgen increased the adhesion of PC-3 cells. Sapino A. et al. found increases in the adhesion plaques of MCF-7 cells treated with estrogen, as well as increases in the number of adhesion-specific structures [58]. Based on the data we obtained, the estrogen with which both breast cancer cell lines were treated had no statistically significant effect on the adhesion of cells.
The level of piRNAs is high in both the normal gamete stem cells and cancer cells. A study by Yang Q. et al. identified 770 known miRNA and 5 new miRNA lines, as well as 20121 piRNA lines in healthy human testes. In this study, common line matches were found between piRNAs and miRNAs by using the advanced sequencing (Next Generation Sequencing—NGS) method. The study by Yang et al. showed that miRNAs and piRNAs act as regulators in spermatogenesis, and it is a useful source for studies in this field [59]. In their study, Kang et al. administered testosterone hormone to the testes of healthy male rats and measured piRNA expressions at certain time intervals. They eventually determined that testosterone increased the expression of the PIWI protein and piRNA in the testis [60].
In their study, Chu H. et al. investigated the single nucleotide polymorphism (SNP) of the sites containing piRNA clusters in the development of colorectal cancer. They determined that individuals that have the polymorphism of piR-01551 and the long non-coding RNA site lnc00964-3 had a high risk of developing colorectal cancer [61]. The results of microarray and other functionality assays of hepatocellular carcinoma revealed that the expression of the gene site piR-Hep1 increased by 46.6% in hepatocellular carcinoma tissue when compared with healthy tissue. In the same study, it was argued that silencing of the gene site piR-Hep1 prevented the cancer cells from proliferating, migrating, and attacking the healthy tissue [62]. Another study on bladder cancer, however, showed that piRNAs play a role in tumor development [63]. Hashim et al suggested that expression of at least three piRNAs (DQ597945, DQ570994 and DQ598651) was found to be significantly different in estrogen positive and negative breast cancer cell lines. In their study, it was also found that eight piRNAs deregulated in the breast cancer tumors’ encode proteins involved in key cancer cell functions. All of their data showed that piRNAs represent a new perspective in the identification of estrogen levels in hormone responsive breast cancer types [64]. In another study, it was suggested that piRNA expression profiles might be evidence supporting a potentially tissue-specific role for piRNAs in both tumor and non-malignant somatic tissues that may be clinically relevant. Furthermore, in this study, it was also shown that piRNA expression can delineate clinical features, such as histological subgroups, stages, and survival. It was indicated that 6260 piRNA transcriptome profiles were summarized according to tumor and non-malignant tissue types from 11 organs (bladder, breast, colon, head and neck, kidney, lung, ovaries, prostate, stomach, thyroid, and uterine corpus) [65].
It is postulated that the piR-651 and piR-823 that we studied take part in the development of various cancers, especially gastric cancer [10,25,27,66]. In their study, Yan H. et al. detected an increase in the expression of piR-823 in the tissue samples and cell lines of patients with multiple myeloma, and it was also observed that silencing of piR-823 in multiple myeloma cell lines resulted in dysfunction of the elements playing roles in the expression of protein associated with apoptosis and in the regulation of the cell cycle. Additionally, it was determined that piR–823 was directly associated with DNMT3A and 3B de novo DNA methyltransferases [67]. As with the data obtained by Yan H. et al. for multiple myeloma, it was found that silencing of piR-823 in the therapeutic xenograft models effectively inhibited tumor growth [67]. Another study, on the other hand, reported that piR-823 expression decreased in gastric cancer and displayed tumor suppressing properties both in vivo and in vitro [27]. The piR-823 expression that displays different properties (heterogenic, tumor suppressing or oncogenic) in different cancer types is thought to play a dual role in tumor formation. The data we obtained shows that androgen increases the piR-823 expression in LNCaP and PC-3 prostate cancer cells. The treatment of MDA-MB–231 cells with estrogen increases the piR–823 expression. Increasing the hormone levels in cancer cells by external administration of the hormones into these cells revealed that the piR-823 expression displayed oncogenic properties in prostate cancer cell lines and malignant estrogen-independent breast cancer lines, and that this property increased with the increase in hormone levels. The treatment of MCF–7 breast cancer cells with estrogen reduces the piR–823 expression. This result shows that piR-823 may have a different kind of expression in benign estrogen-dependent breast cancer cells treated with hormones. It was observed that therapy with the external administration of estrogen hormone had a positive effect in benign estrogen-dependent MCF-7 cancer cells and caused a decrease in the normally high piR-823 expression.
In our study, the piRNA site is thought to be the one that plays a role in piR-651 cancer development. Previous studies showed that the piR-651 expression increased in various tumors, including gastric, colon, lung and breast tumors, when compared to healthy tissues [10]. Additionally, there is clinical data supporting the notion that increased expression of the piR-651 and PIWI protein during cancer development may play a role in a variety of cancer types, including gastric, pancreas and oesophageal cancer [68–70]. A recent study argued that piR–651 may play a part in the metastasis of gastric tumor cells to the lymph nodes [10]. It was observed that cancer cell growth decreased when piR-651 was silenced in gastric cancer [25]. These studies are among the studies carried out to determine piRNA expression levels in cancer [71]. Moreover, it was also investigated whether the piR-651 expression increased in mesothelioma, hepatocellular, and cervical cancer [10].
Upon the determination of the concentrations at which the externally administered hormones increased cell proliferation the most, piRNA expressions were evaluated in these cells. It was observed that the viability was also at its highest level at the highest proliferation concentrations of the externally administered hormones while adhesion was reduced. On the contrary, a decrease in adhesion was detected in MDA-MB-231 cells. While it was observed that the expression of piR-651 and piR-823 increased upon administration, only the piR-823 expression was detected to have decreased in the estrogen-dependent breast cancer cell line MCF-7.
This study showed that piRNAs, which are becoming increasingly important today, may be affected by hormones. In general, it was observed that cell proliferation and viability increased with androgen or estrogen treatment and that cell adhesion decreased in cells other than PC-3 cells. It was detected that the piR-651 expression increased in LNCaP and PC-3 prostate cancer cells treated with androgen and MCF-7 and MDA-MB-231 breast cancer cells treated with estrogen. The results of this study show that the increase in the expression of piR-651 and piR-823 during gonad development and in cancer may be associated with hormone levels in some cancer cells, and that these piRNA expressions vary depending on the hormone level and cancer cell type in the microenvironment of the cancer. We think that our study will contribute greatly to future studies on piRNAs and may be a source for the consideration of these properties in hormone therapies for some cancer types. More molecular studies are needed in order to obtain detailed information about certain results for the mechanisms of piRNAs in cancer.
Supporting Information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
Çağrı ÖNER: corresponding and first author, took part in editing the study, experimental steps (cell culture, hormone treatment, cell viability, proliferation, adhesion assays, total RNA isolation, reverse transcription, RT-PCR piRNA expression) and the evaluation of the results.
Didem TURGUT COŞAN: author, took part in editing the study, experimental steps (cell culture, hormone treatment, cell viability, proliferation, adhesion assays, total RNA isolation, reverse transcription, RT-PCR piRNA expression) and the evaluation of the results.
Ertuğrul ÇOLAK: took part in the statistical evaluation of the results.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by Eskisehir Osmangazi University Scientific Research Project Commission (Project Number: 2014-576).
References
- 1.Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, Imai Y, et al. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 2011;20 (1): 119–131. 10.1016/j.ccr.2011.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldman BJ, Feldman D. The Development of Androgen-independent Prostate Cancer. Nat Rev Cancer. 2001;1 (1): 34–45. [DOI] [PubMed] [Google Scholar]
- 3.Palmer CP, Mycielska ME, Burcu H, Osman K, Collins T, Beckerman R, et al. Single cell adhesion measuring apparatus (SCAMA): application to cancer cell lines of different metastatic potential and voltage-gated Na+ channel expression. Eur Biophys J. 2008;37 (4): 359–368. [DOI] [PubMed] [Google Scholar]
- 4.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122 (4): 553–563. [DOI] [PubMed] [Google Scholar]
- 5.Litvinov IV, De Marzo AM, Isaacs JT. Is the Achilles’ heel for prostate cancer therapy a gain of function in androgen receptor signaling? J Clin Endocrinol Metab. 2003;88: 2972–2982. [DOI] [PubMed] [Google Scholar]
- 6.Kierszenbaum AL (2006) Histology and Cell Biology—An Introduction to Pathology. Demir R, translator. Maryland, USA: Mosby. [Google Scholar]
- 7.Horoszewicz JS. LNCaP Model of Human Prostatic Carcinoma. Cancer Research. 1983;43: 1809–1818. [PubMed] [Google Scholar]
- 8.Zhou H, Hu H, Lai M. Non-coding RNAs and their epigenetic regulatory mechanisms. Biol Cell. 2010;102: 645–655. 10.1042/BC20100029 [DOI] [PubMed] [Google Scholar]
- 9.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12 (12): 861–874. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- 10.Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, et al. PiRNA, the new non-coding RNA is aberrantly expressed in human cancer cells,. Clinica Chimica Acta. 2011;412 (17–18): 1621–1625. [DOI] [PubMed] [Google Scholar]
- 11.Mei Y, Clark D, Mao L. Novel dimensions of piRNAs in cancer. Cancer Lett. 2013;336 (1): 46–52. 10.1016/j.canlet.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aravin AA, Bourc'his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22 (8): 970–975. 10.1101/gad.1669408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31: 785–799. 10.1016/j.molcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332 (6031): 848–852. 10.1126/science.1203919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442 (7099): 203–207. [DOI] [PubMed] [Google Scholar]
- 16.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germlinespecific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442: 199–202. [DOI] [PubMed] [Google Scholar]
- 17.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20 (13): 1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316 (5825): 744–747. [DOI] [PubMed] [Google Scholar]
- 19.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & Development. 2008;22 (7): 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116 (6): 817–829. [DOI] [PubMed] [Google Scholar]
- 21.Carmell MA, Girard A, van de Kant HJG, Bourc'his D, Bestor TH, de Rooij DG, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental Cell. 2007;12 (4): 503–514. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Pane A, Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17 (7): 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Q, Ma Q, Shehadeh LA, Wilson A, Xia L, Yu H, et al. Expression of the Argonaute protein PiwiL2 and piRNAs in adult mouse mesenchymal stem cells. Biochem Biophys Res Commun. 2010;396 (4): 915–920. 10.1016/j.bbrc.2010.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes & Development. 2012;26 (21): 2361–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui L, Lou YR, Zhang XJ, Zhou H, Deng HX, Song HJ, et al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clinical Biochemistry. 2011;44 (13): 1050–1057. 10.1016/j.clinbiochem.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 26.Pandya GM, Ramani UV, Janmeda M, Dangar NS, Tyagi K, Brahmkshtri BP, et al. piRNA: Basics and their Association with PIWI proteins. Current Trends in Biotechnology and Pharmacy. 2014;8 (3): 303–308. [Google Scholar]
- 27.Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, et al. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315 (1): 12–17. 10.1016/j.canlet.2011.10.004 [DOI] [PubMed] [Google Scholar]
- 28.Lee C, Sutkowski DM, Sensibar JA, Zelner D, Kim I, Amsel I, et al. Regulation of proliferation and production of prostate-specific antigen in androgen-sensitive prostatic cancer cells, LNCaP, by dihydrotestosterone. Endocrinology. 1995;136 (2): 796–803. [DOI] [PubMed] [Google Scholar]
- 29.Henttu P, Liao SS, Vihko P. Androgens up-regulate the human prostate-specific antigen messenger ribonucleic acid (mRNA), but down-regulate the prostatic acid phosphatase mRNA in the LNCaP cell line. Endocrinology. 1992;130 (2): 766–772. [DOI] [PubMed] [Google Scholar]
- 30.Zhao XY, Peehl DM, Navone NM, Feldman D. 1alpha,25-dihydroxyvitamin D3 inhibits prostate cancer cell growth by androgen-dependent and androgen-independent mechanisms. Endocrinology. 2000;141 (7): 2548–2556. [DOI] [PubMed] [Google Scholar]
- 31.Wolf DA, Herzinger T, Hermeking H, Blaschke D, Horz W. Transcriptional and posttranscriptional regulation of human androgen receptor expression by androgen. Mol Endocrinol. 1993;7 (7): 924–936. [DOI] [PubMed] [Google Scholar]
- 32.Falany JL, Falany CN. Expression of Cytosolic Sulfotransferases in Normal Mammary Epithelial Cells and Breast Cancer Cell Lines. Cancer Res. 1996;56 (7): 1551–1555. [PubMed] [Google Scholar]
- 33.Furuya Y, Kohno N, Fujiwara Y, Saitoh Y. Mechanisms of Estrogen Action on the Proliferation of MCF-7 Human Breast Cancer Cells in an Improved Culture Medium. Cancer Res. 1989;49 (23): 6670–6674. [PubMed] [Google Scholar]
- 34.Gierthy JF, Lincoln DW, Gillespie MB, Seeger JI, Martinez HL, Dickerman HW, et al. Suppression of Estrogen-regulated Extracellular Tissue Plasminogen Activator Activity of MCF-7 Cells by 2,3,7,8-Tetrachlorodibenzo-p-dioxin. Cancer Res. 1987;47 (23): 6198–6203. [PubMed] [Google Scholar]
- 35.Le Bail JC, Varnat F, Nicolas JC, Habrioux G. Estrogenic and antiproliferative activities on MCF-7 human breast cancer cells by flavonoids. Cancer Lett. 1998;130 (1–2): 209–216. [DOI] [PubMed] [Google Scholar]
- 36.Tsai EM, Wang SC, Lee JN, Hung MC. Akt activation by estrogen in estrogen receptor-negative breast cancer cells. Cancer Res. 2001;61 (23): 8390–8392. [PubMed] [Google Scholar]
- 37.Wiese TE, Kral LG, Dennis KE, Butler WB, Brooks SC. Optimization of estrogen growth response in MCF-7 cells. In Vitro Cell Dev Biol. 1992;28A (9–10): 595–602. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi Y, Seino Y, Takei H, Kurosumi M, Hayashi S. Detection of estrogen-independent growth-stimulating activity in breast cancer tissues: implication for tumor aggressiveness. Cancer Microenviron. 2014;7 (1–2): 23–31. 10.1007/s12307-013-0139-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan S, Meng Q, Gao B, Grossman J, Yadegari M, Goldberg I, et al. Alcohol stimulatesestrogen receptor signaling in human breast cancer cell lines. Cancer Res. 2000;60: 5635–5639. [PubMed] [Google Scholar]
- 40.Ginsberg E. Estrogen, alcohol and breast cancer risk. JSteroid Biochem Mol Bio 1999;69: 299–306. [DOI] [PubMed] [Google Scholar]
- 41.Przylipiak A, Rabe T, Hafner J, Przylipiak M, Runnebaum B. Influences of ethanol on in vitro growth of human mammary carcinoma call line MCF-7. Arch Gynecol Obstet,. 1996;258 (137–140): [DOI] [PubMed] [Google Scholar]
- 42.Simanowski UA, Seitz HK, Baier B, Kommerell B, Schmidt-Gayk H, Wright NA. Chronic ethanol consumption selectively stimulates rectal cell proliferation in the rat. Gut. 1986;27 (3): 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singletary KW, Frey RS, Yan W. Effect of ethanol on proliferation and estrogen receptor-alpha expression in human breast cancer cells. Cancer Lett. 2001;165 (2): 131–137. [DOI] [PubMed] [Google Scholar]
- 44.Momma T, Hamblin MR, Hasan T. Hormonal modulation of the accumulation of 5-aminolevulinic acid-induced protoporphyrin and phototoxicity in prostate cancer cells. Int J Cancer. 1997;72 (6): 1062–1069. [DOI] [PubMed] [Google Scholar]
- 45.Trapman J, Brinkmann AO. The androgen receptor in prostate cancer. Pathol Res Pract. 1996;192 (7): 752–760. [DOI] [PubMed] [Google Scholar]
- 46.Tai S, Sun Y, Squires JM, Zhang H, Oh WK, Liang CZ, et al. PC3 is a cell line characteristic of prostatic small cell carcinoma. Prostate. 2011;71 (15): 1668–1679. 10.1002/pros.21383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karey KP, Sirbasku DA. Differential Responsiveness of Human Breast Cancer Cell Lines MCF-7 and T47D to Growth Factors and 170-Estradiol. CANCER RESEARCH. 1988;48: 4083–4092. [PubMed] [Google Scholar]
- 48.Burow ME, Tang Y, Collins-Burow BM, Krajewski S, Reed JC, McLachlan JA, et al. Effects of environmental estrogens on tumor necrosis factor alpha-mediated apoptosis in MCF-7 cells. Carcinogenesis. 1999;20 (11): 2057–2061. [DOI] [PubMed] [Google Scholar]
- 49.Harrington WR, Kim SH, Funk CC, Madak-Erdogan Z, Schiff R, Katzenellenbogen JA, et al. Estrogen dendrimer conjugates that preferentially activate extranuclear, nongenomic versus genomic pathways of estrogen action. Mol Endocrinol 2006;20 (3): 491–502. [DOI] [PubMed] [Google Scholar]
- 50.Thompson EW, Katz D, Shima TB, Wakeling AE, Lippman ME, Dickson RB. ICI 164,384, a pure antagonist of estrogen-stimulated MCF-7 cell proliferation and invasiveness. Cancer Res. 1989;49 (24 Pt 1): 6929–6934. [PubMed] [Google Scholar]
- 51.Wang TT, Phang JM. Effects of estrogen on apoptotic pathways in human breast cancer cell line MCF-7. Cancer Res. 1995;55 (12): 2487–2489. [PubMed] [Google Scholar]
- 52.Zhu ML, Kyprianou N. Role of androgens and the androgen receptor in epithelial-mesenchymal transition and invasion of prostate cancer cells. FASEB J. 2010;24 (3): 769–777. 10.1096/fj.09-136994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evangelou A, Letarte M, Marks A, Brown TJ. Androgen modulation of adhesion and antiadhesion molecules in PC-3 prostate cancer cells expressing androgen receptor. Endocrinology. 2002;143 (10): 3897–3904. [DOI] [PubMed] [Google Scholar]
- 54.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest Urol. 1979;17 (1): 16–23. [PubMed] [Google Scholar]
- 55.Heneweer M, Muusse M, Dingemans M, de Jong PC, van den Berg M, Sanderson JT. Co-culture of primary human mammary fibroblasts and MCF-7 cells as an in vitro breast cancer model. Toxicol Sci. 2005;83 (2): 257–263. [DOI] [PubMed] [Google Scholar]
- 56.Teixeira C, Reed JC, Pratt MA. Estrogen promotes chemotherapeutic drug resistance by a mechanism involving Bcl-2 proto-oncogene expression in human breast cancer cells. Cancer Res. 1995;55 (17): 3902–3907. [PubMed] [Google Scholar]
- 57.Rajah TT, Peine KJ, Du N, Serret CA, Drews NR. Physiological concentrations of genistein and 17β-estradiol inhibit MDA-MB-231 breast cancer cell growth by increasing BAX/BCL-2 and reducing pERK1/2. Anticancer Res. 2012;32 (4): 1181–1191. [PubMed] [Google Scholar]
- 58.Sapino A, Pietribiasi F, Bussolati G, Marchisio PC. Estrogen- and tamoxifen-induced rearrangement of cytoskeletal and adhesion structures in breast cancer MCF-7 cells. Cancer Res. 1986;46 (5): 2526–2531. [PubMed] [Google Scholar]
- 59.Yang Q, Hua J, Wang L, Xu B, Zhang H, Ye N, et al. MicroRNA and piRNA profiles in normal human testis detected by next generation sequencing. PLoS One. 2013;8 (6): e66809 10.1371/journal.pone.0066809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang HJ, Moon MJ, Lee HY, Han SW. Testosterone alters testis function through regulation of piRNA expression in rats. Mol Biol Rep. 2014;41 (10): 6729–6735. 10.1007/s11033-014-3558-y [DOI] [PubMed] [Google Scholar]
- 61.Chu H, Xia L, Qiu X, Gu D, Zhu L, Jin J, et al. Genetic Variants in Noncoding PIWI-Interacting RNA and Colorectal Cancer Risk. Cancer. 2015;0 (12): 2044–2052. [DOI] [PubMed] [Google Scholar]
- 62.Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, et al. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58 (6): 1165–1173. 10.1016/j.jhep.2013.01.032 [DOI] [PubMed] [Google Scholar]
- 63.Chu H, Hui G, Yuan L, Shi D, Wang Y, Du M, et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356 (2 Pt B): 561–567. 10.1016/j.canlet.2014.10.004 [DOI] [PubMed] [Google Scholar]
- 64.Hashim A, Rizzo F, Marchese G, Ravo M, Tarallo R, Nassa G, et al. RNA sequencing identifies spesific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget. 2014;5 (20): 9901–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez VD, Vucic EA, Thu KL, Hubaux R, Enfield KS, Pikor LA, et al. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep. 2015;5 (10423): 10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491 (7423): 279–283. 10.1038/nature11502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L, et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29 (1): 196–206. 10.1038/leu.2014.135 [DOI] [PubMed] [Google Scholar]
- 68.Grochola LF, Greither T, Taubert H, Moller P, Knippschild U, Udelnow A, et al. The stem cell-associated Hiwi gene in human adenocarcinoma of the pancreas: expression and risk of tumour-related death. Br J Cancer. 2008;99 (7): 1083–1088. 10.1038/sj.bjc.6604653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He W, Wang Z, Wang Q, Fan Q, Shou C, Wang J. Expression of HIWI in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. BMC Cancer. 2009;9: 426 10.1186/1471-2407-9-426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y, Liu Y, Shen X, Zhang X, Chen X, Yang C. The PIWI protein acts as a predictive marker for human gastric cancer. Int J Clin Exp Pathol. 2012;5: 315–325. [PMC free article] [PubMed] [Google Scholar]
- 71.Siddiqi S, Terry M, Matushansky I. Hiwi mediated tumorigenesis is associated with DNA hypermethylation. PLoS One. 2012;7: 33711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.