Abstract
Single-stranded DNA binding (SSB) proteins coordinate DNA replication, repair, and recombination and are critical for maintaining genomic integrity. SSB binds to single-stranded DNA (ssDNA) rapidly and with very high affinity making it a useful molecular tool to detect free ssDNA in solution. We have labeled SSB from Plasmodium falciparum (Pf-SSB) with the MDCC (7-diethylamino-3-((((2-maleimidyl)ethyl)amino)-carbonyl)coumarin) fluorophore which yields a four-fold increase in fluorescence upon binding to ssDNA. Pf-SSBMDCC binding to DNA is unaffected by NaCl or Mg2+ concentration and does not display salt-dependent changes in DNA binding modes or cooperative binding on long DNA substrates. These features are unique to Pf-SSB, making it an ideal tool to probe the presence of free ssDNA in any biochemical reaction. Using this Pf-SSBMDCC probe as a sensor for free ssDNA, we have investigated the clearing of preformed yeast Rad51 nucleoprotein filaments by the Srs2 helicase during HR. Our studies provide a rate for the disassembly of the Rad51 filament by full length Srs2 on long ssDNA substrates. Mutations in the conserved 2B domain in the homologous bacterial UvrD, Rep and PcrA helicases show an enhancement of DNA unwinding activity, but similar mutations in Srs2 do not affect its DNA unwinding or Rad51 clearing properties. These studies showcase the utility of the Pf-SSB probe in mechanistic investigation of enzymes that function in DNA metabolism.
Introduction
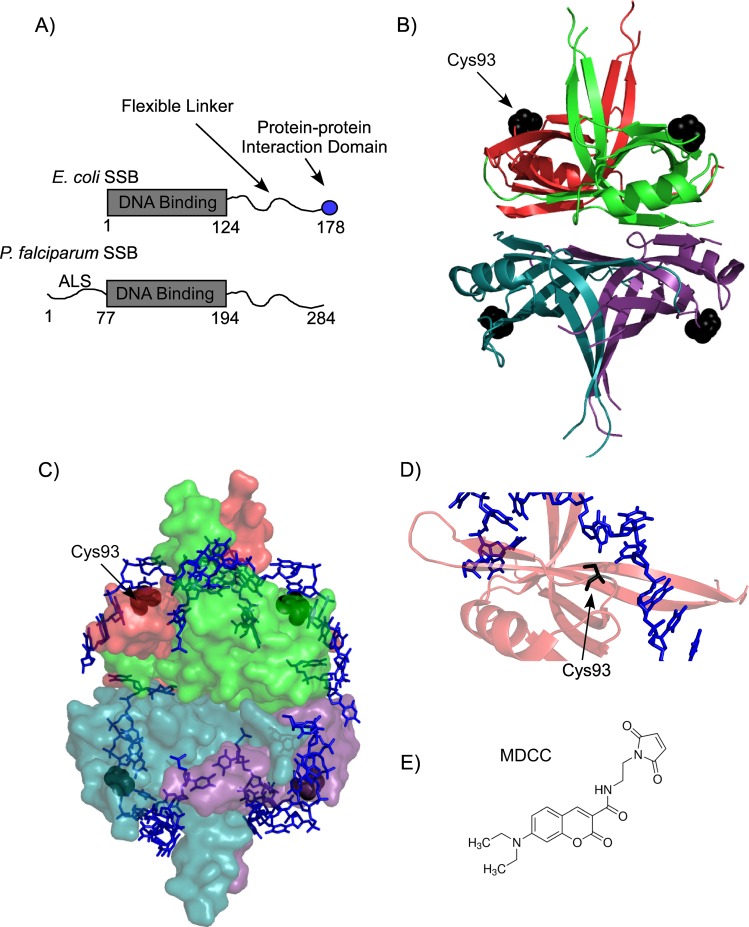
Single-stranded DNA binding (SSB) proteins play essential roles in DNA replication, repair, recombination and replication restart, and are found in all domains of life[1]. They bind to single-stranded DNA (ssDNA) with very high selectivity and affinity, and in the cell they protect ssDNA from nucleolytic attacks[2–4]. In addition, they act as a central hub to coordinate various processes essential for maintaining genomic integrity by binding and recruiting a host of DNA replication and repair proteins[5]. The E. coli SSB (Ec-SSB) protein has been shown to mediate protein-protein interactions with up to fourteen different proteins[5]. Ec-SSB is the most characterized representative in this class of SSBs and functions as a homotetramer[6]. Each subunit contains a conserved DNA binding domain and a C-terminal 9 amino acid tip that mediates the various protein-protein interactions[7,8]. These two functional domains are connected by a non-conserved intrinsically disordered linker (Fig 1A)[9]. Slight variations to this structural organization are observed in the organelle-based eukaryotic homotetrameric SSB proteins. Mitochondrial SSB (mtSSB) for example contains only the conserved DNA binding domain, whereas SSBs found in the apicoplast of eukaryotic parasites such as Plasmodium falciparum and Toxoplasma gondii, have the DNA binding core and the disordered linker, but do not contain the protein-protein interaction tip (Fig 1A)[10,11].
Fig 1. Domain organization of Pf-SSB.
(A) Schematic representation of the DNA binding, protein-protein interaction and linker regions of E. coli and P. falciparum SSB. Pf-SSB also has an apicoplast localization signal (ALS), which is not required for its DNA binding function. The numbers denote positions of the amino acids at the beginning and end of each domain. (B) Individual subunits of the homotetrameric DNA binding domain are depicted as cartoon representation in the crystal structure of Pf-SSB. The Cys93 residues used for attachment of the fluorophore are shown as black spheres. (C) Pf-SSB is shown as surface representation with two (dT)35 DNA molecules (blue stick representation) wrapped around the homotetramer. (D) The proximity of Cys93 (black stick) to the bound DNA in Pf-SSB is highlighted. (E) Structure of the MDCC (7-diethylamino-3-((((2-maleimidyl)ethyl)amino)-carbonyl)coumarin) fluorophore used to label Pf-SSB. Images of the Pf-SSB structure were generated using PDB ID: 3ULP.
Crystal structures of homotetrameric SSB proteins from various organisms show a remarkable similarity in the organization of the DNA binding cores, suggesting a common mechanism of interaction with ssDNA[7,11–15]. Ec-SSB binds to ssDNA in multiple DNA binding modes that differ in the number of nucleotides occluded by the tetramer. (SSB)35 and (SSB)65 are two such modes where the subscript denotes the average number of ssDNA nucleotides engaged by the tetramer[16]. These binding modes are observed when the NaCl or Mg2+ concentration is varied in solution. At high salt concentrations (e.g., 200 mM NaCl) the (SSB)65 mode is favored where all four subunits in the tetramer bind to DNA. In this mode, Ec-SSB has been shown to bind as beads-on-a-string on long ssDNA substrates due to limited cooperativity between the tetramers[17–20]. Interestingly, under these conditions, SSB has been shown to perform numerous activities associated with DNA repair and recombination including, diffusion along ssDNA[21,22], unwinding of hairpins[23] and to promote the formation of RecA nucleoprotein filaments[21]. At lower NaCl concentrations (e.g., 20 mM NaCl), the (SSB)35 mode is observed where only two of the four subunits are bound to DNA. SSB tetramers bind with high cooperativity on long DNA substrates, but within each tetramer binding of DNA to two subunits exerts negative cooperativity on the other two subunits[18,20,24]. The complex (SSB)35 mode is thought to promote SSB binding and function during DNA replication[25].
SSB from the malarial parasite Plasmodium falciparum (Pf-SSB) shares a high degree of structural homology with Ec-SSB in the homotetrameric DNA binding core[10,11,26]. However, it does not contain the conserved protein-protein interaction tip. The intrinsically disordered linker in Ec-SSB tends to adopt more globular conformations, whereas in Pf-SSB they tend to be more extended random coils in structure[9]. The crystal structure of Pf-SSB shows a remarkable similarity to the Ec-SSB structure with ssDNA wrapped around the DNA binding core (Fig 1C), which occludes ~55–62 nt of ssDNA[10,11]. Pf-SSB binds to ssDNA with very high affinity (Ka >1010 M-1)[10]. More striking are the differences in the DNA binding properties between the two proteins with respect to salt concentrations in the reaction: Pf-SSB displays no evidence of salt-dependent cooperative binding on long DNA molecules[10]. Irrespective of the NaCl concentration in the reaction, Pf-SSB binds to two (dT)35 or one (dT)70 molecule with similar affinity[10,11]. This feature is unique to Pf-SSB and is not found in any other bacterial SSBs tested to date. In addition, similar to Ec-SSB, Pf-SSB can be purified with relative ease and is stable in a wide assortment of buffers. While biochemical explorations of Pf-SSB have shed insight on its DNA binding properties, its precise function in the apicoplast is unknown. A role in the replication of apicoplast DNA has been inferred[26]. Whether Pf-SSB coordinates DNA repair and recombination in the apicoplast remains to be ascertained; but is not the subject of investigation in this report.
Fluorescently labeled versions of Ec-SSB and Bacillus subtilis SSB have been developed as real-time probes for ssDNA in biochemical assays[22,27,28]. However, one has to take into account the buffer-dependent variations and cooperative behavior of these SSBs to adequately interpret results. The unique NaCl concentration-resistant DNA binding properties of Pf-SSB along with the loss of all the positive and negative cooperative binding features make it an attractive target for ssDNA probe development.
Here, we describe the development of a MDCC-labeled Pf-SSB protein (Pf-SSBMDCC) as a probe for ssDNA. Using this probe, we have captured the interplay between Srs2 and Rad51 in homologous recombination (HR). Rad51 is the central engine for HR and functions by forming a nucleoprotein filament on the resected ssDNA[29,30]. Srs2 is a superfamily-1 (SF1) helicase that functions as an anti-recombinase by disassembling Rad51 molecules from DNA, a process termed filament clearing[31–33]. During filament clearing, Srs2 physically interacts with Rad51, stimulating ATP hydrolysis within Rad51 and causing it to dissociate from the DNA [33,34]. Earlier studies of filament clearing by Srs2 were carried out on short DNA oligonucleotide substrates using fluorescently end-labeled DNA substrates[33,35]. When Rad51 binds to these DNA substrates, a change in fluorescence occurs corresponding to filament formation. Disappearance of this signal in the presence of Srs2 was corroborated with filament disassembly. An inherent limitation to these assays is the length of the DNA (< 120 nt in length). HR often occurs on DNA ~1–2 kb in length and the experimental ability to measure nucleoprotein filament dynamics on long DNA substrates would be immensely helpful to study events in HR[36,37]. Using Pf-SSBMDCC as a reporter for free ssDNA in the reaction, here we report an assay to monitor the disassembly of Rad51 by Srs2 on long (6400 nt in length), circular m13ssDNA substrates. Using this assay, we report the filament clearing activity of full length Srs2 and a Srs2DK-AA variant carrying mutations in its 2B domain. Our studies reveal that the 2B domain in Srs2 serves functions that appear distinct than in UvrD, PcrA and Rep, the prokaryotic homologs of Srs2.
Results
Generation of the fluorescent Pf-SSBMDCC probe
Pf-SSB contains an N-terminal apicoplast localization signal (ALS), a DNA binding core and an unstructured C-terminal region (Fig 1A)[26]. The ALS is not required for DNA binding and also prevents proper expression in E. coli overexpression systems, and hence is not part of the protein investigated here[11]. The DNA binding core is structurally conserved in the homotetrameric class of SSBs and in the case of Pf-SSB contains a single Cys residue at position 93 in each subunit (Fig 1B). Cys93 is surface exposed in the crystal structure and is positioned adjacent to the bound DNA (Fig 1C and 1D)[11]. Purified Pf-SSB was labeled with MDCC (7-diethylamino-3-((((2-maleimidyl)ethyl)amino)-carbonyl)coumarin) fluorophore (Fig 1E) with >95% labeling efficiency (Fig 2A). MDCC-labeled Pf-SSB protein (Pf-SSBMDCC) shows a fluorescence profile with an excitation and emission maxima of 430 nm and 482 nm, respectively (Fig 2B). The same labeling efficiency was also observed when Pf-SSB was labeled with Alexa-555 (not shown). These results suggest that Cys93 is solvent exposed and accessible to complete labeling with maleimide chemistry. MDCC is a relatively small dye and has been shown to be a sensitive fluorophore when tethered onto protein-based probes to detect the binding of ADP[38,39], inorganic phosphate[40,41] and nucleotides[42]. While data obtained with MDCC-labeled Pf-SSB are reported here, similar data are observed when Pf-SSB is labeled with Alexa-555 (data not shown).
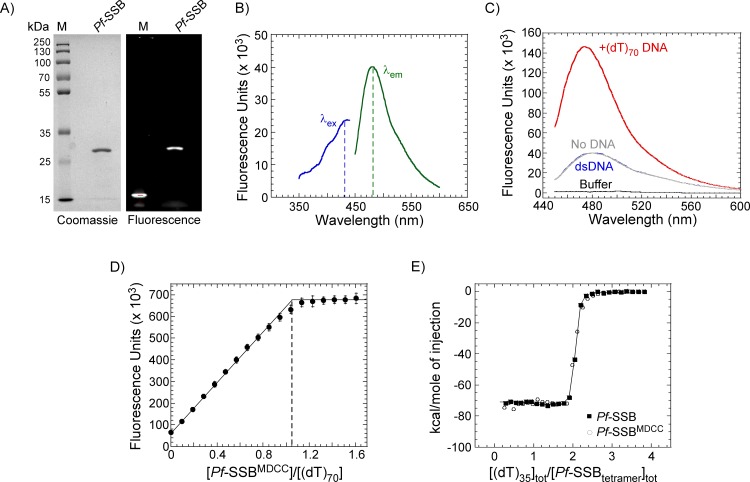
Fig 2. DNA binding properties of Pf-SSBMDCC.
(A) Purified Pf-SSBMDCC was analyzed on a 12% SDS-PAGE gel and imaged after staining with coommassie dye or detected using fluorescence imaging. M denotes the protein ladder. (B) Excitation (blue, λex) and emission (green, λem) spectra of 1 μM Pf-SSBMDCC are shown. The dotted lines correspond to an excitation and emission maxima of 430 nm and 482 nm, respectively. (C) 1 μM Pf-SSBMDCC was excited at 430 nm and emission spectra were measured in the absence of DNA (grey) and in the presence of a 125 bp dsDNA (blue) or an oligo-dT 70 nt ssDNA (red). A four-fold increase in Pf-SSBMDCC fluorescence is observed in the presence of the (dT)70 ssDNA oligonucleotide. (D) Fluorescence titration of Pf-SSBMDCC with increasing concentrations of ssDNA [(dT)70]. Pf-SSBMDCC binds stoichiometrically, with one SSB tetramer binding to one (dT)70 oligonucleotide as denoted by the dotted line. The mean values and standard errors from three independent experiments are shown. (E) Isothermal calorimetric measurement of changes in enthalpy associated with binding of two (dT)35 molecules to Pf-SSB and Pf-SSBMDCC are shown. Both proteins bind stoichiometrically with similar observed heat changes ΔHobs = -73.1±0.2 kcal mol-1 and -71.8±0.2 kcal mol-1 for Pf-SSB and Pf-SSBMDCC, respectively. The mean values and standard errors from three independent experiments are shown.
Pf-SSBMDCC binds to DNA with very high affinity and is not influenced by NaCl concentration
Pf-SSB has an occluded site size of ~55–62 nt and a (dT)70 oligonucleotide completely wraps around the homotetrameric DNA binding core[10,11]. Pf-SSBMDCC binds to ((dT)70) which results in a ~4-fold increase in fluorescence of the attached fluorophore (Fig 2C). Control experiments show that the binding of Pf-SSBMDCC is specific to ssDNA. Addition of double-stranded DNA (dsDNA) to Pf-SSBMDCC does not elicit a change in fluorescence (Fig 2C). Unlabeled Pf-SSB binds to ssDNA stoichiometrically to a (dT)70 oligonucleotide with high affinity (Kobs >1010 M-1)[10]. Pf-SSBMDCC binds to a (dT)70 substrate stoichiometrically with each tetramer binding to one (dT)70 molecule (Fig 2D). In addition, ITC measurements of ssDNA binding to both the unlabeled and labeled Pf-SSB yield similar changes in heat capacity, ΔHobs = -73.1±0.2 kcal mol-1 and -71.8±0.2 kcal mol-1 for Pf-SSB and Pf-SSBMDCC, respectively. These data are consistent with DNA binding parameters previously reported for Pf-SSB[10] and suggest that the fluorophore does not interfere with the DNA binding properties of Pf-SSB, and can be used as a reliable reporter of free ssDNA in a reaction.
Since Pf-SSB is relatively refractory to changes in NaCl concentration in the reaction, we next tested if the binding of Pf-SSBMDCC to ssDNA is affected by [NaCl]. Binding of Pf-SSBMDCC to a (dT)70 ssDNA substrate was measured at 0.02, 0.1, 0.5 or 1 M NaCl in the reaction. In each of these experiments, one Pf-SSBMDCC molecule stoichiometrically binds to one (dT)70 ssDNA molecule (Fig 3A and 3B). These results show that the fluorescently labeled Pf-SSBMDCC protein is also refractory to changes in [NaCl]. A small decrease in total fluorescence is observed as the [NaCl] is increased in the reaction, suggesting that the quantum yield of the fluorophore is slightly sensitive to buffer conditions, but the ssDNA binding properties are relatively unaffected (Fig 3A). Similar results were observed when the [MgCl2] concentration was varied from 0–10 mM in the reaction (not shown). These results showcase the versatility of Pf-SSBMDCC as a probe for ssDNA, and in this respect, is better suited for probe development than E. coli SSB and SSB proteins from other bacterial species tested thus far.
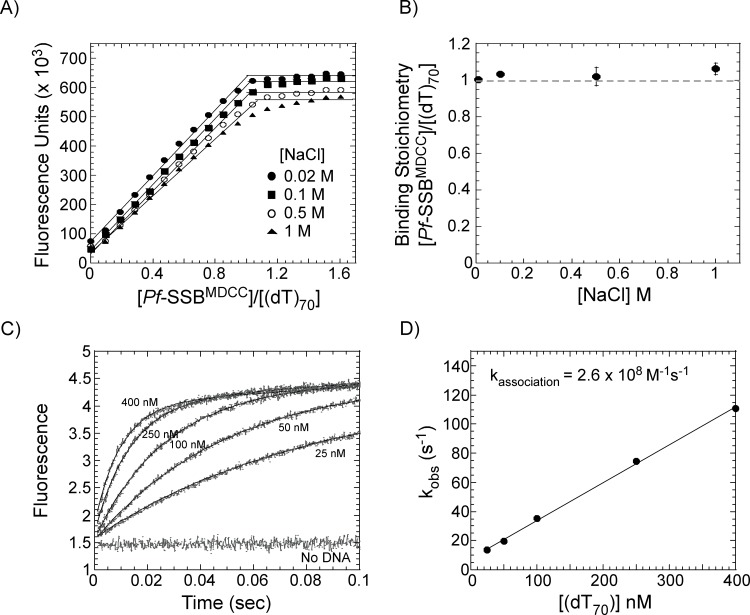
Fig 3. Pf-SSBMDCC binds stoichiometrically to ssDNA over a wide range of NaCl concentrations.
(A) Fluorescence titration of Pf-SSBMDCC with increasing concentrations of ssDNA [(dT)70] in the presence of increasing concentrations of NaCl. Experiments were performed in 20 mM Tris-Cl, pH 8, 0.1 mM EDTA, and 1 mM TCEP with either 0.02 (●), 0.1 (■), 0.5 (○) or 1M (▲) NaCl in the reaction. (B) Stoichiometry of [Pf-SSB bound]/[(dT)70] under various NaCl conditions from A is plotted as a function of [NaCl] and shows no significant change in binding stoichiometry over a wide range of NaCl concentrations. The mean values and standard errors from three independent experiments are shown. (C) Stopped-flow analysis of Pf-SSBMDCC binding to (dT)70 ssDNA. Rapid binding of Pf-SSBMDCC to ssDNA is observed as increasing concentrations of (dT)70 are mixed with a fixed concentration of Pf-SSBMDCC (20 nM). Data were fit to a single exponential equation and (D) the kobs (s-1) from the fits were plotted as a function of DNA concentration yielding an apparent association rate constant 2.6 x 108 M-1s-1.
Pf-SSBMDCC binds rapidly to ssDNA
A probe for ssDNA should possess rapid binding capability. Pf-SSB has been shown to possess fast ssDNA binding kinetics. To test whether the attachment of the fluorophore affects the DNA binding kinetics, we mixed increasing concentrations of ssDNA with Pf-SSBMDCC in a stopped-flow instrument and monitored the change in fluorescence (Fig 3C). Pf-SSBMDCC rapidly binds to DNA as observed by the saturation of the fluorescence signal well before 100 msec. The data are fit to a single exponential function and analysis of the kobs as a function of [(dT)70] yields an apparent association rate constant of 2.6 x 108 M-1s-1 (Fig 3D). These results suggest that Pf-SSBMDCC binding to ssDNA is diffusion limited and is ideal as a probe for free ssDNA in any reaction.
Preformed Rad51 filaments are unaffected by Pf-SSBMDCC
Rad51 binds to ssDNA and forms the nucleoprotein filament during HR[33,37,43]. Mediator proteins, such as the Srs2 helicase, disassemble Rad51 molecules during HR thereby creating transient gaps in ssDNA[32,33]. In an in vitro reaction looking at Rad51 binding and dissociation, these open ssDNA intermediates provide a binding opportunity for a secondary reporter such as Pf-SSBMDCC. The change in fluorescence upon Pf-SSBMDCC binding to ssDNA can be utilized as a measure of Rad51 DNA binding/dissociation kinetics. We have previously reported an apparent rate for filament clearing by a truncated version of Srs2 (lacking a portion of the C-terminal tail—Srs21-898) on short ssDNA oligonucleotides (<125 nt)[33]. Since HR often occurs on DNA ~1 to 2 kb in length, the experimental ability to measure nucleoprotein filament dynamics on longer DNA substrates would be immensely helpful[36,37]. SSB can bind and completely sequester long DNA substrates. To establish the utility of Pf-SSBMDCC as a probe for long DNA substrates, we tested the ability of Pf-SSBMDCC to bind a 6.4 kb circular m13ssDNA substrate. Pf-SSBMDCC binds rapidly to free circular m13ssDNA and displays a robust change in the fluorescence signal (Fig 4A).
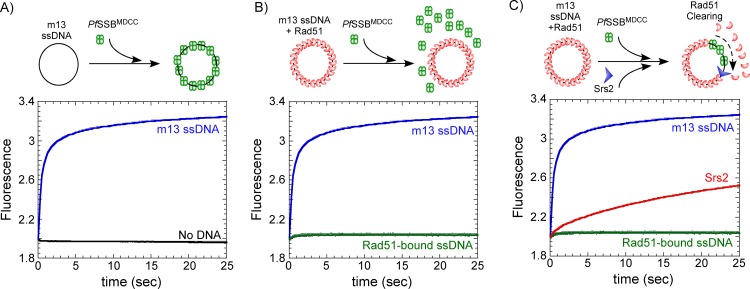
Fig 4. Rad51 filament clearing by Srs2 captured by Pf-SSBMDCC.
(A) Stopped-flow measurement of Pf-SSBMDCC (80 nM) binding to m13 circular ssDNA (3 μM nucleotides) is shown by a rapid increase in fluorescence (blue trace). No change in fluorescence is observed in the absence of ssDNA (black trace). (B) When m13ssDNA (3 μM nucleotides) is pre-coated with Rad51 (3 μM)in the presence of ATP (3 mM) and then mixed with Pf-SSBMDCC (80 nM), no significant change in fluorescence is observed (green trace) suggesting that Pf-SSBMDCC does not gain access to ssDNA when it is completely bound by Rad51 in the form of a nucleoprotein filament. (C) Challenging the Rad51 nucleoprotein filament on m13ssDNA with Pf-SSBMDCC in the presence of full length Srs2 (25 nM) results in a gradual increase in the fluorescence signal (red). Srs2 clears the Rad51 from the ssDNA yielding free ssDNA for the rapid and tight binding of Pf-SSBMDCC. Models for the reaction mixing schemes are presented above each data panel.
To investigate Rad51 filament dynamics on long ssDNA substrates using the Pf-SSBMDCC, we first preformed the Rad51 filament on m13ssDNA. Yeast Rad51 forms a stable nucleoprotein filament in the presence of ATP. This feature is unique to S. cerevisiae Rad51 as both human Rad51 and the bacterial homolog RecA have been shown to be in equilibrium between the bound and unbound states in the presence of ATP[44–48]. We used 80 nM Pf-SSBMDCC (tetramer concentration) to saturate all the potential ssDNA binding sites when 3 μM total nucleotides of the m13ssDNA were used in the reaction. We used a binding site size of ~60 nt bound/SSB tetramer as a rubric for the reaction (50 nM total SSB binding sites), and complete saturation of the Pf-SSBMDCC fluorescence is observed under these conditions (Fig 4A).
In order to be a usable reporter of Rad51 dynamics, Pf-SSBMDCC should not interfere with preformed Rad51 filaments. To test this, Pf-SSBMDCC was mixed with preformed Rad51 filaments (Rad51 + ATP) in a stopped-flow, and the change in fluorescence was monitored (Fig 4B). 2 to 3 μM Rad51 was sufficient to occupy all the ssDNA in the reaction and we used a site size of ~3 nt/Rad51 (1 μM total Rad51 binding sites) to formulate the conditions for filament formation[49–51]. No change in fluorescence is observed suggesting that Pf-SSBMDCC did not displace Rad51and no free ssDNA is available in this scenario for Pf-SSBMDCC to bind in the reaction (Fig 4B). In the control experiments without Rad51, Pf-SSBMDCC rapidly binds to free ssDNA and an exponential rise in fluorescence signal is observed (Fig 4A and 4B).
Srs2 rapidly clears Rad51 molecules off ssDNA
Using Pf-SSBMDCC and Rad51 nucleoprotein filaments formed on circular m13ssDNA, we tested whether full length Srs2 will clear long nucleoproteins filaments. Preformed Rad51 nucleoproteins were first formed as described above by incubating Rad51, m13ssDNA, ATP and an ATP regenerating system as described in the methods section. These filaments were mixed with Srs2 in a stopped-flow and the change in fluorescence was monitored. A gradual increase in fluorescence is observed in the presence of Srs2 (Fig 4C). Srs2 disassembles Rad51 molecules creating gaps in the ssDNA, which are bound by the Pf-SSBMDCC molecules. Srs2 could initiate Rad51 disassembly from multiple positions on the filament and hence a complete model to fit the data cannot be accurately generated. These results show that the Pf-SSBMDCC probe can be used as a reporter for measurement of Rad51 clearing during HR. This assay will be useful to measure the effect of other mediator proteins such as the SHU complex on Rad51 filament stability and to investigate the interplay between Srs2 and other mediator proteins on long nucleoprotein filaments[30,43].
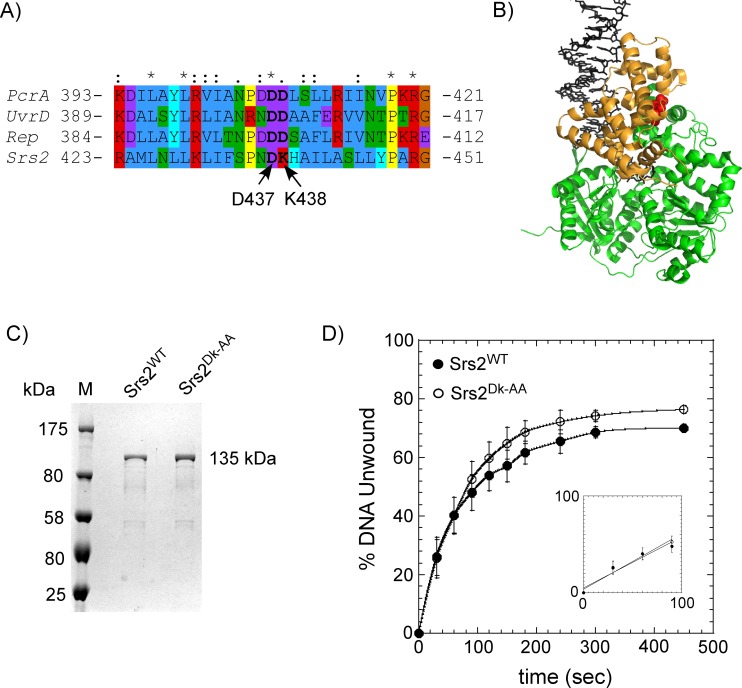
Mutations in the 2B domain of Srs2 do not enhance its DNA unwinding or filament clearing activities
Srs2 is a SF1 helicase capable of unwinding dsDNA[52]. Srs2 is homologous to the bacterial UvrD, Rep and PcrA helicases[53]. The 2B domain in UvrD, Rep and PcrA have been shown to be inhibitory for the DNA unwinding activity of these enzymes[54–60]. Mutations or deletions in the 2B domain stimulate their DNA unwinding activity[58,60]. The role of the 2B domain of Srs2 in DNA unwinding or nucleoprotein filament clearing have not been explored.
In uvrd303, a hyperactive helicase variant, two aspartate residues (D403, D404) are mutated to alanine (Fig 5A)[61–63]. They reside in the 2B domain and are thought to participate in controlling the transition between the ‘closed’ and ‘open’ conformations of this domain (Fig 5B)[61]. The two aspartates are conserved in UvrD, Rep and PcrA, but only the first aspartate residue is conserved in Srs2 (D437). The second aspartate is replaced with a Lysine in S. cerevisiae Srs2 (Fig 5A). To test the significance of these differences in this region, we generated a Srs2 variant by substituting D437 and K438 with two alanine residues and purified the Srs2DK-AA variant (Fig 5C). We next tested the difference in DNA unwinding rates of the full length Srs2WT and Srs2DK-AA proteins on a DNA substrate with a 25bp dsDNA with a 16 nt 3' ssDNA overhang, as previously described[35]. Both proteins unwind DNA with similar kinetics (Fig 5D), unwinding the DNA substrate at ~0.025 s-1. These results suggest that these residues in the 2B domain of Srs2 do not play a role in DNA unwinding compared to the reported observations of enhanced DNA unwinding for the bacterial homologs UvrD, PcrA and Rep[54,57,58]. Other fluorescent SSB probes have been used as a tool to measure DNA unwinding kinetics of helicases[27], and Pf-SSBMDCC can also be used to measure the DNA unwinding kinetics of Srs2 (Davenport et. al., unpublished data).
Fig 5. 2B domain mutations in Srs2 have no effect on its DNA unwinding capabilities.
(A) Alignment of the region in the 2B domains from UvrD, Rep, PcrA and Srs2. D437 and K438 in Srs2 align with D403 and D404 in UvrD, which are mutated in the uvrD303 phenotype, a hyperactive helicase mutant of UvrD. Amino acids are colored according to their physicochemical properties. (B) Crystal structure of the UvrD (PDB ID:2IS4; the bacterial Srs2 homolog) is shown with the 2B domain colored gold. The 2B domain is in the ‘closed conformation’ when bound to the unwinding DNA substrate. The DNA is shown as sticks (black) and the D403-D404 residues are shown as red spheres. (C) SDS-PAGE analysis of the purified full length Srs2WT and Srs2DK-AA proteins. (D) Unwinding kinetics of a DNA substrate (25bp dsDNA with a 16 nt 3' ssDNA overhang) by Srs2WT and Srs2DK-AA. No discernable difference in unwinding kinetics is observable between the two proteins. Fitting the linear portion of the data (insert) yield unwinding rates of 0.026 s-1 and 0.028 s-1 for the Srs2WT and Srs2DK-AA proteins, respectively. The mean values and standard errors from three independent experiments are shown.
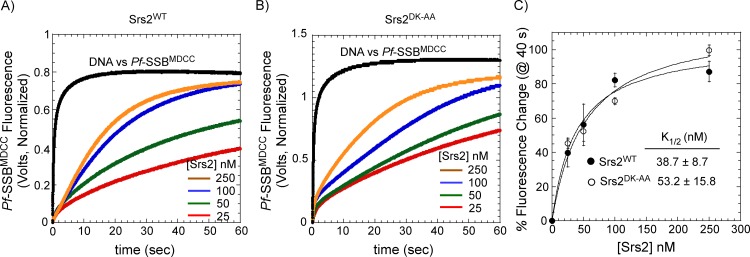
We next tested whether these residues in the 2B domain play a role in the filament clearing activity of Srs2. Using Pf-SSBMDCC as a probe to monitor Rad51 nucleoprotein filament disassembly, we measured the kinetics of filament clearing of Srs2WT and Srs2DK-AA. Filament clearing was measured by challenging preformed nucleoprotein filaments on circular m13ssDNA with increasing concentrations of Srs2 and Pf-SSBMDCC, and the change in fluorescence was measured in a stop flow instrument. To measure the Rad51 filament clearing activity, the total fluorescence from a specific time point in the reaction (e.g., at 40 sec; Fig 6) was plotted as a function of [Srs2] and an apparent K1/2 for clearing was calculated by fitting the data to a Menton hyperbola (Fig 6C). Srs2WT and Srs2DK-AA both cleared long Rad51 nucleoprotein filaments with relatively similar efficiency: K1/2 = 38.7±8.7 and 53.2±15.8 for Srs2WT and Srs2DK-AA respectively. These results suggest that these residues in the 2B domain of Srs2 do not play a key role in the Rad51 clearing mechanism.
Fig 6. Nucleoprotein filament clearing activity of Srs2 is unaffected by mutations in its 2B domain.
Stopped-flow analysis of Rad51 nucleoprotein filament clearing with increasing concentrations of (A) Srs2WT and (B) Srs2DK-AA were measured using Pf-SSBMDCC as a reporter for free ssDNA in the reaction. (C) The percent change in Pf-SSBMDCC fluorescence at time = 40 sec from A and B are plotted against Srs2 concentration and shows a hyperbolic relationship. The K1/2 for Rad51 filament clearing was obtained by fitting the data to a hyperbola and shows that both proteins clear Rad51 filaments with similar efficiency. The mean values and standard errors from three independent experiments are shown.
Discussion
The ability to monitor activity of DNA binding proteins on long DNA substrates is useful in mimicking reaction substrates that are encountered in the cell. Pf-SSBMDCC is well suited as a tool to report the presence of free ssDNA of varying lengths in a reaction irrespective of the buffer conditions used. The binding of Pf-SSBMDCC to DNA is not affected by [NaCl] or [Mg2+] in the reaction. In addition, the salt dependent variations in intra-subunit and inter-subunit cooperativity between the tetramers observed for Ec-SSB[16,19] are not applicable to Pf-SSB[10,11]. Hence, Pf-SSB serves as a more versatile probe for ssDNA compared to SSB proteins from E. coli or SSB proteins from other bacterial origins. Pf-SSBMDCC rapidly binds to ssDNA with very high affinity making it an ideal reporter for real-time kinetics of enzyme activity. Here, we have used Pf-SSBMDCC to monitor an activity that occurs during homologous recombination.
Formation of the Rad51 filament drives HR and its dissociation hinders HR. Pro-HR mediators promote HR by stabilizing the Rad51 filament whereas anti-HR mediators, such as Srs2, prevent inopportune HR events by removing Rad51 molecules from DNA[43]. Ensemble experiments looking at Rad51 filament disassembly were limited to short DNA substrates end-labeled with fluorophores[33,64]. Recent single molecule experiments using DNA curtains have been able to capture Rad51 dynamics on long DNA substrates[65], but are limited by the need for expensive and custom-built total internal reflection microscopy based instrumentation. Pf-SSBMDCC serves as a feasible probe to monitor the disassembly of S. cerevisiae Rad51 nucleoprotein filaments. We have captured disassembly of Rad51 filaments by full length Srs2 on a 6.4 kb long m13ssDNA circular substrate. Our earlier investigation of Rad51 filament clearing by a truncated version of Srs2 (Srs21-898) yielded a K1/2 = 380 nM[33]. Here, we observe a K1/2 = 38.7 nM for filament clearing by full length Srs2 on long DNA substrates (Fig 6C). In the earlier study, Rad51 would dissociate and rebind to the free ssDNA and occluded the true efficiency of filament clearing. In the current assay, Pf-SSBMDCC serves as a trap for free ssDNA and prevents the rebinding of Rad51, thereby providing a more accurate measure of the filament clearing by Srs2. During HR, the RPA protein (the eukaryotic functional homolog of SSB) serves the role of sequestering free DNA as Rad51 is displaced by Srs2[32,66].
The Pf-SSBMDCC based assay for filament clearing was also used to test the role of the Srs2 2B domain. In UvrD and Rep, the 2B domain adopts two specific conformations–an ‘open conformation’ where the 2B domain is situated above the 2A domain, and a ‘closed conformation’ where the 2B domain moves over the 1A domain[54,67]. In the presence of DNA, the 2B domain adopts the closed conformation (Fig 5B)[67–69]. Deletion of the 2B domain stimulates the DNA unwinding activity of both UvrD and Rep suggesting that the 2B domain is inhibitory to unwinding activity[58,60,61]. uvrD303 is a hyperactive DNA unwinding variant of UvrD carrying two mutations in the 2B domain. D403 and D404 are mutated to alanine residues in this UvrD variant[61] and these residues are highly conserved in the Srs2 homologs (Fig 5A). Since only the first aspartate residue is conserved in Srs2, we generated the Srs2DK-AA variant to investigate the role of the 2B domain. We see no differences in either the DNA unwinding or Rad51 clearing kinetics between Srs2WT and Srs2DK-AA (Fig 6), suggesting that these residues in the 2B domain might be serving a different role in Srs2 and warrants more investigation.
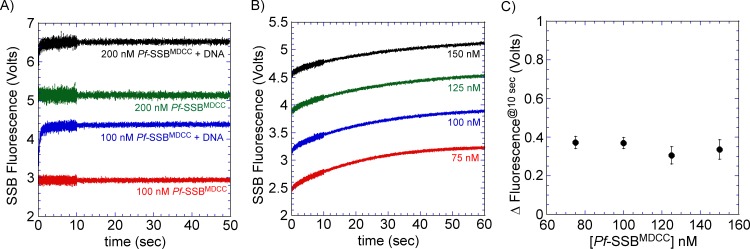
While SSB proteins are attractive tools as probes for free ssDNA in the reaction, one needs to control for inadvertent effects of SSB on their protein/system of investigation. Performing the experiments with multiple concentrations of probe-SSB and being certain that the measured reaction rates/behavior do not change would address such concerns. An example of such control experiments is presented (Fig 7). Increasing the concentration of the Pf-SSBMDCC probe in the reaction results in the enhancement of the total fluorescence signal (Fig 7A). However, the kinetics of filament clearing by Srs2 is independent of probe concentration (Fig 7B and 7C) and hence, in this case, is a true reporter of free ssDNA in the reaction.
Fig 7. Pf-SSBMDCC does not influence the activity of Srs2.
(A) Change in fluorescence upon mixing varying concentrations of Pf-SSBMDCC (100 or 200 nM) with buffer in the presence or absence of m13ssDNA (3 μM nucleotides). (B) Rad51 filament clearing by Srs2 was measured in the presence of increasing concentrations of Pf-SSBMDCC. Preformed Rad51 filaments were rapidly mixed with varying amounts of Pf-SSBMDCC (75, 100, 125 or 150 nM) and Srs2 (100 nm) and the change in fluorescence was measured over time. Data were collected over a split time period with 5000 points each assigned to the first 10 sec and remaining 50 sec, respectively. An average of 10 independent traces is shown. (C) The normalized change in fluorescence at time = 10 sec was subtracted from time = 0.01 sec and the Δfluorescence@10sec values plotted as a function of [Pf-SSBMDCC]. No significant change in fluorescence is observed. The mean values and standard errors from three independent experiments are shown.
In summary, Pf-SSBMDCC serves as a robust tool to measure the presence of free ssDNA in a reaction. In addition to being a tool to monitor DNA unwinding by DNA helicases, it can be tailored to study events such as the dissociation of Rad51 during homologous recombination. This Pf-SSBMDCC probe should be amenable as a tool to monitor the dynamics of other DNA binding enzymes.
Materials and Methods
Chemicals and Reagents
All standard laboratory chemicals, phosphocreatine and creatine phosphokinase were purchased from Sigma-Aldrich (St. Louis, MO). MDCC (7-diethylamino-3-((((2-maleimidyl)ethyl)amino)-carbonyl)coumarin) was purchased from Thermo Fisher Scientific (Grand Island, NY).
DNA
Oligonucleotides used in this study were synthesized by Integrated DNA Technologies (Coralville, IA). Phage m13mp18ssDNA was purchased from New England Biolabs (Ipswich, MA). All ssDNA concentrations were determined spectrophotometrically using the extinction coefficient ε260 = 8.1 x 103 M-1 (nucleotide) cm-1 for oligo(dT) and ε259 = 7370 M-1 cm-1 for m13ssDNA in 20 mM Tris-Cl, pH 8.0.
Proteins
Pf-SSB, Saccharomyces cerevisiae Srs2 (full length) and Rad51 proteins were purified as described[10,11,33]. Concentration of Pf-SSB was determined using extinction coefficient: ε280 = 9.58 ×104 M-1 cm-1 (Pf-SSB tetramer). All SSB concentrations denoted in this study are for the tetramer. Concentration of Srs2 and Rad51 were determined spectroscopically using extinctions coefficients ε280 = 82,670 M-1cm-1 and 12,420 M-1cm-1 respectively. The Srs2DK-AA mutation was generated using the Q5 site directed mutagenesis kit (NEB, Ipswich, MA) and purified similar to the wild type Srs2 protein.
Labeling of Pf-SSB
750 μl of 400 μM Pf-SSB (monomer concentration) was first dialyzed into labeling buffer (50 mM potassium phosphate, pH 7.0, 100 mM NaCl, and 5% v/v glycerol) at 4°C. A 16-fold molar excess of freshly made TCEP was added to the SSB protein prior to labeling. The MDCC fluorophore was resuspended in DMSO and added to the protein at a final molar ratio of 2:1 [MDCC:SSBmonomer]. The reaction was incubated overnight at 4°C and quenched by adding a 5-fold excess of 2-mercaptoethanol. Excess dye from the reaction was removed by purifying the labeled protein on a Biogel P-4 polyacrylamide gel size exclusion column (Bio Rad Laboratories, Hercules, CA) at 4°C using labeling buffer. Fractions containing labeled SSB were pooled and dialyzed into SSB storage buffer (20 mM Tris-Cl, pH 8.3, 1 mM Na3EDTA, 500 mM NaCl, 5 mM 2-mercaptoethanol, and 50%(v/v) glycerol). SSB concentration was measured using ε280 = 95,800 M-1 cm-1 (Pf-SSB tetramer) and MDCC concentration was measured using ε430 = 49,300 M-1 cm-1. A correction factor was applied to the final protein concentration to account for the MDCC absorbance at 280 nm: [SSB]-([SSB]*0.3634), yields the correct concentration of Pf-SSBMDCC. This labeling procedure routinely yielded ~95–99% MDCC-labeled Pf-SSB protein. Proteins were stored at -20°C for up to 12 months with no loss in DNA binding activity.
DNA binding measurements
Binding of Pf-SSB to (dT)70, was examined in buffer (20 mM Tris-Cl, pH 8, 0.1 mM EDTA, 100 mM NaCl and 1 mM TCEP) by monitoring the change in Pf-SSBMDCC fluorescence upon titration with DNA at 25°C (ISS PC1 spectrofluorometer, Champaign, IL) [λex = 430 nm (2 nm band-pass), and λem = 482nm (2–5 nm band-pass)].
Stopped-flow DNA binding and Rad51 clearing experiments
All the stopped-flow experiments were carried out using an Auto SF-120 Stopped-Flow instrument (Kintek Corp., Snow Shoe, PA) at 25°C or a SX20 Stopped-Flow instrument from Applied Photophysics (Surrey, UK). To monitor the fluorescence changes arising from Pf-SSBMDCC, samples were excited at 430 nm and emission was monitored using a 450 nm long pass cut-off filter (Newport Inc., Irvine, CA). Pf-SSBMDCC binding to ssDNA [(dT)70] was measured in buffer (20 mM Tris-Cl, pH 8, 0.1 mM EDTA, 100 mM NaCl and 1 mM TCEP) at 25°C and by rapidly mixing 20 nM Pf-SSBMDCC against varying concentrations of (dT)70 = 0–400 nM. Data were fit to single exponential equation and the rates were plotted against DNA concentration to obtain an association rate constant. For the filament clearing experiments, Rad51 nucleoprotein filaments were first formed on m13ssDNA by incubating 3 μM Rad51 with m13ssDNA (3 μM nucleotides) in filament buffer (25 mM Tris-Cl, pH 7.7, 1 mM TCEP, 5% v/v glycerol, 10 mM MgCl2 an 50 mM NaCl) with 3 mM ATP and an ATP regenerating system (30 mM phosphocreatine and 0.2 mg/ml creatine phosphokinase). These filaments were mixed with reactions containing 80 nM Pf-SSBMDCC in the presence or absence of 25–250 nM Srs2. All concentrations noted here are ‘post-mixing’ in the stop flow setup. The fluorescence intensities at a given time point (e.g. 40 sec; Fig 6A and 6B) were normalized against the total fluorescence change for Pf-SSBMDCC binding to free m13ssDNA to yield the percent change in fluorescence. These values were then plotted against [Srs2] and the data were fitted to a hyperbola to yield apparent K1/2 values for filament clearing. The effect of total [Pf-SSBMDCC] on filament clearing by Srs2 was measured similarly, but the concentration of SSB was increasing in each experiment as denoted (75, 100, 125 or 150 nM). The change in fluorescence at 10 sec (Δfluorescence@10sec) was calculated by subtracting the fluorescence value at t = 10 sec with the value measured at t = 0.01 sec for each trace observed in the dataset.
Isothermal calorimetric measurement of Pf-SSB binding to ssDNA was performed using a VP-ITC microcalorimeter (GE Inc., Piscataway, NJ) as described previously[10]. Briefly, 1 μM of Pf-SSB or Pf-SSBMDCC was incubated in the cell in buffer (20 mM Tris-Cl, pH 8, 0.1 mM EDTA, 200 mM NaCl and 1 mM TCEP) and titrated with (dT)35 (24 μM concentrations in the syringe). The reference heats of dilutions were determined by titrating DNA solution into the buffer in the cell. Proteins and DNA were dialyzed extensively into the reaction buffer (4 buffer changes) prior to the experiments. The ITC data for Pf-SSB binding to two molecules of (dT)35 were analyzed as described[10].
DNA unwinding assay
The substrate for DNA unwinding was prepared by annealing EA-T: 5′-CGATCGTCC-TCTAGACAGCTTACGC-3′ labeled with γ-32P-ATP (Perkin Elmer) and polynucleotide kinase (NEB) and EA-B: 5′-GCGTAAGCTGTCTAGAGGACGATCG[T]16 yielding a 25 bp dsDNA with a 16 nt 3′ ssDNA overhang. DNA unwinding by Srs2 was measured by the incubating 10 nM of the DNA with 250 nM Srs2 and monitoring the displacement of the labeled EA-T strand. Reactions were performed in DNA unwinding buffer [20 mM KHPO4 (pH 8.0), 120 mM NaCl, 10 mM MgCl2, 100 μg/ml bovine serum albumin and 0.2 mM β-mercaptoethanol] at 30°C and initiated by adding ATP (2 mM) to the preformed Srs2-DNA complex. Samples were removed at the denoted time points and quenched with stop buffer [100 mM EDTA (pH 8.0), 20% v/v glycerol and 0.4% SDS]. The dsDNA substrate and dissociated ssDNA product strand were resolved by EMSA in a 10% TBE (Tris–borate–EDTA) acrylamide gel (100 V at 25°C in 1× TBE), dried and quantified using a phosphor imager.
Acknowledgments
This work was funded by a grant from the NIH-NIGMS (R15GM110671) and start-up support from Marquette University to EA. EPD was supported by an Undergraduate Research and Creative Opportunities (URCO) grant from Utah State University. We thank Tawny Moss for generation of the Srs2DK-AA mutation and undergraduate students—Mark Soffe, Cason Worley, Danny Morris and Jeff VonGermeten for their assistance with protein purification. We also acknowledge the members of the Antony Lab for critical reading of the manuscript and sharing ideas for using the Pf-SSB probe.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was funded by a grant from the NIH-NIGMS (R15GM110671) and start-up support from Marquette University to EA. EPD was supported by an Undergraduate Research and Creative Opportunities (URCO) grant from Utah State University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chase JW, Williams KR (1986) Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem 55: 103–136. [DOI] [PubMed] [Google Scholar]
- 2.Meyer RR, Laine PS (1990) The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev 54: 342–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohman TM, Ferrari ME (1994) Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem 63: 527–570. [DOI] [PubMed] [Google Scholar]
- 4.Wold MS (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66: 61–92. [DOI] [PubMed] [Google Scholar]
- 5.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL (2008) SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol 43: 289–318. 10.1080/10409230802341296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohman TM, Bujalowski W, Overman LB (1988) E. coli single strand binding protein: a new look at helix-destabilizing proteins. Trends Biochem Sci 13: 250–255. [PubMed] [Google Scholar]
- 7.Raghunathan S, Kozlov AG, Lohman TM, Waksman G (2000) Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol 7: 648–652. [DOI] [PubMed] [Google Scholar]
- 8.Antony E, Weiland E, Yuan Q, Manhart CM, Nguyen B, Kozlov AG, et al. (2013) Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J Mol Biol 425: 4802–4819. 10.1016/j.jmb.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozlov AG, Weiland E, Mittal A, Waldman V, Antony E, Fazio N, et al. (2015) Intrinsically disordered C-terminal tails of E. coli single-stranded DNA binding protein regulate cooperative binding to single-stranded DNA. J Mol Biol 427: 763–774. 10.1016/j.jmb.2014.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antony E, Kozlov AG, Nguyen B, Lohman TM (2012) Plasmodium falciparum SSB tetramer binds single-stranded DNA only in a fully wrapped mode. J Mol Biol 420: 284–295. 10.1016/j.jmb.2012.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antony E, Weiland EA, Korolev S, Lohman TM (2012) Plasmodium falciparum SSB tetramer wraps single-stranded DNA with similar topology but opposite polarity to E. coli SSB. J Mol Biol 420: 269–283. 10.1016/j.jmb.2012.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein DA, Eggington JM, Killoran MP, Misic AM, Cox MM, Keck JL (2004) Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc Natl Acad Sci U S A 101: 8575–8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saikrishnan K, Manjunath GP, Singh P, Jeyakanthan J, Dauter Z, Sekar K, et al. (2005) Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association. Acta Crystallogr D Biol Crystallogr 61: 1140–1148. [DOI] [PubMed] [Google Scholar]
- 14.Saikrishnan K, Jeyakanthan J, Venkatesh J, Acharya N, Sekar K, Varshney U, et al. (2003) Structure of Mycobacterium tuberculosis single-stranded DNA-binding protein. Variability in quaternary structure and its implications. J Mol Biol 331: 385–393. [DOI] [PubMed] [Google Scholar]
- 15.Yang C, Curth U, Urbanke C, Kang C (1997) Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nat Struct Biol 4: 153–157. [DOI] [PubMed] [Google Scholar]
- 16.Bujalowski W, Lohman TM (1986) Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry 25: 7799–7802. [DOI] [PubMed] [Google Scholar]
- 17.Overman LB, Bujalowski W, Lohman TM (1988) Equilibrium binding of Escherichia coli single-strand binding protein to single-stranded nucleic acids in the (SSB)65 binding mode. Cation and anion effects and polynucleotide specificity. Biochemistry 27: 456–471. [DOI] [PubMed] [Google Scholar]
- 18.Bujalowski W, Overman LB, Lohman TM (1988) Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. J Biol Chem 263: 4629–4640. [PubMed] [Google Scholar]
- 19.Bujalowski W, Lohman TM (1989) Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. I. Evidence and a quantitative model. J Mol Biol 207: 249–268. [DOI] [PubMed] [Google Scholar]
- 20.Bujalowski W, Lohman TM (1989) Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. II. Salt, temperature and oligonucleotide length effects. J Mol Biol 207: 269–288. [DOI] [PubMed] [Google Scholar]
- 21.Roy R, Kozlov AG, Lohman TM, Ha T (2009) SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 461: 1092–1097. 10.1038/nature08442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy R, Kozlov AG, Lohman TM, Ha T (2007) Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J Mol Biol 369: 1244–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou R, Kozlov AG, Roy R, Zhang J, Korolev S, Lohman TM, et al. (2011) SSB functions as a sliding platform that migrates on DNA via reptation. Cell 146: 222–232. 10.1016/j.cell.2011.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bujalowski W, Lohman TM (1987) Limited co-operativity in protein-nucleic acid interactions. A thermodynamic model for the interactions of Escherichia coli single strand binding protein with single-stranded nucleic acids in the "beaded", (SSB)65 mode. J Mol Biol 195: 897–907. [DOI] [PubMed] [Google Scholar]
- 25.Kozlov AG, Lohman TM (2002) Kinetic mechanism of direct transfer of Escherichia coli SSB tetramers between single-stranded DNA molecules. Biochemistry 41: 11611–11627. [DOI] [PubMed] [Google Scholar]
- 26.Prusty D, Dar A, Priya R, Sharma A, Dana S, Choudhury NR, et al. (2010) Single-stranded DNA binding protein from human malarial parasite Plasmodium falciparum is encoded in the nucleus and targeted to the apicoplast. Nucleic Acids Res 38: 7037–7053. 10.1093/nar/gkq565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green M, Gilhooly NS, Abedeen S, Scott DJ, Dillingham MS, Soultanas P (2014) Engineering a reagentless biosensor for single-stranded DNA to measure real-time helicase activity in Bacillus. Biosens Bioelectron 61: 579–586. 10.1016/j.bios.2014.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bianco PR, Stanenas AJ, Liu J, Cohan CS (2012) Fluorescent single-stranded DNA-binding proteins enable in vitro and in vivo studies. Methods Mol Biol 922: 235–244. 10.1007/978-1-62703-032-8_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daley JM, Gaines WA, Kwon Y, Sung P (2014) Regulation of DNA pairing in homologous recombination. Cold Spring Harb Perspect Biol 6: a017954 10.1101/cshperspect.a017954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.San Filippo J, Sung P, Klein H (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257. 10.1146/annurev.biochem.77.061306.125255 [DOI] [PubMed] [Google Scholar]
- 31.Macris MA, Sung P (2005) Multifaceted role of the Saccharomyces cerevisiae Srs2 helicase in homologous recombination regulation. Biochem Soc Trans 33: 1447–1450. [DOI] [PubMed] [Google Scholar]
- 32.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, et al. (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309. [DOI] [PubMed] [Google Scholar]
- 33.Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T (2009) Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell 35: 105–115. 10.1016/j.molcel.2009.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colavito S, Macris-Kiss M, Seong C, Gleeson O, Greene EC, Klein HL, et al. (2009) Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res 37: 6754–6764. 10.1093/nar/gkp748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lytle AK, Origanti SS, Qiu Y, VonGermeten J, Myong S, Antony E (2014) Context-dependent remodeling of Rad51-DNA complexes by Srs2 is mediated by a specific protein-protein interaction. J Mol Biol 426: 1883–1897. 10.1016/j.jmb.2014.02.014 [DOI] [PubMed] [Google Scholar]
- 36.Heyer WD (2015) Regulation of recombination and genomic maintenance. Cold Spring Harb Perspect Biol 7: a016501 10.1101/cshperspect.a016501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Ehmsen KT, Heyer WD, Morrical SW (2011) Presynaptic filament dynamics in homologous recombination and DNA repair. Crit Rev Biochem Mol Biol 46: 240–270. 10.3109/10409238.2011.576007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunzelmann S, Webb MR (2009) A biosensor for fluorescent determination of ADP with high time resolution. J Biol Chem 284: 33130–33138. 10.1074/jbc.M109.047118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webb MR, Corrie JE (2001) Fluorescent coumarin-labeled nucleotides to measure ADP release from actomyosin. Biophys J 81: 1562–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brune M, Hunter JL, Corrie JE, Webb MR (1994) Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 33: 8262–8271. [DOI] [PubMed] [Google Scholar]
- 41.Brune M, Hunter JL, Howell SA, Martin SR, Hazlett TL, Corrie JE, et al. (1998) Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli. Biochemistry 37: 10370–10380. [DOI] [PubMed] [Google Scholar]
- 42.Walsh MT, Roller EE, Ko KS, Huang X (2015) Measurement of DNA Polymerase Incorporation Kinetics of Dye-Labeled Nucleotides Using Total Internal Reflection Fluorescence Microscopy. Biochemistry 54: 4019–4021. 10.1021/acs.biochem.5b00269 [DOI] [PubMed] [Google Scholar]
- 43.Sung P, Krejci L, Van Komen S, Sehorn MG (2003) Rad51 recombinase and recombination mediators. J Biol Chem 278: 42729–42732. [DOI] [PubMed] [Google Scholar]
- 44.Bianco PR, Tracy RB, Kowalczykowski SC (1998) DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci 3: D570–603. [DOI] [PubMed] [Google Scholar]
- 45.Eggleston AK, Kowalczykowski SC (1991) An overview of homologous pairing and DNA strand exchange proteins. Biochimie 73: 163–176. [DOI] [PubMed] [Google Scholar]
- 46.Kowalczykowski SC, Clow J, Krupp RA (1987) Properties of the duplex DNA-dependent ATPase activity of Escherichia coli RecA protein and its role in branch migration. Proc Natl Acad Sci U S A 84: 3127–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Menetski JP, Kowalczykowski SC (1985) Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J Mol Biol 181: 281–295. [DOI] [PubMed] [Google Scholar]
- 48.Zaitseva EM, Zaitsev EN, Kowalczykowski SC (1999) The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J Biol Chem 274: 2907–2915. [DOI] [PubMed] [Google Scholar]
- 49.Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJ, et al. (2007) Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure 15: 599–609. [DOI] [PubMed] [Google Scholar]
- 50.Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, et al. (2005) Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res 33: 3292–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Z, Yang H, Pavletich NP (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453: 489–484. 10.1038/nature06971 [DOI] [PubMed] [Google Scholar]
- 52.Rong L, Klein HL (1993) Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J Biol Chem 268: 1252–1259. [PubMed] [Google Scholar]
- 53.Lohman TM, Tomko EJ, Wu CG (2008) Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol 9: 391–401. 10.1038/nrm2394 [DOI] [PubMed] [Google Scholar]
- 54.Jia H, Korolev S, Niedziela-Majka A, Maluf NK, Gauss GH, Myong S, et al. (2011) Rotations of the 2B sub-domain of E. coli UvrD helicase/translocase coupled to nucleotide and DNA binding. J Mol Biol 411: 633–648. 10.1016/j.jmb.2011.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balci H, Arslan S, Myong S, Lohman TM, Ha T (2011) Single-molecule nanopositioning: structural transitions of a helicase-DNA complex during ATP hydrolysis. Biophys J 101: 976–984. 10.1016/j.bpj.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomko EJ, Jia H, Park J, Maluf NK, Ha T, Lohman TM (2010) 5'-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocase. EMBO J 29: 3826–3839. 10.1038/emboj.2010.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, et al. (2010) PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell 142: 544–555. 10.1016/j.cell.2010.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM (2005) Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A 102: 10076–10081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rasnik I, Myong S, Cheng W, Lohman TM, Ha T (2004) DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes. J Mol Biol 336: 395–408. [DOI] [PubMed] [Google Scholar]
- 60.Cheng W, Brendza KM, Gauss GH, Korolev S, Waksman G, Lohman TM (2002) The 2B domain of the Escherichia coli Rep protein is not required for DNA helicase activity. Proc Natl Acad Sci U S A 99: 16006–16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meiners MJ, Tahmaseb K, Matson SW (2014) The UvrD303 hyper-helicase exhibits increased processivity. J Biol Chem 289: 17100–17110. 10.1074/jbc.M114.565309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Centore RC, Leeson MC, Sandler SJ (2009) UvrD303, a hyperhelicase mutant that antagonizes RecA-dependent SOS expression by a mechanism that depends on its C terminus. J Bacteriol 191: 1429–1438. 10.1128/JB.01415-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang G, Deng E, Baugh L, Kushner SR (1998) Identification and characterization of Escherichia coli DNA helicase II mutants that exhibit increased unwinding efficiency. J Bacteriol 180: 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiu Y, Antony E, Doganay S, Koh HR, Lohman TM, Myong S (2013) Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat Commun 4: 2281 10.1038/ncomms3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robertson RB, Moses DN, Kwon Y, Chan P, Zhao W, Chi P, et al. (2009) Visualizing the disassembly of S. cerevisiae Rad51 nucleoprotein filaments. J Mol Biol 388: 703–720. 10.1016/j.jmb.2009.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312. [DOI] [PubMed] [Google Scholar]
- 67.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G (1997) Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell 90: 635–647. [DOI] [PubMed] [Google Scholar]
- 68.Lee JY, Yang W (2006) UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127: 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Korolev S, Yao N, Lohman TM, Weber PC, Waksman G (1998) Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci 7: 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.