Abstract
The mechanisms by which exercise mediates its multiple cardiac benefits are only partly understood. Prior comprehensive analyses of the cardiac transcriptional components and microRNAs dynamically regulated by exercise suggest that the CBP/p300-interacting protein CITED4 is a downstream effector in both networks. While CITED4 has documented functional consequences in neonatal cardiomyocytes in vitro, nothing is known about its effects in the adult heart. To investigate the impact of cardiac CITED4 expression in adult animals, we generated transgenic mice with regulated, cardiomyocyte-specific CITED4 expression. Cardiac CITED4 expression in adult mice was sufficient to induce an increase in heart weight and cardiomyocyte size with normal systolic function, similar to the effects of endurance exercise training. After ischemia-reperfusion, CITED4 expression did not change initial infarct size but mediated substantial functional recovery while reducing ventricular dilation and fibrosis. Forced cardiac expression of CITED4 also induced robust activation of the mTORC1 pathway after ischemic injury. Moreover, pharmacological inhibition of mTORC1 abrogated CITED4’s effects in vitro and in vivo. Together, these data establish CITED4 as a regulator of mTOR signaling that is sufficient to induce physiologic hypertrophy at baseline and mitigate adverse ventricular remodeling after ischemic injury.
The exercise-induced factor CITED4 is sufficient to induce physiological cardiac hypertrophy and promote functional recovery after ischemic injury through activation of mTOR signaling.
Introduction
Multiple lines of evidence suggest exercise has benefits for primary and secondary prevention of cardiovascular disease (1). Exercise induces important systemic changes affecting the cardiovascular system through alterations in metabolism, peripheral vessels, and skeletal muscle. However, studies in animal models suggest that exercise also induces changes intrinsic to the heart itself, which contribute to its benefits (2). Improved cardiovascular performance, enhanced metabolic efficiency, and cardiac growth in the form of physiologic hypertrophy are all adaptive responses to exercise (3).
Cardiac hypertrophy secondary to exercise can result in increases in left ventricular mass of 20% or more (4). While physiologic hypertrophy can grossly appear similar to pathological hypertrophy, the underlying processes are distinct at both the cellular and molecular levels (5). Although increased cardiomyocyte size underlies both physiologic and pathologic hypertrophy, the former is associated with proportional increases in both the length and width of cardiomyocytes, while the latter often demonstrates disproportionate increases in cardiomyocyte length that are thought to be maladaptive (6). In addition to a distinct morphology, exercise leads to improved cardiomyocyte calcium sensitivity (7), T-tubule organization (8), mitochondrial biosynthesis (9), and enhanced contractility (10). Exercise training reduces ischemic injury (11) but also independently mitigates adverse remodeling when initiated after infarction by attenuating fibrosis, ventricular dilation, and cardiac dysfunction (1, 12, 13).
Previous collaborative work from our laboratories profiled all known transcriptional components in hearts from mice exposed to swim stress in comparison with sedentary controls and with mice subjected to pressure-overload to identify genes differentially expressed in exercise (2). Among these, CITED4 (Creb binding protein [CBP]/p300–interacting transactivator with ED-rich carboxy-terminal domain-4) was increased in exercised hearts. CITED4 was originally cloned as a protein interacting with CBP/p300 and as a coactivator of the AP-2 family of transcription factors (14). More recent analyses of cardiac microRNA pathways regulated in 2 distinct models of exercise identified miR-222 as necessary for exercise-induced cardiac and cardiomyocyte growth and demonstrated that targets of miR-222 are negative regulators of CITED4 in neonatal cardiomyocytes (13). Thus, CITED4 has been implicated as a downstream effector of exercise-related transcriptional and microRNA pathways in neonatal cardiomyocytes.
Initial functional studies in vitro demonstrated that CITED4 induces hypertrophy and hyperplasia in neonatal cardiomyocytes (2). A subsequent high-throughput phenotypic screen of hypertrophic agonists on cardiomyocyte gene expression and morphology found that neuregulin-1 (NRG1), a physiological stimulus increased in exercise, was most closely linked to increased CITED4 expression (15). Interestingly, CITED4 expression attenuated cardiomyocyte elongation, promoting a more physiological growth pattern (15). However, the effects of CITED4 expression in vivo, particularly in the adult heart, are completely unknown.
To determine whether cardiac CITED4 expression is sufficient to recapitulate the effects of exercise, we developed an inducible, cardiac-specific transgenic model and characterized the effects of CITED4 expression at baseline and after ischemic injury.
Results
Creation of a cardiac-specific CITED4 transgenic.
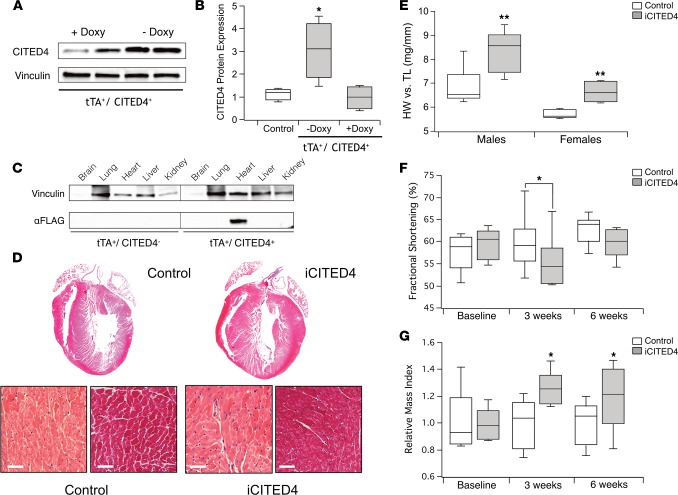
To examine the effects of CITED4 expression in vivo, we generated inducible, cardiac-specific transgenic mice — referred to throughout as iCITED4 — in which transgene expression is suppressed by doxycycline (16). Removal of doxycycline from chow in 8- to 10-week-old mice resulted in robust expression of the FLAG-tagged CITED4 transgene in double-positive mice (tTA+/CITED4+) (Figure 1A). The induction of CITED4 by transgene activation culminated in a 3-fold increase in CITED4 protein (P < 0.01 by 1-way ANOVA) compared with iCITED4 mice still under doxycycline suppression or single-positive (tTA+ or CITED4+) controls (Figure 1B). There was full expression of the transgene just 6 days after removal of doxycycline, and expression persisted at similar levels over the ensuing weeks (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/jci.insight.85904DS1). Examination of multiple tissues confirmed that expression of the FLAG-tagged CITED4 transgene was cardiac specific (Figure 1C) and had minimal expression when not induced (Supplemental Figure 1B). Quantitative PCR (qPCR) performed on cardiomyocytes and noncardiomyocytes isolated from adult iCITED4 mice demonstrated that the CITED4 transgene was induced only in cardiomyocytes (Supplemental Figure 2, A and B).
Figure 1. Cardiac expression of CITED4 produces physiological cardiac hypertrophy.
(A) Western blot demonstrating CITED4 from whole-heart lysates after 3 weeks of expression by removal of doxycycline (Doxy) in transgenic mice expressing tTA and CITED4-FLAG. (B) Quantification of CITED4 expression as compared with control mice in iCITED4 without doxycycline (3.04- ± 0.63-fold) or with doxycycline (1.13- ± 0.13-fold); n = 4 mice per group, *P < 0.05 by 1-way ANOVA for serial comparisons. (C) Restricted cardiac expression of CITED4-Flag in the double-positive (tTA+/CITED4+) transgenics as demonstrated by Western blot from lysates from multiple tissues. (D) Gross and microscopic histology of representative samples, including H&E (left panels) and MTS for fibrosis (right panels). Scale bars: 50 μm. (E) Increases in heart weight to tibial length for males (8.36 ± 0.35 mg/mm vs. 6.84 ± 0.32 mg/mm) and for females (6.62 ± 0.22 mg/mm vs. 5.72 ± 0.08 mg/mm). n = at least 5 animals per group. (F) Fractional shortening before and after induction of iCITED4 for 3 weeks and 6 weeks, respectively; n = 8–12 males per group. (G) Increase in relative mass index after 3 weeks of induction in CITED4 transgenics remains stable for at least 6 weeks of expression. Relative increase in LV mass for CITED4 transgenics as compared with controls: 27.7% ± 5.2% at 3 weeks and 25.1% ± 7% at 6 weeks. For all graphs: significance determined by the Student’s t test unless otherwise specified; *P < 0.05, **P < 0.01.
Cardiac expression of CITED4 induces physiologic hypertrophy.
Three weeks after doxycycline withdrawal, heart weight in iCITED4 mice increased 15%–20% in both male and female mice compared with littermate controls (Figure 1E). There was no difference in lung weight, demonstrating iCITED4 mice do not develop pulmonary congestion suggestive of heart failure (Supplemental Figure 2C). Histological examination by H&E staining and Masson’s Trichrome stain (MTS) confirmed concentric hypertrophy without an increase in fibrosis (Figure 1D). Echocardiographic analyses before and 3 weeks after induction of CITED4 demonstrated a small but statistically significant decrease in fractional shortening in iCITED4 mice, as compared with single-positive controls (Figure 1F). With continued expression up to 6 weeks, there was no difference in fractional shortening between iCITED4 animals and controls, suggesting the initial small decrease in ventricular function was a transient effect that did not lead to progressive decline (Figure 1F and Supplemental Figure 3). Consistent with the gravimetric data, the relative mass index determined by echocardiography (17) at both 3 and 6 weeks after CITED4 expression demonstrated a stable, approximately 25% increase in left-ventricular mass over the entire period (Figure 1G). These data demonstrate that cardiac-specific expression of CITED4 induces stable cardiac hypertrophy with normal cardiac function. This is similar to what is seen after endurance training in mice.
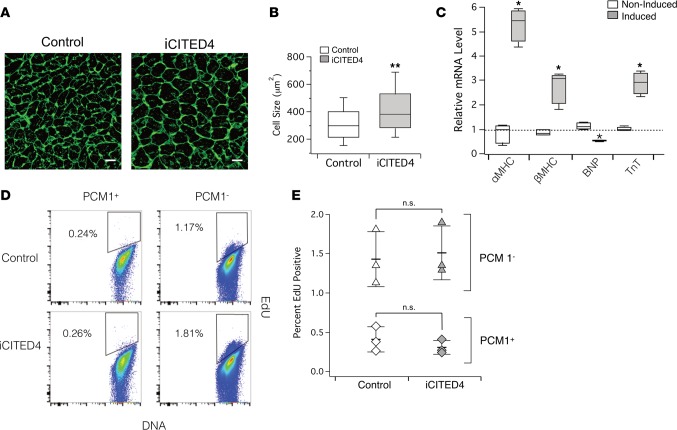
Histological analyses of Wheat Germ Agglutinin–stained (WGA-stained) cardiac sections demonstrated a 24.5% increase (P < 0.01) in cardiomyocyte cross-sectional area in CITED4-expressing transgenics compared with matched controls (Figure 2, A and B), suggesting that the change in cardiomyocyte size is sufficient to account for the increased heart mass. qPCR revealed increases in the ratio of αMHC/βMHC and expression of Troponin T (TnT), as well as a decrease in brain natriuretic peptide (BNP) (Figure 2C). These changes are characteristic of physiological hypertrophy. Exercise induces markers of proliferation in adult hearts (2, 13, 18), and forced expression of CITED4 increased proliferation of neonatal rat ventricular myocytes (NRVMs) (2, 15). To examine whether cardiomyocyte proliferation contributed to the increased heart size seen in adult hearts with short-term CITED4 expression, we administrated the thymidine analogue EdU for the last 2 weeks of induction. Since unambiguous identification of cardiomyocyte nuclei is challenging in fixed heart sections (19), we isolated nuclei and stained them for PCM1 to identify cardiomyocyte nuclei (20). The percent EdU-positive PCM1-positive cardiomyocyte nuclei as determined by flow cytometry was consistent with the low basal rate of DNA synthesis reported by others (21) and was not altered by CITED4 expression (Figure 2, D and E). Taken together, these data suggest cardiomyocyte-specific expression of CITED4 in the adult heart is sufficient to induce physiological cardiac hypertrophy similar to that seen with endurance exercise training (22). However, CITED4 does not appear to induce baseline cardiomyocyte proliferation in the adult mouse heart, at least as reflected in PCM1-positive nuclei over the time frames examined.
Figure 2. CITED4 expression for 2 weeks increases cell size but does not induce EdU incorporation of PCM1-positive cardiomyocytes at baseline.
(A) Representative images of cardiac sections stained with WGA demonstrate increased cardiomyocyte cell size in iCITED4 transgenics. (B) Quantification of increased cell size (343 ± 4.6 μm2 vs. 427 ± 6.4 μm2, n = ~800 cells per group, 3 hearts per group, **P < 0.001). Scale bar: 30 μm. (C) Quantitative PCR (qPCR) demonstrates induction of mRNA transcripts consistent with physiologic cardiac hypertrophy gene profile. (D) Flow cytometry of adult cardiomyocyte nuclei stained for the marker PCM1 demonstrates no increase in EdU positivity in CITED4 transgenics. (E) Quantification of flow cytometry data demonstrates no difference in the amount of DNA synthesis as a marker for proliferation in cardiac nuclei from iCITED4 transgenics. For all graphs: significance determined by the Student’s t test unless otherwise specified; *P < 0.05, **P < 0.01.
CITED4 improves cardiac remodeling and survival after ischemic injury.
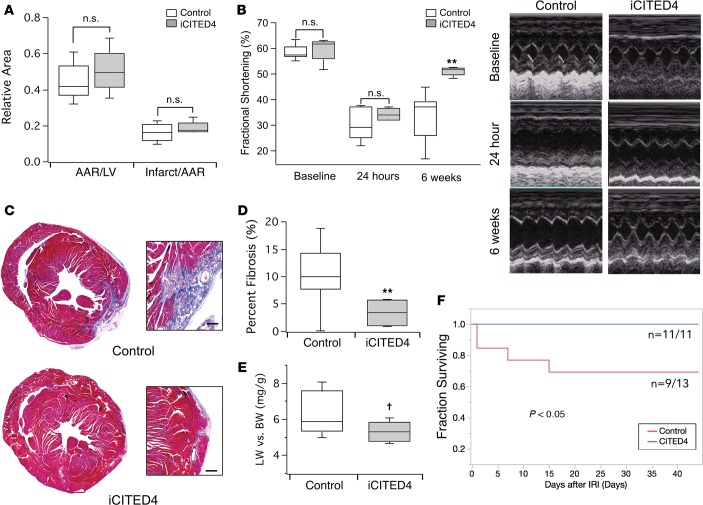
Exercise reduces initial ischemic injury (23). However, exercise also reduces adverse remodeling, attenuating fibrosis, ventricular dilatation, and cardiac dysfunction in both animal models (24) and humans (25). Thus, we examined whether CITED4 expression is sufficient to mediate the effects of exercise on the response to ischemic injury. We subjected iCITED4 mice or controls to ischemia-reperfusion injury (IRI) induced by 30 minutes of left coronary artery ligation followed by reperfusion. CITED4 expression had no effect on the initial infarct size, as determined by triphenyltetrazolium chloride (TTC) staining at 24 hours (Figure 3A). Consistent with this, echocardiography showed a similar degree of dysfunction in CITED4-expressing transgenics and control mice at 24 hours (Figure 3B). However, 6 weeks after ischemic injury, ventricular function in CITED4-expressing mice recovered to near normal levels, while there was little to no recovery in littermate controls (Figure 3B). Correspondingly, there was a 3.4-fold reduction (P < 0.05) in cardiac fibrosis in iCITED4 mice as compared with controls (Figure 3, C and D). There was also a nonsignificant trend toward lower lung weights (P = 0.06), suggestive of protection against heart failure (Figure 3E). Finally, there was a 30% reduction in mortality (P < 0.05) in iCITED4 transgenics as compared with controls 6 weeks after cardiac injury (Figure 3F). These data demonstrate that, while CITED4 expression does not change initial infarct size after ischemic injury, it is sufficient to promote functional recovery, dramatically reducing adverse cardiac remodeling, late fibrosis, and mortality.
Figure 3. CITED4 improves cardiac remodeling after ischemia/reperfusion injury.
(A) No change in infarct size at 24 hours after ischemia reperfusion injury (IRI) from transient ligation of the left anterior descending artery (LAD) as demonstrated by TTC staining. (B) Noninvasive assessment of ventricular function demonstrates substantial dysfunction at 24 hours after IRI that improves significantly over time in the iCITED4 transgenics but not controls. n ≥ 5 mice per group per time point for analysis. **P < 0.01. (C) Representative micrographs of cardiac sections stained with MTS demonstrating a reduction of fibrosis in iCITED4 transgenics 4 weeks after IRI. High-magnification inlays demonstrate area of fibrosis and myocyte loss. Scale bar: 200 μm. (D) Quantification of fibrosis as a percentage of the total area (Control: 11.9% ± 2.6%, iCITED4: 3.5% ± 1.2%, n = 5 per group). **P < 0.01. (E) Trend to reduced lung weights consistent with a decreased degree of heart failure in CITED4 expressing mice, †P = 0.06 by Student’s t test. (F) Improvement in mortality in iCITED4 transgenics as compared with controls; n = 13 (control), 11 (iCITED4) at the start of the experiment; P < 0.05 by Log-Rank parametric test. For all graphs: significance determined by the Student’s t test unless otherwise specified; *P < 0.05, **P < 0.01.
CITED4 regulates autophagy and apoptosis after ischemic injury.
Autophagy is a self-degradative process that plays important roles in homeostasis through removal of defective proteins and organelles. In the heart, autophagy alters the response to ischemia and reperfusion. However, autophagy can have either beneficial or detrimental effects in cardiac IRI depending on the context (26). After reperfusion, autophagosomes accumulate in the heart due to impaired clearance and lead to increases in ROS, mitochondrial permeability, and cardiomyocyte death (27). Because exercise activates basal autophagy in both skeletal and cardiac muscle (28), we examined whether CITED4 expression alters autophagy at baseline or after IRI.
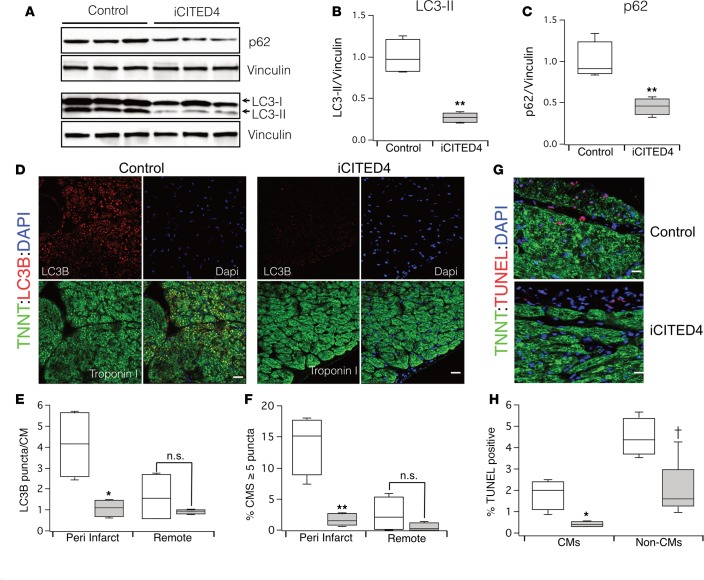
CITED4 expression did not significantly alter basal autophagy in the heart, as indicated by levels of LC3-II, a marker of autophagosomes (data not shown). We next examined hearts from iCITED4 transgenics and controls 1 week after IRI, when ventricular function begins to diverge between the groups as the iCITED4 mice recover (Supplemental Figure 4). Whole-heart lysates from CITED4 transgenics demonstrated a substantial reduction in LC3-II levels 1 week after IRI compared with controls (Figure 4, A and B). To determine the cell lineage involved, we quantified the number of LC3-II puncta on histological sections and found a dramatic reduction in the number of puncta in cardiomyocytes (Figure 4, D–F). LC3-II levels and the number of autophagosomes reflect both changes in activation of autophagy and flux through the pathway (29). Interestingly, cardiac levels of p62, a scaffolding protein that accumulates when flux is impaired, were reduced > 2-fold in CITED4 transgenics compared with controls (Figure 4, A and C). Thus, although CITED4 expression does not regulate basal autophagy in the heart, it reduces the accumulation of autophagosomes in cardiomyocytes after reperfusion, which could reflect either reduced formation or enhanced elimination.
Figure 4. Modulation of autophagy with CITED4 expression 1 week after IRI.
(A) Western blot demonstrating accumulation of LC3-II and p62 in control samples as compared with iCITED4 transgenics. (B) Quantification of LC3-II (relative reduction of 71% ± 3.1% in iCITED4 mice) and (C) quantification of p62 (Relative reduction of 54% ± 8% in iCITED4 transgenics). (D) Representative micrographs of cardiac sections demonstrating accumulation of LC3B dots in cardiomyocytes (CMs). Sections were stained with antibodies for Cardiac Troponin T (green), LC3B (red), and DAPI (blue). Scale bars: 50 μm. (E) Quantification of LC3B puncta per cardiomyocyte and (F) percentage of cardiomyocyte with greater than 5 LC3B puncta in both the peri-infarct area and remote to the area of injury. n = 5 mice per group and at least 20 high power fields (hpf) per section. (G) Micrographs of TUNEL staining demonstrate a reduction in apoptosis in iCITED4 transgenics. Sections were stained with antibodies for Cardiac Troponin T (green), TUNEL (red), and DAPI (blue). Scale bars: 50 μm. (H) Quantification of TUNEL-positive cells both CMs and noncardiomyocytes (Non-CMs). †P = 0.06 for Non-CMs by Student’s t test. For all graphs: significance determined by the Student’s t test unless otherwise specified; *P < 0.05, **P < 0.01.
As noted above, reperfusion-induced accumulation of autophagosomes can contribute to cardiomyocyte loss (30). Consistent with the reduction in autophagosome accumulation in iCITED4 hearts, TUNEL staining 1 week after IRI also demonstrated a reduction in cardiomyocyte apoptosis (Figure 4, G and H). To determine whether CITED4 also has direct antiapoptotic effects in cardiomyocytes, we expressed CITED4 in NRVMs that were subjected to hypoxia-reoxygenation. CITED4 expression substantially reduced cardiomyocyte apoptosis after hypoxia-reoxygenation in vitro to an extent comparable with that seen with pharmacological IGF-I treatment (Supplemental Figure 5A). Expression profiling in NRVMs revealed that CITED4 expression in NRVMs upregulated several antiapoptotic genes, including BCL2 (data not shown) — which was confirmed by qPCR (Supplemental Figure 5B) — likely accounting for the observed anti-apoptotic effect. We concluded that CITED4 expression is sufficient to reduce apoptosis of cardiomyocytes both in vitro and in vivo after an ischemic injury.
mTORC1 signaling is activated by CITED4 expression and necessary for physiologic hypertrophy and improved cardiac remodeling after injury.
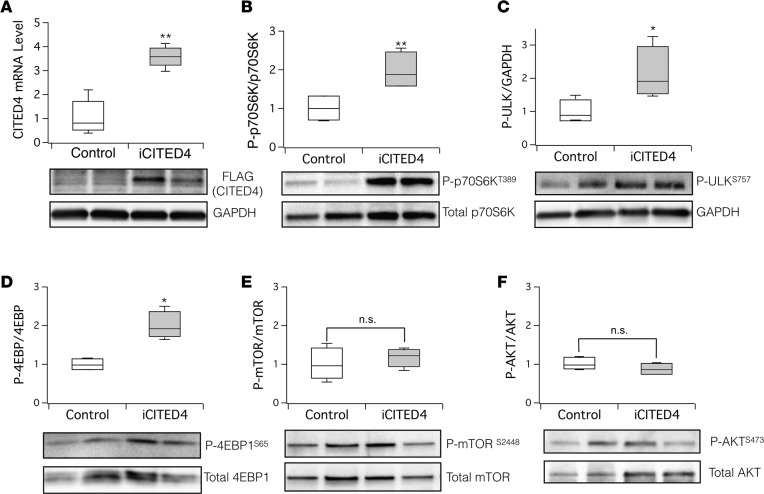
The serine/theorine kinase mTOR is a central regulator of cell growth and autophagy through phosphorylation of downstream substrates including ULK1/2 and Atg13 (31). We interrogated mTOR signaling in hearts from iCITED4 transgenics and controls 1 week after IRI (Figure 5A), since this is when cardiac function is clearly recovering in iCITED4 mice but not controls (Supplemental Figure 4). mTOR signals via 2 distinct complexes, mTORC1 and mTORC2, which are distinguished by their associated components, their specific downstream signaling targets, and their sensitivity to rapamycin (32). mTORC2 is less sensitive to rapamycin and mediates the phosphorylation of serine 473 (S473) on AKT1 and its subsequent activation (33). Immunoblotting did not reveal any increase in phosphorylation of mTOR (Figure 5E) or of AKT1 (S473; Figure 5F) in iCITED4 transgenics, suggesting that mTORC2 is not affected by CITED4 expression. In contrast, phosphorylation of targets of mTORC1 — including P70 S6 kinase (p70S6K) (Figure 5B), ULK1/2 (Figure 5C), and the regulator of protein translation 4EBP1 (Figure 5D) — was consistently increased by CITED4 expression. AMP kinase is a multisubunit protein that regulates multiple metabolic pathways including mTOR and can directly activate autophagy through phosphorylation of ULK1 (34). Phosphorylation of AMPK (T172) normalized to total AMPK was also not altered by CITED4 expression (Supplemental Figure 6). Taken together, these data demonstrate that CITED4 expression leads to mTORC1 activation in vivo without activation of either AMPK or AKT1.
Figure 5. CITED4 expression activates mTORC1 signaling targets in the heart after ischemic injury.
Increased CITED4 expression in hearts 1 week after IRI activates targets of mTORC1 including p70S6K, ULK, and 4EBP1 without affecting AKT or mTOR phosphorylation. (A) Quantitative PCR (qPCR) from whole-heart mRNA (upper panel) and Western blotting of FLAG-tagged CITED4 from whole-heart lysates (lower panel), demonstrating increased CITED4 expression in the hearts of iCITED4 transgenics 1 week after ischemic injury. Western blotting of whole-heart lysates with phospho-specific antibodies at the indicated phosphorylation site (lower panels) and quantification by densitometry (upper panels) reveals a significant increase in the downstream targets of mTORC1, including p70S6K (B), ULK (C), and 4EPB1 (D), without any significant change in the phosphorylation of mTOR at serine 2448 (E) or AKT at serine 473 (F). n = 4 mice for each group. For all graphs: significance determined by the Student’s t test unless otherwise specified; *P < 0.05, **P < 0.01.
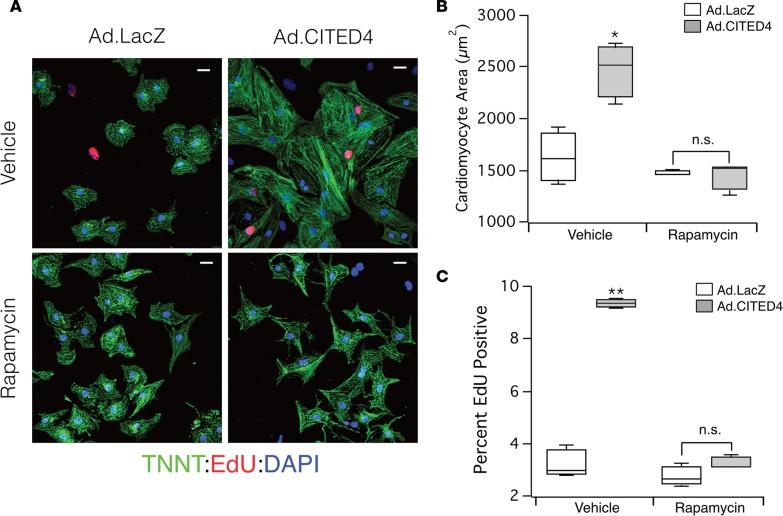
To examine the functional contribution of mTOR activation to the phenotypes induced by CITED4 expression, we examined the effects of the mTOR inhibition by rapamycin. As previously described, adenoviral expression of CITED4 in NRVMs increases both cell size (hypertrophy) and DNA synthesis (proliferation marker), as determined by incorporation of the thymidine analogue (EdU) (2, 15). Expression of CITED4 increased cardiomyocyte size (Figure 6, A and B), and this effect was completely blocked by rapamycin. CITED4 also increased NRVM EdU incorporation almost 3-fold (LacZ: 3.26 ± 0.44% vs. CITED4: 9.35 ± 0.11%, P < 0.001); EdU incorporation was also inhibited by rapamycin (LacZ: 2.74 ± 0.31% vs. CITED4: 3.31 ± 0.15% P = 0.4) (Figure 6, A and C). Notably, rapamycin only inhibited the increase in cell size and proliferation induced by CITED4 expression but did not reduce baseline NRVM size or EdU incorporation. Thus, CITED4-driven activation of mTORC1 appears necessary for its effects on NRVM hypertrophy and proliferation in vitro.
Figure 6. Inhibition of mTOR with rapamycin blocks effects of CITED4 expression in vitro.
(A) Representative micrographs demonstrating increased cell size and EdU uptake with adenoviral expression of CITED4 that is blocked by addition of rapamycin. Staining antibodies: Cardiac Troponin T (green), EdU (red), and DAPI (blue). Scale bars: 25 μm. (B) CITED4 expression increases cell size with vehicle (Ad.LacZ: 1,631 ± 202 μm2 vs. Ad.CITED4: 2,459 ± 212 μm2, P < 0.05) but is blocked with addition of 10 μM of rapamycin (Ad.LacZ: 1,473 ± 173 μm2 vs. Ad.CITED4: 1,434 ± 108 μm2). Analysis performed on 400–800 cells per sample and 3 samples per condition. (C) Stereotypical increases in proliferation observed with CITED4 expression in NRVMs as compared with LacZ controls is completely inhibited by treatment with rapamycin. For all graphs: significance determined by the Student’s t test unless otherwise specified; *P < 0.05, **P < 0.01.
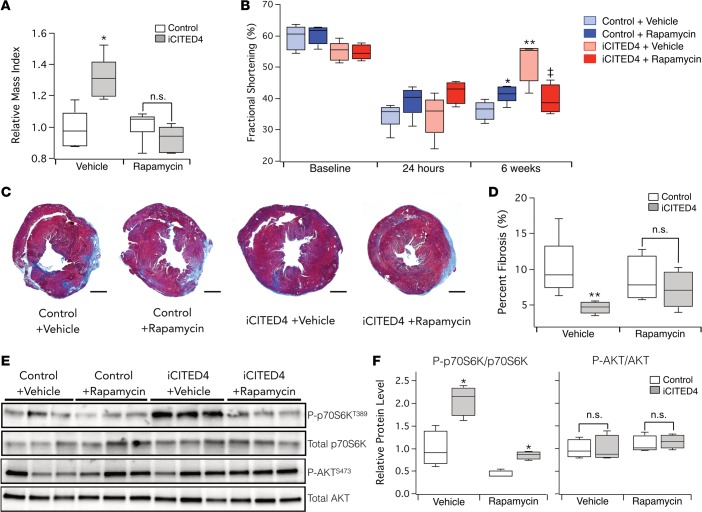
To determine if the phenotypes seen with CITED4 expression in vivo are also mediated by mTORC1, we treated iCITED4 transgenics and controls with rapamycin before and after IRI. The increased left ventricle (LV) mass evident on echocardiography with CITED4 expression was completely blocked by rapamycin (Figure 7A), suggesting that in vivo, as in vitro, CITED4-induced cardiac growth is mTOR dependent.
Figure 7. Treatment with rapamycin inhibits the development of physiologic cardiac hypertrophy and cardioprotection from ischemic injury induced by CITED4.
(A) Measure of relative mass by echocardiography demonstrates an increase at baseline with CITED4 expression, which is inhibited by daily rapamycin (2 mg/kg) treatment. (B) Echocardiography demonstrates a nonsignificant trend toward improved function in animals receiving rapamycin versus those injected with vehicle (controls: with vehicle fractional shortening (FS) = 34.5% ± 1.6% vs. with rapamycin FS = 39.2% ± 2.1%, P = 0.1; iCITED4: with vehicle FS = 34.6% ± 3% vs. with rapamycin FS = 42.2% ± 1.8%, P = 0.08). Repeat echocardiography at 6 weeks after IRI demonstrated substantial recovery in CITED4 transgenics that was blocked by rapamycin treatment, **P < 0.01 by 1-way ANOVA. (iCITED4: with vehicle FS = 51.6% ± 2.8% vs. with rapamycin FS = 42.2% ± 1.8%, ‡P < 0.01) Comparison of control mice treated with rapamycin demonstrated recovery compared with mice receiving vehicle (controls: with vehicle % FS = 36.3% ± 1.2%, vs. with rapamycin FS = 41.5% ± 1.2%, *P = 0.01), n = 4–6 animals per group. (C) Representative images of cardiac sections stained with MTS demonstrated a reduction in the fibrosis in iCITED4 mice receiving vehicle. Scale bars: 1 mm. (D) Quantification of fibrosis 6 weeks after IRI revealed reduced fibrosis in CITED4 transgenics that was inhibited by rapamycin treatment (vehicle, controls: 9.9% ± 1.6% vs. iCITED4: 4.6% ± 0.4%, **P = 0.01; rapamycin, controls: 8.5% ± 1.6% vs. iCITED4: 7.1% ± 1.3%; n.s. designates P = 0.48). n = 4–6 animals per group. (E) Protein lysates from hearts 6 weeks after IRI demonstrate increased phosphorylation of S6 kinase in iCITED4 mice receiving vehicle, which was suppressed by administration of rapamycin, without a significant change in AKT1 activation. (F) Quantification of Western blotting reveals increased S6 kinase activation with CITED4 expression; *P < 0.05 by 1-way ANOVA. Rapamycin inhibits S6 kinase phosphorylation in control mice administered rapamycin by ratio of phosphorylated p70S6K to total p70S6K (P-p70S6K/p70S6K) (vehicle: 1 ± 0.26 vs. rapamycin: 0.44 ± 0.05, *P = 0.05) and blocks activation of S6 kinase by CITED4 (vehicle: 2.06 ± 0.22 vs. rapamycin: 0.85 ± 0.06, *P < 0.01). n.s. denotes no significant difference between CITED4 transgenics given rapamycin and controls. Student’s t test was used in paired groups unless otherwise specified.
We then performed IRI in iCITED4 transgenics and controls receiving vehicle or rapamycin and followed cardiac function by echocardiography. In WT mice, rapamycin has an acute cardioprotective effect thought to be mediated by PI-3 kinase and/or JAK2-STAT3, but it also mitigates adverse remodeling — possibly through inhibition of negative feedback to IRS-1 (35). We hypothesized that the favorable remodeling seen with CITED4 expression is dependent on mTOR activation and is similar to that observed with transgenic mTOR expression in the heart (36). Therefore, rapamycin should specifically exacerbate remodeling in CITED4 transgenics as well as mediate an acute cardioprotective effect in both control and iCITED4 mice. As seen previously, at 24 hours, there was no significant difference in the fractional shortening between controls and iCITED4 transgenics receiving vehicle (Figure 7B). There was a trend toward improvement in ventricular function in both the iCITED4 transgenics and controls treated with rapamycin as compared with those receiving vehicle (P = 0.08 and 0.1, respectively) (Figure 7B), consistent with the acute cardioprotective effects of rapamycin (37). Six weeks after IRI, control mice receiving rapamycin demonstrated modest functional improvement compared with controls receiving vehicle, likely reflecting the initial cardioprotective effect and smaller infarcts compared with vehicle-treated animals. In contrast, the functional recovery seen again in iCITED4 transgenics treated with vehicle was completely blocked by rapamycin (Figure 7B). Correspondingly, late fibrosis in controls improved while fibrosis in iCITED4 transgenics worsened in response to rapamycin (P = 0.01) (Figure 7, C and D). Thus, in all respects, iCITED4 transgenics treated with rapamycin behave indistinguishably from littermate controls after IRI. The increased S6 kinase phosphorylation at threonine 389 indicative of mTORC1 activation in iCITED4 transgenics was also suppressed by rapamycin treatment (Figure 7, E and F). As before, there was no increase in AKT1 (S473) phosphorylation with CITED4 expression, suggesting an absence of mTORC2 activation. Thus, the combination of the functional, anatomic, and signaling data in controls and iCITED4 transgenics with or without rapamycin treatment all strongly suggest that CITED4 induction of both physiologic hypertrophy and improved cardiac remodeling after IRI in vivo is mediated by activation of mTORC1.
Discussion
To examine the effects of CITED4 in the adult heart in vivo, we generated an inducible transgenic mouse line that results in regulated expression of CITED4 specifically in cardiomyocytes. Transgene induction in adult animals culminated in an approximately 3-fold increase in CITED4 protein levels, comparable with increases seen in exercise (2). Cardiac CITED4 expression was sufficient to recapitulate many but not all of the phenotypes seen with endurance exercise training. Heart and cardiomyocyte size increased without fibrosis, and cardiac function remained normal with induction of physiologic hypertrophy genes. Although prior studies suggested that exercise induces an increase in cardiac proliferation markers (2, 13, 18) and CITED4 induces proliferation in NRVMs (Figure 7, A and C), we did not see an increase in EdU incorporation in PCM1-positive cardiomyocyte nuclei in iCITED4 transgenics during 2 weeks of observation. This suggests CITED4 expression is not sufficient to induce this phenotype in PCM1-positive cardiomyocyte nuclei in adult hearts in vivo over these time frames. Whether CITED4 expression can enhance cardiomyogenesis over longer time frames and/or be induced by other pathophysiological stimuli — or whether there could be a PCM1-negative subset of cardiomyocytes undetected in these assays — are interesting questions for future investigation.
Using a model of myocardial IRI, we found that CITED4 expression did not significantly affect initial infarct size at 24 hours but substantially enhanced functional recovery at 6 weeks, in association with a 3-fold reduction in late fibrosis, a trend toward lower lung weights, and improved survival. Taken together, these data demonstrate that, although CITED4 expression does not confer acute cardioprotection, it does markedly mitigate adverse remodeling after ischemic injury. Since exercise training has independent beneficial effects on both initial infarct size and late adverse remodeling (38), we infer that CITED4-independent pathways mediate the former, while CITED4-dependent pathways are likely major contributors to the latter. Adverse remodeling is an important cause of human heart failure that may be more amenable to therapeutic targeting than initial infarct size, given the unanticipated nature of acute coronary syndromes. Therefore, finding ways to induce or activate CITED4 warrants further investigation as a potentially feasible strategy to promote functional recovery and lessen adverse remodeling after ischemic injury.
CITED4 expression decreased both the accumulation of autophagosomes and cardiomyocyte apoptosis after IRI. Although ischemia induces autophagy, reperfusion appears to impair autophagic flux, leading to accumulation of both LC3-II and p62 thought to contribute to cardiomyocyte apoptosis (26). While we did not directly examine autophagic flux, our results are consistent with these studies in demonstrating autophagosome accumulation with increased LC3-II and p62 that were mitigated by CITED4 expression. This CITED4-induced reduction in autophagosome accumulation also correlated with a decrease in cardiomyocyte apoptosis demonstrated in vivo. The reduction in autophagosome accumulation may seem paradoxical, given data that exercise induces autophagy in both skeletal and cardiac muscle (39). However, it is possible that the reduction in autophagosomes after reperfusion reflects enhanced autophagic flux or that the effects of exercise-related pathways on autophagy are different in settings such as IRI.
These phenotypes are most consistent with increased activation of mTORC1 and its downstream targets in iCITED4 transgenics. Inhibition of mTORC1 with rapamycin revealed that both the cardiomyocyte hypertrophy and proliferation induced by CITED4 in vitro and the increase in LV mass observed in vivo were mTORC1 dependent. While the relative contribution of mTORC2 appears to be negligible, given both the lack of altered AKT phosphorylation and the sensitivity to rapamycin, other signaling mechanisms may predominate in different cellular contexts. Interestingly, the initial (24 hour) dysfunction after IRI in both CITED4-expressing mice and controls was slightly improved by rapamycin treatment, consistent with the previously described acute cardioprotective effect of rapamycin (40). In contrast, while rapamycin-treated controls demonstrated better function at 6 weeks compared with vehicle-treated controls, consistent with smaller initial infarcts, the iCITED4 transgenics treated with rapamycin did substantially worse than vehicle-treated iCITED4 mice, bringing them back to control levels of dysfunction. These divergent responses to rapamycin of control and iCITED4 mice underscore the specificity of the observations and support the role of mTOR in mediating the beneficial effects of CITED4 on ventricular remodeling. These data implicate CITED4 as a regulator of mTOR signaling that is sufficient to induce physiologic hypertrophy similar to that seen with endurance exercise training and demonstrate that mTORC1 activation is necessary for these effects.
In summary, we examined the effects in the adult heart of expressing CITED4, a common downstream effector in cardiac transcriptional and microRNA pathways differentially regulated in the heart in response to exercise. We found that CITED4 expression was sufficient to induce physiological heart growth, similar to that seen with endurance exercise training. CITED4 did not alter the initial infarct size or cardiac dysfunction after IRI, but it induced a remarkable functional recovery over the ensuing weeks, associated with reduced cardiomyocyte autophagosome accumulation and apoptosis, a dramatic reduction in fibrosis, more favorable LV remodeling, and improved survival. These phenotypes appear dependent on CITED4-induced activation of mTORC1. Given these findings, learning to manipulate CITED4 expression or activity in vivo warrants further investigation as a therapeutic strategy to promote functional recovery and mitigate adverse ventricular remodeling after ischemia injury.
Methods
Generation of iCITED4 transgenic mice.
A tetracycline-off binary α-MHC transgene system was used as previously described (16). For the responder mouse line, the full-length CITED4 transcript was amplified by PCR with a C-terminal FLAG tag from a mouse library and subcloned into a vector generously given by Jeffrey Robbins (Cincinnati Children’s Hospital, Cincinnati, Ohio, USA) (16). A Not I fragment was microinjected into FVB/NJ oocytes and transferred to pseudopregnant mice. After confirmation of stable Mendelian transmission, positive mice were bred to the appropriate tTA driver line, and cardiac-specific, doxycycline-regulated CITED4 expression was confirmed in line 9, which was used for all the experiments presented. Double-positive mice (tTA+ and CITED4+) are referred to as iCITED4 mice throughout the text unless otherwise specified. Doxycycline was administered in the food using a special diet formulated by Purina (200 mg/kg in pellets). To induce CITED4 expression, 8- to 10-week-old mice were fed normal chow without doxycycline for the indicated interval, usually 3 weeks prior to experiments or analysis. tTA and CITED4 lines were always maintained as single-positive lines and crossed for experiments to ensure consistent copy number and expression levels of the transgene. Male mice were used throughout the study unless indicated otherwise. The parent strain was FVB, and at least 4 backcrosses were performed prior to the experiments presented in this study.
Ischemia reperfusion model and analyses.
Transgenic iCITED mice and controls were subjected to ischemia reperfusion as previously described (41). Briefly, the left anterior descending artery (LAD) was ligated with 7-0 silk (Ethicon). Five minutes into ischemia, 50 μl of fluorescent microspheres (10 μm FluoSpheres, Molecular Probes) were injected into the LV cavity to delineate the perfused area and nonperfused area-at-risk (AAR). Following 30 minutes of LAD occlusion, the LAD ligature was released, and reperfusion was confirmed visually. After 24 hours, 1 week, or 6 weeks of reperfusion, mice were sacrificed and hearts were collected for analyses. To determine the myocardial infarct size, hearts were stained with 2,3,5-TTC (Sigma-Aldrich) as reported previously (42). In animals euthanized 6 weeks after IRI, 2-chamber cardiac sections were made and stained with MTS to identify fibrosis. All surgeries and analyses were performed by investigators blinded to genotype or treatment. For the rapamycin experiments, animals were treated with rapamycin at 2 mg/kg or vehicle by daily i.p. injections starting 3 weeks before ischemic injury for a total period of 9 weeks.
Cardiomyocyte isolation, culture, and transfection.
Primary NRVMs were prepared as described (43). Isolated NRVMs were purified by preplating. Before treatment, NRVMs were synchronized and cultured in serum-free media 24 hours after plating. EdU was added to the media 18 hours prior to fixation with 4% paraformaldehyde. EdU detection was performed according to the manufacturer’s protocol, and cells were counter-stained with an antibody to TnT (Abcam, catalog ab56357).
Cardiomyocytes and noncardiomyocytes were isolated from adult mice as described previously (13). Briefly, left ventricles were harvested from mice and Langendorff perfused with collagenase. Digested hearts were than dissected into small pieces and dissociated into single cells. After filtering, the cell solution was allowed to settle for several minutes in a Falcon tube (Falcon). The cell pellet and supernatant were transferred to individual Falcon tubes for further separation. The cell pellet was resuspended in transfer buffer and settled to precipitation. After the second precipitation, the cell pellet was checked for typical rod-shaped morphology before RNA and microRNA extraction to confirm expression of cardiomyocyte markers. The initial supernatant was centrifuged first at 50 g (3 minutes) and then 300 g (5 minutes) before confirmation of cardiomyocyte versus noncardiomyocyte identity of pelleted cells by qPCR for fibroblast and cardiomyocyte markers.
Determination of EdU Incorporation by flow cytometry.
Isolation and staining of adult cardiomyocyte nuclei was performed as previously described (20). In brief, control and iCITED4 animals were given i.p. injections of the thymidine analogue EdU (1 mg) every other day for 2 weeks after induction of CITED4 expression by doxycycline removal. Hearts were removed and homogenized using a handheld tissue homogenizer, and cells were then incubated with a hypotonic solution and mechanically lysed with a dounce homogenizer. Nuclei were isolated by sucrose gradient centrifugation and stained with anti-PCM1 antibody (Sigma) to identify the nuclear envelope of adult cardiomyocyte nuclei. A secondary stain with FITC-labeled secondary antibody was performed, and the nuclei were separated by FACS using the S3e Cell Sorter (Propell Labs). Sample purity was then determined by analysis of the sorted population on a fluorescent microscope.
Microscopy, confocal microscopy, and image quantification.
Images of cultured cells were taken with Leica DM 5000 B microscope. For fluorescent imaging, heart sections were imaged on an inverted Zeiss LSM 510 Meta confocal microscope using standard techniques. At least 30 random images were obtained from each group. For quantification of autophagy, frozen cardiac sections were fixed with 4% paraformaldehyde and stained with antibodies for Cardiac Troponin (TNNT) and LC3B (Abcam, catalog ab56357, Cell Signaling Technologies, catalog 2775S, respectively). For quantification of fibrosis, H&E and MTS slides were imaged automatically with a VS120 (Olympus) slide scanner. Images were then quantified using ImageJ in a blinded fashion.
Echocardiography.
Echocardiography was performed on conscious mice using a GE Vivid7 with i13L probe (14 MHZ) as described previously (41). Briefly, parasternal long-axis views, short-axis views, and 2D guided M-mode images of short axes at the papillary muscle level were recorded. The average of at least 6 measurements was used for every data point from each mouse. Analyses were performed in a blinded fashion to genotype or treatment.
Western blotting.
For Western blot analysis, protein lysates were separated by SDS-PAGE using precast gels (Bio-Rad) and transferred to nitro-cellulose membranes using a semi-dry transfer method for 45 minutes at 15 V. Membranes were blocked with 5% milk for 1 hour and incubated with primary antibodies either for 3 hours at room temperature or overnight at 4 degrees with agitation. Species-specific secondary antibodies conjugated to horseradish perioxidase (Cell Signaling Technologies) were used for detection. Imaging and quantification was performed with a digital imager (Bio-Rad) using commercial software. All antibodies used in are described in Supplemental Table 1.
qPCR.
For analysis of relative mRNA levels, total RNA was isolated from either whole hearts or isolated cells by inorganic extraction with Trizol and subsequent ethanol purification. After normalization, we created total cDNA by reverse transcription (Applied Biosystems) following the manufacturer instructions. Using specific primers for each target gene, real-time PCR reactions were carried out using SYBR-green and standard amplification protocols. All target genes were compared with at least 2 housekeeping genes (TBP and GAPDH) and calculated as the relative fold change from the house keeping gene and the relevant control (ΔΔ). At least 3 biologic replicates were averaged for each experiment. Primer pairs for BNP, αMHC, βMHC, SMA1, and TNT were all from PrimerBank (https://pga.mgh.harvard.edu/primerbank/). For CITED4, the following primers were used: forward - CCTGGCATACGGCTCCTTC, reverse - AGACTGCAGGTGCGTGCTAC. Standard melting curves and separation on a DNA gel demonstrated a single band of the appropriate size representing amplification of CITED4.
Statistics.
Data are presented as box/whisker plots, with the center line representing the mean and the upper and lower bounds of the box corresponding to the 25th and 75th percentiles of the averaged data. The whiskers denote the 10th and 90th percentile. Unpaired, 2-tailed Student’s t tests were used as indicated, with P < 0.05 considered significant, n.s. designates nonsignificance and a P value greater than 0.1. When assessing multiple groups, 1-way ANOVA was utilized with Tukey’s post hoc test. Kaplan-Meier survival analysis was performed with JMP software using the Log Rank test for significance. All other statistical analyses were performed in IGOR Pro.
Study approval.
All animal protocols were approved by the Institutional Animal Care and Use Committee at the Beth Israel Deaconess Medical Center. Periodic review of procedures was performed, and amendments were made as needed.
Author contributions
VJB and AR conceived and designed the study. VJB and CP conducted the experiments. CL, YC, KP, and NLO assisted with the experiments. CX performed all the surgeries. NM created the CITED4 virus and the inducible transgenic mouse line. VB and AR wrote and edited the paper. BMS assisted with critical review and editing of the paper. AR secured funding and directed the project as a whole.
Supplementary Material
Acknowledgments
This research was supported by grants from the NIH (A. Rosenzweig [1UH2TR000901, R01HL110733, R01 R01HL122987], C. Platt [T32GM007226], and V.J. Bezzerides [T32HL007572-27A1]). V.J. Bezzerides was also supported by a LaDue Fellowship Award from Harvard Medical School. K. Paruchuri was supported by a fellowship from the Sarnoff Foundation. C. Lerchenmüller was supported by the German Research Foundation (DFG Le 3257/1-1). A. Rosenzweig is a principal faculty member of the Harvard Stem Cell Institute.
Footnotes
V.J. Bezzerides’s present address is: Cardiovascular Division of Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
C. Platt, C. Lerchenmüller, C. Xiao, Y. Cao, and A. Rosenzweig’s present address is: Cardiovascular Division of Massachusetts General Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2016;1(9):e85904. doi:10.1172/jci.insight.85904.
Contributor Information
Colin Platt, Email: platt.colin@gmail.com.
Carolin Lerchenmüller, Email: carolin.lerchenmueller@gmail.com.
Kaavya Paruchuri, Email: kaavyaparuchuri@gmail.com.
Chunyang Xiao, Email: CXIAO@PARTNERS.ORG.
Yunshan Cao, Email: yunshancao@126.com.
Nina Mann, Email: Nina.Mann@childrens.harvard.edu.
Bruce M. Spiegelman, Email: bruce_spiegelman@dfci.harvard.edu.
Anthony Rosenzweig, Email: arosenzweig@partners.org.
References
- 1.Wei X, Liu X, Rosenzweig A. What do we know about the cardiac benefits of exercise? Trends Cardiovasc Med. 2015;25(6):529–536. doi: 10.1016/j.tcm.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boström P, et al. C/EBPβ controls exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell. 2010;143(7):1072–1083. doi: 10.1016/j.cell.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Platt C, Houstis N, Rosenzweig A. Using exercise to measure and modify cardiac function. Cell Metab. 2015;21(2):227–236. doi: 10.1016/j.cmet.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeMaria AN, Neumann A, Lee G, Fowler W, Mason DT. Alterations in ventricular mass and performance induced by exercise training in man evaluated by echocardiography. Circulation. 1978;57(2):237–244. doi: 10.1161/01.CIR.57.2.237. [DOI] [PubMed] [Google Scholar]
- 5.Bernardo BC, Weeks KL, Pretorius L, McMullen JR. Molecular distinction between physiological and pathological cardiac hypertrophy: experimental findings and therapeutic strategies. Pharmacol Ther. 2010;128(1):191–227. doi: 10.1016/j.pharmthera.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Gerdes AM, et al. Structural remodeling of cardiac myocytes in patients with ischemic cardiomyopathy. Circulation. 1992;86(2):426–430. doi: 10.1161/01.CIR.86.2.426. [DOI] [PubMed] [Google Scholar]
- 7.Wisløff U, et al. Increased contractility and calcium sensitivity in cardiac myocytes isolated from endurance trained rats. Cardiovasc Res. 2001;50(3):495–508. doi: 10.1016/S0008-6363(01)00210-3. [DOI] [PubMed] [Google Scholar]
- 8.Kemi OJ, et al. The effect of exercise training on transverse tubules in normal, remodeled, and reverse remodeled hearts. J Cell Physiol. 2011;226(9):2235–2243. doi: 10.1002/jcp.22559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vettor R, et al. Exercise training boosts eNOS-dependent mitochondrial biogenesis in mouse heart: role in adaptation of glucose metabolism. Am J Physiol Endocrinol Metab. 2014;306(5):E519–E528. doi: 10.1152/ajpendo.00617.2013. [DOI] [PubMed] [Google Scholar]
- 10.Kemi OJ, Wisløff U. Mechanisms of exercise-induced improvements in the contractile apparatus of the mammalian myocardium. Acta Physiol (Oxf) 2010;199(4):425–439. doi: 10.1111/j.1748-1716.2010.02132.x. [DOI] [PubMed] [Google Scholar]
- 11.Calvert JW, et al. Exercise protects against myocardial ischemia-reperfusion injury via stimulation of β(3)-adrenergic receptors and increased nitric oxide signaling: role of nitrite and nitrosothiols. Circ Res. 2011;108(12):1448–1458. doi: 10.1161/CIRCRESAHA.111.241117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French JP, et al. Ischemia-reperfusion-induced calpain activation and SERCA2a degradation are attenuated by exercise training and calpain inhibition. Am J Physiol Heart Circ Physiol. 2006;290(1):H128–H136. doi: 10.1152/ajpheart.00739.2005. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, et al. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015;21(4):584–595. doi: 10.1016/j.cmet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bragança J, et al. Human CREB-binding protein/p300-interacting transactivator with ED-rich tail (CITED) 4, a new member of the CITED family, functions as a co-activator for transcription factor AP-2. J Biol Chem. 2002;277(10):8559–8565. doi: 10.1074/jbc.M110850200. [DOI] [PubMed] [Google Scholar]
- 15.Ryall KA, Bezzerides VJ, Rosenzweig A, Saucerman JJ. Phenotypic screen quantifying differential regulation of cardiac myocyte hypertrophy identifies CITED4 regulation of myocyte elongation. J Mol Cell Cardiol. 2014;72:74–84. doi: 10.1016/j.yjmcc.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanbe A, Gulick J, Hanks MC, Liang Q, Osinska H, Robbins J. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ Res. 2003;92(6):609–616. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- 17.Collins KA, et al. Accuracy of echocardiographic estimates of left ventricular mass in mice. Am J Physiol Heart Circ Physiol. 2001;280(5):H1954–H1962. doi: 10.1152/ajpheart.2001.280.5.H1954. [DOI] [PubMed] [Google Scholar]
- 18.Waring CD, et al. The adult heart responds to increased workload with physiologic hypertrophy, cardiac stem cell activation, and new myocyte formation. Eur Heart J. 2014;35(39):2722–2731. doi: 10.1093/eurheartj/ehs338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ang KL, et al. Limitations of conventional approaches to identify myocyte nuclei in histologic sections of the heart. Am J Physiol Cell Physiol. 2010;298(6):C1603–C1609. doi: 10.1152/ajpcell.00435.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergmann O, Jovinge S. Isolation of cardiomyocyte nuclei from post-mortem tissue. J Vis Exp. 2012;(65):e85904. doi: 10.3791/4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senyo SE, Lee RT, Kühn B. Cardiac regeneration based on mechanisms of cardiomyocyte proliferation and differentiation. Stem Cell Res. 2014;13(3 pt B):532–541. doi: 10.1016/j.scr.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins KA, Korcarz CE, Lang RM. Use of echocardiography for the phenotypic assessment of genetically altered mice. Physiol Genomics. 2003;13(3):227–239. doi: 10.1152/physiolgenomics.00005.2003. [DOI] [PubMed] [Google Scholar]
- 23.Abergel E, et al. Serial left ventricular adaptations in world-class professional cyclists: implications for disease screening and follow-up. J Am Coll Cardiol. 2004;44(1):144–149. doi: 10.1016/j.jacc.2004.02.057. [DOI] [PubMed] [Google Scholar]
- 24.Giannuzzi P, Temporelli PL, Corrà U, Gattone M, Giordano A, Tavazzi L. Attenuation of unfavorable remodeling by exercise training in postinfarction patients with left ventricular dysfunction: results of the Exercise in Left Ventricular Dysfunction (ELVD) trial. Circulation. 1997;96(6):1790–1797. doi: 10.1161/01.CIR.96.6.1790. [DOI] [PubMed] [Google Scholar]
- 25.Haykowsky M, et al. A meta-analysis of the effects of exercise training on left ventricular remodeling following myocardial infarction: start early and go longer for greatest exercise benefits on remodeling. Trials. 2011;12:e85904. doi: 10.1186/1745-6215-12-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Diwan A. Autophagy is impaired in cardiac ischemia-reperfusion injury. Autophagy. 2012;8(9):1394–1396. doi: 10.4161/auto.21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma X, et al. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125(25):3170–3181. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He C, Sumpter R., Jr and Levine B. Exercise induces autophagy in peripheral tissues and in the brain. Autophagy. 2012;8(10):1548–1551. doi: 10.4161/auto.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishida K, Kyoi S, Yamaguchi O, Sadoshima J, Otsu K. The role of autophagy in the heart. Cell Death Differ. 2009;16(1):31–38. doi: 10.1038/cdd.2008.163. [DOI] [PubMed] [Google Scholar]
- 30.Wu X, et al. Impaired autophagy contributes to adverse cardiac remodeling in acute myocardial infarction. PLoS One. 2014;9(11):e85904. doi: 10.1371/journal.pone.0112891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sciarretta S, Volpe M, Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res. 2014;114(3):549–564. doi: 10.1161/CIRCRESAHA.114.302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chong E, et al. Resveratrol, a red wine antioxidant, reduces atrial fibrillation susceptibility in the failing heart by PI3K/AKT/eNOS signaling pathway activation. Heart Rhythm. 2015;12(5):1046–1056. doi: 10.1016/j.hrthm.2015.01.044. [DOI] [PubMed] [Google Scholar]
- 34.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu YB, et al. Mechanisms mediating the cardioprotective effects of rapamycin in ischaemia-reperfusion injury. Clin Exp Pharmacol Physiol. 2011;38(1):77–83. doi: 10.1111/j.1440-1681.2010.05467.x. [DOI] [PubMed] [Google Scholar]
- 36.Aoyagi T, et al. Cardiac mTOR protects the heart against ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2012;303(1):H75–H85. doi: 10.1152/ajpheart.00241.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buss SJ, et al. Beneficial effects of Mammalian target of rapamycin inhibition on left ventricular remodeling after myocardial infarction. J Am Coll Cardiol. 2009;54(25):2435–2446. doi: 10.1016/j.jacc.2009.08.031. [DOI] [PubMed] [Google Scholar]
- 38.Chen T, et al. Aerobic exercise inhibits sympathetic nerve sprouting and restores β-adrenergic receptor balance in rats with myocardial infarction. PLoS One. 2014;9(5):e85904. doi: 10.1371/journal.pone.0097810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481(7382):511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khan S, Salloum F, Das A, Xi L, Vetrovec GW, Kukreja RC. Rapamycin confers preconditioning-like protection against ischemia-reperfusion injury in isolated mouse heart and cardiomyocytes. J Mol Cell Cardiol. 2006;41(2):256–264. doi: 10.1016/j.yjmcc.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 41.Das S, et al. Pathological role of serum- and glucocorticoid-regulated kinase 1 in adverse ventricular remodeling. Circulation. 2012;126(18):2208–2219. doi: 10.1161/CIRCULATIONAHA.112.115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagoshi T, et al. PI3K rescues the detrimental effects of chronic Akt activation in the heart during ischemia/reperfusion injury. J Clin Invest. 2005;115(8):2128–2138. doi: 10.1172/JCI23073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsui T, Nagoshi T, Rosenzweig A. Akt and PI 3-kinase signaling in cardiomyocyte hypertrophy and survival. Cell Cycle. 2003;2(3):220–223. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.