Phosphatidylethanolamine (PE) and cardiolipin have specific roles in the activity and assembly of the mitochondrial respiratory chain supercomplexes, respectively, whereas phosphatidylcholine is redundant. Nonmitochondrial PE can be transported into mitochondria, where it can fully substitute for the lack of mitochondrial PE biosynthesis.
Abstract
Mitochondrial membrane phospholipid composition affects mitochondrial function by influencing the assembly of the mitochondrial respiratory chain (MRC) complexes into supercomplexes. For example, the loss of cardiolipin (CL), a signature non–bilayer-forming phospholipid of mitochondria, results in disruption of MRC supercomplexes. However, the functions of the most abundant mitochondrial phospholipids, bilayer-forming phosphatidylcholine (PC) and non–bilayer-forming phosphatidylethanolamine (PE), are not clearly defined. Using yeast mutants of PE and PC biosynthetic pathways, we show a specific requirement for mitochondrial PE in MRC complex III and IV activities but not for their formation, whereas loss of PC does not affect MRC function or formation. Unlike CL, mitochondrial PE or PC is not required for MRC supercomplex formation, emphasizing the specific requirement of CL in supercomplex assembly. Of interest, PE biosynthesized in the endoplasmic reticulum (ER) can functionally substitute for the lack of mitochondrial PE biosynthesis, suggesting the existence of PE transport pathway from ER to mitochondria. To understand the mechanism of PE transport, we disrupted ER–mitochondrial contact sites formed by the ERMES complex and found that, although not essential for PE transport, ERMES facilitates the efficient rescue of mitochondrial PE deficiency. Our work highlights specific roles of non–bilayer-forming phospholipids in MRC function and formation.
INTRODUCTION
The mitochondrial respiratory chain (MRC) consists of four multimeric protein complexes (complexes I–IV) that generate an electrochemical proton gradient across the mitochondrial inner membrane driving ATP synthesis via ATP synthase (complex V). MRC complexes are embedded within the inner mitochondrial membrane, where they interact in different stoichiometries and assemble into supramolecular structures known as supercomplexes (Schagger and Pfeiffer, 2000; Dudkina et al., 2011). Supercomplex formation increases the efficiency of electron transfer by spatial restriction of electron carrier diffusion (Acín-Pérez et al., 2008). Disruption in the function or formation of MRC complexes or supercomplexes perturbs mitochondrial bioenergetics, manifesting clinically in a wide range of metabolic disorders (Koopman et al., 2012; Nunnari and Suomalainen, 2012; Vafai and Mootha, 2012). Thus manipulating the function of the MRC complexes has been proposed as a possible therapeutic avenue for the treatment and prevention of these diseases (Walters et al., 2012; Weinberg and Chandel, 2015).
Traditionally, the functions of MRC complexes have been studied in isolation from their native membrane environment, which often overlooks the contribution of membrane lipids (Gohil and Greenberg, 2009). Phospholipids constitute a large proportion of mitochondrial membrane lipid milieu, where they specify MRC function by influencing physical properties of the membrane and by specific phospholipid:protein interactions (Horvath and Daum, 2013; Ren et al., 2014). Mitochondrial membranes contain all of the major classes of phospholipids found in cell membranes, including phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol, phosphatidylserine (PS), and phosphatidic acid (PA), as well as cardiolipin (CL), which is predominantly, if not exclusively, found in the mitochondria (Horvath and Daum, 2013). PC is the most abundant mitochondrial phospholipid, accounting for almost 50% of total phospholipids (Horvath and Daum, 2013). The cylindrical shape of PC promotes bilayer formation. PE is the second-most-abundant mitochondrial phospholipid and has a cone-shape structure that imposes negative curvature stress on the membrane, increasing its tendency to form nonbilayer structures (Holthuis and Menon, 2014). Like PE, CL is the other major cone-shaped mitochondrial phospholipid with the tendency to induce negative membrane curvature to the lipid bilayer. These “non–bilayer-forming” phospholipids are proposed to facilitate membrane fusion and influence the binding and activity of membrane proteins (van Meer et al., 2008). The ratio of bilayer to non–bilayer-forming phospholipids in the mitochondria is roughly equal and highly conserved, suggesting both coordinate regulation of their biosynthesis and their importance in mitochondrial function (Daum, 1985).
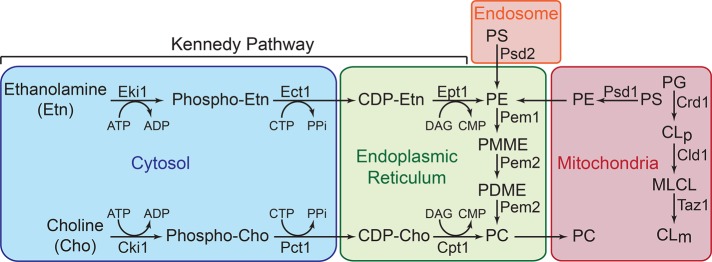
Phospholipid biosynthesis in eukaryotes is compartmentalized into various subcellular organelles, including the endoplasmic reticulum (ER), endosome, and mitochondria. Mitochondria are able to synthesize PE and CL in situ, whereas all of the other phospholipids are imported (Gohil and Greenberg, 2009; Figure 1). CL is synthesized from phosphatidylglycerol and cytidine diphosphate–diacylglycerol in the inner mitochondrial membrane by CL synthase, Crd1. The acyl chain composition of newly formed CL species is remodeled by the sequential action of cardiolipin-specific deacylase, Cld1, and a transacylase, Taz1, to generate mature CL. PE is synthesized by decarboxylation of PS via phosphatidylserine decarboxylase 1, Psd1, which is localized to the inner mitochondrial membrane and is the primary source of cellular PE (Bürgermeister et al., 2004a). The yeast-specific enzyme Psd2 is localized to an endosomal compartment (Gulshan et al., 2010) and contributes to the cellular PE content to a lesser extent (Bürgermeister et al., 2004a). The de novo biosynthesis of PC proceeds through three sequential methylations of PE catalyzed by phosphatidylethanolamine methyltransferases, Pem1 and Pem2. In addition to these de novo biosynthetic pathways, PE and PC can be synthesized from their precursors ethanolamine (Etn) or choline, respectively, through the nonmitochondrial Kennedy pathway (Figure 1). The functional contribution of the Kennedy pathway phospholipids to mitochondrial membranes is not clear. Results from Bürgermeister et al. (2004a, b) are consistent with equilibrium transport of PE and PC between mitochondrial and microsomal membranes. However, subsequent studies showed that PE synthesized by the Kennedy pathway is not able to restore mitochondrial respiration (Chan and McQuibban, 2012; Wang et al., 2014), implying that either PE transport from ER to mitochondria is insufficient to functionally compensate for the lack of mitochondrial PE biosynthesis or that the nonmitochondrial PE species cannot substitute for mitochondrial PE species.
FIGURE 1:
Aminoglycerophospholipids and CL biosynthetic pathways in yeast. PE biosynthesis in yeast is accomplished by three major pathways: 1) Psd1-catalyzed decarboxylation of PS in the mitochondria, 2) Psd2-catalyzed decarboxylation of PS in endosomal compartments, and 3) incorporation of Etn via the cytosolic/ER Kennedy pathway. PC is produced by two major pathways: 1) formation of Pem1 and Pem2 PC by successive methylation of PE, and 2) incorporation of choline via the Kennedy pathway. CL biosynthesis occurs exclusively in the mitochondria, where premature cardiolipin (CLp) is synthesized from phosphatidylglycerol (PG) by Crd1. The resulting CLp is deacylated by the phospholipase Cld1 to produce monolysocardiolipin (MLCL) and reacylated by Taz1 to form mature cardiolipin (CLm). CDP, cytidine diphosphate; CTP, cytidine triphosphate; CMP, cytidine monophosphate; DAG, diacylglycerol; PA, phosphatidic acid; PI, phosphatidylinositol; PDME, phosphatidyldimethylethanolamine; PPi, inorganic pyrophosphate.
Although the role of CL in MRC function and formation has been extensively studied (Joshi et al., 2009; Mileykovskaya and Dowhan, 2014), the roles of more abundant mitochondrial phospholipids, including PE and PC, are incompletely understood. Depletion of mitochondrial PE in mammalian cells was shown to result in reduced respiratory capacity, ATP production, and destabilization of complex IV–containing supercomplexes (Tasseva et al., 2013). In contrast, yeast cells lacking mitochondrial PE biosynthesis were shown to contain higher-order supercomplexes (Bottinger et al., 2012). Although these studies suggest conflicting roles of PE in MRC assembly, the role of PC in mitochondrial bioenergetics has not been investigated at all. Therefore it is not clear whether MRC defects are exclusive to alterations in nonbilayer mitochondrial phospholipids like PE and CL or simply the result of any perturbation in mitochondrial phospholipid composition.
To address these gaps in our knowledge, we used isogenic yeast mutants of PE and PC biosynthetic enzymes to systematically dissect their roles in MRC biogenesis. We show that the disruption of mitochondrial PE biosynthesis causes reduced activities of fully assembled MRC complex III and IV containing supercomplexes. Surprisingly, the most abundant mitochondrial phospholipid, PC, is not required for MRC function or formation. In contrast to the prevailing model of interorganelle phospholipid trafficking (Osman et al., 2011; Lahiri et al., 2015), we demonstrate that PE synthesized in the ER via the Kennedy pathway can be transported into mitochondria, where it can fully substitute for the loss of mitochondrial PE biosynthesis. Our work thus identifies specific roles of the two most abundant mitochondrial phospholipids in MRC biogenesis and provides a means for manipulating mitochondrial phospholipid composition by stimulating the nonmitochondrial Kennedy pathway.
RESULTS
Deletion of Psd1 and Pem2 dramatically alters mitochondrial PE/PC ratio
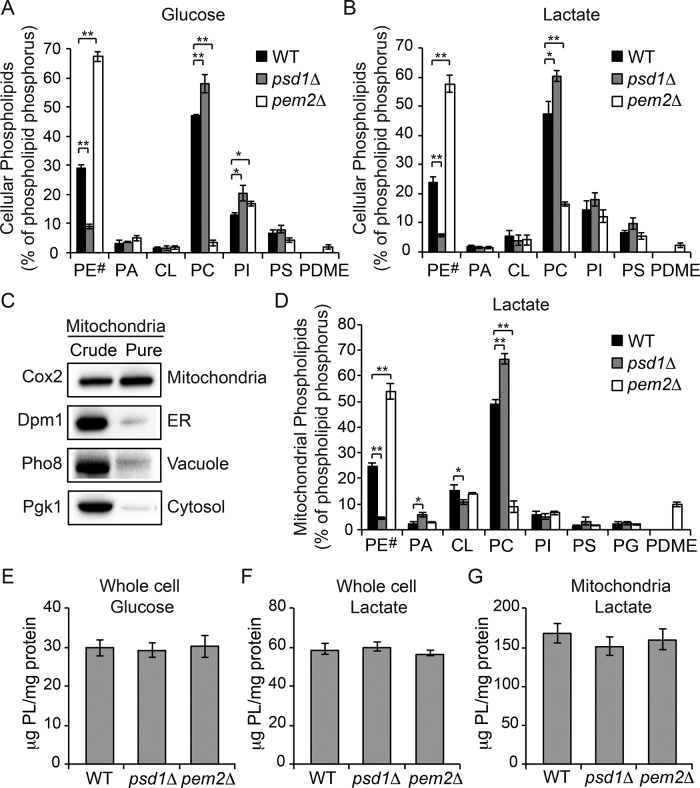
To dissect the roles of the two most abundant mitochondrial phospholipids, PE and PC, in MRC function and formation in vivo, we focused on psd1Δ and pem2Δ cells, which lack key enzymes for PE and PC biosynthesis, respectively (Figure 1). Previous studies showed that phospholipid composition and mitochondrial biogenesis is dependent upon the carbon source used in the growth medium (Tuller et al., 1999). Therefore we analyzed the phospholipid composition of yeast cells grown in glucose-containing, fermentable (SC glucose) or lactate-containing, nonfermentable (SC lactate) carbon sources. Whole-cell phospholipid analysis of glucose-grown psd1Δ cells revealed a threefold reduction in PE with a concomitant increase in PC (Figure 2A). Conversely, pem2Δ cells showed a 15-fold reduction in PC, with a twofold increase in PE and its precursor, phosphatidylmonomethylethanolamine (PMME), which could not be separated by the TLC used in this study (Figure 2A). Similar PE and PC changes were observed when psd1Δ and pem2Δ cells were grown in a medium containing lactate (Figure 2B). To analyze mitochondrial phospholipid composition, we obtained highly purified mitochondria with minimal contamination from other cellular organelles (Figure 2C). Consistent with whole-cell phospholipid composition, the levels of PE decreased by approximately sixfold in psd1Δ mitochondria (Figure 2D). The decrease in PE in psd1Δ mitochondria was accompanied by alterations in other phospholipids, including a significant increase in PC and PA and a decrease in CL (Figure 2D). In pem2Δ mitochondria, PC levels decreased fivefold, whereas the PE/PMME content doubled (Figure 2D). We also observed a significant accumulation of phosphatidyldimethylethanolamine in pem2Δ mitochondria (Figure 2D). In both glucose- and lactate-containing media, the absolute phospholipid levels in whole cells and in isolated mitochondria did not change in either mutant relative to the wild-type (WT) cells (Figure 2, E–G). These results suggest that in yeast cells, a homeostatic mechanism exists that buffers cells against the loss of the absolute amount of membrane phospholipids such that the depletion in PE is compensated by an increase in PC and vice versa. Therefore psd1Δ and pem2Δ cells have a significantly altered PE/PC ratio without any change in their absolute amount of membrane phospholipids. The mitochondrial PE/PC ratio in WT was 0.503, which was reduced to 0.065 in psd1Δ cells and increased to 6.02 in pem2Δ cells. Despite these dramatic deviations in the mitochondrial PE/PC ratios in psd1Δ and pem2Δ cells, the gross cellular and mitochondrial morphology was unaltered, with only a small reduction in the average length of mitochondrial cristae and outer membrane in psd1Δ cells (Supplemental Figure S1). There was no change in mitochondrial cristae length of pem2Δ cells, but we did observe a slight reduction in the average outer membrane length (Supplemental Figure S1). Collectively these results demonstrate that yeast cells can tolerate extensive alteration in mitochondrial PE/PC ratios and that a decrease in the PE level is countered by an increase in PC and vice versa.
FIGURE 2:
Cellular and mitochondrial phospholipid composition of psd1Δ and pem2Δ cells. The whole-cell phospholipid composition of WT, psd1Δ, and pem2Δ cells grown in (A) SC glucose and (B) SC lactate. Phospholipid levels are expressed as the percentage of total phospholipid phosphorus in each phospholipid class. PE# represents the sum of PE and PMME in pem2Δ cells. Data are expressed as mean ± SD (n = 3); **p < 0.005, *p < 0.05. (C) Western blot analysis of crude and sucrose-gradient purified mitochondria from WT cells. Cox2, Dpm1, Pho8, and Pgk1 are used as markers of the yeast mitochondria, ER, vacuole, and cytoplasm, respectively. (D) Phospholipid composition of sucrose gradient–purified mitochondria from WT, psd1Δ, and pem2Δ cells grown in SC lactate. Data are expressed as mean ± SD (n = 3); **p < 0.005, *p < 0.05. (E, F) Total phospholipid content of whole-cell homogenates of WT, psd1Δ, and pem2Δ cells grown in (E) SC glucose or (F) SC lactate. (G) Total phospholipid content of mitochondria from SC lactate–grown cells. Data are expressed as mean ± SD (n = 3).
Decreased mitochondrial PE/PC ratio results in reduced respiration and ATP levels
To dissect the specific roles of PC and PE in MRC function, we performed extensive growth characterization of psd1Δ and pem2Δ cells in different carbon sources. The growth of psd1Δ and pem2Δ cells in fermentable SC glucose medium was comparable to that for WT cells (Figure 3A and Supplemental Figure S2A). Consistent with previous work (Birner et al., 2001), the growth of psd1Δ cells in nonfermentable SC lactate medium was severely compromised, whereas pem2Δ cells were able to grow, albeit at a slightly reduced rate (Figure 3B and Supplemental Figure S2B). The growth defects in nonfermentable medium suggested respiratory deficiency. To directly assess respiration, we measured oxygen consumption in WT, psd1Δ, and pem2Δ cells. Consistent with the severely diminished respiratory growth, the psd1Δ cells had a ∼60% reduction in oxygen consumption compared with WT cells (Figure 3, C and D). Oxygen consumption in pem2Δ cells was comparable to WT cells in SC lactate and even slightly elevated in SC glucose medium (Figure 3, C and D). The reduced growth of pem2Δ cells could be due to defects in mitochondrial protein import machinery, as reported recently (Schuler et al., 2015), and not to defects in respiration per se. In accordance with reduced respiration in psd1Δ cells, we observed a significant 50% reduction in ATP levels, whereas the ATP levels in the respiratory competent pem2Δ cells were unaltered (Figure 3, E and F). These results suggest that mitochondrial PE, but not PC, is essential for maintaining normal respiration.
FIGURE 3:
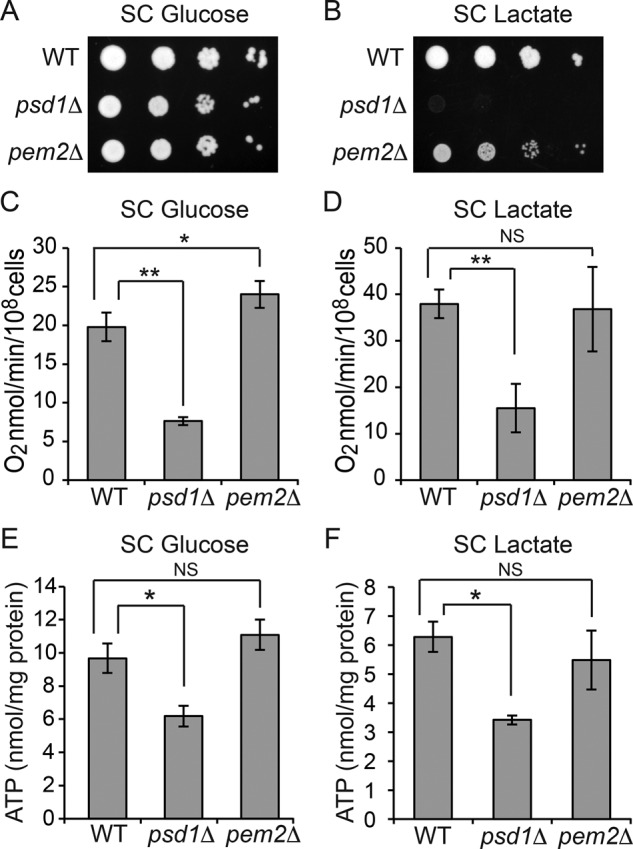
Mitochondrial respiration is dependent on PE but not PC levels. Tenfold serial dilutions of WT, psd1Δ, and pem2Δ cells were spotted onto (A) SC glucose and (B) SC lactate plates, and images were captured after 2 (SC glucose) or 5 d (SC lactate) of growth at 30°C. Data are representative of at least three independent experiments. (C, D) WT, psd1Δ, and pem2Δ cells were grown in (C) SC glucose or (D) SC lactate to late log phase, and the rate of oxygen consumption was measured. Data are expressed as mean ± SD (n = 6); *p < 0.05, **p < 0.005. (E, F) Cellular ATP levels of WT, psd1Δ, and pem2Δ cells cultured in (E) SC glucose or (F) SC lactate. Data are expressed as mean ± SD (n = 3); *p < 0.05.
Decreased mitochondrial PE/PC ratio reduces MRC supercomplex activities without affecting supercomplex formation
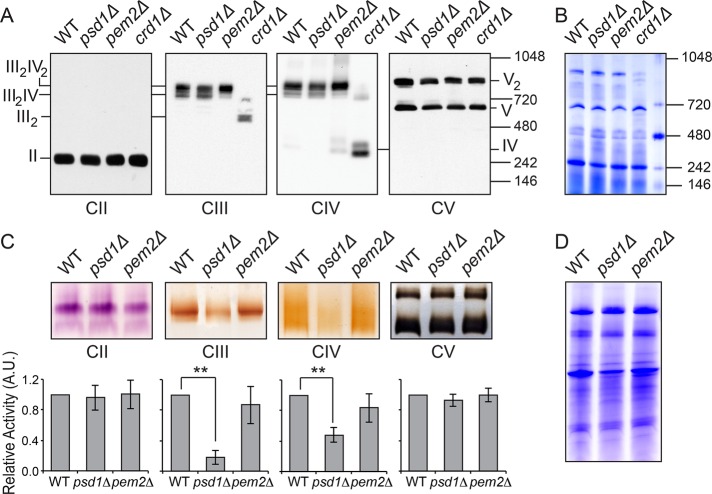
To ascertain the biochemical basis for reduced respiration in PE-depleted cells, we analyzed the levels of native and denatured MRC complexes in the mitochondrial lysate from WT, psd1Δ, and pem2Δ cells grown in SC lactate medium. There was no change in the steady-state levels of individual MRC subunits (Supplemental Figure S3) or their incorporation into fully assembled MRC complexes in either of the mutant cells (Figure 4, A and B). As reported previously, MRC supercomplexes containing complexes III and IV were disrupted in CL-lacking crd1Δ cells (Zhang et al., 2002; Pfeiffer et al., 2003). These results imply that reduced PE and accompanying 33% decrease in CL levels in psd1Δ are insufficient to disrupt supercomplex formation. We noticed that pem2Δ cells, which exhibit an increased PE/PC ratio, showed an enhanced formation of a larger supercomplex (III2IV2) at the expense of the smaller supercomplex (III2IV; Figure 4A). The lack of alterations in the amount and assembly of the MRC complexes cannot explain the respiratory deficiency of psd1Δ cells, suggesting that the reduced respiration could be due to a decrease in MRC activity. Therefore we measured the enzymatic activities of MRC complexes and observed four- and 2.5-fold reductions in complex III and IV activities, respectively, in psd1Δ cells (Figure 4, C and D). The specific reduction in complex III and IV activities could, in part, explain the respiratory defects observed in psd1Δ cells. The activities of MRC complexes were comparable in WT and pem2Δ cells (Figure 4, C and D), which is consistent with the normal respiratory phenotype of pem2Δ cells. Together these results demonstrate specific requirement of CL for MRC supercomplex formation and PE for MRC complex III and IV activities, whereas PC is redundant for these functions.
FIGURE 4:
Mitochondrial PE is required for MRC complex III and IV activities but not MRC supercomplex formation. (A) Mitochondria from SC lactate–grown cells were solubilized by 1% digitonin and subjected to BN–PAGE/Western blot, and complexes II–V were detected by Sdh2, Rip1, Cox2, and Atp2 antibodies, respectively. Mitochondria from CL-deficient crd1Δ cells were used as positive control to demonstrate loss of supercomplexes (III2IV2, large supercomplex; III2IV, small supercomplex) under identical conditions. (B) Samples from A were stained with Coomassie blue to demonstrate equal loading. (C) Digitonin-solubilized mitochondrial complexes from WT, psd1Δ, and pem2Δ cells were separated by CN-PAGE, followed by in-gel activity staining for complexes II–V. In-gel activities of MRC complexes were quantified by densitometric analysis, and relative activities were plotted for complexes II–V. Data were normalized to WT cells and expressed as mean ± SD (n = 3); **p < 0.005. (D) Samples from C were stained with Coomassie blue, and total protein, quantified using densitometric analysis, was used to normalize activity staining.
Depletion of PE results in a specific loss of mitochondrial DNA–encoded MRC subunits
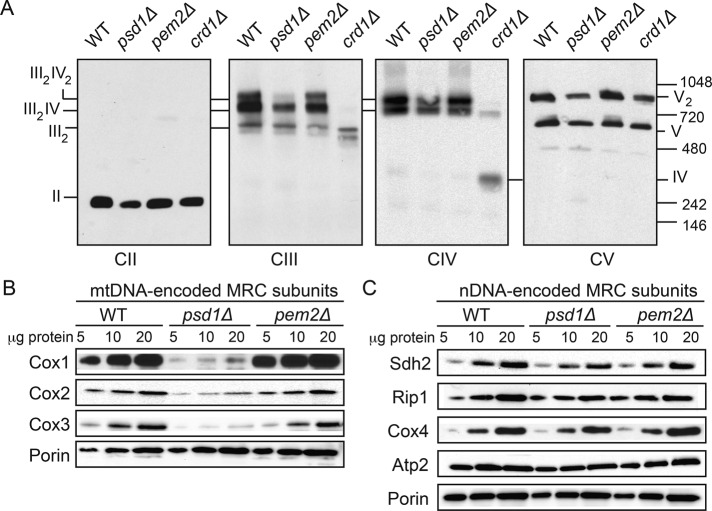
In contrast to previous studies (Bottinger et al., 2012; Tasseva et al., 2013), which reported aberrant formation of MRC supercomplexes in PE depleted cells, we did not find any alterations in the MRC supercomplexes in psd1Δ cells. We reasoned that this discrepancy could be related to use of different growth conditions and carbon sources in these studies. Indeed, we found reduced levels of MRC supercomplexes (III2IV2 and III2IV) in glucose-grown psd1Δ cells (Figure 5A). These carbon source–dependent differences in MRC supercomplex assembly might be due to increased petite formation in psd1Δ cells, as reported previously (Birner et al., 2001). The petite phenotype results from mutations in the mitochondrial genome or loss of mitochondrial DNA (mtDNA), which leads to the loss of mtDNA-encoded MRC subunits. Consistent with the previous report, we found a significant increase in the number of petite colonies in the psd1Δ mutant (Supplemental Figure S4). Accordingly, SDS–PAGE/Western blot analysis of MRC subunits showed a specific decrease in the steady-state levels of mtDNA-encoded subunits Cox1, Cox2, and Cox3 (Figure 5B) without affecting the levels of the nuclear-encoded subunits Sdh2, Rip1, Cox4, and Atp2 (Figure 5C). Unlike PE-depleted psd1Δ cells, loss of PC in pem2Δ cells did not result in any alterations in the assembly or steady-state levels of MRC complexes or petite formation (Figure 5 and Supplemental Figure S4). Collectively these results suggest that the decrease in the MRC supercomplex levels in glucose-grown psd1Δ cells results from the loss of mtDNA-encoded subunits.
FIGURE 5:
Depletion of mitochondrial PE in glucose-grown psd1Δ cells results in a specific loss of mtDNA-encoded MRC subunits. (A) Digitonin-solubilized mitochondria from SC glucose–grown WT, psd1Δ, and pem2Δ cells were subjected to BN–PAGE/Western blot. Complexes II–V were detected by Sdh2, Rip1, Cox2, and Atp2 antibodies, respectively. Data are representative of at least three independent experiments. (B) Mitochondria from SC glucose–grown WT, psd1Δ, and pem2Δ cells were subjected to SDS–PAGE, and mtDNA-encoded subunits were probed using Cox1, Cox2, and Cox3 antibodies. (C) Nuclear-encoded subunits were probed using Sdh2, Rip1, Cox4, and Atp2. Porin was used as a loading control. Data are representative of at least three independent experiments.
PE synthesized via Kennedy pathway completely rescues respiratory defects of psd1Δ cells by restoring mitochondrial PE levels
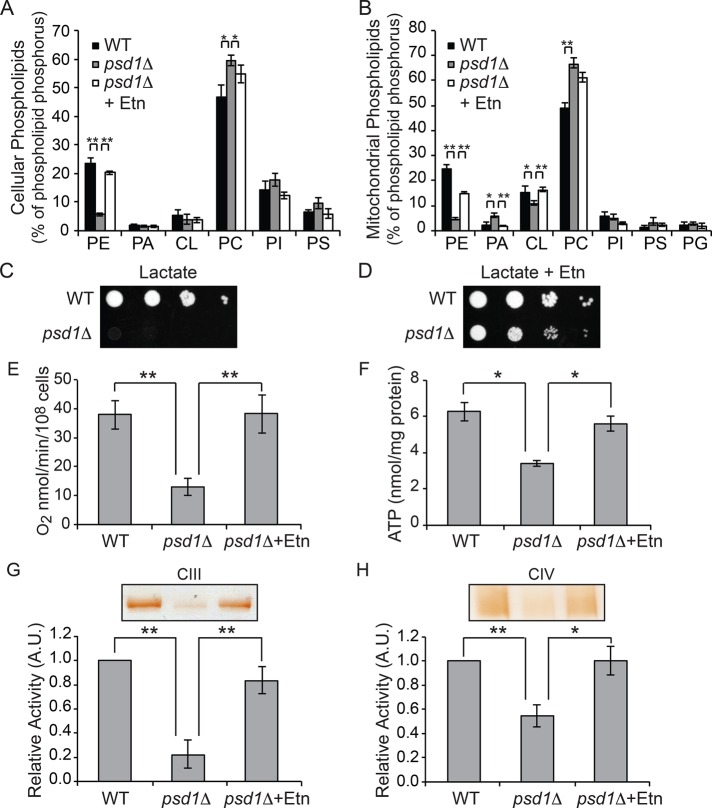
To determine whether PE synthesized in ER by the cytidine diphosphate–Etn branch of the Kennedy pathway could compensate for the loss of mitochondrial PE, we grew psd1Δ cells in the presence of Etn and measured cellular and mitochondrial phospholipids. Etn supplementation in psd1Δ cells completely restored cellular PE and significantly restored mitochondrial PE levels (Figure 6, A and B). Of interest, supplementation of Etn not only rescued mitochondrial PE levels, but it also restored PA and CL levels in psd1Δ mitochondria (Figure 6B), implying that a yet-unidentified homeostatic mechanism regulates the precise proportion of individual phospholipids in mitochondrial membranes. Next we asked whether the partial restoration of mitochondrial PE through exogenous Etn supplementation could restore the respiratory defects observed in psd1Δ cells. Indeed, Etn supplementation rescued the respiratory growth defect of psd1Δ cells in SC lactate medium (Figure 6, C and D). Consistent with the rescue of respiratory growth, Etn supplementation restored oxygen consumption (Figure 6E) and cellular ATP content (Figure 6F) in psd1Δ cells to WT levels. To investigate the mechanism by which Etn rescued respiration, we measured MRC complex III and IV activities and found that they were restored to WT levels in Etn-supplemented psd1Δ cells (Figure 6, G and H). Etn supplementation not only restored respiratory function in SC lactate, but it also rescued petite formation and cellular ATP levels in SC glucose medium (Supplemental Figure S5, A and B). Taken together, these results show that PE synthesized in ER by the Kennedy pathway can replenish mitochondrial PE and restore MRC complex III and IV activities in psd1Δ cells.
FIGURE 6:
Ethanolamine supplementation rescues respiratory defects of psd1Δ cells by restoring mitochondrial PE levels. (A) Cellular and (B) mitochondrial phospholipid composition of WT cells grown in SC lactate and psd1Δ cells grown in SC lactate with and without 2 mM Etn. Phospholipid levels are expressed as percentage of total phospholipid phosphorus in each phospholipid class. Data are expressed as mean ± SD (n = 3); **p < 0.005, *p < 0.05. Tenfold serial dilutions of WT and psd1Δ cells were spotted onto (C) SC lactate and (D) SC lactate + 2 mM Etn plates, and images were captured after 4 d of growth at 30°C. Data are representative of at least three independent trials. (E) Rate of oxygen consumption and (F) total cellular ATP levels of WT and psd1Δ cells grown in SC lactate ± 2 mM Etn to late logarithmic phase were quantified. Data are expressed as mean ± SD (n = 3); *p < 0.05, **p < 0.005 (G, H) Digitonin- solubilized mitochondrial complexes from WT and psd1Δ cells grown in SC lactate ± 2 mM Etn were separated by CN-PAGE, followed by in-gel activity staining for (G) complex III and (H) complex IV. Densitometric quantifications of relative in-gel activities for complexes III and IV. Data were normalized to WT cells and are expressed as mean ± SD (n = 3); **p < 0.005, *p < 0.05. A.U., arbitrary units.
Endoplasmic reticulum–mitochondria encounter structure facilitates Etn-dependent rescue of mitochondrial PE deficiency
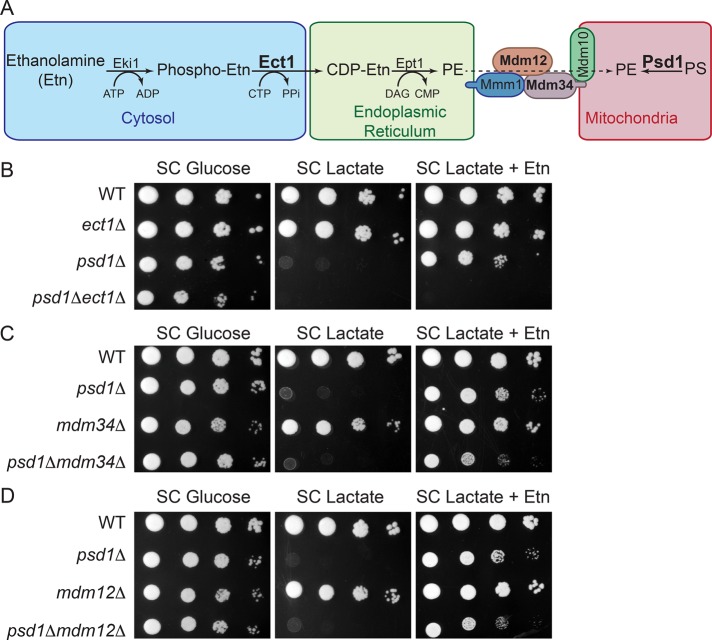
The complete rescue of mitochondrial bioenergetic phenotypes and respiratory growth of psd1Δ cells by Etn supplementation implies efficient transport of PE from ER to mitochondria (Figure 7A). To understand the molecular basis of PE import to mitochondria, we focused on the endoplasmic reticulum–mitochondria encounter structure (ERMES) complex, a mitochondria–ER tethering structure proposed to be involved in trafficking phospholipids between ER and mitochondria (Kornmann et al., 2009). First, to rule out the possibility that Etn itself or one of its metabolites is responsible for the psd1Δ rescue, we deleted the Kennedy pathway enzyme Ect1 in psd1Δ cells and showed that the rescue of psd1Δ cells by Etn is completely abrogated (Figure 7B). This result clearly demonstrates that PE synthesized via the Kennedy pathway is essential for psd1Δ rescue. Next we deleted two ERMES subunits, Mdm34 or Mdm12, both of which contain the synaptotagmin-like mitochondrial lipid-binding protein domain (SMP; AhYoung et al., 2015), in psd1Δ cells and found that Etn rescue is reduced in the double mutants (Figure 7, C and D). To reveal the involvement of the ERMES complex in PE transport, we measured the phospholipid levels in mitochondria of psd1Δmdm34Δ cells with and without Etn supplementation. We did not observe any significant decrease in the steady levels of mitochondrial PE in the double mutant compared with psd1Δ single mutant after Etn supplementation (Supplemental Figure S6). These results suggest that ERMES is not essential for the import of nonmitochondrial PE to mitochondria but might only indirectly facilitate Etn-mediated rescue of psd1Δ cells.
FIGURE 7:
PE synthesized by the Kennedy pathway requires ERMES for complete rescue of mitochondrial PE deficiency. (A) Schematic representation of the Kennedy pathway of PE biosynthesis and the ERMES complex. The Kennedy pathway enzyme Ect1, mitochondrial Psd1, and Mdm34 and Mdm12 of the ERMES complex are depicted in boldface to indicate that these genes are targeted to construct double-knockout strains. Tenfold serial dilutions of (B) WT, ect1Δ, psd1Δ, and psd1Δect1Δ, (C) WT, psd1Δ, mdm34Δ, and psd1Δmdm34Δ, and (D) WT, psd1Δ, mdm12Δ, and psd1Δmdm12Δ cells were spotted onto SC glucose and SC lactate ± Etn plates, and images were captured after 2 (SC glucose) or 5 d (SC lactate ± Etn) of growth at 30°C. Data are representative of at least three independent experiments.
DISCUSSION
Perturbations in mitochondrial membrane phospholipid composition have been shown to cause MRC dysfunction (Lu and Claypool, 2015; Chicco and Sparagna, 2007). Therefore delineating the specific roles of the individual phospholipids of mitochondrial membranes is critical to understanding MRC function. In this study, we used yeast mutants of PE and PC biosynthetic pathways to show a specific requirement of mitochondrial PE for MRC complex III and IV activities, whereas loss of PC was well tolerated. Furthermore, we showed that PE synthesized in the ER via the Kennedy pathway can be efficiently transported to the mitochondrial membranes, where it can completely substitute for the lack of mitochondrial PE biosynthesis. Our results suggest a specific requirement of non–bilayer-promoting phospholipids PE and CL in MRC function and formation.
Analysis of the phospholipid profiles of psd1Δ and pem2Δ cells revealed a cellular homeostatic mechanism that maintained absolute phospholipid concentrations. For example, depletion of PE is always accompanied by an increase in the level of PC and vice versa (Figure 2, A, B, and D). As a result, the mutants harboring PSD1 or PEM2 deletions had altered PE/PC ratios rather than a change in the absolute amount of one particular phospholipid (Figure 2, E–G). Therefore it is important to interpret results in terms of alterations in relative levels of different classes of phospholipids rather than a single phospholipid. We do not know the molecular basis for homeostatic mechanisms that strictly maintain total cellular and mitochondrial phospholipid levels. We predict that such homeostatic mechanisms are likely mediated by feedback inhibition of phospholipid biosynthetic enzymes and cross-pathway regulations by phospholipid precursors, a prediction supported by previous studies (Carman and Han, 2011). Apart from the overall mitochondrial membrane phospholipid content, MRC assembly and activity have been linked to cristae shape (Cogliati et al., 2013). Therefore we analyzed mitochondrial morphology of psd1Δ and pem2Δ mutants by transmission electron microscopy and found that the gross mitochondrial structure and overall cristae morphology were unaffected in PE- and PC-deficient mitochondria (Supplemental Figure S1), with only a small reduction in the average mitochondrial cristae length in psd1Δ mutants (Supplemental Figure S1E).
Earlier studies focused on defining the role of PE in mitochondrial functions were performed in different growth media, resulting in conflicting phenotypes (Birner et al., 2001; Chan and McQuibban, 2012). This is not surprising because it has been long known that phospholipid composition is influenced by the growth conditions of yeast cells (Storey et al., 2001; Gohil et al., 2005). For example, the reduction in respiratory growth of psd1Δ cells is much more pronounced in synthetic lactate medium devoid of Etn (Birner et al., 2001) compared with complex lactate medium (Bottinger et al., 2012), which presumably contains enough Etn to drive PE synthesis by the Kennedy pathway (Storey et al., 2001). Therefore all of the experiments in the present study were performed in the synthetic defined medium to reproducibly control phospholipid levels. The psd1Δ cells cultured in synthetic medium showed a dramatic decrease in the mitochondrial PE/PC ratio (Figure 2D), which reduced MRC function (Figure 3). These bioenergetic deficits could be partly attributed to the reduced activities of the MRC complexes III and IV (Figure 4C). Decreased ATP and respiration in psd1Δ cells could also be due to the reduced mitochondrial membrane potential and protein import, as previously described (Bottinger et al., 2012). Thus our findings on the role of PE in mitochondrial bioenergetics are partly consistent with previous studies in yeast (Bottinger et al., 2012) and mammalian cell lines (Tasseva et al., 2013), but, unlike these reports, we did not find any perturbations in MRC supercomplexes in PE-depleted mitochondria (Figure 4A). This discrepancy could be due to use of different growth conditions. Indeed, we observed reduced levels of MRC supercomplexes in psd1Δ cells cultured in glucose-containing medium (Figure 5A). Thus our results obtained from isogenic yeast mutants grown under identical conditions clearly demonstrate that of the three most abundant mitochondrial phospholipids (PC, PE, and CL) only CL is essential for MRC supercomplex formation (Figure 4, A and B).
Although previous studies provided insights into the specific role of CL in MRC biogenesis (Wenz et al., 2009; Bazán et al., 2013), they did not address whether the mitochondrial phospholipid requirement is specific for non–bilayer-forming phospholipids or whether any perturbation in phospholipid composition will result in MRC defects in vivo. We found that, unlike non–bilayer-forming PE and CL, depletion in bilayer-forming PC did not alter mitochondrial bioenergetic parameters (Figures 3–5). A fivefold depletion in mitochondrial PC had no effect on MRC function or formation, which is surprising, considering that PC constitutes almost half of all the mitochondrial membrane phospholipids. Three previous reports provide possible explanations for this observation. First, the loss of PC in yeast is accompanied by the remodeling of fatty acyl species of PE into more of a bilayer-forming phospholipid, thus maintaining the ratio of non–bilayer- to bilayer-forming phospholipids (Boumann et al., 2006). Second, 90% of bacteria do not contain PC in the cellular membranes that harbor their oxidative phosphorylation machinery (Aktas et al., 2010). Third, elevation of nonbilayer phospholipids such as PE and phosphatidylpropanolamine in PC-lacking cells was able to support growth in nonfermentable medium, suggesting that bilayer-forming PC is not essential for respiratory growth of yeast Saccharomyces cerevisiae (Choi et al., 2004). Collectively these studies indicate that MRC activity or assembly is more resistant to perturbation in PC levels but is very sensitive to changes in nonbilayer phospholipids. Of interest, we observed that depletion of PC with a concomitant increase in PE plus PMME resulted in enhanced formation of the large supercomplex III2IV2 and free complex IV at the expense of the small supercomplex III2IV (Figure 4A), suggesting that formation of MRC supercomplexes can be modulated by altering the bulk properties of membranes in addition to specific molecular interactions with CL. Further, PC-depleted cells showed an increased rate of oxygen consumption (Figure 3C), consistent with a recent study in mammalian cells lacking a mammalian homologue of yeast Pem2 (van der Veen et al., 2014). Thus our findings in yeast cells are likely applicable to mammalian cells.
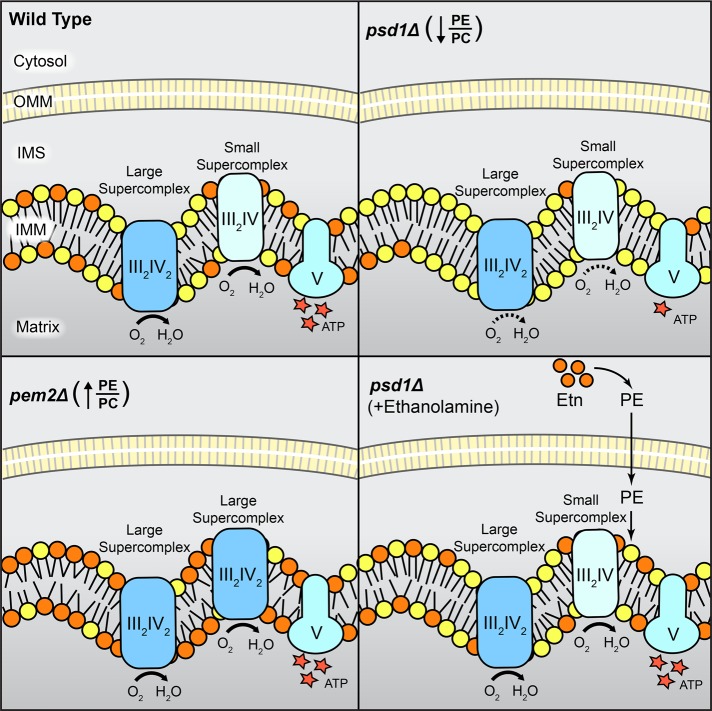
Next we asked whether PE synthesized in ER could be transported to mitochondria to compensate for the lack of mitochondrial PE biosynthesis. Previous reports paint a conflicting picture. One line of evidence shows that the reduced respiratory growth of psd1Δ cells is rescued by Etn supplementation (Birner et al., 2001; Riekhof and Voelker, 2006), whereas other studies show that PE synthesized by the Kennedy pathway does not meet the requirements for respiration in psd1Δ cells (Chan and McQuibban, 2012; Wang et al., 2014). Here we show that exogenous Etn supplementation not only restores cellular PE levels, but it also significantly increases mitochondrial PE levels in psd1Δ cells and fully rescues all mitochondrial bioenergetic defects observed in psd1Δ cells (Figure 6). These results imply that mitochondrial PE import pathway(s) exist and that nonmitochondrial PE can functionally compensate for the lack of mitochondrial PE biosynthesis. The requirement of ERMES complex for efficient rescue suggested that the membrane contact sites between ER and mitochondria may facilitate PE import into mitochondria. However, steady-state levels of mitochondrial PE were not reduced upon Etn supplementation in psd1Δ cells lacking the ERMES complex (Supplemental Figure S6) implying that ERMES is not essential for PE transport but might only play an indirect role in Etn-mediated rescue (Figure 7). In summary, as depicted in our model (Figure 8), this work identified critical roles of nonbilayer phospholipids in MRC activity and assembly, respectively, and showed that nonmitochondrial PE can be transported to mitochondria, where it can functionally substitute for PE deficiency.
FIGURE 8:
Model depicting the specific roles of mitochondrial PE and PC in MRC complex activity and assembly. Reduced PE/PC ratio in mitochondrial PE–depleted psd1Δ cells leads to decreased MRC complex III and IV activities without affecting supercomplex formation. Increased PE/PC ratio in mitochondrial PC–depleted pem2Δ cells results in enhanced formation of the larger supercomplex (III2IV2) without altering the activities of complexes. PE synthesized in ER by exogenous supplementation of Etn is transported into mitochondria and completely restores MRC supercomplex activities in PE-deficient psd1Δ cells. IMM, inner mitochondrial membrane; IMS, intermembrane space; OMM, outer mitochondrial membrane.
MATERIALS AND METHODS
Yeast strains, growth medium composition, and culture conditions
S. cerevisiae strains used in this study are listed in Supplemental Table S1. For growth in liquid media, strains were precultured in YPLac medium (1% yeast extract, 2% peptone, and 2% lactate, pH 5.5) and inoculated into SC media (0.2% dropout mix containing amino acid and other supplements, as previously described (Amberg et al., 2005), 0.17% yeast nitrogen base without amino acids and ammonium sulfate, and 0.5% ammonium sulfate) containing either 2% glucose or 2% lactate (pH 5.5) and grown to late logarithmic phase. Solid media were prepared by the addition of 2% agar. Liquid SC glucose and SC lactate were inoculated with WT or the indicated strains to a starting A600 of 0.1, and growth was monitored for up to 30 h (SC glucose) or 60 h (SC lactate) at 30°C. For growth on solid media, 10-fold serial dilutions of overnight precultures were spotted on SC glucose or SC lactate plates and incubated at 30°C for 2 and 4–5 d, respectively. For Etn supplementation experiments, 2 mM Etn was added to SC growth medium. Petite formation was determined by spreading glucose-grown cells (200 cells/100 μl) onto YPD and YPlac plates. Petite formation was calculated based on the number of colonies that grew on YPD (total viable cells) compared with YPlac (respiratory-competent) media. Single- and double-knockout yeast strains (Supplemental Table S1) were constructed by one-step gene disruption using hygromycin cassette (Janke et al., 2004).
Mitochondrial isolation
Isolation of crude and pure mitochondria was performed as described previously (Meisinger et al., 2006). Mitochondria were isolated from yeast cells grown to late logarithmic phase and subsequently used for Western blot analysis, as well as for in-gel activity assays. To obtain pure mitochondrial fractions, crude mitochondria were loaded onto a sucrose step gradient (60, 32, 23, and 15%) and centrifuged at 134,000 × g for 1 h. The intact mitochondria recovered from the gradient interface (60 and 32%) were washed in isotonic buffer, pelleted at 10,000 × g, and subsequently used for mitochondrial phospholipid quantification. Protein concentrations were determined by the bicinchoninic acid (BCA) assay (Thermo Scientific, Rockford, IL).
Cellular and mitochondrial phospholipid measurements
For the quantification of cellular and mitochondrial phospholipids, lipids were extracted from cells (1 g of wet weight) or pure mitochondria (1.5 mg of protein) using the Folch method (Folch et al., 1957), and individual phospholipids were separated by two-dimensional TLC using the following solvent systems: chloroform/methanol/ammonium hydroxide (65:35:5) in the first dimension, followed by chloroform/acetic acid/methanol/water (75:25:5:2.2) in the second dimension (Storey et al., 2001). Phospholipids were visualized with iodine vapor and scraped into glass tubes, and Pi was quantified. The quantification of total cellular phospholipids was performed as previously described (Horvath et al., 2011). Briefly, 100 mg of SC glucose– or SC lactate–grown cells were digested with Zymolyase and homogenized, and the cells debris was pelleted (3000 × g, 5 min). Phospholipids were extracted from the cell-free homogenate using the Folch method (Folch et al., 1957), and the total phospholipid phosphate was quantified using the Bartlett method (Bartlett, 1959). The mitochondrial phospholipid phosphate was extracted from 1.5 mg of pure mitochondria and quantified using the Bartlett method (Bartlett, 1959).
Electron microscopy
Yeast cells cultured in SC lactate medium were harvested and fixed in 3% glutaraldehyde contained in 0.1 M sodium cacodylate, pH 7.4, 5 mM CaCl2, 5 mM MgCl2, and 2.5% (wt/vol) sucrose for 1 h at room temperature with gentle agitation, spheroplasted, embedded in 2% ultralow-temperature agarose (prepared in water), cooled, and subsequently cut into small pieces (∼1 mm3). The cells were then postfixed in 1% OsO4 and 1% potassium ferrocyanide contained in 0.1 M sodium cacodylate for 1 h at room temperature. The blocks were washed thoroughly three times with double-distilled H2O, transferred to 1% thiocarbohydrazide at room temperature for 3 min, washed in double-distilled H2O twice, and transferred to 1% OsO4 and 1% potassium ferrocyanide in 0.1 M sodium cacodylate for an additional 3 min at room temperature. The cells were washed three times with double-distilled H2O and stained in Kellenberger’s uranyl acetate at room temperature overnight. Cells were then dehydrated through a graded series of cold ethanol, followed by room temperature 100% ethanol exchanges, washed twice in propylene oxide, left overnight in propylene oxide/Epon with gentle agitation, and subsequently embedded in Epon resin. Sections were cut on a Leica Ultracut UCT ultramicrotome and observed on a Philips EM420 transmission electron microscope at 100 kV. Images were recorded with a Soft Imaging System Megaview III digital camera, and figures were assembled in Adobe Photoshop with only linear adjustments in contrast and brightness.
Measurement of oxygen consumption and ATP levels
For measurements of respiration rates, cells were grown to late log phase in SC glucose or SC lactate medium and resuspended in fresh medium at 108 cells/ml (SC glucose) or 2 × 107 cells/ml (SC lactate), and the rate of oxygen consumption was measured at 30°C using Oxytherm (Hansatech, Norfolk, UK). Cyanide-sensitive respiration was calculated after the addition of 1 mM KCN, and the cyanide-insensitive respiration was subtracted from the total respiration. For cellular ATP quantification, cells were seeded at 250,000 cells/well in a 96-well plate, and ATP was measured using the BacTiter-Glo Microbial Cell Viability Assay (Promega, Madison, WI) kit according to the manufacturer’s instructions. All data were normalized to the protein concentration, determined by the BCA assay (Thermo Scientific).
SDS–blue native PAGE and immunoblotting
SDS–PAGE was performed on mitochondrial samples solubilized in lysis buffer (150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with protease inhibitor cocktail (Roche Diagnostic, Indianapolis, IN). Protein extracts were separated on NuPAGE 4–12% Bis-Tris gels (Life Technologies, Carlsbad, CA) and transferred on polyvinylidene fluoride membranes using a Trans-Blot transfer cell (Bio-Rad, Hercules, CA). Membranes were blocked in 5% fatty acid–free bovine serum albumin dissolved in Tris-buffered saline with 0.1% Tween-20 and probed with antibodies as indicated. Blue native PAGE (BN–PAGE) was performed to separate native MRC protein complexes as previously described (Wittig et al., 2006). Briefly, yeast mitochondria were solubilized in buffer containing 1% digitonin (Life Technologies) and incubated for 15 min at 4°C. After a clarifying spin at 20,000 × g (30 min, 4°C), 50x G-250 sample additive was added to the supernatant, and 20 μg of protein was loaded on a 3–12% gradient of native PAGE Bis-Tris gel (Life Technologies). Western blot was performed using a Mini-PROTEAN Tetra cell (Bio-Rad). Membrane was blocked in 5% nonfat milk in Tris-buffered saline with 0.1% Tween-20 and probed with antibodies as indicated. Primary antibodies for yeast proteins used were Cox1, 1:5000 (110 270; Abcam, Cambridge, MA); Cox2, 1:50,000 (110 271; Abcam); Cox3, 1:5000 (110 259; Abcam); Cox4, 1:5000 (110 272; Abcam); Sdh2, 1:5000 (from Dennis Winge); Rip1, 1:100,000 (from Vincenzo Zara); Atp2, 1:40,000 (from Sharon Ackerman); porin, 1:50,000 (110 326; Abcam); Dpm1, 1:250 (113 686; Abcam); Pho8, 1:250 (113 688; Abcam); and Pgk1, 1:5000 (113 687; Abcam). Secondary antibodies (1:5000) were incubated for 1 h at room temperature, and membranes were developed using Western Lightning Plus-ECL (PerkinElmer, Waltham, MA).
In-gel activity measurements
In-gel activities for mitochondrial respiratory chain complexes were performed as described previously (Wittig et al., 2007). Clear native (CN)–PAGE was used to avoid interference of Coomassie blue with activity measurements. Briefly, mitochondria solubilized in 1% digitonin were resolved on a 4–16% gradient native PAGE Bis-Tris gel (Life Technologies) using the following additions to the cathode buffer: for complex II, 0.01% n-dodecyl β-d-maltoside (DDM) and 0.05% sodium deoxycholate (DOC) were added; for complex III, there was no addition; for complex IV, 0.05% DDM and 0.05% DOC were added; and for complex V, 0.01% DDM and 0.05% DOC were added. Gels were loaded with 30 μg of protein for complex II, 90 μg of protein for complexes III and IV, and 20 μg of protein for complex V and incubated in activity staining solutions for complex II, III, IV, or V as reported previously (Wittig et al., 2007). Band intensity quantification was done using the gel analysis method in ImageJ (National Institutes of Health, Bethesda, MD). Equal loading was determined by Coomassie blue stain, and total protein and band intensity quantification was done using the gel analysis method in ImageJ.
Supplementary Material
Acknowledgments
We thank Miriam L. Greenberg (Wayne State University) for yeast strains and Sharon Ackerman (Wayne State University School of Medicine), Dennis Winge (University of Utah), and Vincenzo Zara (Università del Salento) for their generous gift of antibodies. We also thank Craig Kaplan, Steve Lockless, Jean-Philippe Pellois, and the members of the Gohil lab for valuable discussions and comments, as well as Mary Casillas and Shrishiv Timbalia for artwork and John Neff for technical help. Research reported here was supported by Welch Foundation Grant A-1810 and National Institutes of Health Award R01GM111672 to V.M.G. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used:
- CL
cardiolipin
- ER
endoplasmic reticulum
- ERMES
endoplasmic reticulum–mitochondria encounter structure
- Etn
ethanolamine
- MRC
mitochondrial respiratory chain
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PMME
phosphatidylmonomethylethanolamine
- PS
phosphatidylserine.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-12-0865) on May 25, 2016.
REFERENCES
- Acín-Pérez R, Fernández-Silva P, Peleato ML, Pérez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- AhYoung AP, Jiang J, Zhang J, Khoi Dang X, Loo JA, Zhou ZH, Egea PF. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc Natl Acad Sci USA. 2015;112:3179–3188. doi: 10.1073/pnas.1422363112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktas M, Wessel M, Hacker S, Klusener S, Gleichenhagen J, Narberhaus F. Phosphatidylcholine biosynthesis and its significance in bacteria interacting with eukaryotic cells. Eur J Cell Biol. 2010;89:888–894. doi: 10.1016/j.ejcb.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics. A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Bartlett GR. Phosphorus assay in column chromatography. J Biol Chem. 1959;234:466–468. [PubMed] [Google Scholar]
- Bazán S, Mileykovskaya E, Mallampalli VK, Heacock P, Sparagna GC, Dowhan W. Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J Biol Chem. 2013;288:401–411. doi: 10.1074/jbc.M112.425876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner R, Burgermeister M, Schneiter R, Daum G. Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol Biol Cell. 2001;12:997–1007. doi: 10.1091/mbc.12.4.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottinger L, Horvath SE, Kleinschroth T, Hunte C, Daum G, Pfanner N, Becker T. Phosphatidylethanolamine and cardiolipin differentially affect the stability of mitochondrial respiratory chain supercomplexes. J Mol Biol. 2012;423:677–686. doi: 10.1016/j.jmb.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumann HA, Gubbens J, Koorengevel MC, Oh CS, Martin CE, Heck AJ, Patton-Vogt J, Henry SA, de Kruijff B, de Kroon AI. Depletion of phosphatidylcholine in yeast induces shortening and increased saturation of the lipid acyl chains: evidence for regulation of intrinsic membrane curvature in a eukaryote. Mol Biol Cell. 2006;17:1006–1017. doi: 10.1091/mbc.E05-04-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürgermeister M, Birner-Grünberger R, Heyn M, Daum G. Contribution of different biosynthetic pathways to species selectivity of aminoglycerophospholipids assembled into mitochondrial membranes of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2004a;1686:148–160. doi: 10.1016/j.bbalip.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Bürgermeister M, Birner-Grunberger R, Nebauer R, Daum G. Contribution of different pathways to the supply of phosphatidylethanolamine and phosphatidylcholine to mitochondrial membranes of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2004b;1686:161–168. doi: 10.1016/j.bbalip.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Carman GM, Han GS. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu Rev Biochem. 2011;80:859–883. doi: 10.1146/annurev-biochem-060409-092229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, McQuibban GA. Phosphatidylserine decarboxylase 1 (Psd1) promotes mitochondrial fusion by regulating the biophysical properties of the mitochondrial membrane and alternative topogenesis of mitochondrial genome maintenance protein 1 (Mgm1) J Biol Chem. 2012;287:40131–40139. doi: 10.1074/jbc.M112.399428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol Cell Physiol. 2007;292:33–44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- Choi JY, Martin WE, Murphy RC, Voelker DR. Phosphatidylcholine and N-methylated phospholipids are nonessential in Saccharomyces cerevisiae. J Biol Chem. 2004;279:42321–42330. doi: 10.1074/jbc.M405074200. [DOI] [PubMed] [Google Scholar]
- Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC, et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155:160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G. Lipids of mitochondria. Biochim Biophys Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc Natl Acad Sci USA. 2011;108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Gohil VM, Greenberg ML. Mitochondrial membrane biogenesis: phospholipids and proteins go hand in hand. J Cell Biol. 2009;184:469–472. doi: 10.1083/jcb.200901127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohil VM, Thompson MN, Greenberg ML. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J Biol Chem. 2005;280:35410–35416. doi: 10.1074/jbc.M505478200. [DOI] [PubMed] [Google Scholar]
- Gulshan K, Shahi P, Moye-Rowley WS. Compartment-specific synthesis of phosphatidylethanolamine is required for normal heavy metal resistance. Mol Biol Cell. 2010;21:443–455. doi: 10.1091/mbc.E09-06-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510:48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- Horvath SE, Wagner A, Steyrer E, Daum G. Metabolic link between phosphatidylethanolamine and triacylglycerol metabolism in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2011;1811:1030–1037. doi: 10.1016/j.bbalip.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath SE, Daum G. Lipids of mitochondria. Prog Lipid Res. 2013;52:590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Joshi AS, Zhou J, Gohil VM, Chen S, Greenberg ML. Cellular functions of cardiolipin in yeast. Biochim Biophys Acta. 2009;1793:212–218. doi: 10.1016/j.bbamcr.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman WJ, Willems PH, Smeitink JA. Monogenic mitochondrial disorders. N Engl J Med. 2012;366:1132–1141. doi: 10.1056/NEJMra1012478. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S, Toulmay A, Prinz WA. Membrane contact sites, gateways for lipid homeostasis. Curr Opin Cell Biol. 2015;33:82–87. doi: 10.1016/j.ceb.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YW, Claypool SM. Disorders of phospholipid metabolism: an emerging class of mitochondrial disease due to defects in nuclear genes. Front Genet. 2015;6:1–27. doi: 10.3389/fgene.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem Phys Lipids. 2014;179:42–48. doi: 10.1016/j.chemphyslip.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Voelker DR, Langer T. Making heads or tails of phospholipids in mitochondria. J Cell Biol. 2011;192:7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- Ren M, Phoon CK, Schlame M. Metabolism and function of mitochondrial cardiolipin. Prog Lipid Res. 2014;55:1–16. doi: 10.1016/j.plipres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Riekhof WR, Voelker DR. Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J Biol Chem. 2006;281:36588–36596. doi: 10.1074/jbc.M608851200. [DOI] [PubMed] [Google Scholar]
- Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MH, Di Bartolomeo F, Böttinger L, Horvath SE, Wenz LS, Daum G, Becker T. Phosphatidylcholine affects the role of the sorting and assembly machinery in the biogenesis of mitochondrial β-barrel proteins. J Biol Chem. 2015;290:26523–26532. doi: 10.1074/jbc.M115.687921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey MK, Clay KL, Kutateladze T, Murphy RC, Overduin M, Voelker DR. Phosphatidylethanolamine has an essential role in Saccharomyces cerevisiae that is independent of its ability to form hexagonal phase structures. J Biol Chem. 2001;276:48539–48548. doi: 10.1074/jbc.M109043200. [DOI] [PubMed] [Google Scholar]
- Tasseva G, Bai HD, Davidescu M, Haromy A, Michelakis E, Vance JE. Phosphatidylethanolamine deficiency in mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J Biol Chem. 2013;288:4158–4173. doi: 10.1074/jbc.M112.434183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller G, Nemec T, Hrastnik C, Daum G. Lipid composition of subcellular membranes of an FY1679-derived haploid yeast wild-type strain grown on different carbon sources. Yeast. 1999;15:1555–1564. doi: 10.1002/(SICI)1097-0061(199910)15:14<1555::AID-YEA479>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- van der Veen JN, Lingrell S, da Silva RP, Jacobs RL, Vance DE. The concentration of phosphatidylethanolamine in mitochondria can modulate ATP production and glucose metabolism in mice. Diabetes. 2014;63:2620–2630. doi: 10.2337/db13-0993. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters AM, Porter GA, Jr, Brookes PS. Mitochondria as a drug target in ischemic heart disease and cardiomyopathy. Circ Res. 2012;111:1222–1236. doi: 10.1161/CIRCRESAHA.112.265660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang S, Liou LC, Ren Q, Zhang Z, Caldwell GA, Caldwell KA, Witt SN. Phosphatidylethanolamine deficiency disrupts α-synuclein homeostasis in yeast and worm models of Parkinson disease. Proc Natl Acad Sci USA. 2014;111:3976-E3985. doi: 10.1073/pnas.1411694111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Hielscher R, Hellwig P, Schägger H, Richers S, Hunte C. Role of phospholipids in respiratory cytochrome bc(1) complex catalysis and supercomplex formation. Biochim Biophys Acta. 2009;1787:609–616. doi: 10.1016/j.bbabio.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Wittig I, Karas M, Schagger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics. 2007;6:1215–1225. doi: 10.1074/mcp.M700076-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.