Cdc5 associates with centromeric chromatin during mitosis. Cdc5 plays a critical role in the differential removal of cohesin from centromeric chromatin compared to chromosome arms.
Abstract
Sister chromatid cohesion is essential for tension-sensing mechanisms that monitor bipolar attachment of replicated chromatids in metaphase. Cohesion is mediated by the association of cohesins along the length of sister chromatid arms. In contrast, centromeric cohesin generates intrastrand cohesion and sister centromeres, while highly cohesin enriched, are separated by >800 nm at metaphase in yeast. Removal of cohesin is necessary for sister chromatid separation during anaphase, and this is regulated by evolutionarily conserved polo-like kinase (Cdc5 in yeast, Plk1 in humans). Here we address how high levels of cohesins at centromeric chromatin are removed. Cdc5 associates with centromeric chromatin and cohesin-associated regions. Maximum enrichment of Cdc5 in centromeric chromatin occurs during the metaphase-to-anaphase transition and coincides with the removal of chromosome-associated cohesin. Cdc5 interacts with cohesin in vivo, and cohesin is required for association of Cdc5 at centromeric chromatin. Cohesin removal from centromeric chromatin requires Cdc5 but removal at distal chromosomal arm sites does not. Our results define a novel role for Cdc5 in regulating removal of centromeric cohesins and faithful chromosome segregation.
INTRODUCTION
Accurate chromosome segregation is important for the maintenance of genome stability during growth and proliferation of organisms (Lengauer et al., 1998; Maddox et al., 2012). Errors in chromosome segregation result in aneuploidy, which has been linked with several human diseases, such as developmental disorders and many cancers (Santaguida and Amon, 2015; Singh and Gerton, 2015). The kinetochore, composed of centromeric DNA (CEN) and associated proteins, is an essential component of the chromosomal segregation machinery (Burrack and Berman, 2012; Choy et al., 2012; Maddox et al., 2012). The assembly of kinetochore proteins at budding yeast CEN (core centromere composed of ∼125 base pairs of defined DNA sequences) results in a highly ordered, unique, and topologically distinct chromatin structure in the chromosomes (Clarke and Carbon, 1980; Bloom and Carbon, 1982; Bloom et al., 1984; Saunders et al., 1990), the structural integrity of which is required for accurate chromosome segregation (Newlon, 1988; Verdaasdonk and Bloom, 2011; Haase et al., 2013; Mishra et al., 2013; Mishra et al., 2015). Budding yeast centromeric chromatin (CEN and pericentromeric DNA that extends ∼30–50 kb around the CEN) exhibits a cruciform structure referred to as centromere loop (C-loop), which is analogous to that proposed in the looping model for vertebrate centromeres (Yeh et al., 2008; Verdaasdonk et al., 2012). The intramolecular linkages in the C-loop and the intermolecular linkages along the length of the chromosomes promote the biorientation of kinetochores and sister chromatids toward opposite spindle pole bodies for faithful chromosome segregation.
Cohesion along the length of the chromosomes is facilitated by chromosomal association of cohesins (Hornig and Uhlmann, 2004; Gerton, 2007; Brooker and Berkowitz, 2014). The cohesin complex is composed of four subunits: Scc1/Mcd1, Scc3, Smc1, and Smc3 (Hartman et al., 2000; Laloraya et al., 2000; Nasmyth, 2002; Glynn et al., 2004; Weber et al., 2004; Gerton, 2005; Bose and Gerton, 2010; Rossio et al., 2010; Marston, 2014; Roccuzzo et al., 2015). An enrichment of cohesins is seen at cohesin-associated regions (CARs) and at centromeric chromatin (Blat and Kleckner, 1999; Megee et al., 1999; Tanaka et al., 1999). Removal of cohesins from chromosomes facilitates separation of sister chromatids at anaphase and cell cycle progression (Hartman et al., 2000; Marston, 2014; Guacci et al., 2015). Evolutionarily conserved Cdc5 has been shown to regulate several aspects of mitosis (Zitouni et al., 2014; Archambault et al., 2015), including sister chromatid separation, by phosphorylating Scc1/Mcd1 (Alexandru et al., 2001) to promote its proteolytic cleavage by separase (Uhlmann et al., 2000). Mutation of phosphorylatable serines to alanines in Scc1/Mcd1 or depletion of Cdc5 results in anaphase delay (Alexandru et al., 2001; Hornig and Uhlmann, 2004). It has been proposed that preferential phosphorylation of chromatin-bound Scc1/Mcd1 by Cdc5 accelerates its cleavage compared with soluble cohesin (Hornig and Uhlmann, 2004); however, not all Scc1/Mcd1 is cleaved by separase in anaphase (Uhlmann et al., 1999).
Here we report that Cdc5 associates with centromeric chromatin and CARs in a cell cycle–dependent manner. Enrichment of Cdc5 in mitosis overlaps with the timing of removal of cohesin from chromosomes. Cohesin proteins interact with Cdc5 in vivo and are required for the enrichment of Cdc5 at centromeric chromatin during mitosis. We demonstrate the persistence of centromeric cohesin in anaphase cells of a cdc5 mutant. Our results show that Cdc5 associates with centromeric chromatin to regulate the removal of cohesin from this region and promote faithful chromosome segregation.
RESULTS
Cdc5 associates with centromeric chromatin in a cell cycle–dependent manner
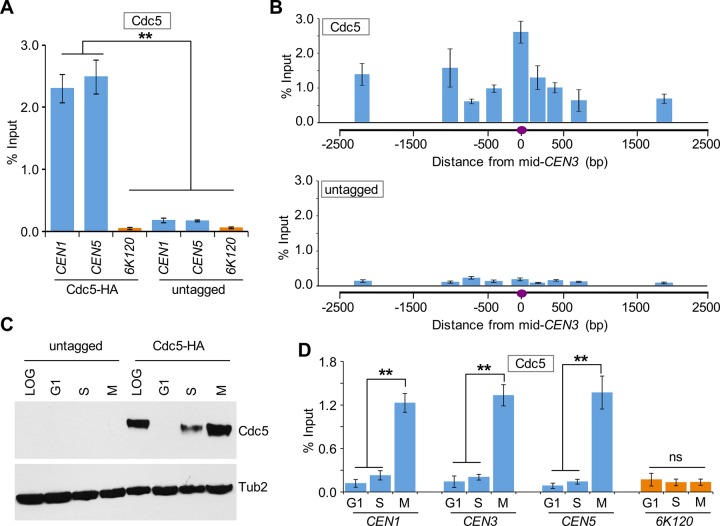
To determine whether Cdc5 associates with centromeric chromatin, we performed chromatin immunoprecipitation (ChIP) experiments using a wild-type strain carrying hemagglutinin (HA)-tagged Cdc5 expressed from its own promoter. ChIP–quantitative PCR (qPCR) showed that Cdc5 associates with core CEN (CEN1 and CEN5) in logarithmically growing cells (Figure 1A). No significant enrichment of Cdc5 was observed at the negative control region 6K120 (Rossio et al., 2010) or in an untagged wild-type strain (Figure 1A). We next examined Cdc5 enrichment in chromatin regions flanking CEN. ChIP-qPCR showed that Cdc5 is strongly enriched at the central core region of CEN3 and flanking chromatin (Figure 1B). No significant enrichment was detected in an untagged wild-type strain used as a control (Figure 1, A and B).
FIGURE 1:
Cdc5 associates with centromeric chromatin in a cell cycle–dependent manner. (A) Cdc5 associates with core CEN DNA. ChIP was performed for Cdc5 using a wild-type strain containing Cdc5-HA (YMB9264) and an untagged control strain (YMB9263). Strains were grown to logarithmic phase at 30°C. ChIP was performed using α-HA antibodies. Enrichment of Cdc5 to CEN1 and CEN5 as determined by qPCR is shown as percentage input. The negative control region 6K120 was used as a background control. Average from three biological replicates ± SE. **p <0.01, Student’s t test. (B) Cdc5 is enriched at the CEN and peri-CEN chromatin. ChIP was performed as in A. Average from three biological replicates ± SE. (C) Western blotting showing the protein expression levels of Cdc5-HA in wild-type strain containing Cdc5-HA (YMB9264) and untagged control (YMB9263). Strains were in logarithmic phase of growth (LOG) or arrested in G1-phase (α-factor), S-phase (hydroxyurea), or M-phase (nocodazole) stages of the cell cycle. Antibodies used in Western blotting were α-HA (Cdc5) and α-Tub2 (loading control). (D) Association of Cdc5 with CEN DNA increases in mitotic cells. ChIP-qPCR shows the levels of CEN-associated Cdc5 in a wild-type strain containing Cdc5-HA (YMB9264) and untagged control (YMB9263) grown to logarithmic phase at 30°C or arrested at various stages of the cell cycle as in C. Enrichment at CEN (CEN1, CEN3, and CEN5) and a negative control region 6K120 was determined by qPCR and is shown as percentage input. Average from three biological replicates ± SE. **p < 0.01; ns, not statistically significant, Student’s t test.
To examine whether association of Cdc5 with CEN varies during the cell cycle, we performed ChIP experiments using a wild-type strain arrested in the G1 (α-factor treatment), S (hydroxyurea treatment), or M (nocodazole treatment) phases of the cell cycle. The cell cycle arrest was confirmed by fluorescence-activated cell sorting (FACS) assay (Supplemental Figure S1). Western blot analysis revealed a cell cycle–dependent expression pattern of Cdc5, with maximum protein levels observed in M-phase cells, only low levels detected in S-phase cells, and no detectable expression in G1 cells (Figure 1C), as previously observed (Hardy and Pautz, 1996; Charles et al., 1998). The CEN enrichment of Cdc5 was significantly higher in mitotic (M) cells (Figure 1D). No significant enrichment of Cdc5 at CEN was detected in G1- and S-phase cells; the ChIP signals in G1- and S-phase cells were largely similar to those detected at a negative control region, 6K120 (Figure 1D). Control experiments performed with an untagged wild-type strain showed no significant enrichment of Cdc5 at either CEN or negative control region 6K120 (Supplemental Figure S2).
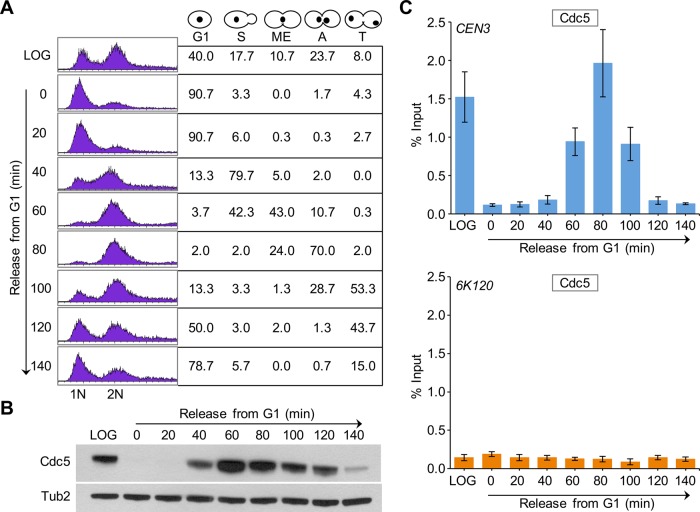
Next we examined the CEN association of Cdc5 during cell cycle progression using wild-type cells arrested in G1 phase by α-factor treatment and at various times after release from G1 arrest (Figure 2A). On the basis of cell and nuclear morphology, we categorized cells as G1, S, metaphase (ME), anaphase (A), and telophase (T) following procedures described previously (Calvert and Lannigan, 2010). In agreement with previous results (Figure 1B), Western blot analysis revealed cell cycle–regulated expression of Cdc5, with increased levels observed in metaphase and anaphase cells (Figure 2B). ChIP results showed that CEN association of Cdc5 is low in cells soon after release from α-factor arrest (40 min), increasing as cells enter mitosis (60 min), maximum in anaphase (80 min), and decreasing as cells exit mitosis (100–120 min; Figure 2, A and C). We did not detect Cdc5 enrichment at a negative control region, 6K120 (Figure 2C bottom). On the basis of these results, we conclude that CEN association of Cdc5 is cell cycle dependent, with maximum enrichment in mitosis (metaphase and anaphase).
FIGURE 2:
CEN association of Cdc5 is cell cycle regulated, with maximum enrichment in cells undergoing mitosis. A wild-type strain containing Cdc5-HA (YMB9264) was grown at 30°C, arrested in G1 with α-factor, washed, and released into pheromone-free medium. α-Factor was added again 80 min after release to block the cells at the subsequent G1. Samples were taken at various time points (minutes) after G1 release. (A) FACS analysis revealed cell cycle progression. Cell cycle stages were determined based on cell morphology and nuclear position by microscopic examination of 100 cells for each time point. Different stages of the cell cycle: G1, S phase (S), metaphase (ME), anaphase (A), and telophase (T). (B) Expression of Cdc5 is cell cycle regulated. Western blot analysis was carried out on whole-cell protein extracts prepared from samples taken at various times after release from G1 arrest as in A. Blots were probed with α-HA (Cdc5) and α-Tub2 (loading control) antibodies. (C) Centromeric levels of Cdc5 are highest in mitotic cells. ChIP for Cdc5 was carried out using α-HA antibodies. Enrichment of Cdc5 at CEN3 (top) and a negative control region 6K120 (bottom) determined by qPCR and shown as percentage input. Average from three biological replicates ± SE.
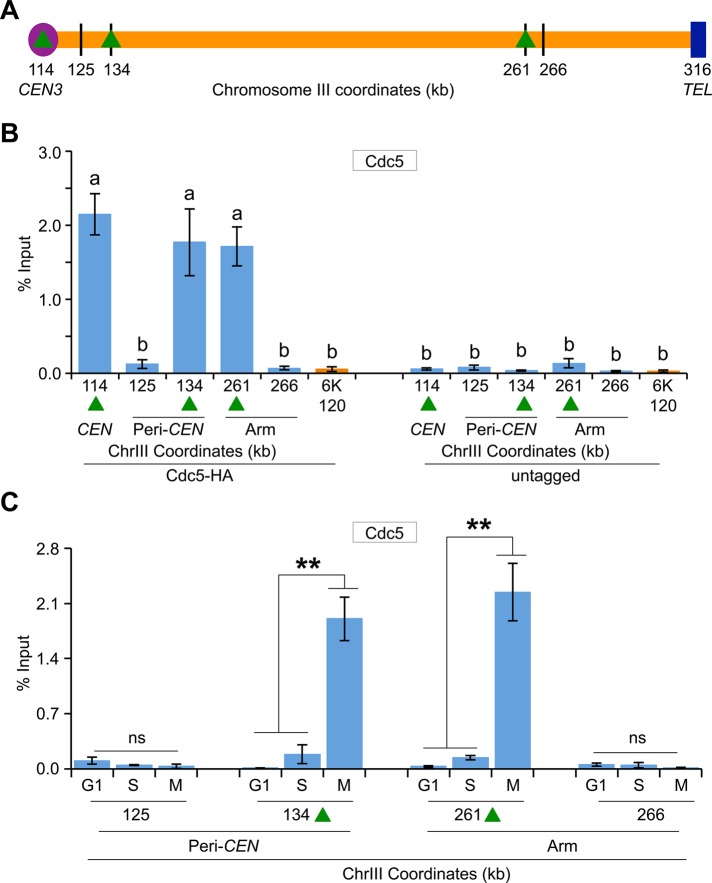
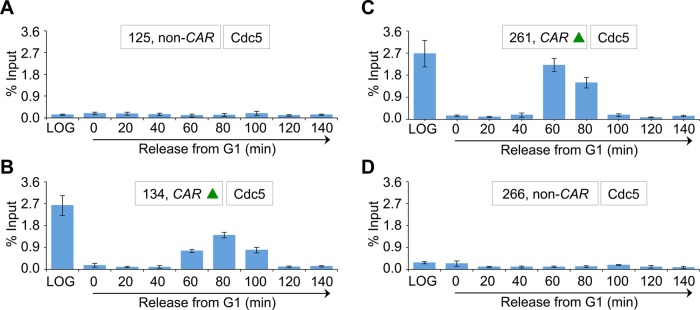
Because a previous study showed enrichment of Cdc5 at CARs (Rossio et al., 2010), we investigated whether Cdc5 localization at CARs is cell cycle dependent. We examined three CARs on chromosome III (Figure 3A, green triangles), and two non-CARs (Figure 3A, black vertical lines) previously reported to either associate or fail to associate with cohesin components, respectively (Eckert et al., 2007; Ng et al., 2009). The CARs are 114 (CEN3), 134 (peri-CEN, 20 kb from CEN3), and 261 (chromosome arm, 147 kb from CEN3), and non-CARs are 125 (peri-CEN, 11 kb from CEN3) and 266 (chromosome arm, 152 kb from CEN3; Figure 3A). Experiments performed with an untagged wild-type strain showed no significant enrichment of Cdc5 at any of the regions tested (Figure 3B). In logarithmically growing cells, Cdc5 associates specifically with CARs (Figure 3B). Cdc5 enrichment at CARs was significantly higher in mitotic cells (M) than in S-phase cells and was barely detectable in G1 (Figure 3C). The increased levels of Cdc5 at CARs in mitosis were verified by assaying cells synchronized in G1 and released into pheromone-free medium. Consistent with the increased enrichment of Cdc5 at peri-CEN and arm CARs (134 and 261, respectively) in nocodazole-treated cells (Figure 3C), we observed higher levels of Cdc5 at these regions in cells undergoing mitosis (60–100 min postrelease; Figure 4). Taken together, these results show that Cdc5 enrichment at CEN and CARs is cell cycle dependent, with maximum enrichment during mitosis.
FIGURE 3:
Cdc5 associates with CARs in a cell cycle–dependent manner, with maximum enrichment in mitosis. (A) Schematic showing CARs and non-CARs on chromosome III. Cohesins have been shown to either associate (CARs) or fail to associate (non-CARs) at these regions in previous studies (Eckert et al., 2007; Ng et al., 2009). Centromere (CEN3, violet circle), CAR (green triangle), and non-CAR (black vertical line) sites. CEN is located at 114 kb; peri-CEN CAR 134 (20 kb from CEN), chromosome arm CAR 261 (147 kb from CEN), peri-CEN non-CAR 125 (11 kb from CEN), and chromosome arm non-CAR 266 (152 kb from CEN) were examined. (B) Cdc5 associates with CARs. ChIP was performed for Cdc5 in a wild-type strain tagged with Cdc5-HA (YMB9264) and an untagged control strain (YMB9263). Cells were grown to logarithmic phase at 30°C. Enrichment of Cdc5 to CEN (114), CAR (134 and 261), and non-CAR control regions (125 and 266) on chromosome III was determined by qPCR and is shown as percentage input. Average values of three biological replicates ± SE of the mean. Values sharing the same letter (a, b) are not significantly different at the 5% level based on analysis of variance. (C) Enrichment of Cdc5 at CARs is higher in mitotic cells. ChIP-qPCR was performed with wild-type strain YMB9264 (Cdc5-HA) using cells prepared as in Figure 1C. Enrichment at CAR (134 and 261) and non-CAR control (125 and 266) regions is shown as percentage input. Average from three biological replicates ± SE. **p < 0.01; ns, not statistically significant, Student’s t test.
FIGURE 4:
CAR association of Cdc5 is cell cycle regulated, with maximum enrichment in cells undergoing mitosis. A wild-type strain containing Cdc5-HA (YMB9264) was grown at 30°C, arrested in G1 with α-factor, washed, and released into pheromone-free medium. α-Factor was added again 80 min after release to block the cells at the subsequent G1. Samples were taken at various time points (minutes) after G1 release. ChIP-qPCR was performed to determine the enrichment pattern of Cdc5 at CARs and non-CARs on chromosome III throughout the cell cycle. The CAR (green triangles) and non-CAR (black vertical lines) examined are shown schematically in Figure 3A. Cdc5 enrichment is shown as percentage input. Average from three biological replicates ± SE for each time point. (A) Enrichment levels of Cdc5 at non-CAR (125) located at peri-CEN3. (B) Enrichment levels of Cdc5 at CAR (134) located at peri-CEN3. (C) Enrichment levels of Cdc5 at CAR (261) located at the chromosomal arm. (D) Enrichment levels of Cdc5 at non-CAR (266) located at the chromosomal arm.
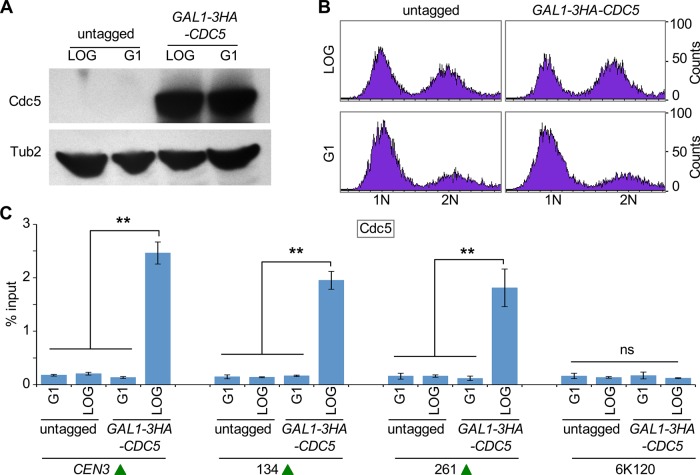
Induced expression of Cdc5 in G1 does not result in its recruitment to centromeric chromatin and CARs
The mitotic enrichment of Cdc5 at CEN and CAR sites correlates with the protein expression levels of Cdc5, and no significant enrichment of Cdc5 was observed at these sites in G1, when protein expression of Cdc5 was not detected (Figure 1, C and D). Therefore we sought to determine whether recruitment of Cdc5 at CEN and CAR sites is regulated by the cell cycle or is an outcome of its cell cycle–dependent expression pattern. We constructed a strain expressing HA-tagged Cdc5 from a galactose-inducible promoter (GAL1-3HA-CDC5) and performed ChIP experiments using cells growing logarithmically and cells arrested in G1 with α-factor treatment. The galactose-induced expression of Cdc5 in logarithmically growing and G1 cells was determined by Western blotting (Figure 5A). FACS analysis confirmed the synchronization of cells in G1 (Figure 5B). ChIP-qPCR revealed significant enrichment of Cdc5 at CEN3 and CARs (134, 261) in logarithmically growing cells, but no significant enrichment of Cdc5 was detected at these regions in G1 cells (Figure 5C). The ChIP signals in G1 cells were largely similar to those detected at the negative control region 6K120 or at CEN3, CARs, and 6K120 region in an untagged wild-type strain (Figure 5C). On the basis of these results, we conclude that the enrichment of Cdc5 at CEN and CARs is cell cycle regulated.
FIGURE 5:
Induced expression of Cdc5 in G1 does not result in its recruitment to CEN and CARs. A wild-type strain containing GAL1-3HA-CDC5 (YMB9441) and untagged control (JG595) were grown at 30°C in yeast extract/peptone with 2% galactose and raffinose and arrested in G1 with α-factor for 2 h. Samples were collected for protein extraction, DNA content, and ChIP analyses. (A) Induced expression of Cdc5 (GAL1-3HA-CDC5) in G1. Western blot analysis was carried out on whole-cell protein extracts prepared from cultures growing logarithmically or arrested in G1 with α-factor treatment. Blots were probed with α-HA (Cdc5) and α-Tub2 (loading control) antibodies. (B) FACS analysis revealed synchronization of cells in G1. (C) Cdc5 does not associate with CEN and CARs in G1. ChIP for Cdc5 (GAL1-3HA-CDC5) was carried out using α-HA antibodies. Enrichment of Cdc5 at CEN3, CARs (134, 261), and a negative control region 6K120 was determined by qPCR and is shown as percentage input. Average from three biological replicates ± SE. **p < 0.01; ns, not statistically significant, Student’s t test.
Cohesin proteins interact with Cdc5 and regulate the mitotic enrichment of Cdc5 at centromeric chromatin and CARs
The association of Cdc5 at CEN and CARs (Figures 1–4) coincides with the high levels of cohesins shown to associate at these regions (Megee et al., 1999; Laloraya et al., 2000; Glynn et al., 2004; Weber et al., 2004; Eckert et al., 2007); hence we asked whether cohesin interacts with Cdc5 in vivo and investigated its role in Cdc5 centromeric association. In budding yeast, the cohesin complex is composed of four subunits: Scc1/Mcd1, Scc3, Smc1, and Smc3 (Nasmyth, 2002). We constructed strains expressing Cdc5 and TAP-tagged Mcd1 or Cdc5 and green fluorescent protein (GFP)–tagged Smc3 from their endogenous promoters. Immunoprecipitation experiments showed in vivo interaction of Cdc5 with Mcd1 and Smc3 (Figure 6). No interaction was observed in control experiments using a strain with untagged Mcd1 or Smc3 (Figure 6).
FIGURE 6:
Cdc5 interacts with cohesin proteins Mcd1 and Smc3 in vivo. (A) Cdc5 interacts in vivo with Mcd1. Wild-type strain (MAY8878) expressing TAP-tagged Mcd1 (CDC5 MCD1-TAP) and a control strain (BY4741) expressing untagged Mcd1 (CDC5 MCD1) from their native promoters were grown at 30°C. Cell extracts were prepared and used in immunoprecipitation experiments using α-Cdc5 monoclonal and α-TAP antibodies. Eluted proteins were analyzed by Western blotting with α-Cdc5, α-TAP (Mcd1), and α-Tub2 (loading control) antibodies. IN, input; IP, immunoprecipitated samples. (B) Cdc5 interacts in vivo with Smc3. Wild-type strain (YMB9431) expressing GFP-tagged Smc3 (CDC5 SMC3-GFP) and a control strain (YMB8702) expressing untagged Smc3 (CDC5 SMC3) from their native promoters were grown at 30°C. Cell extracts were prepared and used in immunoprecipitation experiments using α-Cdc5 monoclonal and α-GFP antibodies. Eluted proteins were analyzed by Western blotting with α-Cdc5, α-GFP (Smc3), and α-Tub2 (loading control) antibodies.
Next we examined whether Mcd1 is required for enrichment of Cdc5 at CEN and CARs. Because cohesin subunits are essential for cell viability, we used mcd1-1, a well-characterized temperature-sensitive mutant that arrests in M phase at the nonpermissive temperature (37°C) and exhibits defects in establishment and maintenance of cohesion (Guacci et al., 1997; Noble et al., 2006; Skibbens et al., 2010; Eng et al., 2014, 2015). The expression of Cdc5 was similar in wild-type and mcd1-1 strains grown at 25 and 37°C, as measured by Western blot analysis (Figure 7A). The enrichment of Cdc5 at CEN and CARs in nocodazole-treated (M-phase arrest) wild-type and mcd1-1 cells grown at 25°C and after shift to 37°C was assayed by ChIP. The cell cycle arrest was verified by FACS assay (Figure 7B). ChIP-qPCR showed that Cdc5 levels at CEN3, as well as CARs located at peri-CEN (134) and the chromosomal arm (261), are reduced threefold to fourfold specifically in the mcd1-1 strain at 37°C (Figure 7C). There was no significant enrichment of Cdc5 at negative control region 6K120 under any condition (Figure 7C). We conclude that cohesin is required for enrichment of Cdc5 at CEN and CARs.
FIGURE 7:
Mitotic enrichment of Cdc5 at CEN and CARs requires cohesin. Wild type (WT; MAY8775), mcd1-1 (MAY8851), and untagged control (YPH499) expressing Cdc5-13Myc were grown in YPD with nocodazole (20 μg/ml) to arrest cells in M phase at permissive temperature (25°C) and shifted to nonpermissive temperature (37°C) for 2.5 h. (A) Western blotting showing the protein expression levels of Cdc5 in WT, mcd1-1, and untagged control strains. Antibodies used were α-Myc (Cdc5) and α-Tub2 (loading control). (B) FACS profile showing synchronization of WT, mcd1-1, and untagged control strains in M phase of the cell cycle after nocodazole treatment. (C) Cdc5 levels are reduced at CEN and CARs in mcd1-1 strains. ChIP was performed using chromatin extracts from WT, mcd1-1, and untagged control strains with α-Myc antibodies. Enrichment of Cdc5 at the CEN3 (114), peri-CEN CAR (134), and chromosomal arm CAR (261) was determined by qPCR and is shown as percentage input. CARs are marked (green triangles). The negative control region 6K120 was used as a background control. Average from three biological replicates ± SE. **p < 0.01; ns, not statistically significant, Student’s t test.
cdc5-99 strains exhibit persistence of cohesin at CEN and peri-CEN regions in anaphase
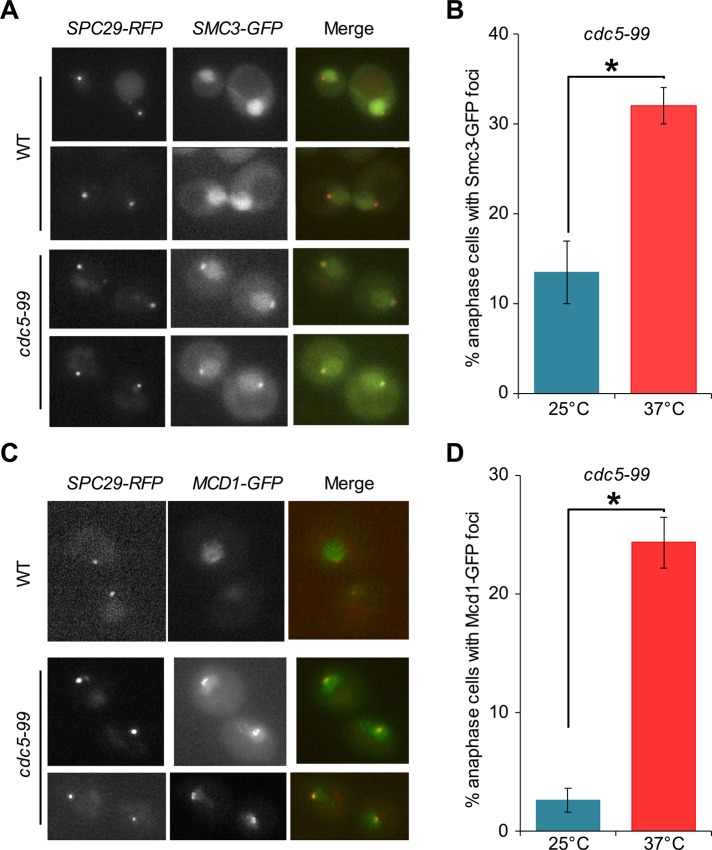
Given our findings that association of Cdc5 at centromeric chromatin and CARs is maximal in cells undergoing the metaphase-to-anaphase transition (Figures 2 and 4), we posited that Cdc5 might be required for the removal of chromatin-bound cohesin. In wild-type cells, cohesin is lost from both centromeric chromatin and chromosomal arms at anaphase onset (Marston, 2014). To characterize the Cdc5 dependence of cohesin loss, we monitored the localization of Smc3-GFP in anaphase cells of wild-type and cdc5-99 strains grown at permissive temperature (25°C) and after a 2.5-h shift to the nonpermissive temperature of 37°C. The well-characterized temperature-sensitive cdc5-99 mutant exhibits a defect in mitotic exit at the nonpermissive temperature of 37°C (St-Pierre et al., 2009). Spindle pole body (SPB) component Spc29–red fluorescent protein (RFP) was used as a marker for centromeric chromatin due to the close physical proximity of the SPB to the kinetochores in budding yeast. Using the distance between SPBs as a measure for spindle length, we quantitated Smc3-GFP foci in anaphase cells with a spindle length of 3–8 μm. In addition to the diffuse Smc3-GFP fluorescence observed over the separated chromatin masses in both wild-type and cdc5-99 cells, distinct Smc3-GFP foci were observed in the cdc5-99 mutant (Figure 8A). No detectable Smc3-GFP foci were ever observed in wild-type cells in anaphase. Quantification of the cdc5-99 cells with Smc3-GFP foci at one or both of the spindle poles was significantly higher (32 vs. 13%, p < 0.05) at 37°C (n = 144) than at 25°C (n = 125; Figure 8B). We next examined the localization pattern of another cohesin component, Mcd1-GFP, in anaphase cells of wild-type and cdc5-99 strains using growth conditions and procedure as described earlier. The persistence of Mcd1-GFP foci was observed in anaphase cells of cdc5-99 strain, whereas no detectable Mcd1-GFP foci were observed in wild-type cells (Figure 8C). The number of cdc5-99 cells with Mcd1-GFP foci at one or both of the spindle poles was significantly higher (24 vs. 3%, p < 0.05) at 37°C (n = 197) than at 25°C (n = 94; Figure 8D).
FIGURE 8:
Persistence of Smc3 and Mcd1 close to spindle pole bodies in anaphase cells of cdc5-99 strains. (A) Wild-type (WT, KBY6098-1) and cdc5-99 (KBY6097-1) strains containing SMC3-GFP and SPC29-RFP were grown at permissive temperature (25°C) and shifted to the nonpermissive temperature (37°C) for 2.5 h. Cells with spindle length 3–8 μm were considered to be in anaphase and were used for imaging. Spc29-RFP was used as a spindle pole marker. Representative images showing the persistence of Smc3-GFP signals in anaphase cells of cdc5-99 strain (KBY6097-1). (B) Percentage of Smc3-GFP foci proximal to the spindle poles was determined in cdc5-99 strain (KBY6097-1) at permissive temperature (25°C, n = 125) and 2.5 h after shift to nonpermissive temperature (37°C, n = 144). Standard weighted-means analysis was used to calculate averages, and significance was determined by one-way analysis of variance. Bar diagram represents average ± SE of the mean. *p < 0.05. (C) WT (YMB9695) and cdc5-99 (YMB9696) strains containing MCD1-GFP and SPC29-RFP were used. Strain VG3507-2A carrying MCD1-GFP (a gift from Vincent Guacci, University of California, Berkeley, CA) was used to construct strains YMB9695 and YMB9696. Strains were grown at permissive temperature (25°C) and shifted to the nonpermissive temperature (37°C) for 3 h. Cells with spindle length 3–8 μm were considered to be in anaphase and were used for imaging. Spc29-RFP was used as a spindle pole marker. Representative images showing the persistence of Mcd1-GFP signals in anaphase cells of cdc5-99 strain (YMB9696). (D) Percentage of Mcd1-GFP foci proximal to the spindle poles was determined in cdc5-99 strain (YMB9696) at permissive temperature (25°C, n = 94) and 3 h after shift to nonpermissive temperature (37°C, n = 197). Standard weighted-means analysis was used to calculate averages, and significance was determined by one-way analysis of variance. Bar diagram represents average ± SE of the mean. *p < 0.05.
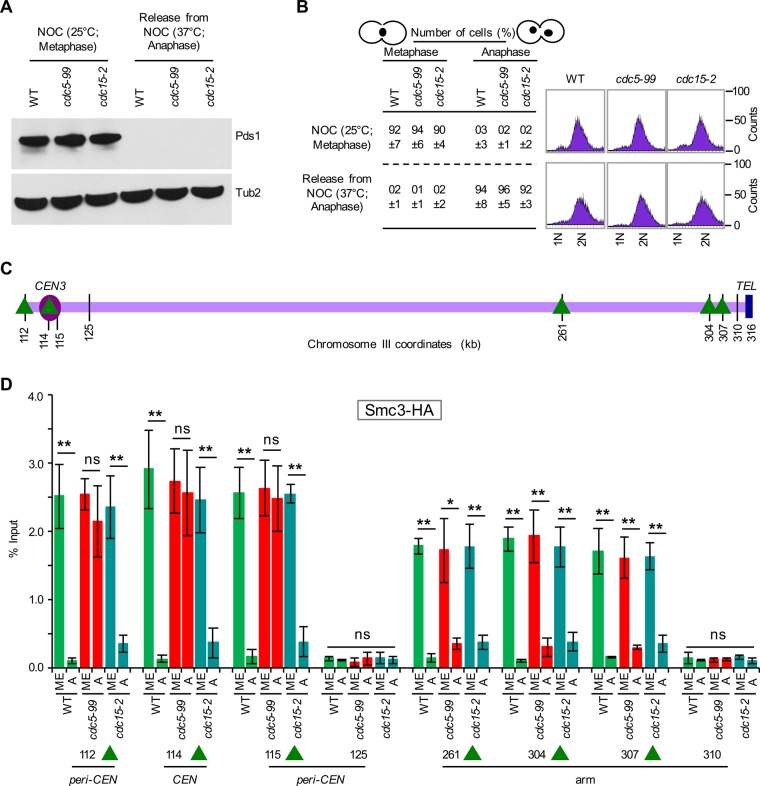
The proximity of Smc3-GFP and Mcd1-GFP foci to Spc29-RFP in cdc5-99 strains suggested the persistence of cohesin in centromeric chromatin; hence we used ChIP to assay the association of Smc3-HA at CARs close to (CEN) or distant from the centromere (arm), using either metaphase or anaphase cells from wild-type, cdc5-99, and cdc15-2 strains. Metaphase cells were obtained by growing strains at 25°C in nocodazole, and anaphase cells were obtained after release from nocodazole at the nonpermissive temperature of 37°C (Figure 9). We used cdc15-2 as a control because this strain arrests at a stage similar to cdc5 mutants during mitosis (Surana et al., 1993; St-Pierre et al., 2009). The cell cycle stage was determined by monitoring Pds1 levels (Cohen-Fix et al., 1996) by Western blotting, DNA content by FACS, and nuclear morphology by microscopy (Calvert and Lannigan, 2010). High levels of Pds1 are present in metaphase; however, levels of Pds1 are undetectable in anaphase, as degradation of Pds1 is required for anaphase onset (Cohen-Fix et al., 1996; Cohen-Fix and Koshland, 1999). Consistent with this, high levels of Pds1 were observed in metaphase but not in anaphase cells of wild-type, cdc5-99, and cdc15-2 strains (Figure 9, A and B). Large-budded cells with the nucleus at the neck or separated nuclei indicate metaphase and anaphase cells, respectively (Figure 9B). ChIP-qPCR showed enrichment of Smc3-HA at CEN (114), CARs flanking the centromere (112, 115), and the arms (261, 304, 307) in metaphase cells of wild-type, cdc5-99, and cdc15-2 strains (Figure 9, C and D). Smc3-HA levels were significantly reduced at both the centromeric chromatin (CEN [114], peri-CEN CARs [112, 115]) and the chromosomal arms (261, 304, 307) in anaphase cells of wild-type and cdc15-2 strains, and these levels were largely similar to those at non-CARs (125, 310; Figure 9D). In contrast, in anaphase cells of the cdc5-99 strain, Smc3-HA levels were significantly reduced only at CARs at chromosomal arms (261, 304, 307) and remained high at centromeric chromatin (CEN [114] and peri-CEN CARs [112, 115]). The levels of Smc3-HA at CEN and peri-CEN CARs in anaphase cells of cdc5-99 strains were not statistically different from those observed at these chromosomal regions in metaphase cells (Figure 9D). No significant enrichment of Scm3-HA was detected at non-CARs (125, 310) at either metaphase or anaphase (Figure 9D). These results suggest that Cdc5 is required for the removal of cohesin from centromeric chromatin during mitosis.
FIGURE 9:
Cdc5 is required for the removal of Smc3 from centromeric chromatin. WT (YMB9693), cdc5-99 (YMB9709), and cdc15-2 (YMB9703) expressing Smc3-6HA and Pds1-18Myc from their endogenous promoters were used. Smc3-6HA was an internally tagged version of Smc3 (six copies of HA inserted at amino acid residue 607) using plasmid pVG466 (a gift from Vincent Guacci). Strains were grown in YPD at permissive temperature (25°C) with nocodazole (NOC, 20 μg/ml) for 2 h to synchronize cells in metaphase. Cells were then shifted to nonpermissive temperature (37°C) for 1 h, followed by release from NOC into YPD at 37°C for 45 min to enrich cells in anaphase. Samples were collected for Western blot analysis for Pds1, nuclear morphology, DNA content, and ChIP analyses. Three biological replicates were performed for each strain. (A) Western blot analysis showing high levels of Pds1 only in metaphase cells of WT, cdc5-99, and cdc15-2 strains. Antibodies used were α-Myc (Pds1) and α-Tub2 (loading control). (B) Cell cycle stages (metaphase and anaphase) were determined based on nuclear position and cell morphology by microscopic examination of 100 cells for each strain per replicate as described previously (Calvert and Lannigan, 2010; Mishra et al., 2011). DNA content was determined by FACS. (C) Schematic representation of CARs and non-CARs on chromosome III. The schematic is derived from Figure 3A with additional CARs and non-CARs. Centromere (CEN3, violet circle), CAR (green triangle), and non-CAR (black vertical line) sites. CEN is located at 114 kb; peri-CEN CARs 115 (1 kb from CEN) and 112 (2 kb from CEN), chromosome arm CAR 261 (147 kb from CEN), 304 (190 kb from CEN), 307 (193 kb from CEN), peri-CEN non-CAR 125 (11 kb from CEN), and chromosome arm non-CAR 310 (196 kb from CEN) were examined. (D) Smc3 persists at CEN and peri-CEN chromatin in cdc5-99 strains. ChIP was performed with chromatin prepared from metaphase (ME) or anaphase (A) with strains as described using α-HA antibodies. Enrichment of Smc3 at CEN3 (114), peri-CEN CARs (112, 115), and chromosomal arm CARs (261, 304, 307) were determined by qPCR and is shown as percentage input. The non-CAR peri-CEN loci 125 and 310 in chromosome arms were used as a negative control. Average from three biological replicates ± SE. **p <0.01; *p <0.05; ns, not significant, Student’s t test.
A previous study showed that SMC subunits of cohesin complex (e.g., Smc1) remain associated with chromosomes for ∼10 min after the degradation of Scc1/Mcd1 in anaphase (Tanaka et al., 1999). Hence we investigated whether Cdc5 is also required for removal of Mcd1 from centromeric chromatin. We examined the association of Mcd1-HA at CEN and CARs in cdc5-99 and cdc15-2 grown at 25°C in nocodazole (metaphase) and after release from nocodazole at 37°C (anaphase). We detected high levels of Pds1 in metaphase but not in anaphase cells of cdc5-99, and cdc15-2 strains (Figure 10, A and B). The levels of Pds1 were slightly lower in cdc15-2 than in the cdc5-99 strain (Figure 10A). ChIP-qPCR showed enrichment of Mcd1-HA at CEN (114), CARs flanking the centromere (112, 115), and the arms (261, 304, 307) in metaphase cells of cdc5-99 and cdc15-2 strains (Figure 10, C and D). Mcd1-HA levels were significantly reduced at CEN (114), peri-CEN CARs (112, 115), and chromosomal arms CARs (261, 304, 307) in anaphase cells of cdc15-2, and these levels were largely similar to those at non-CARs (125, 31; Figure 10D). Consistent with the results for Smc3-HA, significantly reduced levels of Mcd1-HA were observed at CARs in chromosomal arms (261, 304, 307), whereas Mcd1-HA levels remained high at centromeric chromatin (CEN [114] and peri-CEN CARs [112, 115]) in anaphase cells of cdc5-99 strains. The levels of Mcd1-HA at CEN and peri-CEN CARs in anaphase cells of cdc5-99 strains were statistically not different from those observed at these chromosomal regions in metaphase cells (Figure 10D). No significant enrichment of Mcd1-HA was detected at non-CARs (125, 310) at either metaphase or anaphase (Figure 10D). The persistence of both Smc3 and Mcd1 in CEN and peri-CEN chromatin in anaphase cells of the cdc5-99 strain supports a role for Cdc5 in the removal of cohesin from centromeric chromatin.
FIGURE 10:
Cdc5 is required for the removal of Mcd1 from centromeric chromatin. cdc5-99 (YMB9711), and cdc15-2 (YMB9712) strains expressing Mcd1-3HA, and Pds1-18Myc from their endogenous promoters were used. Strains KT046 (Tong and Skibbens, 2014) and D1469 carrying MCD1-3HA were used to construct strains YMB9711 and YMB9712, respectively. Strains were grown in YPD at permissive temperature (25°C) with nocodazole (NOC; 20 μg/ml) for 2 h to synchronize cells in metaphase. Cells were then released into YPD at nonpermissive temperature (37°C) for 2.5 h to enrich cells in anaphase. Samples were collected for Western blot analysis, nuclear morphology, DNA content, and ChIP analyses. Three biological replicates were performed for each strain. (A) Western blotting showing high levels of Pds1 only in metaphase cells of cdc5-99 and cdc15-2 strains. Antibodies used were α-Myc (Pds1) and α-Tub2 (loading control). (B) Cell cycle stages (metaphase and anaphase) were determined based on nuclear position and cell morphology by microscopic examination of 100 cells for each strain per replicate. DNA content was determined by FACS. (C) Schematic representation of CARs and non-CARs on chromosome III as detailed in Figure 9C. (D) Mcd1 persists at CEN and peri-CEN chromatin in cdc5-99 strains. ChIP experiments were performed using chromatin prepared from metaphase (ME) or anaphase (A) cells from strains as described using α-HA antibodies. Enrichment of Mcd1 at CEN3 (114), peri-CEN CARs (112, 115), and chromosomal arm CARs (261, 304, 307) were determined by qPCR and is shown as percentage input. The non-CAR peri-CEN loci 125 and 310 in chromosome arms were used as negative controls. Average from three biological replicates ± SE. **p <0.01; *p < 0.05; ns, not significant, Student’s t test.
Cdc5 is required for faithful chromosome segregation
The cell cycle–regulated centromeric association of Cdc5 and its role in removal of centromeric cohesin led us to examine whether defects in Cdc5 would result in increased chromosome loss. We constructed a cdc5-99 strain carrying a reporter chromosome (RC) and determined the frequency of loss of the RC using a colony color assay as described previously (Spencer et al., 1990). Loss of the RC results in formation of red sectors in an otherwise white colony. Colonies that are at least half red indicate loss of the RC in the first cell division. The frequency of RC loss in cdc5-99 strains was about sixfold higher than in the wild-type strain (Figure 11A), similar to that reported for kinetochore mutants (Kastenmayer et al., 2005; Ma et al., 2012).
FIGURE 11:
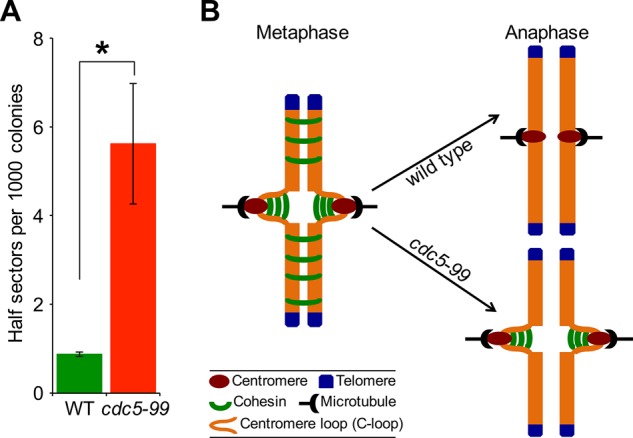
Cdc5 is required for faithful chromosome segregation. (A) Cdc5 is required for faithful chromosome segregation. Frequency of RC loss in wild-type (WT; YPH363) and cdc5-99 (YMB9438) strains was determined as described in Materials and Methods. At least 3000 colonies from three independent transformants were counted. Values are mean ± SE. *p < 0.05, Student’s t test. (B) Schematic model for the role of Cdc5 in cohesin removal from centromeric chromatin during mitosis. In WT cells, cohesin is removed from chromosomal arms and centromeric chromatin during metaphase-to-anaphase transition, whereas in cdc5-99 strains, cohesin is removed from chromosomal arms but persists at centromeric chromatin.
DISCUSSION
Evolutionarily conserved polo-like kinase Cdc5 and its homologues play a range of crucial and conserved roles in regulation of cell cycle events (Alexandru et al., 2001; Lee et al., 2005; St-Pierre et al., 2009; Rossio et al., 2010; Ratsima et al., 2011; Botchkarev et al., 2014; Walters et al., 2014; Zitouni et al., 2014; Archambault et al., 2015). Among its many roles, Cdc5 is required for the phosphorylation and subsequent cleavage of Scc1/Mcd1 in order to promote sister chromatid separation (Alexandru et al., 2001; Hornig and Uhlmann, 2004). Our results provide mechanistic insight into the role of Cdc5 in the removal of centromeric cohesin during mitosis. We show that Cdc5 associates with centromeric chromatin and is required for the efficient removal of cohesin at core CEN and peri-CEN regions during mitosis. These conclusions are based on results showing that Cdc5: 1) associates with CEN and CARs in a cell cycle–dependent manner, with maximum enrichment in mitosis; 2) interacts with cohesins in vivo; 3) requires cohesin for its mitotic enrichment at CEN and CARs; 4) is required for the removal of cohesin from centromeric chromatin in anaphase; and 5) is required for faithful chromosome segregation.
Enrichment of Cdc5 at centromeric chromatin was highest in mitotic cells undergoing the transition from metaphase to anaphase. Even though Cdc5 is expressed in S-phase and anaphase cells, we observed significantly lower levels of chromatin-associated Cdc5 during the S phase of the cell cycle. Moreover, ectopic expression of Cdc5 in G1 does not result in its recruitment to CEN and CARs, suggesting that the binding of Cdc5 to these chromosomal regions is regulated by the cell cycle. Association of Cdc5 flanking CEN6 and CARs was reported in a ChIP-on-chip study evaluating the role of Rsc2 in mitotic exit (Rossio et al., 2010). Our comprehensive ChIP analysis shows that the cell cycle–regulated centromeric enrichment of Cdc5 coincides with its role in removal of cohesin from this region. We discovered that cohesin components Scc1/Mcd1 interact with Cdc5 in vivo, and the interaction is required for enrichment of Cdc5 at CEN and CARs. It is of note that high-throughput proteomic studies also identified cohesin components when Cdc5 was used as bait in affinity-capture mass spectroscopy experiments (Ho et al., 2002; Snead et al., 2007; Breitkreutz et al., 2010). The cohesin-dependent enrichment of Cdc5 at CEN and CARs reflects a dynamic enzyme–substrate relationship between Cdc5 and cohesin components, resulting in preferential removal of chromatin-bound cohesin. Of most importance, our results suggest a role for Cdc5 in removal of centromeric cohesins, most likely from the intramolecular linkages in the C-loop (Yeh et al., 2008; Hu et al., 2011), but not cohesins in the arms of chromosomes (Figure 11B). The C-loop model resolved a paradox in which sister kinetochores can separate up to 800 nm in vivo despite high levels of cohesin at centromeres. The persistence of Smc3 and Mcd1 in anaphase of cdc5-99 cells at the nonpermissive temperature may reflect the presence of intramolecularly linked cohesin in the C-loop, whereas the absence of cohesin in chromosome arms shows that Cdc5 is dispensable for removal of intermolecularly linked cohesins. Cdc5 has been shown to regulate removal of cohesin from chromosome arms for segregation of homologous chromosomes in meiosis I but to be dispensable for meiosis II (Brar et al., 2006; Katis et al., 2010; Attner et al., 2013). Attner et al. (2013) concluded that additional kinases or substrates and/or the differential nature of centromeric versus chromosome arm cohesin regulate the removal of centromeric cohesin in meiosis II. On the basis of our results, we conclude that Cdc5 represents the first example of differential regulation of centromeric versus arm cohesin during mitosis in budding yeast.
The mitotic enrichment of Cdc5 at CEN suggests a role for Cdc5 in kinetochore function because of major changes in spatial geometry and spindle tension that occur at the kinetochore during mitosis. Consistent with this hypothesis, we observed errors in chromosome segregation in the cdc5-99 strain. Because Cdc5 is involved in many biological processes (Archambault and Glover, 2009; St-Pierre et al., 2009; Botchkarev et al., 2014; Walters et al., 2014; Zitouni et al., 2014; Roccuzzo et al., 2015), chromosome segregation errors observed in cdc5-99 strain may not be due to solely defects in cohesin removal. Previous studies showed that Cdc5 interacts with and phosphorylates kinetochore proteins in vivo (e.g., Slk19) to promote proper microtubule attachment and spindle function (Stegmeier et al., 2002; Snead et al., 2007; Park et al., 2008; Liang et al., 2009; Richmond et al., 2013). Plk1, the human homologue of Cdc5, also localizes to the CEN (Arnaud et al., 1998; Kishi et al., 2009), where it likely phosphorylates Rad21 (the human Scc1 homologue; Waizenegger et al., 2000). In addition, Plk1 phosphorylates kinetochore protein Mis18BP1, which in turn licenses the recruitment of new CENP-A to kinetochores in the G1 phase of the cell cycle (McKinley and Cheeseman, 2014). Of note, Cse4 (the budding yeast CENP-A homologue) was identified as an in vivo interacting partner of Cdc5 in a genome-wide proteomic study (Snead et al., 2007). Future studies should help us to address whether Cdc5-mediated phosphorylation of kinetochore proteins affects the removal of centromeric cohesins.
In summary, our studies provide novel insights into the role of Cdc5 in removal of centromeric cohesin but not cohesin from chromosomal arms. Differential regulation of cohesin removal has also been reported in mammalian cells, where removal of cohesin from chromosomal arms in prometaphase is regulated by the cohesin antagonist WAPL, whereas removal of centromeric cohesin at the metaphase-to-anaphase transition by Plk1 is achieved by a separase-dependent mechanism (Waizenegger et al., 2000; Hauf et al., 2001; Gandhi et al., 2006; Kueng et al., 2006; Haarhuis et al., 2013). Mutations and overexpression of Plk1 are observed in many cancers (Davies et al., 2005; Stephens et al., 2005; Kan et al., 2010), displaying chromosomal instability and aneuploidy (Degenhardt and Lampkin, 2010). Hence elucidation of molecular mechanisms by which polo kinases regulate cohesin removal and faithful chromosome segregation might help us identify and develop novel therapeutic targets for cancer therapy.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions
Saccharomyces cerevisiae strains and plasmids used in this study are listed in Table 1. Strains were grown in 1% yeast extract/2% Bacto-peptone/2% glucose (YPD) or in yeast synthetic medium containing 2% glucose and supplements to allow for the selection of plasmids used.
TABLE 1:
Strains and plasmids used in this study.
| Strain name | Genotype | Reference |
|---|---|---|
| YMB9431 | MATa ura3-1 leu2-3112 his3-11,15 trp1-1 ade2-1 can1-100 SMC3-GFP::URA3 | This study |
| YMB9263 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CSE4-13MYC::LEU2 CEN-TRP1 | This study |
| YMB9264 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CSE4-13MYC::LEU2 CEN-CDC5-3HA::TRP1 (p344) | This study |
| JG595 | MATa ura3-1 leu2,3-112 his3-1 trp1-1 ade2-1 can1-100 Δbar1 CSE4-12MYC::URA3 SCM3-3FLAG::kanMX | Camahort et al. (2007) |
| YMB9441 | MATa ura3-1 leu2,3-112 his3-1 trp1-1 ade2-1 can1-100 Δbar1 CSE4-12Myc::URA3 SCM3-3FLAG::kanMX GAL1-3HA-CDC5::TRP1 | This study |
| YMB9693 | MATa ura3-1 leu2-3112 his3-11,15 trp1-1 ade2-1 can1-100 SMC3-N607-6HA::URA3 PDS1-18MYC::LEU2 | This study |
| YMB9695 | MATa MCD1-GFP leu2-3112 ura3-52 his3-11,15 bar1 GAL+ SPC29-RFP::Hyg | This study |
| YMB9696 | MATa MCD1-GFP leu2-3112 ura3-52 his3-11,15 bar1 GAL+ SPC29-RFP::Hyg cdc5-99::HIS3MX6 | This study |
| YMB9703 | MATa ura3-1 leu2-3112 his3-11,15 trp1-1 ade2-1 can1-100 SMC3-N607-6HA::URA3 PDS1-18MYC::LEU2 cdc15-2 | This study |
| YMB9709 | MATa ura3-1 leu2-3112 his3-11,15 trp1-1 ade2-1 can1-100 SMC3-N607-6HA::URA3 PDS1-18MYC::LEU2 cdc5-99::HIS3MX6 | This study |
| YMB9711 | MATa ade2-1 his3-11,15 leu2-3112 trp1-1 ura3-1 can1-100 MCD1-3HA::TRP1 PDS1-18MYC::LEU2 cdc5-99::HIS3MX6 | This study |
| YMB9712 | MATα leu2-3112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 [phi+] rad5-535 MCD1-3HA::HPHMX6 cdc15-2::TRP1 PDS1-18MYC::LEU2 | This study |
| YMB8702 | MATa ura3-1 leu2-3112 his3-11,15 trp1-1 ade2-1 can1-100 | This study |
| KBY6097-1 | MATα ura3-1 leu2-3112 his3-11,15 trp1-1 ade2-1 can1-100 SMC3-GFP::URA3 cdc5-99::HIS3MX6 SPC29-RFP::Hyg | This study |
| KBY6098-1 | MATa ura3-1 leu2-3112 his3-11,15 trp1-1 ade2-1 can1-100 SMC3-GFP::URA3 SPC29-RFP::Hyg | This study |
| YMB9438 | MATα ura3-52 lys2-801 leu2-Δ1 ade2-101 his3-Δ200 [CFIII (CEN3.L) CFVII (RAD2.d) URA3 SUP11] cdc5-99::HIS3 | This study |
| YPH363 | MATα ura3-52 lys2-801 leu2-Δ1 ade2-101 his3-Δ200 [CFIII (CEN3.L) CFVII (RAD2.d) URA3 SUP11] | Spencer et al. (1990) |
| BY4741 | MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 | Open Biosystems, Lafayette, CO |
| MAY8878 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 MCD1-TAP::HIS3 | This study |
| YPH499 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 | Sikorski and Hieter (1989) |
| MAY8775 | MATa ura3-52 ade2-101 his3-11,15 leu2-3112 CDC5-13MYC | This study |
| MAY8851 | MATa ura3-52 ade2-101 his3-11,15 leu2-3112 CDC5-13MYC mcd1-1 | This study |
| VG3507-2A | MATa MCD1-GFP leu2-3112 ura3-52 his3-11,15 bar1 GAL+ | V. Guacci, University of California, Berkeley, CA |
| KT046 | MATa ade2-1 his3-11,15 leu2-3112 trp1-1 ura3-1 can1-100 MCD1-3HA::TRP1 | Tong and Skibbens (2014) |
| D1469 | MATα leu2-3112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 [phi+] rad5-535 MCD1-3HA::HPHMX6 cdc15-2::TRP1 | D. D’Amours, Université de Montréal, Montréal, QC |
| Plasmids | Description | Reference |
| p344 | CEN-CDC5-3HA::TRP1 | D. D’Amours |
| pVG466 | SMC3-N607-6HA::URA3 | V. Guacci |
| pSB205 | PDS1-18MYC::LEU2 | S. Biggins, Fred Hutchinson Cancer Research Center, Seattle, WA |
Assay for the loss of reporter chromosome
Loss of a nonessential RC was measured using a colony color assay in which RC loss results in red sectors in an otherwise white colony (Spencer et al., 1990). Wild-type and cdc5-99 strains were grown to logarithmic phase in selective medium to maintain the RC. Cultures were diluted and plated on complete synthetic medium with limiting adenine at 30ºC. The frequency of chromosome loss was measured by counting the colonies that were at least half red, indicating loss of the RC at the first cell division. At least 3000 colonies of three individual transformants were examined for each strain.
ChIP and qPCR experiments
ChIP assays were performed with three biological replicates as described previously (Mishra et al., 2007, 2011). Antibodies used to capture protein–DNA complexes were α-HA (A2095; Sigma-Aldrich, St. Louis, MO) and α-Myc (A7470; Sigma-Aldrich). ChIP-qPCR was carried out using SYBR Green Master Mix in a 7500 Fast Real Time PCR System (Applied Biosystems, Foster City, CA) following conditions used previously (Mishra et al., 2011). The enrichment values were determined as percentage input using the ΔΔCT method (Livak and Schmittgen, 2001). Primer sequences used in this study are presented in Table 2.
TABLE 2:
Primers used in this study.
| Locus | Forward (5′–3′) | Reverse (5′–3′) | Reference |
|---|---|---|---|
| CEN1 | CTCGATTTGCATAAGTGTGCC | GTGCTTAAGAGTTCTGTACCAC | Choy et al. (2011) |
| CEN3 | GATCAGCGCCAAACAATATGG | AACTTCCACCAGTAAACGTTTC | Choy et al. (2011) |
| CEN5 | AAGAACTATGAATCTGTAAATGACTGATTCAAT | CTTGCACTAAACAAGACTTTATACTACGTTTAG | Choy et al. (2011) |
| 6K120 | AACGTCACTTTTTTTCCAGGG | GCAAAGCTAGCTAACGAACAA | This study |
| CEN3-L1 | ATATTGTTTGGCGCTGATCGCC | TTGATGAACTTTTCAAAGATGAC | Choy et al. (2011) |
| CEN3-L2 | TCATCTTTGAAAAGTTCATCAAGG | GATAACAAAGCATGGTATGGCG | Choy et al. (2011) |
| CEN3-L3 | GCCATACCATGCTTTGTTATCGTC | TATTATGCTCCCCTGGATTTTATGCG | Choy et al. (2011) |
| CEN3-L4 | AGCATTAGAGCCACTGTCATTTC | ATAATTAAGATACGAATGTGTTCGTTG | Mishra et al. (2013) |
| CEN3-R1 | TTTACTGGTGGAAGTTTTGCTCA | GTCAACGAGTCCTCTCTGGCTA | Choy et al. (2011) |
| CEN3-R2 | GAGAGGACTCGTTGACGTAGAA | GAATATGATAATGGTTACACCAGTAGG | Choy et al. (2011) |
| CEN3-R3 | TGTAACCATTATCATATTCATGAC | GATTTAATGCACGTTATGTTTCG | Choy et al. (2011) |
| CEN3-R4 | ACTGACAGCACCATTAATCAATCA | TATCCTCAGTAGAGGGCAAAGTT | Mishra et al. (2013) |
| 125 | TGCCAAGTTGTGCTTTTTAGTTGAG | TTGATGTGTTGATTGTTCGGTCAC | Eckert et al. (2007), Ng et al. (2009) |
| 134 | CCGATGGTTAGGATTTCCAACG | GGTTTTCAGAACAGAATGGGGC | Eckert et al. (2007), Ng et al. (2009) |
| 261 | TTGCCACAGCCACAGATATAACTG | GATGGACAAAGCGTTGTATCCG | Eckert et al. (2007), Ng et al. (2009) |
| 266 | CAGACTTCCTCCAAGCAACAGG | TCCTCCTTAGCCTTTTCTTCAGC | Eckert et al. (2007), Ng et al. (2009) |
| 304 | CCCCTTCCAAAGACCTGACA | CCAGCCGTCGATCCTAAAGA | Laloraya et al. (2000) |
| 307 | GTAGCGCTCTCAACTACCCT | TCGTATACTGTTAGGGTCTGCA | Laloraya et al. (2000) |
| 310 | TCTCGGAATTTATCATGACCCAT | AAACCCTGCACACATTTCGT | Laloraya et al. (2000) |
Immunoprecipitation and Western blotting
Immunoprecipitation assays were performed as described previously (Mishra et al., 2011). Protein extracts were prepared with the trichloroacetic acid procedure and quantified using the Bio-Rad DC protein assay (Bio-Rad Laboratories, Hercules, CA). Equal amounts of protein for each sample were resolved on SDS–polyacrylamide gels and transferred to nitrocellulose membrane. Primary antibodies used were α-HA (clone 12CA5; Roche Molecular Systems, Pleasanton, CA), α-GFP (A11122; Life Technologies, Carlsbad, CA), α-TAP (CAB1001; Thermo Scientific, Waltham, MA), α-Cdc5 (11H12 and 4F10; Medimabs, Montreal, Canada), α-Myc (Z-5, sc-789; Santa Cruz Biotechnology, Dallas, TX), and α-Tub2 (Mishra et al., 2011). Secondary antibodies were horseradish peroxidase (HRP)–conjugated donkey α-rabbit immunoglobulin G (IgG; NA934V, Amersham Biosciences, Amersham, United Kingdom), and HRP-conjugated sheep α-mouse IgG (NA931V; Amersham Biosciences).
Microscopy
Cultures grown to logarithmic phase at permissive temperature (25°C) and after a shift to nonpermissive temperature (37°C) were used for all imaging experiments. Anaphase cells that exhibited a spindle length of 3–8 μm were selected based on the position of spindle pole bodies (Spc29-RFP; Chen et al., 2000; Maddox et al., 2000). Cells were imaged for Smc3-GFP, Mcd1-GFP, and Spc29-RFP signals at room temperature on a Nikon TE-2000E inverted microscope equipped with a 1.4 numerical aperture/100× Plan-Apo objective (Nikon Instruments, New York, NY) as described previously (Haase et al., 2013; Salmon et al., 2013).
Supplementary Material
Acknowledgments
We are highly thankful to Vincent Guacci and Robert Skibbens for strains, plasmids, and helpful suggestions. We thank Sue Biggins for plasmids and Jennifer Gerton for strains, Kathy McKinnon of the National Cancer Institute Vaccine Branch FACS Core for assistance with FACS and the Basrai laboratory for discussions. P.K.M., S.C.Y., W.C.A., L.B., L.E.D., Z.J., T.P., and M.A.B. were supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health; M.A.H. and D.R. by the National Science Foundation; E.Y. and K.B. by the National Institutes of Health (R37 GM32238); and D.D. by the Cancer Research Society and the Canadian Institutes of Health Research (MOP 82912).
Abbreviations used:
- CAR
cohesin-associated region
- CEN
centromere
- ChIP
chromatin immunoprecipitation
- C-loop
centromere loop
- GFP
green fluorescent protein
- qPCR
quantitative PCR
- RC
reporter chromosome
- RFP
red fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-01-0004) on May 25, 2016.
REFERENCES
- Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- Archambault V, Lepine G, Kachaner D. Understanding the Polo kinase machine. Oncogene. 2015;34:4799–4807. doi: 10.1038/onc.2014.451. [DOI] [PubMed] [Google Scholar]
- Arnaud L, Pines J, Nigg EA. GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma. 1998;107:424–429. doi: 10.1007/s004120050326. [DOI] [PubMed] [Google Scholar]
- Attner MA, Miller MP, Ee LS, Elkin SK, Amon A. Polo kinase Cdc5 is a central regulator of meiosis I. Proc Natl Acad Sci USA. 2013;110:14278–14283. doi: 10.1073/pnas.1311845110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Bloom KS, Amaya E, Carbon J, Clarke L, Hill A, Yeh E. Chromatin conformation of yeast centromeres. J Cell Biol. 1984;99:1559–1568. doi: 10.1083/jcb.99.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Bose T, Gerton JL. Cohesinopathies, gene expression, and chromatin organization. J Cell Biol. 2010;189:201–210. doi: 10.1083/jcb.200912129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VV, Jr, Rossio V, Yoshida S. The budding yeast Polo-like kinase Cdc5 is released from the nucleus during anaphase for timely mitotic exit. Cell Cycle. 2014;13:3260–3270. doi: 10.4161/15384101.2014.953882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Kiburz BM, Zhang Y, Kim JE, White F, Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker AS, Berkowitz KM. The roles of cohesins in mitosis, meiosis, and human health and disease. Methods Mol Biol. 2014;1170:229–266. doi: 10.1007/978-1-4939-0888-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack LS, Berman J. Flexibility of centromere and kinetochore structures. Trends Genet. 2012;28:204–212. doi: 10.1016/j.tig.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert ME, Lannigan J. Yeast cell cycle analysis: combining DNA staining with cell and nuclear morphology. Curr Protoc Cytom. 2010;Chapter 9 doi: 10.1002/0471142956.cy0932s52. Unit 9.32.1–16. [DOI] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Acuna R, Au WC, Basrai MA. A role for histone H4K16 hypoacetylation in Saccharomyces cerevisiae kinetochore function. Genetics. 2011;189:11–21. doi: 10.1534/genetics.111.130781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Mishra PK, Au WC, Basrai MA. Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae. Biochim Biophys Acta. 2012;1819:776–783. doi: 10.1016/j.bbagrm.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Koshland D. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev. 1999;13:1950–1959. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- Degenhardt Y, Lampkin T. Targeting Polo-like kinase in cancer therapy. Clin Cancer Res. 2010;16:384–389. doi: 10.1158/1078-0432.CCR-09-1380. [DOI] [PubMed] [Google Scholar]
- Eckert CA, Gravdahl DJ, Megee PC. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21:278–291. doi: 10.1101/gad.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng T, Guacci V, Koshland D. ROCC, a conserved region in cohesin’s Mcd1 subunit, is essential for the proper regulation of the maintenance of cohesion and establishment of condensation. Mol Biol Cell. 2014;25:2351–2364. doi: 10.1091/mbc.E14-04-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng T, Guacci V, Koshland D. Interallelic complementation provides functional evidence for cohesin-cohesin interactions on DNA. Mol Biol Cell. 2015;26:4224–4235. doi: 10.1091/mbc.E15-06-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton J. Chromosome cohesion: a cycle of holding together and falling apart. PLoS Biol. 2005;3:e94. doi: 10.1371/journal.pbio.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL. Enhancing togetherness: kinetochores and cohesion. Genes Dev. 2007;21:238–241. doi: 10.1101/gad.1523107. [DOI] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Stricklin J, Bloom MS, Guo X, Bhatter M, Koshland D. A novel mechanism for the establishment of sister chromatid cohesion by the ECO1 acetyltransferase. Mol Biol Cell. 2015;26:117–133. doi: 10.1091/mbc.E14-08-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JH, Elbatsh AM, van den Broek B, Camps D, Erkan H, Jalink K, Medema RH, Rowland BD. WAPL-mediated removal of cohesin protects against segregation errors and aneuploidy. Curr Biol. 2013;23:2071–2077. doi: 10.1016/j.cub.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Haase J, Mishra PK, Stephens A, Haggerty R, Quammen C, Taylor RM, 2nd, Yeh E, Basrai MA, Bloom K. A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Curr Biol. 2013;23:1939–1944. doi: 10.1016/j.cub.2013.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Pautz A. A novel role for Cdc5p in DNA replication. Mol Cell Biol. 1996;16:6775–6782. doi: 10.1128/mcb.16.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hornig NC, Uhlmann F. Preferential cleavage of chromatin-bound cohesin after targeted phosphorylation by Polo-like kinase. EMBO J. 2004;23:3144–3153. doi: 10.1038/sj.emboj.7600303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Itoh T, Mishra A, Katoh Y, Chan KL, Upcher W, Godlee C, Roig MB, Shirahige K, Nasmyth K. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr Biol. 2011;21:12–24. doi: 10.1016/j.cub.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- Kastenmayer JP, Lee MS, Hong AL, Spencer FA, Basrai MA. The C-terminal half of Saccharomyces cerevisiae Mad1p mediates spindle checkpoint function, chromosome transmission fidelity and CEN association. Genetics. 2005;170:509–517. doi: 10.1534/genetics.105.041426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell. 2010;18:397–409. doi: 10.1016/j.devcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi K, van Vugt MA, Okamoto K, Hayashi Y, Yaffe MB. Functional dynamics of Polo-like kinase 1 at the centrosome. Mol Cell Biol. 2009;29:3134–3150. doi: 10.1128/MCB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Park JE, Asano S, Park CJ. Yeast polo-like kinases: functionally conserved multitask mitotic regulators. Oncogene. 2005;24:217–229. doi: 10.1038/sj.onc.1208271. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Liang F, Jin F, Liu H, Wang Y. The molecular function of the yeast polo-like kinase Cdc5 in Cdc14 release during early anaphase. Mol Biol Cell. 2009;20:3671–3679. doi: 10.1091/mbc.E08-10-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma L, Ho K, Piggott N, Luo Z, Measday V. Interactions between the kinetochore complex and the protein kinase A pathway in Saccharomyces cerevisiae. G3 (Bethesda) 2012;2:831–841. doi: 10.1534/g3.112.002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Corbett KD, Desai A. Structure, assembly and reading of centromeric chromatin. Curr Opin Genet Dev. 2012;22:139–147. doi: 10.1016/j.gde.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL. Chromosome segregation in budding yeast: sister chromatid cohesion and related mechanisms. Genetics. 2014;196:31–63. doi: 10.1534/genetics.112.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee PC, Mistrot C, Guacci V, Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, Foltz DR, Basrai MA. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 2011;7:e1002303. doi: 10.1371/journal.pgen.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK, Baum M, Carbon J. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol Genet Genomics. 2007;278:455–465. doi: 10.1007/s00438-007-0263-8. [DOI] [PubMed] [Google Scholar]
- Mishra PK, Guo J, Dittman LE, Haase J, Yeh E, Bloom K, Basrai MA. Pat1 protects centromere-specific histone H3 variant Cse4 from Psh1-mediated ubiquitination. Mol Biol Cell. 2015;26:2067–2079. doi: 10.1091/mbc.E14-08-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK, Ottmann AR, Basrai MA. Structural integrity of centromeric chromatin and faithful chromosome segregation requires Pat1. Genetics. 2013;195:369–379. doi: 10.1534/genetics.113.155291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Newlon CS. Yeast chromosome replication and segregation. Microbiol Rev. 1988;52:568–601. doi: 10.1128/mr.52.4.568-601.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TM, Waples WG, Lavoie BD, Biggins S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell. 2009;20:3818–3827. doi: 10.1091/mbc.E09-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble D, Kenna MA, Dix M, Skibbens RV, Unal E, Guacci V. Intersection between the regulators of sister chromatid cohesion establishment and maintenance in budding yeast indicates a multi-step mechanism. Cell Cycle. 2006;5:2528–2536. doi: 10.4161/cc.5.21.3405. [DOI] [PubMed] [Google Scholar]
- Park CJ, Park JE, Karpova TS, Soung NK, Yu LR, Song S, Lee KH, Xia X, Kang E, Dabanoglu I, et al. Requirement for the budding yeast polo kinase Cdc5 in proper microtubule growth and dynamics. Eukaryotic Cell. 2008;7:444–453. doi: 10.1128/EC.00283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratsima H, Ladouceur AM, Pascariu M, Sauve V, Salloum Z, Maddox PS, D’Amours D. Independent modulation of the kinase and polo-box activities of Cdc5 protein unravels unique roles in the maintenance of genome stability. Proc Natl Acad Sci USA. 2011;108:E914–E923. doi: 10.1073/pnas.1106448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond D, Rizkallah R, Liang F, Hurt MM, Wang Y. Slk19 clusters kinetochores and facilitates chromosome bipolar attachment. Mol Biol Cell. 2013;24:566–577. doi: 10.1091/mbc.E12-07-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccuzzo M, Visintin C, Tili F, Visintin R. FEAR-mediated activation of Cdc14 is the limiting step for spindle elongation and anaphase progression. Nat Cell Biol. 2015;17:251–261. doi: 10.1038/ncb3105. [DOI] [PubMed] [Google Scholar]
- Rossio V, Galati E, Ferrari M, Pellicioli A, Sutani T, Shirahige K, Lucchini G, Piatti S. The RSC chromatin-remodeling complex influences mitotic exit and adaptation to the spindle assembly checkpoint by controlling the Cdc14 phosphatase. J Cell Biol. 2010;191:981–997. doi: 10.1083/jcb.201007025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ED, Shaw SL, Waters JC, Waterman-Storer CM, Maddox PS, Yeh E, Bloom K. A high-resolution multimode digital microscope system. Methods Cell Biol. 2013;114:179–210. doi: 10.1016/B978-0-12-407761-4.00009-9. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol. 2015;16:473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- Saunders MJ, Yeh E, Grunstein M, Bloom K. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol Cell Biol. 1990;10:5721–5727. doi: 10.1128/mcb.10.11.5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VP, Gerton JL. Cohesin and human disease: lessons from mouse models. Curr Opin Cell Biol. 2015;37:9–17. doi: 10.1016/j.ceb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Marzillier J, Eastman L. Cohesins coordinate gene transcriptions of related function within Saccharomyces cerevisiae. Cell Cycle. 2010;9:1601–1606. doi: 10.4161/cc.9.8.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead JL, Sullivan M, Lowery DM, Cohen MS, Zhang C, Randle DH, Taunton J, Yaffe MB, Morgan DO, Shokat KM. A coupled chemical-genetic and bioinformatic approach to Polo-like kinase pathway exploration. Chem Biol. 2007;14:1261–1272. doi: 10.1016/j.chembiol.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Douziech M, Bazile F, Pascariu M, Bonneil E, Sauve V, Ratsima H, D’Amours D. Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol Cell. 2009;34:416–426. doi: 10.1016/j.molcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Tong K, Skibbens RV. Cohesin without cohesion: a novel role for Pds5 in Saccharomyces cerevisiae. PLoS One. 2014;9:e100470. doi: 10.1371/journal.pone.0100470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Verdaasdonk JS, Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol. 2011;12:320–332. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk JS, Gardner R, Stephens AD, Yeh E, Bloom K. Tension-dependent nucleosome remodeling at the pericentromere in yeast. Mol Biol Cell. 2012;23:2560–2570. doi: 10.1091/mbc.E11-07-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Walters AD, May CK, Dauster ES, Cinquin BP, Smith EA, Robellet X, D’Amours D, Larabell CA, Cohen-Fix O. The yeast polo kinase Cdc5 regulates the shape of the mitotic nucleus. Curr Biol. 2014;24:2861–2867. doi: 10.1016/j.cub.2014.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitouni S, Nabais C, Jana SC, Guerrero A, Bettencourt-Dias M. Polo-like kinases: structural variations lead to multiple functions. Nat Rev Mol Cell Biol. 2014;15:433–452. doi: 10.1038/nrm3819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.