Stimulation of cells with the IL-12–type cytokine IL-12 or IL-23 results in activation of receptor-associated Janus kinases (Jak) and phosphorylation of STAT proteins in target cells. Functional association of IL 12Rβ1 with tyrosine kinase 2 and IL-23R with Jak2 is mandatory for IL-12 and/or IL-23 signaling.
Abstract
The interleukin (IL)-12–type cytokines IL-12 and IL-23 are involved in T-helper (Th) 1 and Th17 immunity, respectively. They share the IL-12 receptor β1 (IL-12Rβ1) as one component of their receptor signaling complexes, with IL-12Rβ2 as second receptor for IL-12 and IL-23R for IL-23 signal transduction. Stimulation with IL-12 and IL-23 results in activation of receptor-associated Janus kinases (Jak) and phosphorylation of STAT proteins in target cells. The Janus kinase tyrosine kinase (Tyk) 2 associates with IL-12Rβ1, whereas Jak2 binds to IL-23R and also to IL-12Rβ2. Receptor association of Jak2 is mediated by Box1 and Box2 motifs located within the intracellular domain of the receptor chains. Here we define the Box1 and Box2 motifs in IL-12Rβ1 and an unusual Jak2-binding site in IL-23R by the use of deletion and site-directed mutagenesis. Our data show that nonfunctional box motifs abolish IL-12– and IL-23–induced STAT3 phosphorylation and cytokine-dependent proliferation of Ba/F3 cells. Coimmunoprecipitation of Tyk2 by IL-12Rβ1 and Jak2 by IL‑23R supported these findings. In addition, our data demonstrate that association of Jak2 with IL-23R is mandatory for IL-12 and/or IL-23 signaling, whereas Tyk2 seems to be dispensable.
INTRODUCTION
The proinflammatory interleukin (IL)-12 family members IL-12 and IL-23 are heterodimeric cytokines composed of the shared p40 subunit and p35 or p19 (Garbers et al., 2012; Floss et al., 2015). IL-12 and IL-23 have structural similarities but different functions in the immune system. IL-12 drives the development of T-helper (Th) 1 cells mainly through the activation of STAT4. Th1 cells that produce interferon-γ (IFN-γ) are crucial for antimicrobial and antitumor responses (Trinchieri et al., 2003). IL-23 is important for the development of Th17 cells, which mediate antimicrobial and antifungal responses and are characterized by the production of the cytokines IL-17A, IL-17F, and IL-21 (Park et al., 2005; Ouyang et al., 2008; Korn et al., 2009). Th1 and Th17 cells are involved in the pathogenesis of autoimmune diseases and chronic inflammatory disorders (Cosmi et al., 2014).
The IL-12 receptor β1 (IL-12Rβ1) is the shared receptor chain of the IL-12 and IL-23 receptor signaling complexes, which consist of IL-12Rβ1/IL-12Rβ2 and IL-12Rβ1/IL-23R, respectively (Presky et al., 1996; Parham et al., 2002). IL-23 signaling is initiated by activation of tyrosine kinase (Tyk) 2 and Janus kinase (Jak) 2, which phosphorylate predominantly STAT3 and to a lesser extent STAT1, STAT4, and STAT5 (Parham et al., 2002). The downstream signaling molecule of IL-12Rβ2 is predominantly STAT4 (Bacon et al., 1995b; Jacobson et al., 1995; Thierfelder et al., 1996) and to a lesser extent STAT1, STAT3, and STAT5 (Collison and Vignali, 2008). Tyk2 associates with IL‑12Rβ1 and Jak2 with IL‑12Rβ2 (Bacon et al., 1995a; Zou et al., 1997). Phosphotyrosine-binding sites for STAT molecules have been found only in the intracellular domains of IL‑12Rβ2 and IL-23R and not in IL‑12Rβ1. Consequently they have been described as the only signal-transducing components of the IL-12 and IL‑23 receptor complexes (Collison and Vignali, 2008). Recently a noncanonical tyrosine-independent STAT3 activation site was identified within IL-23R (Floss et al., 2013). The IL-12Rβ1 was not found to contribute to IL-12/IL-23–induced intracellular signaling and is therefore considered as a ligand-binding receptor (Wu et al., 2000). However, p40 homodimers induce macrophage migration via the IL-12Rβ1 (Ha et al., 1999), suggesting a role of IL-12Rβ1 in signal transduction (Russell et al., 2003). IL-12 signaling is mediated through tyrosine residues within IL-12Rβ2 (Watford et al., 2003). In humans, tyrosine at position 800 is critical for STAT4 activation (Naeger et al., 1999), whereas the tyrosines at positions 757, 804, and 811 are important for STAT4 and tyrosines at positions 737, 804, and 811 for STAT3 activation in mice (Nishikomori et al., 2002). SOCS3 is recruited to human IL-12Rβ2 the interaction with the phosphorylated tyrosine at position 800 and thereby inhibits IL‑12–induced activation of STAT4 (Yamamoto et al., 2003). STAT-initiated signaling via the IL-12Rβ1 is unlikely, since the human IL-12Rβ1 contains no tyrosine residue and the murine IL-12Rβ1 contains only one (Y635), which is not embedded in a classical STAT-binding motif (Chua et al., 1995). Therefore it needs to be investigated whether Y635 is phosphorylated during IL-12/IL-23 signal transduction.
The intracellular domain of the IL-12Rβ1 contains two predicted Box1 and Box2 motifs as docking sites for Tyk2, which are conserved in human and mouse IL-12Rβ1 proteins (Chua et al., 1995). Whereas IL-23R does not contain conserved Box1 and Box2 motifs, the binding site of Jak2 at the IL-23R has been predicted (Pidasheva et al., 2011). Box1 motifs are highly conserved among many cytokine receptors (Ihle and Kerr, 1995; Murakami et al., 1991). These motifs are proline-rich, ∼6–10 amino acid residues long, and located in close proximity to the transmembrane domain. The consensus sequence of Box1 motifs is ΦΦP1X(I/V)P2XP3(E/K) (Φ denotes hydrophobic amino acid; Usacheva et al., 2002), where P1 and P2 are always present in cytokine receptors that associate with Jak2 (Usacheva et al., 2002). The Box2 motif is not exactly defined but is characterized by the consensus sequence (V/L)E(V/L)L (ΦEΦΦ; Usacheva et al., 2002). Box1 is needed for the association of the Janus kinase to the receptor chain (Tanner et al., 1995), whereas Box2 somehow mediates the full activity of the associated Janus kinase (Fukunaga et al., 1991; Colosi et al., 1993; Usacheva et al., 2002). The Janus kinase–binding regions within the IL-12Rβ1 and the IL-23R were not characterized.
Here we define the Box1 and Box2 motifs in murine IL-12Rβ1 and an unusual Jak2-binding site in IL-23R. Functional analysis revealed that association of Jak2 to IL-23R is mandatory for IL-12 and IL-23 signaling, whereas the function of Tyk2 can be taken over by another kinase.
RESULTS
Jak kinase activation upon IL-23 stimulation
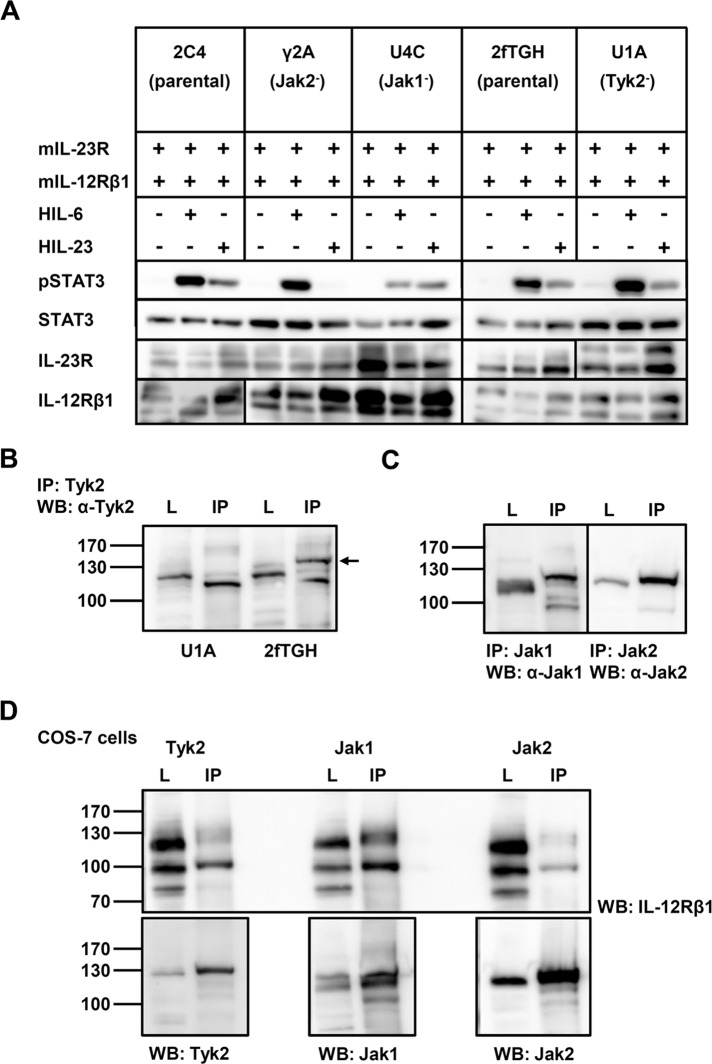
It has been shown that the same spectra of Jak/STAT molecules as in IL-12 signaling are activated by IL-23. IL‑23R, like IL‑12Rβ2, interacts with Jak2 to initiate signaling (Parham et al., 2002). To confirm the requirement for Jak2 and Tyk2 in IL-23 signal transduction, we cotransfected human fibrosarcoma cells (2C4 and 2fTGH) and cells that are deficient in Jak1 (U4C), Jak2 (γ2A), or Tyk2 (U1A; Behrmann et al., 2004) with the cDNAs for murine IL‑23R and IL‑12Rβ1 (Figure 1A). Expression analysis of both receptors performed by Western blotting revealed comparable levels. The cotransfected cells were incubated in the absence or presence of hyper–IL (HIL)-23. IL‑23 signaling was assessed by analysis of STAT3 activation using immunoblotting. HIL‑6 stimulation was included as control. HIL-6 is a fusion protein of IL‑6 and the soluble IL‑6R, which mimics IL‑6 trans-signaling (Fischer et al., 1997). Previous studies demonstrated a crucial role of Jak1 for STAT3 activation in response to IL‑6 (Guschin et al., 1995; Aparicio-Siegmund et al., 2014). Comparable results were obtained in gp130-expressing, kinase-deficient cell lines cotransfected with both IL‑23 receptors. HIL-6 dependent STAT3 phosphorylation was greatly reduced in the absence of Jak1, but loss of Jak2 or Tyk2 had no effect (Figure 1A). Western blot data demonstrated IL-23–dependent tyrosine phosphorylation of STAT3 in cotransfected parental cell lines (2C4, 2fTGH) and Jak1-deficient cells (U4C). No signal was detected in Jak2 deficient fibrosarcoma cells (γ2A). In contrast, Tyk2-deficient U1A and parental 2fTGH cells showed comparable activation of STAT3 upon stimulation with IL‑23 (Figure 1A). To confirm Tyk2 deficiency in U1A cells, we precipitated the kinase with a specific antibody and detected it by Western blotting (Figure 1B). In addition, we found the presence of Jak1 and Jak2 (Figure 1C). Cotransfection of COS-7 cells with cDNAs coding for IL-12Rβ1 and Tyk2, Jak1, or Jak2 indicated that in the absence of Tyk2, Jak1 takes over the kinase function (Figure 1D). These data demonstrate that Jak2 is crucial for IL-23 signaling, whereas Tyk2 is dispensable.
FIGURE 1:
Tyk2 is dispensable in IL-23 signal transduction. (A) Kinase-deficient fibrosarcoma cell lines γ2A (lacking Jak2), U4C (lacking Jak1), and U1A (lacking Tyk2) and the parental cell lines 2C4 and 2fTGH were cotransfected with expression plasmids for IL‑23 receptors. At 30 h after transfection, cells were washed with PBS and starved overnight in serum-free medium. Cells were then stimulated with 0.2% HIL‑6 for 15 min or 0.2% HIL‑23 for 30 min. Cellular lysates were prepared, and 50 μg of total protein (31 μg for HIL-6 stimulation) per lane was loaded on SDS gels, followed by immunoblotting using specific antibodies for phospho-STAT3, STAT3, mIL‑23R, and mIL-12Rβ1. Three independent experiments were performed and one representative experiment is shown. (B) Confirmation of Tyk2 absence in U1A cells. Tyk2 was immunoprecipitated from cellular lysates, and Western blotting was performed to detect the kinase. The arrow highlights the precipitated Tyk2 in 2fTGH cells. One representative experiment out of three is shown. IP, immunoprecipitation; L, lysate. (C) Detection of Jak1 and Jak2 in Tyk2-deficient U1A cells after immunoprecipitation. Cellular lysates were prepared and subjected to immunoprecipitation, followed by Western blotting. One representative experiment is shown. (D) Jak1 associates with IL-12Rβ1 in the absence of Tyk2. COS-7 cells were transiently transfected with cDNAs coding for IL-12Rβ1 and either Tyk2, Jak1 or Jak2. Kinases were immunoprecipitated and Western blotting was performed to detect IL-12Rβ1 and the appropriate kinase. One representative experiment out of three is shown.
The intracellular domain of IL-12Rβ1 spanning amino acid residues 592–656 is crucial for IL‑23–induced cellular proliferation and STAT3 phosphorylation
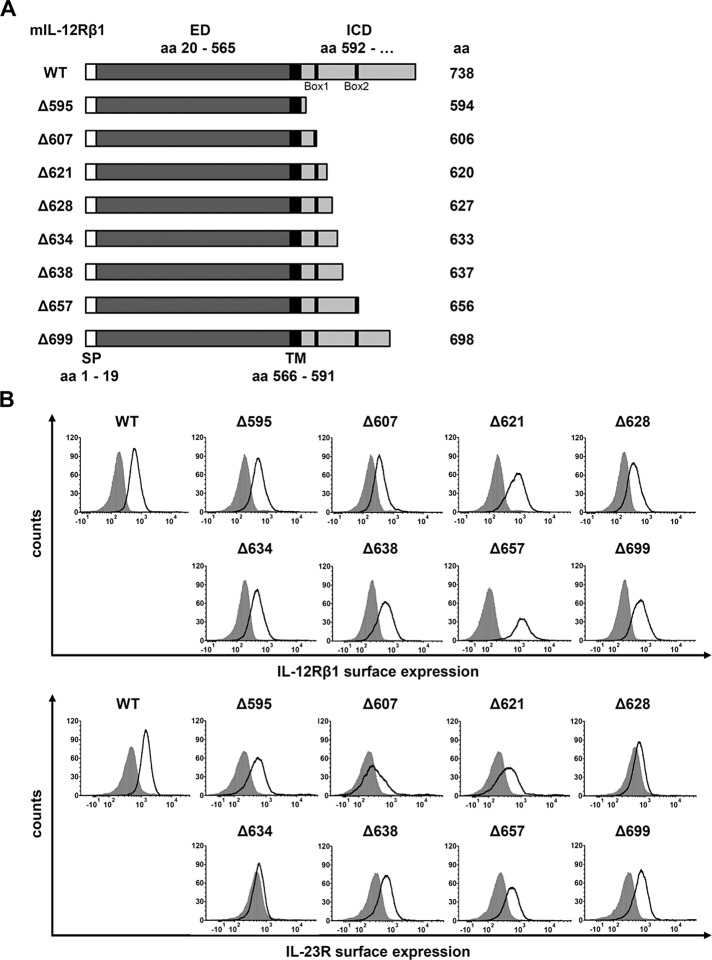
Murine IL-12Rβ1 consists of 738 amino acids. Its transmembrane domain is made up by amino acid residues 566–591 and its intracellular domain (ICD) by amino acid residues 592–738 (Chua et al., 1995; UniProtKB: Q60837). To analyze sequence requirements for IL-23–induced signaling within the intracellular domain of the IL-12Rβ1, we initially generated a set of eight different IL‑12Rβ1 variants containing various deletions of the intracellular domain (Figure 2A). The cDNAs coding for the full-length IL‑12Rβ1 (WT) protein and its deletions were stably transduced into Ba/F3-gp130-IL‑23R cells expressing murine IL‑23R (Floss et al., 2013). Ba/F3 cells are ideal to study cytokine signaling because their proliferation depends on IL-3 and activation of STAT5. After stable transfection with a cDNA coding for gp130, proliferation of Ba/F3-gp130 cells and activation of STAT3 depends on IL-6 and soluble IL-6R or HIL-6. The correct cellular location of the various IL-12Rβ1 variants was shown by cell surface staining followed by flow cytometry analysis (Figure 2B).
FIGURE 2:
Analysis of the intracellular domain of IL-12Rβ1 for signal transduction. (A) The 738–amino acid mouse IL‑12Rβ1 is a transmembrane protein containing Box1 and Box2 motifs within the ICD. Eight deletion variants were generated by PCR, and the first amino acid of the missing part of the IL‑12Rβ1 cytoplasmic domain is noted (e.g., Δ594). The extracellular (ED) and the transmembrane domain (TM) of IL-12Rβ1 were unaffected. (B) Representative histograms of IL‑12Rβ1 (top) and IL‑23R (bottom) surface expression of stably transduced Ba/F3-gp130 cell lines. Gray-shaded areas indicate Ba/F3-gp130 cells (negative control), and dark solid lines are the respective Ba/F3 cell lines as indicated.
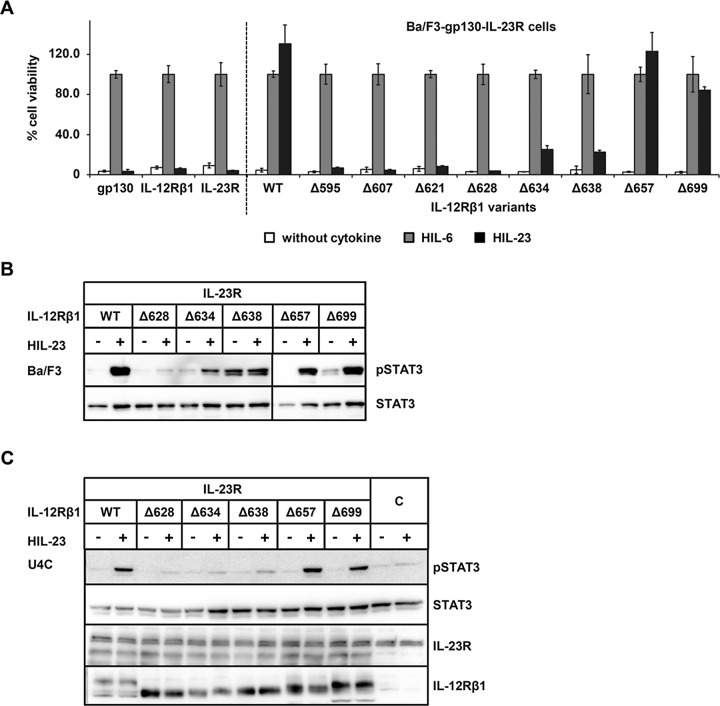
We then analyzed IL-23 signaling by measuring cell proliferation and STAT3 activation. Ba/F3-gp130-IL-23R-IL‑12Rβ1 (WT) cells with WT receptors proliferated in the presence of HIL‑23, whereas Ba/F3-gp130 cells expressing only IL‑23R or IL‑12Rβ1 did not grow (Floss et al., 2013; Figure 3A). Next, we compared IL-23–induced proliferation of Ba/F3-gp130-IL-23R cells, stably transduced with cDNAs coding for each of the eight IL-12Rβ1 variants, with IL-12Rβ1 WT cells. For this purpose, we stimulated cells with saturating concentrations of either HIL-6 or HIL‑23. Given that the cell lines were all independent cell clones and differed slightly in their maximal proliferation speed/capacity, we calculated IL‑23–induced proliferation as percentage of HIL-6–induced proliferation, which was set to 100%. As shown in Figure 3A, we did not detect differences in the proliferation of Ba/F3-gp130-IL‑23R-IL‑12Rβ1 cells (WT) stimulated with either HIL-6 or HIL‑23. However, proliferation was abolished in the Ba/F3-gp130-IL‑23R-IL‑12Rβ1Δ595, -Δ607, -Δ621, and -Δ628 cell lines upon IL-23 stimulation. In contrast, IL-23–induced cellular proliferation of Ba/F3-gp130-IL‑23R-IL‑12Rβ1Δ657 and -Δ699 cells was undistinguishable from that of Ba/F3-gp130-IL‑23R-IL‑12Rβ1 (WT) cells, whereas IL-23–induced proliferation of Ba/F3-gp130-IL‑23R-IL‑12Rβ1Δ634 and -Δ638 cells was about fourfold reduced compared with Ba/F3-gp130-IL‑23R-IL‑12Rβ1 (WT) cells (Figure 3A).
FIGURE 3:
The cytoplasmic domain of mouse IL‑12Rβ1 is important for STAT3 activation. (A) Proliferation of stably cotransduced Ba/F3-gp130 cells with cDNAs coding for murine IL‑23R and murine IL‑12Rβ1 (WT) or appropriate deletion variants. Equal numbers of cells were cultured for 3 d in the presence of 0.2% HIL‑6–conditioned cell culture supernatant or 0.2% HIL‑23 or without cytokine. Parental Ba/F3-gp130 cells were used as control. Proliferation was measured using the colorimetric CellTiter-Blue Cell Viability Assay. HIL‑6–dependent proliferation was set to 100%. One representative experiment is shown. Error bars represent SD for technical replicates. (B) Stably transduced Ba/F3 cells were washed three times, starved, and stimulated with 0.2% HIL‑23 for 30 min. Cellular lysates were prepared, and equal amounts of total protein (50 μg/lane) were loaded on SDS gels, followed by immunoblotting using specific antibodies for phospho-STAT3 and STAT3. Western blot data show one representative experiment out of three. (C) U4C cells were transiently transfected with cDNAs for murine IL‑23R and IL‑12Rβ1 variants. At 30 h after transfection, cells were washed with PBS and starved overnight in serum-free medium. Cells were then stimulated with HIL‑23 for 30 min. Cellular lysates were prepared, and equal amounts of total protein (50 μg/lane) were loaded on SDS gels, followed by immunoblotting using specific antibodies for phospho-STAT3, STAT3, mIL-23R, and mIL-12Rβ1. Nontransfected U4C cells served as negative control (C). Western blot data show one representative experiment out of three.
Analysis of IL-23 signaling by activation of STAT3 was investigated in either stably transduced Ba/F3 cells or transiently transfected U4C cells expressing IL-23R and the IL‑12Rβ1 (WT) or the variant IL‑12Rβ1Δ628, -Δ634, -Δ638, -Δ657, or -Δ699. Consistent with the cellular proliferation results, IL-23–induced STAT3 phosphorylation was observed in cells expressing IL-23R and full-length IL‑12Rβ1 (WT) or IL‑12Rβ1Δ657 or -Δ699. Coexpression of IL-23R and IL‑12Rβ1Δ628, however, did not result in STAT3 phosphorylation. Deletion of the murine IL‑12Rβ1 at amino acid residue 633 or 637 resulted in reduction of STAT3 phosphorylation, which is in agreement with the slight proliferation of these cells. Although Ba/F3-gp130-IL‑23R-IL‑12Rβ1Δ638 cells had a somewhat higher STAT3 phosphorylation in the absence of IL-23 (basal pSTAT3 level), IL-23 stimulation only slightly increased STAT3 phosphorylation (Figure 3B). Expression of IL-23R and IL-12Rβ1 in transiently transfected U4C cells was confirmed by Western blotting (Figure 3C). Taken together, our data demonstrate that amino acid residues 657–738 of IL‑12Rβ1 were not critically required for cellular proliferation and STAT3 activation. Moreover, amino acid residues N592 to P627, which flank the proposed Box1 motif of the IL-12Rβ1, rescue ∼20% of biological activity, whereas the amino acid residues N592 to Q656 were required for full activity. This indicates that the proposed Box1 and Box2 motifs are required for kinase binding and induction of IL-23 signaling.
Verification of the proposed Box1 and Box2 motifs for Tyk2 binding and IL-23/IL-12–induced signaling
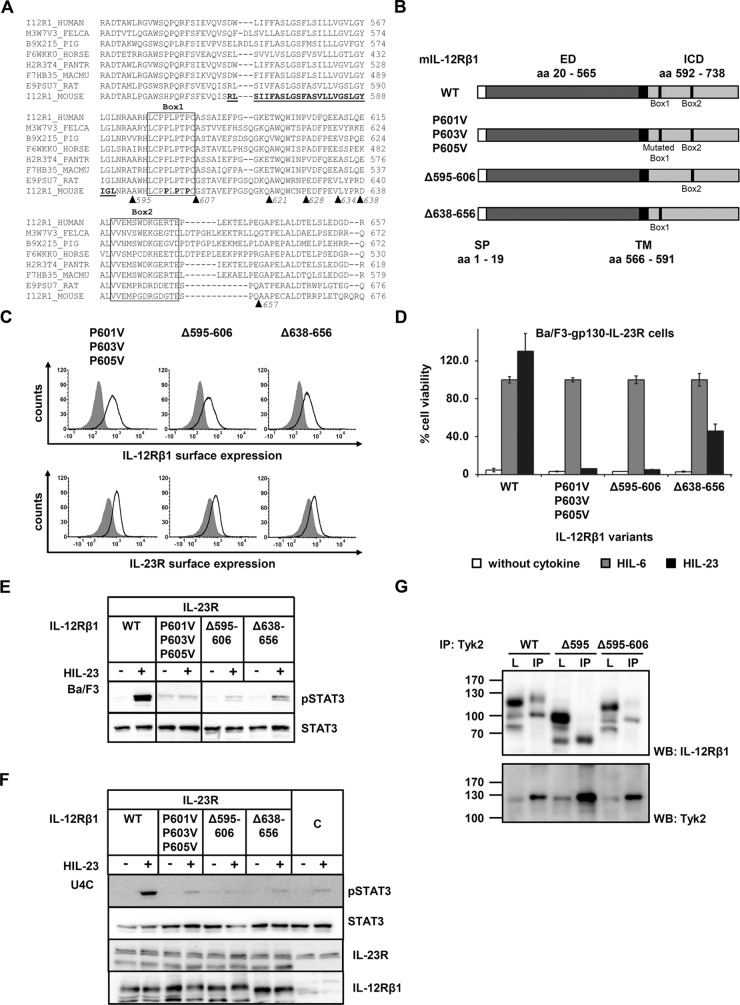
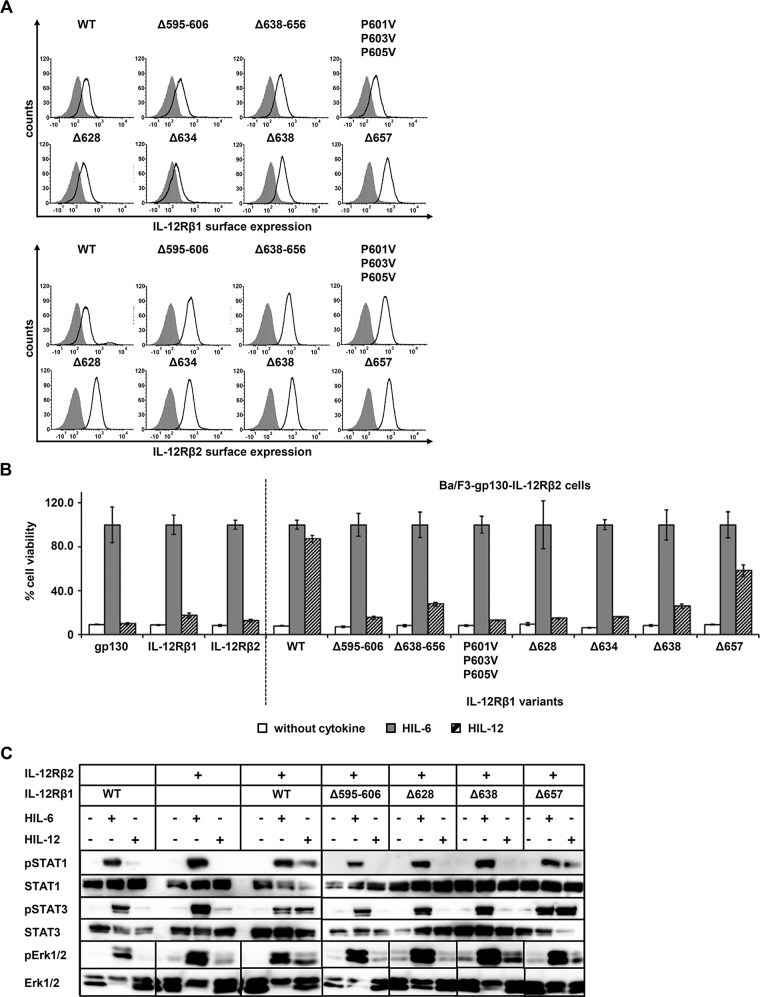
Sequence homology comparison of the intracellular domains of IL-12Rβ1 from different species revealed a highly conserved amino acid residue stretch from murine L598 to C606 (LCPPLPTPC), which shows similarity to the Box1 consensus sequence (φφPX(I/V)PXP(E/K) (Usacheva et al., 2002). A second motif, which is not exactly defined, was found in the murine V641-E653 sequence (VVEMPGDRGDGTE) and might serve as Box2 motif (φEφφ; Usacheva et al., 2002; Figure 4A). To verify the proposed Box1 and Box2 motifs, we generated appropriate amino acid deletions of murine IL‑12Rβ1 (∆Box1: mIL‑12Rβ1Δ595-606, ∆Box2: mIL‑12Rβ1-Δ638-656). In addition, we mutated three proline residues within the Box1 to valine in the mIL‑12Rβ1 variant mIL‑12Rβ1P601V/P603V/P605V (Figure 4B). Expression of the murine IL‑12Rβ1 variants and IL‑23R on the cell surface of generated Ba/F3 cell lines was confirmed by flow cytometry (Figure 4C).
FIGURE 4:
Association of Tyk2 with IL-12Rβ1 via the Box1 motif is mandatory for IL-23 signal transduction. (A) Aligned amino acid sequences of IL‑12Rβ1. The transmembrane domain of the mouse IL‑12Rβ1 is highlighted in bold. Box1 and Box2 motifs are as described in Chua et al. (1995). Deletion variants are depicted by triangles. (B) IL‑12Rβ1 variants with mutated or deleted Box motifs were generated by PCR. The extracellular (ED) and the transmembrane domain (TM) of IL-12Rβ1 are unaffected. (C) Representative histograms of IL‑12Rβ1 (top) and IL‑23R (bottom) surface expression of stably transduced Ba/F3-gp130 cell lines. Gray-shaded areas indicate Ba/F3-gp130 cells (negative control), and dark solid lines are the respective Ba/F3 cell lines as indicated. (D) Proliferation of stably cotransduced Ba/F3-gp130 cells with cDNAs coding for murine IL‑23R and murine IL‑12Rβ1 Box1 or Box2 deletion variants or mutant IL‑12Rβ1. Equal numbers of cells were cultured for 3 d in the presence of 0.2% HIL‑6 or 0.2% HIL‑23 or without cytokine. Ba/F3-gp130 cells expressing murine IL‑23R and IL‑12Rβ1 were used as control. Proliferation was measured using the colorimetric CellTiter-Blue Cell Viability Assay, and HIL‑6–dependent proliferation was set to 100%. Error bars represent SD for technical replicates. (E) Stably transduced Ba/F3 cells were washed three times, starved, and stimulated with 0.2% HIL‑23 for 30 min. Cellular lysates were prepared, and equal amounts of total protein (50 μg/lane) were loaded on SDS gels, followed by immunoblotting using specific antibodies for phospho-STAT3 and STAT3. Western blot data show one representative experiment out of three. (F) U4C cells were transiently transfected with cDNAs for murine IL‑23R and IL‑12Rβ1 deletion and mutant variants. Cotransfected U4C cells expressing wild-type receptors were used as control. At 30 h after transfection, cells were washed with PBS and starved overnight in serum-free medium. Cells were then stimulated with HIL‑23 for 30 min. Cellular lysates were prepared, and 50 μg of total protein per lane was loaded on SDS gels, followed by immunoblotting using specific antibodies for phospho-STAT3, STAT3, mIL-23R, and mIL-12Rβ1. Nontransfected U4C cells served as negative control (C). Western blot data show one representative experiment. (G) The Box1 motif of IL‑12Rβ1 is important for the association of Tyk2. COS-7 cells were cotransfected with cDNAs coding for murine Tyk2 and full-length IL‑12Rβ1, a deletion variant lacking Box1 motif, or an IL‑12Rβ1 variant without the cytoplasmic domain. Tyk2 was immunoprecipitated, and Western blot analysis was performed to detect the appropriate IL-12Rβ1 variant and Tyk2. Three independent experiments were performed, and one representative experiment is shown. IP, Tyk2 coimmunoprecipitation; L, lysate.
We analyzed IL-23–dependent proliferation of stably transduced Ba/F3-gp130-IL‑23R-IL‑12Rβ1Δ595-606, -Δ638-656, and -P601V/P603V/P605V cells. Whereas deletion or mutation of the Box1 motif completely abolished IL-23–induced proliferation, deletion of Box2 resulted in a markedly reduced but detectable cellular proliferation (Figure 4D). Analysis of IL-23–induced STAT3 phosphorylation in either stably transduced Ba/F3 or transiently transfected U4C cells supported the results of the cellular proliferation. STAT3 phosphorylation was undetectable in IL‑12Rβ1 variants with a deleted or mutated Box1 motif and slightly detectable in the IL‑12Rβ1 variant with the deleted Box2 motif. No STAT3 phosphorylation was detected in IL-23–stimulated, cotransfected U4C cells containing IL-23R and the IL-12Rβ1 Box2 deletion variant (Δ638-656; Figure 4, E and F). These results show that Box1 is mandatory for IL-23–induced signaling, whereas Box2 is not essential but needed to achieve full activity.
The kinases Tyk2 and Jak2 are involved in IL-12 signaling (Watford et al., 2003), and direct association of Jak2 with IL-12Rβ2 and Tyk2 with IL-12Rβ1 has been shown (Zou et al., 1997). We investigated the binding of Tyk2 to mIL‑12Rβ1 variants by coimmunoprecipitation approaches. As expected, IL‑12Rβ1 was coimmunoprecipitated with Tyk2, whereas binding of IL‑12Rβ1Δ595 (deletion of Box1 and Box2) and IL-12Rβ1Δ595-606 (deletion Box1) to Tyk2 was greatly diminished (Figure 4G). Unspecific bands of lower molecular weight were detected in lysates and immunoprecipitates of each variant. However, bands with the appropriate molecular weight for the IL-12Rβ1 deletion variants were significantly reduced in the coimmunoprecipitates, indicating that the Box1 motif is essential for binding of the Janus kinase Tyk2 to IL-12Rβ1.
IL‑12Rβ1 is required not only for IL-23–induced signal transduction but also for IL-12 signaling. To verify our results for IL-12 signaling, we generated Ba/F3-gp130 cells stably transduced with murine IL‑12Rβ1 (and variants thereof) and the murine IL‑12Rβ2, making these cells responsive to IL-12. Correct location of the receptors on the cell surface of stably transduced Ba/F3 cells was confirmed by flow cytometry (Figure 5A). As shown in Figure 5B, proliferation of Ba/F3-gp130-IL‑12Rβ2-IL‑12Rβ1 (WT) was dependent on HIL-12, whereas single transduced cells with IL‑12Rβ1 or IL‑12Rβ2 and Ba/F3-gp130 cells did not proliferate in the presence of the cytokine. As expected, Ba/F3-gp130-IL‑12Rβ2 expressing the IL‑12Rβ1 variants ‑Δ628, -Δ634, -Δ595-656, and -P601V/P603V/P605V did not proliferate in the presence of HIL-12. As for IL-23–induced cellular proliferation, Ba/F3-gp130-IL‑12Rβ2 cells expressing mIL‑12Rβ1 variants -Δ638-656 and -Δ638 showed reduced IL-12–dependent proliferation, whereas IL‑12–induced proliferation of Ba/F3-gp130-IL‑12Rβ2-IL‑12Rβ1Δ657 cells was only slightly reduced and more comparable to that of WT Ba/F3-gp130-IL‑12Rβ2-IL‑12Rβ1 cells (Figure 5B).
FIGURE 5:
IL-12Rβ1 plays a central role in IL-12 signal transduction. (A) Selected IL‑12Rβ1 variants were retrovirally transduced in Ba/F3-gp130-IL‑12Rβ2 cells. Representative histograms of IL‑12Rβ1 (top) and IL‑12Rβ2 (bottom) surface expression of resulting transduced Ba/F3-gp130 cell lines. Gray-shaded areas indicate Ba/F3-gp130 cells (negative control), and dark solid lines are the respective Ba/F3 cell lines as indicated. (B) Proliferation of stably transduced Ba/F3-gp130-IL-12Rβ2 cells with cDNAs coding for murine IL‑12Rβ1 (WT) or appropriate variants. Equal numbers of cells were cultured for 3 d in the presence of 0.2% HIL‑6–conditioned cell culture supernatant or 0.4% HIL‑12 or without cytokine. Parental Ba/F3-gp130, Ba/F3-gp130-mIL-12Rβ1, and Ba/F3-gp130-mIL‑12Rβ2 cells were used as control. Proliferation was measured using the colorimetric CellTiter-Blue Cell Viability Assay. HIL‑6–dependent proliferation was set to 100%. Error bars represent SD for technical replicates. (C) Stably transduced Ba/F3 cells were washed three times, starved, and stimulated with 0.2% HIL‑6 or 0.4% HIL‑12 for 15 or 30 min. Cellular lysates were prepared, and 50 μg of total protein per lane was loaded on SDS gels, followed by immunoblotting using specific antibodies for phospho-STAT3, STAT3, phospho-STAT1, STAT1, phospho-Erk1/2, and Erk1/2. Ba/F3-gp130-mIL‑12Rβ1 and Ba/F3-gp130-mIL-12Rβ2 cells were used as control. Western blot data show one representative experiment.
IL-12 induces STAT4, STAT1, and STAT3 phosphorylation but apparently not Erk1/2 activation (Zou et al., 1997; Watford et al., 2003). Due to very low levels of STAT4 protein in Ba/F3 cells (Zou et al., 1997), our analysis was focused on activation of STAT1 and 3. Western blot analysis of IL-12–induced STAT1/3 phosphorylation was performed with stably transduced Ba/F3 cell lines. Only double transduction of the cDNAs coding for IL‑12Rβ1 and IL‑12Rβ2 led to IL-12–induced STAT1/3 phosphorylation (Figure 5C). Cotransduction of IL‑12Rβ2 and IL‑12Rβ1Δ595-606, -Δ628, and -Δ638 did not lead to IL-12–induced STAT1/3 phosphorylation. However, IL-12–stimulated Ba/F3 cells expressing IL-12Rβ2 and IL‑12Rβ1Δ657 induced STAT1/3 phosphorylation comparable to that of WT cells (Figure 5C), which is consistent with IL‑12–dependent proliferation (Figure 5B). We also investigated these stable cell lines for their ability to phosphorylate Erk1/2. In contrast to the assumption of Watford et al. (2003), clearly induced activation of Erk1/2 was detected for Ba/F3 cells expressing IL‑12Rβ2 and WT IL‑12Rβ1 upon IL-12 stimulation. Slight phosphorylation of Erk1/2 was also shown for the IL-12Rβ2/IL‑12β1Δ657 variant (Figure 5C). As already shown for IL-23–induced STAT3 phosphorylation, the amino acid residues (634–656) near the Box2 motif (642–653) of IL‑12Rβ1 are also important for the activation of the Erk1/2 pathway (Figure 5C).
Taken together, our data indicate that the identified Box1 motif in the murine IL‑12Rβ1 from 595 to 606 (LCPPLPTPC) is absolutely mandatory for IL-12– and IL-23–induced signal transduction. The identified Box2 motif is involved in STAT3 phosphorylation, but its deletion did not completely abolish signal transduction and cellular proliferation.
Characterization of the binding site of Jak2 within the intracellular domain of the IL‑23R
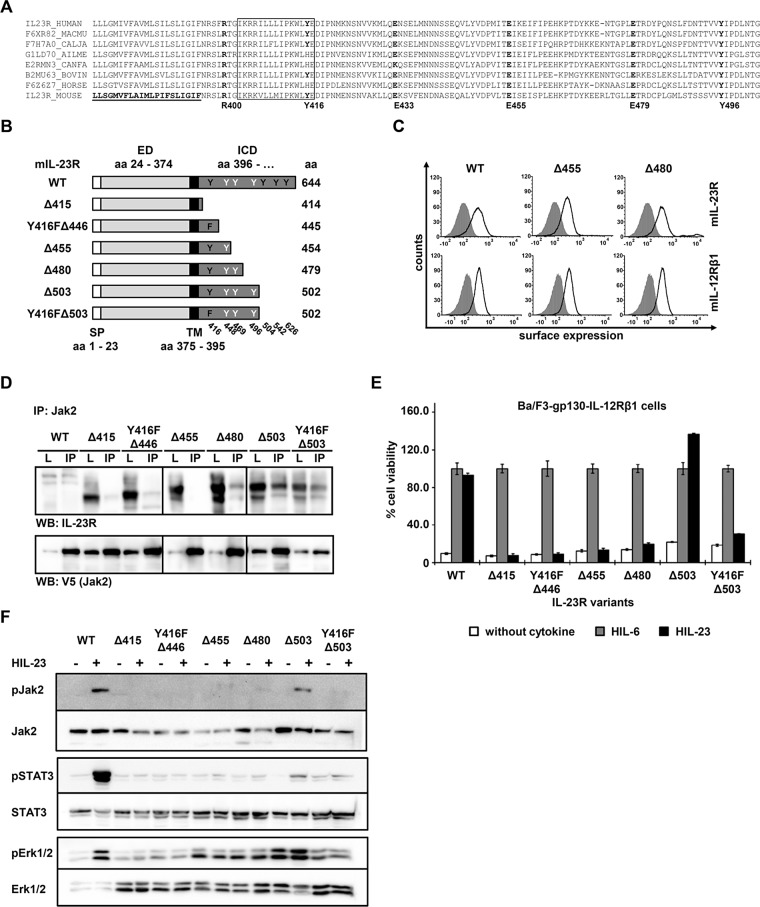
So far, consensus sequence homology comparisons within the intracellular domain of IL‑23R did not lead to the identification of Box1 and Box2 motifs (Figure 6A). However, sequence alignments of IL-23R intracellular chains from different species suggested a conserved region from I403 to E417 in murine IL‑23R as a potential Jak2-binding site (Pidasheva et al., 2011). Polymorphisms in the IL‑23R chain may influence IL‑23 responses, and a nonsynonymous nucleotide substitution in the human IL23R gene, which results in the R381Q variant, showed a protective role against autoimmune diseases (Di Cesare et al., 2009). The R381Q polymorphism is conserved across different species (mouse R400Q) and is located between the transmembrane domain and the postulated Jak2 binding site. In vitro–polarized Th17 cells from IL‑23R R381Q donors failed to induce IL-23–mediated IL-17A production and STAT3 phosphorylation, possibly due to impaired association of Jak2 with the cytoplasmic domain of the receptor (Pidasheva et al., 2011). However, this hypothesis has not been functionally proven. A series of IL‑23R deletion and mutation variants (‑Δ415, ‑Y416FΔ446, ‑Δ503, -Y416FΔ503) was characterized according to their ability to activate STAT3 (Floss et al., 2013).
FIGURE 6:
The membrane-proximal region of IL‑23R is important for association with Jak2. (A) Aligned amino acid sequences of IL‑23R. The transmembrane domain of the mouse IL‑23R is underlined. The hypothetical Jak2 binding motif as postulated by Pidasheva et al. (2011) is highlighted and highly conserved among different species. Mouse SNP R400Q, Y416, E433, E455, and Y496 are shown in bold. (B) Schematic representation of mIL-23R deletion variants. Six deletion variants were generated, and the first amino acid of the missing part of the IL‑23R cytoplasmic domain (CD) is indicated (e.g., Δ415). The extracellular (ED) and the transmembrane domain (TM) of IL-23R are unaffected. Tyrosines within the CD are highlighted in black (Y416, Y504, Y542, and Y626, involved in IL-23 signaling) and white (Y448, Y469, and Y496). Y416F is indicated as F. (C) Representative histograms of IL‑23R (top) and IL‑12Rβ1 (bottom) surface expression of transduced Ba/F3-gp130 cell lines. Gray-shaded areas indicate Ba/F3-gp130 cells (negative control), and dark solid lines are the respective Ba/F3 cell lines as indicated. (D) Amino acid sequence from 455 to 479 is important for the association of Jak2. COS-7 cells were cotransfected with cDNAs coding for murine Jak2 (V5 tagged) and full-length IL‑23R or a set of deletion variants (Floss et al., 2013). Jak2 was immunoprecipitated, and Western blot analysis was performed to detect the appropriate IL‑23R variant and Jak2. Three independent experiments were performed, and one representative experiment is shown. IP, Jak2 coimmunoprecipitation; L, lysate. (E) Proliferation of stably transduced Ba/F3-gp130-IL-12Rβ1 cells with cDNAs coding for murine IL‑23R (WT) or appropriate variants. Equal numbers of cells were cultured for 3 d in the presence of 0.2% HIL‑6–conditioned cell culture supernatant or 0.2% HIL‑23 or without cytokine. Proliferation was measured using the colorimetric CellTiter-Blue Cell Viability Assay. HIL‑6–dependent proliferation was set to 100%. Error bars represent SD for technical replicates. (F) Jak2 association does not implement kinase activation. Stably transduced Ba/F3 cells were washed three times, starved, and stimulated with HIL‑23. Cellular lysates were prepared, and 50 μg of total protein per lane were loaded on SDS gels, followed by immunoblotting using specific antibodies for phospho-Jak2, Jak2, phospho-STAT3, STAT3, phospho-Erk1/2, and Erk1/2. Western blot data show one representative experiment out of three.
In this study, we tested two additional variants (‑Δ455 and ‑Δ480), whose cell surface expression has been verified by flow cytometry (Figure 6, B and C). To investigate the interaction of Jak2 and IL-23R and variants thereof, we verified the interaction of Jak2 with full-length IL-23R (WT) and the deletion variants -Δ480, -Δ503, and -Y416F/Δ503 by coimmunoprecipitation experiments (Figure 6D). Although cotransfection of COS-7 cells with the WT receptor and Jak2 was not as efficient as for the other variants, we detected weak signals for the precipitated receptor and Jak2 in Western blots. In contrast, we detected no interaction of Jak2 and IL-23R for the IL-23RΔ415, -Y416FΔ446, and -Δ455 variants (Figure 6D). Consistent with these results, the corresponding Ba/F3-gp130-IL‑12Rβ1-IL-23RΔ415, -IL-23R-Y416FΔ446, and –IL-23R∆455 cells did not proliferate in the presence of HIL‑23. However, Ba/F3-gp130-IL‑12Rβ1-IL‑23RΔ503 showed IL‑23–dependent proliferation comparable to that of wild-type cells, whereas Y416FΔ503 failed to induce proliferation of the respective Ba/F3 cells (Figure 6E). Despite the interaction of Jak2 and IL-23R∆480, Ba/F3 cells expressing IL-12Rβ1 and the latter variant failed to proliferate in the presence of IL-23. Analysis of IL-23–dependent Jak2 phosphorylation by Western blotting clearly demonstrated activation of the kinase exclusively in Ba/F3-gp130-IL-12Rβ1 cells expressing IL-23R WT or -∆503 (Figure 6F) but not in -∆415, -Y416F∆415, -∆455, -∆480, and -Y416F∆503 variants. We previously showed that STAT3 phosphorylation is not mandatory for the proliferation of Ba/F3-gp130-IL‑12Rβ1-IL‑23RΔ503 cells, which is maintained by activation of Erk1/2 and/or PI3K/Akt (Floss et al., 2013). IL-23–dependent STAT3 phosphorylation was exclusively detected in Ba/F3-gp130-IL‑12Rβ1-IL‑23R WT cells, whereas activation of Erk1/2 occurs also in cells expressing the IL-23R∆503 variant (Figure 6F).
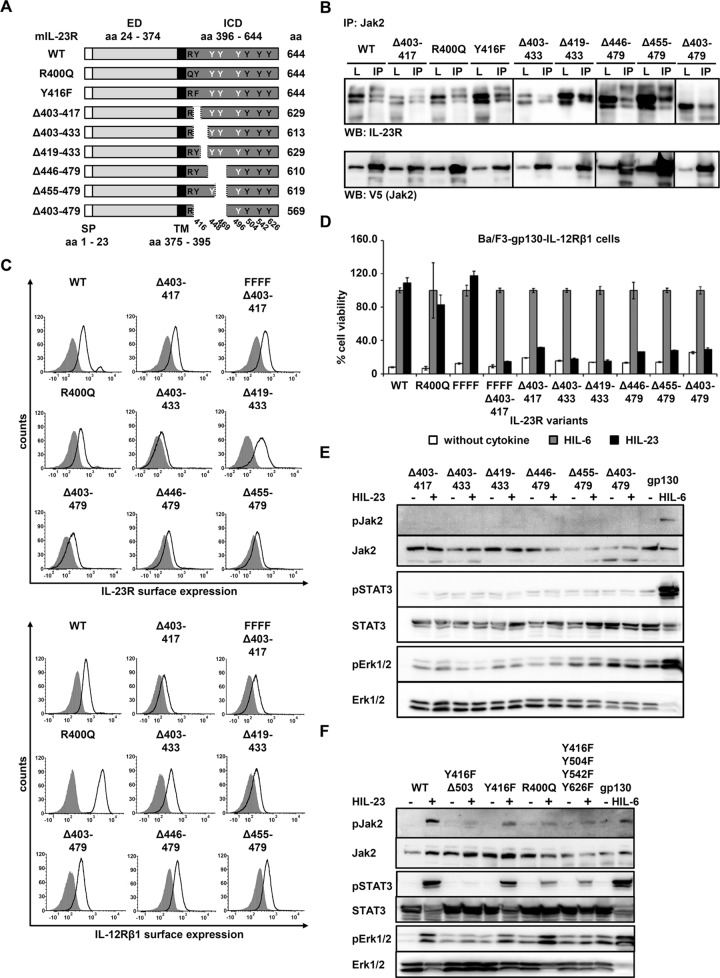
The foregoing results indicated that association of Jak2 with the IL-23R receptor does not necessarily implicate kinase activation upon stimulation with IL-23. In fact, our data suggest that the ICD spanning amino acid residues N396 to E479 of the IL-23R is important for activation of Jak2. To identify the region within the ICD required for kinase association/activation, we generated a second set of IL-23R variants (Figure 7A) and tested their interaction with Jak2. Surprisingly, interaction of Jak2 and IL-23R was detected for all IL-23R variants, including those that carry a deletion in the proposed Jak2-binding site (Δ403-417; Pidasheva et al., 2011), and two IL‑23R mutants that contain amino acid changes located next to (single nucleotide polymorphism [SNP] R400Q) or within (Y416F) the postulated Jak2-binding site (Figure 7B). Ba/F3-gp130-IL-12Rβ1-IL-23R cells were analyzed with regard to the cell surface expression of the receptors (Figure 7C) and their proliferation in the presence of HIL-23. Deletion of the postulated Jak2-binding site was further done in the IL-23R variant with Y → F substitutions of four tyrosines (FFFF; Y416F/Y504F/Y542F/Y626F). Cell viability assays revealed that deletion of the putative Jak2- binding site from Δ403-417 and all other deletions abolished IL-23–dependent cellular proliferation. In contrast, IL-23–induced cellular proliferation of Ba/F3-gp130-IL‑12Rβ1-IL‑23R-R400Q cells was comparable to that for Ba/F3-gp130-IL‑12Rβ1-IL‑23R cells (Figure 7D). Analysis of Jak2, STAT3, and Erk1/2 phosphorylation confirmed the results of the cell viability assay (Figure 7E). No STAT3 and Erk1/2 activation was observed for all deletion variants, demonstrating that the amino acid residues 403–479 are indispensable for signal transduction. For the R400Q variant and the IL-23R-Y416F/Y504F/Y542F/Y626F variant, an apparently weaker STAT3 phosphorylation was detected than with Ba/F3-gp130-IL‑12Rβ1-IL‑23R cells (Figure 7F). We previously showed that this receptor variant possess altered kinetics of STAT3 activation (Floss et al., 2013). Consistent with STAT3 activation, phosphorylation of Jak2 was reduced in these cells. The IL-23R-Y416F and -R400Q variants activated Erk upon stimulation with IL-23, in contrast to cells with Y416F-mutated IL-23R lacking 120 amino acids of the intracellular part (-Y416F ∆503) or harboring mutations of signal-transducing tyrosines (-Y416F/Y504F/Y542F/Y626F; Figure 7F). We previously showed that the predicted SHP2-binding site Y416EDI and an additional motif within the C-terminal part of the IL-23R are needed for phosphorylation of Erk1/2 (Floss et al., 2013).
FIGURE 7:
Mutations and deletions of IL-23R ICD cannot disrupt Jak2 association. (A) Schematic representation of mIL-23R variants with mutations and deletions within the cytoplasmic domain (CD). Two mutation (R400Q and Y416F) and six deletion variants were generated. The extracellular (ED) and transmembrane (TM) domains of IL-23R are unaffected. Tyrosines within the CD are highlighted in black (Y416, Y504, Y542, and Y626, involved in IL-23 signaling) and white (Y448, Y469, and Y496). Y416F and R400Q are indicated. (B) COS-7 cells were cotransfected with cDNAs coding for murine Jak2 (V5 tagged) and full-length IL‑23R or a deletion/mutant variant. Jak2 was immunoprecipitated, and Western blot analysis was performed to detect the appropriate IL‑23R variant and Jak2. Three independent experiments were performed, and one representative experiment is shown. IP, Jak2 coimmunoprecipitation; L, lysate. (C) Representative histograms of IL‑23R (top) and IL‑12Rβ1 (bottom) surface expression of transduced Ba/F3-gp130 cell lines. Gray-shaded areas indicate Ba/F3-gp130 cells (negative control), and dark solid lines are the respective Ba/F3 cell lines as indicated. (D) Proliferation of stably transduced Ba/F3-gp130-mIL-12Rβ1 cells with cDNAs coding for murine IL‑23R (WT) or appropriate variants. Equal numbers of cells were cultured for 3 d in the presence of 0.2% HIL‑6–conditioned cell culture supernatant or 0.2% HIL‑23 or without cytokine. Proliferation was measured using the colorimetric CellTiter-Blue Cell Viability Assay. HIL‑6–dependent proliferation was set to 100%. Error bars represent SD for technical replicates. (E, F) Stably transduced Ba/F3 cells were washed three times, starved, and stimulated with HIL‑23. Cellular lysates were prepared, and 50 μg total protein per lane was loaded on SDS gels, followed by immunoblotting using specific antibodies for phospho-Jak2, Jak2, phospho-STAT3, STAT3, phospho-Erk1/2, and Erk1/2. Western blot data show one representative experiment out of three.
Taken together, our results indicate that the amino acid sequence spanning E455 to E479 is critical for Jak2 binding. To identify the region required for Jak2 binding, we analyzed additional deletion variants, -Δ455-479, -Δ446-479, and -Δ403-479. We included a variant lacking the postulated Jak2-binding site (-Δ403-433) and one IL-23R protein with the lack of 12 amino acids after the postulated site (-Δ419-433). However, all of these receptor deletion variants, including Δ403-479, coimmunoprecipitate with Jak2 (Figure 7B). IL-23 receptors in which parts of the intracellular domain have been removed (Δ403-433, Δ419-433, Δ446-479, Δ455-479, and Δ403-479) showed a strong reduction in cell viability (Figure 7D). These results indicate that despite Jak2 association with the IL-23R receptor, activation of IL-23 signaling did not occur. Our data clearly show that association of Jak2 with the IL-23R is not mediated via classical Box1/Box2 motifs and that the amino acid sequence E455 to E479 is important but not sufficient for Jak2 activation. Considering the lack of IL-23–dependent proliferation of Ba/F3-IL-12Rβ1 cells expressing IL-23R variants with deletions of the previously proposed Jak2- binding site (Δ403-417, Δ403-433, Δ419-433, Δ446-479, Δ455-479, Δ403-479; Figure 7D), it is likely that other receptor sequences contribute to the regulation of the kinase activity.
DISCUSSION
Two members of the Janus kinase family are involved in signaling of IL-12–type cytokines: Jak2 and Tyk2 for IL-12 and IL-23, and Jak2 and Jak1 for IL-27 and IL-35 (Vignali and Kuchroo, 2012). Here we characterized the Tyk2- and Jak2-binding sites in IL-12Rβ1 and IL-23R, respectively. Tyk2 binding to IL-12Rβ1 was mediated by canonical Box1 and Box2 motifs for IL-12– and IL-23–induced receptor complexes. In contrast, IL-23R does not contain Box1 and Box2 motifs. Furthermore, the recently proposed Jak2-binding site I403 to E417 in the close vicinity of the plasma membrane was not responsible for Jak2 binding. Instead, Jak2 binds to the amino acid sequence E455 to E479 of IL-23R. Surprisingly, E455 is ∼60 amino acid residues away from the plasma membrane. Taken together, our data indicate that Jak2 association with IL-23R is mandatory for IL-23 signaling. However, despite binding of Jak2 to the IL-23R kinase, activation occurs only in IL-23R variants with intact cellular domains spanning N396 to T503 (WT, ∆503). Phosphorylation of Jak2 in Ba/F3-IL-12Rβ1 cells expressing these variants induced activation of STAT proteins and/or Erk, leading to cell proliferation.
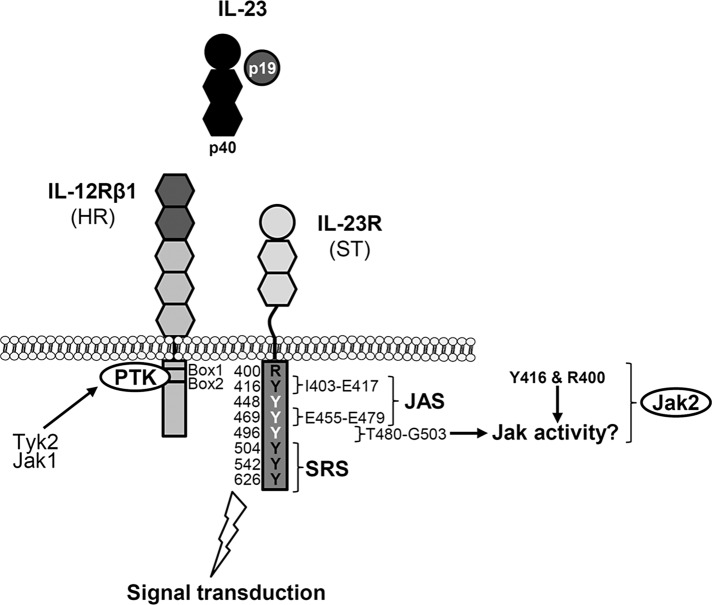
Biological effects of IL-12 and IL-23 are mediated by nonreceptor protein tyrosine kinases, which phosphorylate tyrosine residues on the intracellular domains of the receptor chains. The signal-transducing component is either IL‑12Rβ2 or IL-23R. Specific interactions were detected between Tyk2 and IL-12Rβ1 and between Jak2 and IL-12Rβ2, using IL-12 receptor chimeras containing the extracellular domain of epidermal growth factor receptor, as well as the transmembrane and cytoplasmic domains of either IL-12Rβ1 or IL-12Rβ2 (Zou et al., 1997). Accordingly, IL-23R associates with Jak2 (Parham et al., 2002). We demonstrated that association and activation of Tyk2 are essential for IL-12– and IL-23–induced signal transduction. Our data showed that IL-12Rβ1 is not only a bystander receptor for binding of the cytokine. The intracellular part of IL-12Rβ1 is important for signal transduction of IL-12 and IL-23, as visualized by receptor deletion variants. Analysis of kinase-deficient fibrosarcoma cells showed that Jak2 is indispensable for IL-23 signal transduction. Initial studies showed that generation of Jak2-knockout mice led to embryonic lethality due to defects in erythropoiesis (Neubauer et al., 1998; Parganas et al., 1998). In contrast, Tyk2-deficient mice are viable but show impaired cytokine responses (Karaghiosoff et al., 2000; Shimoda et al., 2000). Neither IL‑12 nor IL-23 induced STAT4 activation in T-cells from a Tyk2-deficient patient (Minegishi et al., 2006). However, STAT3 phosphorylation upon IL‑23 stimulation was not analyzed in this study. Because STAT4 activation is the main signaling event after IL-12 stimulation, STAT3 phosphorylation seems to be dominated by IL‑23 (Watford et al., 2004). We examined Tyk2-deficient fibrosarcoma cells expressing both IL-23 receptors in regard to activation of STAT3 upon stimulation with IL‑23. Tyk2 deficiency did not abolish IL-23 signaling, indicating that STAT3 was activated by another kinase, which may substitute for Tyk2. Coimmunoprecipitation of IL-12Rβ1 from COS-7 cells transiently transfected with cDNAs coding for IL-12Rβ1 and either Tyk2, Jak1, or Jak2 suggests the binding of Jak1 in the absence of Tyk2. Substitution of Janus kinases within the Jak-STAT pathway was shown by analysis of chimeric human IFN-γR2 receptors (Kotenko et al., 1996). Consequently a model was hypothesized in which signal- transducing receptor chains can be divided into two classes: 1) the actual signal transducers (ST) with STAT (or SH2 domain–containing proteins) recruitment sites (SRS) and Jak association sites (JAS), and 2) helper receptors (HR) containing only JAS and no SRS (Kotenko et al., 1996). In this model, IL-12Rβ1 represents the so-called helper receptor, which does not contain functional tyrosine residues but has additional protein tyrosine kinase activity upon IL-12 or IL-23 binding. Kotenko et al. (1996) hypothesized that the intracellular domains of HR can be associated with any Jak isoform and thus do not provide any specificity for signal transduction. Only the extracellular domains of helper receptors are specific for particular ligand–receptor complexes. This hypothesis can be transferred to the IL-12 and IL-23 signaling complexes (Figure 8). In this case, the signal transducers for IL-12 or IL-23 signaling are IL‑12Rβ2 and IL‑23R, respectively, and tyrosines involved in IL-12 and IL-23 signaling have been identified (Watford et al., 2004; Floss et al., 2013). This assumption is supported by the fact that removal of Jak association sites within the helper receptor IL-12Rβ1 completely prevents IL-23 and IL-12 signal transduction.
FIGURE 8:
Overview of Jak-binding sites within the IL-23 receptor complex. Formation of the IL-23 receptor complex consisting of p19, p40, IL-23R, and IL-12Rβ1 results in the activation of signaling pathways (Floss et al., 2015), and interaction of Janus kinases with the intracellular domains of the receptors initiates signaling cascades. According to Kotenko et al. (1996), IL-23R represents the signal transducer (ST) with a Janus kinase association site (JAS) and STAT recruitment sites (SRS). IL-12Rβ1 is regarded as a helper receptor (HR), which contains Box1 and Box2 motifs for association of the protein tyrosine kinase (PTK) Tyk2 or Jak1 and no SRS. Tyrosines within the IL-23R cytoplasmic domain are highlighted in black (Y416, Y504, Y542, and Y626, involved in IL-23 signaling) and white (Y448, Y469, and Y496). Postulated JAS I403-E417 is indicated (Pidasheva et al., 2011). Experimentally validated JAS E455-E479 is highlighted. Amino acid sequences that might influence kinase activity are designated.
Association of Tyk2 and IL-12Rβ1 is mediated by a Box1 motif, which is located within the intracellular part of the receptor. We demonstrated that deletion or mutation of this motif clearly inhibited cytokine signaling. Deletion of the second motif (Box2) reduced but did not abolish IL-23 and IL-12 signal transduction. Early studies of GM-CSFRβ and GH receptor revealed that Box2 motifs positively modulate signaling for these two receptors but are not absolutely required for signaling (Sakamaki et al., 1992; Colosi et al., 1993). Based on the hypothesized signal transducer/helper receptor model, IL-23R should also contain at least one association site for Jak2. However, classical Box1 and Box2 motifs (Usacheva et al., 2002) within the intracellular domain of the receptor have not been identified by amino acid sequence comparison. Recently a putative Jak2-binding site was predicted without any functional analysis (Pidasheva et al., 2011). Ba/F3-IL-12Rβ1 cells expressing IL‑23R, which lack this motif, did not activate STAT3 and did not proliferate upon stimulation with IL-23. However, Jak2 is still associated with the IL-23R deletion variant. Coprecipitation of Jak2 and IL-23R was obtained for all analyzed receptor variants except three deletion variants, -Δ415, -Y416FΔ446, and -Δ455. Activation of Jak2 was exclusively found in cells expressing IL-12Rβ1 and either IL-23R WT or -Δ503. We demonstrated that Ba/F3-IL-12Rβ1 cells expressing either the Δ415 or Y416FΔ446 mutant fail to activate STAT3 and cellular proliferation upon IL-23 stimulation (Floss et al., 2013). Wallweber et al. (2014) identified an unexpected receptor-binding mode in the Tyk2-IFNAR1 interface and described a glutamate residue (E497) in IFNAR1 that is required for Tyk2 binding. Of note, a classical proline-rich Box1 motif does not occur in IFNAR1. However, this glutamate, which is ∼40 amino acids away from the transmembrane domain, is in close vicinity to a dileucine motif important for the Tyk2 FERM-SH2 domain stability. They speculated that the dileucine interaction site might represent a Box1-binding motif for Jak kinase(s). Our results indicate that the amino acid residues E455 to E479 of murine IL-23R are important for association with Jak2. However, the proximal IL-23R cytoplasmic tail is also essential for IL-23 signaling. The polymorphism R381Q, which occurs at a frequency of up to 17%, depending on the population, confers protection against inflammatory bowel diseases, psoriasis, ankylosing spondylitis, and graft-versus-host disease (De Paus et al., 2008). Several studies considered that R381Q IL-23R might result in a loss of function. CD8+ T-cells from healthy R381Q IL-23R carriers showed decreased IL-23–dependent IL-17 and IL-22 production compared with WT individuals. Furthermore, Sarin et al. (2011) found decreased IL-23–dependent expansion and STAT3 activation of CD8+ T-cells from R381Q individuals compared with WT cells. In parallel, Di Meglio et al. (2013) showed that the IL23R R381Q gene variant had no major effect on Th17 cell differentiation, but IL-23–mediated Th17 cell effector function was impaired. Th17 cells from R381Q carriers had significantly reduced IL‑23–induced IL-17A production and STAT3 phosphorylation compared with WT carriers (Di Meglio et al., 2011). Coexpression of IL-12Rβ1 and IL-23R or the R381Q variant in HeLa cells resulted in decreased STAT3 activation upon IL-23 stimulation (Sarin et al., 2011). These results coincide with our data for the corresponding murine SNP R400Q. Ba/F3-IL-12Rβ1 cells expressing IL-23R R400Q proliferate in the presence of IL-23 but have reduced STAT3 activation, which is similar to behavior of an IL-23R (FFFF) containing mutations of STAT3 tyrosine residues (Floss et al., 2013). However, cotransduction of T-cell blasts from a patient with a null mutation of IL-12Rβ1 with IL-12Rβ1 and IL-23R WT or R381Q showed a similar activation of STAT1, STAT3, and STAT4 upon IL-23 stimulation. Proliferation of these cells also showed no significant differences (De Paus et al., 2008).
Of interest, the identified Jak2-binding site in the IL-23R from E455 to E479 shows no homology to Box1 and Box2 motifs and is located ∼60 amino acids from the plasma membrane. The question arises of why the functional Jak2 binding site in the IL-23R is so far from the membrane, whereas all known Box1 motifs are in close proximity to the transmembrane domain. One explanation might be that Jak2 is redirected to the membrane by a special tertiary structure of the intracellular domain of the receptor, thereby facilitating Jak2 activation after cytokine stimulation. Alternatively, Jak2 binding to IL-23R is an exception, and the far location of Jak2 still enables receptor activation. Of interest, deletion of the proposed Jak2-binding site from I403 to E417 also disrupted receptor activation but not Jak2 binding. Folding of the intracellular domain and functional location of Jak2 in a near-membrane position might be part of the explanation of why the deletion of the IL-23R amino acids I403 to E417 is crucial for IL-23–induced signal transduction and cellular proliferation.
MATERIALS AND METHODS
Cells and reagents
COS-7 (ACC-60) cells were purchased from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). Human fibrosarcoma cell lines 2C4, U4C, γ2A, 2fTGH, and U1A have been described (Behrmann et al., 2004). G418 (Genaxxon Biosciences, Ulm, Germany; 400 μg/ml) was added to the medium of 2C4, U4C, and γ2A cells, and 2fTGH and U1A cells were supplemented with 125 or 250 μg/ml hygromycin B (Carl Roth GmbH, Karlsruhe, Germany). Murine Ba/F3-gp130 cells transduced with human gp130 were provided by Immunex (Seattle, WA) (Gearing et al., 1994). The packaging cell line Phoenix-Eco was described previously (Ketteler et al., 2002). Ba/F3-gp130 cell lines with murine IL-12Rβ1 and murine IL-23R (WT), deletion (Δ415, Y416FΔ446, Δ503, Y416FΔ503), and mutation variants (Y416F, FFFF) were described previously (Floss et al., 2013). All cell lines were grown in DMEM high-glucose culture medium (GIBCO, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal calf serum (GIBCO, Thermo Fisher Scientific), 60 mg/l penicillin, and 100 mg/l streptomycin (Genaxxon Bioscience) at 37°C with 5% CO2 in a water-saturated atmosphere. Ba/F3-gp130 cells were maintained in the presence of HIL-6, a fusion protein of IL‑6 and the soluble IL‑6R, which mimics IL‑6 trans-signaling (Fischer et al., 1997). Either recombinant protein (10 ng/ml) or 0.2% of conditioned cell culture medium from a stable CHO-K1 clone secreting HIL-6 (final concentration 20 ng/ml as determined by enzyme-linked immunosorbent assay [ELISA]) was used to supplement the growth medium. Ba/F3-gp130-IL-23R-IL‑12Rβ1 cells expressing murine IL‑23R and murine IL‑12Rβ1 or variants thereof were stimulated with 0.2% of conditioned cell culture medium from a stable CHO-K1 clone secreting HIL‑23 (a fusion of murine p40 and murine p19) in a final concentration of 10 ng/ml, as determined by ELISA (unpublished data; Floss et al., 2013). Accordingly, Ba/F3-gp130-IL‑12Rβ2-IL‑12Rβ1 cells containing the murine IL‑12Rβ2 and murine IL‑12Rβ1 or variants were stimulated with HIL‑12. HIL‑12 is a fusion protein of murine p40 followed by a synthetic linker (RGGGGSGGGGSVE) and murine p35 with an N-terminal Flag and a C-terminal histidine (His) tag. A stable CHO-K1 clone was selected, and 0.4% of conditioned cell culture medium from this clone was used for cell culture and proliferation assays in a final concentration of 4 ng/ml, as determined by ELISA (unpublished data). Phospho-STAT3 (Tyr-705) (D3A7), STAT3 (124H6), phospho-STAT1 (Tyr-701), STAT1, phospho-p44/42 MAPK (Erk1/2) (Thr-202/Tyr-204;D13.14.4E), p44/42 MAPK (Erk1/2) antibody, phospho-Jak2 (Tyr-1007/1008), and Jak2 (D2E12) monoclonal antibodies (mAbs) were obtained from Cell Signaling Technology (Frankfurt, Germany). V5 mAb was purchased from Invitrogen, Thermo Fisher Scientific. Tyk2 (C-20) and Jak1 (HR-785) polyclonal antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX). The Tyk2 rabbit polyclonal antibody was raised against an N-terminal peptide of murine Tyk2 (Prchal-Murphy et al., 2012). Peroxidase-conjugated secondary mAbs were obtained from Pierce (Thermo Fisher Scientific). Biotinylated mIL‑23R (BAF1686) and mIL‑12Rβ1 (BAF1998) mAbs, streptavidin–horseradish peroxidase (HRP), phycoerythrin (PE)-conjugated mIL‑12Rβ1, and mIL‑23R mAbs were from R&D Systems (Minneapolis, MN). Purified hamster mIL‑12Rβ2 and PE mouse Armenian and Syrian hamster immunoglobulin G (IgG) cocktail were purchased from BD Biosciences (Heidelberg, Germany). Alexa Fluor 647–conjugated Fab goat anti-rat IgG was obtained from Dianova (Hamburg, Germany).
Cloning of IL-12Rβ1, IL-12Rβ2, and IL-23R variants
Generation of the eukaryotic expression vector p409 (Althoff et al., 2001) and the retroviral plasmid pMOWS-hygro (Suthaus et al., 2010) containing the cDNA encoding murine IL‑12Rβ1 were described elsewhere (Floss et al., 2013). cDNA coding for murine IL‑12Rβ2 (gene ID 16162) was amplified from total RNA extracts derived from mouse T-cells and cloned into p409 expression vector and the pMOWS-puro retroviral plasmid (Ketteler et al., 2002). A C-terminal c‑myc tag was added. IL‑12Rβ1 variants with truncated intracellular domains were cloned using standard PCR. Mutations of proline to valine in the Box1 motif of IL‑12Rβ1 were introduced by PCR using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific), followed by DpnI digestion of methylated template DNA. Box1 and Box2 deletion variants were generated by splicing with overlap extension PCR (SOE-PCR). Eukaryotic p409 expression plasmids for murine IL-23R (WT) and deletion (-Δ415, -Y416FΔ446, -Δ503, -Y416FΔ503) and mutation variants (-Y416F, -FFFF) were described in Floss et al. (2013). The p409 expression vector containing the cDNA for the murine IL‑23R was used as template for the generation of receptor variants with deletions of and within the membrane-proximal region by standard and SOE-PCR. Mutation of arginine 400 to glutamine was generated by PCR using Phusion high-fidelity DNA polymerase, followed by DpnI digestion of methylated template DNA. Resulting p409 expression vectors were verified by sequencing and used for transient transfection of U4C or COS-7 cells. The cDNA variants of mIL‑12Rβ1 and mIL‑23R variants were transferred into pMOWS-hygro or pMOWS-puro for retroviral transduction of Ba/F3-gp130 cells. The expression plasmid pEFmTyk2 was described in Prchal-Murphy et al. (2012). The pEF/V5-His-mJak2 plasmid was kindly provided by Eric Keil, David Finkenstädt, and Klaus Pfeffer (Institute of Medical Microbiology, Heinrich-Heine-University, Düsseldorf, Germany). The pSVL-mJak1 was a generous gift from Iris Behrmann (Life Sciences Research Unit, University of Luxembourg, Luxembourg).
Transfection, transduction, and selection of cells
COS-7 and human fibrosarcoma cells (2C4, γ2A, U4C, 2fTGH, U1A) were transiently transfected with TurboFect (Thermo Fisher Scientific) according to the manufacturer’s instructions. Ba/F3-gp130 cells were retrovirally transduced with the pMOWS expression plasmids coding for the various IL-23R and IL‑12Rβ1 variants as described in Floss et al. (2013). In addition, Ba/F3-gp130-mIL-12Rβ2 cells were generated using the pMOWS-puro-IL-12Rβ2 vector. Transduced cells were grown in standard DMEM medium as described, supplemented with 10 ng/ml HIL‑6. Selection of transduced Ba/F3 cells was performed with puromycin (1.5 μg/ml), hygromycin B (1 mg/ml; Carl Roth GmbH), or both for at least 2 wk. Then HIL‑6 was washed away, and the generated Ba/F3-gp130 cell lines were selected for HIL‑23– or HIL‑12–dependent growth.
Cell viability assay
To remove the cytokines, Ba/F3-gp130 cell lines were washed three times with sterile phosphate-buffered saline (PBS). We suspended 5 × 103 cells in DMEM supplemented with 10% fetal calf serum, 60 mg/l penicillin, and 100 mg/l streptomycin and cultured them for 3 d in a final volume of 100 μl with or without cytokines as indicated. The CellTiter-Blue Cell Viability Assay (Promega, Karlsruhe, Germany) was used to estimate the number of viable cells by recording the fluorescence (excitation, 560 nm; emission, 590 nm) using the Infinite M200 PRO plate reader (Tecan, Crailsheim, Germany) immediately after adding 20 μl of reagent per well (time point 0) and up to 1 h after incubation under standard cell culture conditions. All of the values were measured in triplicate per experiment. Fluorescence values were normalized by subtraction of values at time point 0. For direct comparison of the individual cell lines, proliferation in the presence of HIL-6 was defined as 100%. All experiments were performed at least three times, and one representative experiment was selected.
Stimulation assays
For analysis of STAT3 activation in cotransfected fibrosarcoma cell lines, cells were starved for 16 h in serum-free medium. This was followed by stimulation with cytokines as indicated. Subsequently cells were harvested and lysed in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1 mM NaF, 1 mM Na3VO4, 1% Nonidet P‑40, and 1% Triton X-100 supplemented with complete protease inhibitor cocktail tablets (Roche Diagnostics, Mannheim, Germany). Ba/F3-gp130 cell lines were washed three times with sterile PBS and incubated in serum-free DMEM for at least 4 h. Cells were stimulated with HIL‑23, HIL‑12, or HIL-6 as indicated, harvested, frozen in liquid nitrogen, and lysed as described. For analysis of phospho-Jak2, cells were lysed in 10 mM Tris-HCl, pH 7.8, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 1 mM sodium vanadate, and 10 mM MgCl2 supplemented with complete protease inhibitor cocktail tablets. Protein concentration of cell lysates was determined by bicinchoninic acid protein assay (Thermo Fisher Scientific) according to the manufacturer’s instructions. Analysis of Jak2, STAT1/3, and Erk1/2 activation was done by immunoblotting using 50 μg of proteins from total cell lysates and using phospho-Jak2, phospho-STAT1, phospho-STAT3, or phospho-Erk1/2 mAbs for detection.
Immunoprecipitation
Cotransfected COS-7 cells were lysed in 20 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1 mM sodium vanadate, 10 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, and 1% Brij 97 supplemented with complete protease inhibitor cocktail tablets (Roche Diagnostics), and fibrosarcoma cells in 2fTGH and U1A in 20 mM Tris-HCl, pH 7.8, 130 mM NaCl, 0.5% NP-40, 0.5 mM EDTA, 1 mM sodium vanadate, and protease inhibitors (Haan and Haan, 2013). Cell debris was removed by centrifugation. Whole-cell extracts were incubated with α-Tyk2 (2 μg/ml rabbit polyclonal Ab or 1:50 C-20), α-Jak1 (1:50), or α-Jak2 (1:1000) mAb at 4°C overnight. We added 50 μl of Protein A-Agarose (Roche Diagnostics) and incubated the samples at 4°C for 2 h under slow agitation. The samples were then washed three times with lysis buffer, suspended in 50 μl of 2.5× Laemmli sample buffer, and incubated for 10 min at 95°C. Analysis of interaction between cytokine receptor and respective Janus kinase was done by Western blotting.
Western blotting
Defined amounts of proteins from cell lysates or coimmunoprecipitations were loaded per lane, separated by SDS‑PAGE under reducing conditions, and transferred to polyvinylidene fluoride membranes. The membranes were blocked in 5% fat-free dried skimmed milk in TBS‑T (10 mM Tris‑HCl, pH 7.6, 150 mM NaCl, 1% Tween-20) and probed with the indicated primary antibodies in 5% fat-free dried skimmed milk in TBS‑T (α-STAT3, α-Jak2) or 5% bovine serum albumin (BSA) in TBS‑T (phospho-Jak2, phospho-STAT3, phospho-STAT1, STAT1, phospho-Erk1/2, Erk1/2, mIL‑23R, mIL‑12Rβ1, V5, α-Tyk2, α-Jak1) at 4°C overnight. After washing, the membranes were incubated with streptavidin-HRP or secondary peroxidase-conjugated antibodies diluted in 5% BSA or 5% fat-free dried skimmed milk in TBS‑T for 1 h at room temperature. The ECL Prime Western Blotting Detection Reagent (GE Healthcare, Freiburg, Germany) and the ChemoCam Imager (INTAS Science Imaging Instruments, Göttingen, Germany) were used for signal detection. For reprobing with another primary antibody, the membranes were stripped in 62.5 mM Tris-HCl, pH 6.8, 2% SDS, and 0.1% β‑mercaptoethanol for 30 min at 60°C and blocked again.
Cell surface detection of cytokine receptors
To detect cell surface expression of the cytokine receptors mIL‑12Rβ2 and mIL‑23R, stably transduced Ba/F3-gp130 cells were washed with FACS buffer (PBS containing 1% BSA) and incubated at 5 × 105 cells/100 μl FACS buffer supplemented with 2.5 μg of mIL‑23R mAbs (R&D Systems) or 1 μg of purified hamster mIL‑12Rβ2 mAbs (BD Biosciences) for 2 h on ice. After a single wash with FACS buffer, cells were incubated in 100 μl of FACS buffer containing a 1:100 dilution of Alexa Fluor 647–conjugated Fab goat anti-rat IgG (Dianova) or 0.25 μg PE mouse Armenian and Syrian hamster IgG cocktail (BD Biosciences) for 1 h at 4°C. Finally, cells were washed once with FACS buffer, suspended in 500 μl FACS buffer, and analyzed by flow cytometry (BD FACSCanto II flow cytometer; BD Biosciences). Data were evaluated using the FCS Express software (De Novo Software, Los Angeles, CA). Detection of surface expression of mIL‑12Rβ1 was achieved by incubating cells prepared as described with 100 μl of FACS buffer containing 10 μl of PE-conjugated IL‑12Rβ1 mAbs (R&D Systems) for 2 h at 4°C.
Acknowledgments
We thank Eric Keil, David Finkenstädt, and Klaus Pfeffer (Institute of Medical Microbiology, Medical Faculty, Heinrich-Heine-University, Düsseldorf, Germany) for providing the plasmid pEF/V5-His-mJak2. We are grateful to Iris Behrmann for the plasmid coding for murine Jak1 and to Daniela S. Floss and Roland Piekorz for critical reading of the manuscript. This work was funded by grants from the Deutsche Forschungsgemeinschaft (SCHE 907/3-1 to J.S.) and the RCMF of the University Düsseldorf (to T.K., J.S., and D.M.F.). B.S. is supported by the Austrian Science Fund (Grants SFB-F28 and P25642) and H.H. by the Deutsche Forschungsgemeinschaft (SFB688 TPA20).
Abbreviations used:
- HIL
hyper-interleukin
- IL
interleukin
- Jak
Janus kinase
- Tyk
tyrosine kinase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-12-1645) on May 18, 2016.
REFERENCES
- Althoff K, Müllberg J, Aasland D, Voltz N, Kallen K, Grötzinger J, Rose-John S. Recognition sequences and structural elements contribute to shedding susceptibility of membrane proteins. Biochem J. 2001;353:663–672. doi: 10.1042/0264-6021:3530663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio-Siegmund S, Moll JM, Lokau J, Grusdat M, Schröder J, Plöhn S, Rose-John S, Grötzinger J, Lang PA, Scheller J, et al. Inhibition of protein kinase II (CK2) prevents induced signal transducer and activator of transcription (STAT) 1/3 and constitutive STAT3 activation. Oncotarget. 2014;5:2131–2148. doi: 10.18632/oncotarget.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon CM, McVicar DW, Ortaldo JR, Rees RC, O’Shea JJ, Johnston JA. Interleukin 12 (IL-12) induces tyrosine phosphorylation of JAK2 and TYK2: differential use of Janus family tyrosine kinases by IL-2 and IL-12. J Exp Med. 1995a;181:399–404. doi: 10.1084/jem.181.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon CM, Petricoin EF, Ortaldo JR, Rees RC, Larner AC, Johnston JA, O’Shea JJ. Interleukin 12 induces tyrosine phosphorylation and activation of STAT4 in human lymphocytes. Proc Natl Acad Sci USA. 1995b;92:7307–7311. doi: 10.1073/pnas.92.16.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrmann I, Smyczek T, Heinrich PC, Schmitz-Van de Leur H, Komyod W, Giese B, Muller-Newen G, Haan S, Haan C. Janus kinase (Jak) subcellular localization revisited: the exclusive membrane localization of endogenous Janus kinase 1 by cytokine receptor interaction uncovers the Jak receptor complex to be equivalent to a receptor tyrosine kinase. J Biol Chem. 2004;279:35486–35493. doi: 10.1074/jbc.M404202200. [DOI] [PubMed] [Google Scholar]
- Chua A, Wilkinson V, Presky D, Gubler U. Cloning and characterization of a mouse IL-12 receptor-beta component. J Immunol. 1995;155:4286–4294. [PubMed] [Google Scholar]
- Collison L, Vignali DA. Interleukin-35: odd one out or part of the family. Immunol Rev. 2008;226:248–262. doi: 10.1111/j.1600-065X.2008.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosi P, Wong K, Leong SR, Wood WI. Mutational analysis of the intracellular domain of the human growth hormone receptor. J Biol Chem. 1993;268:12617–12623. [PubMed] [Google Scholar]
- Cosmi L, Maggi L, Santarlasci V, Liotta F, Annunziato F. T helper cells plasticity in inflammation. Cytometry A. 2014;85:36–42. doi: 10.1002/cyto.a.22348. [DOI] [PubMed] [Google Scholar]
- De Paus R, van de Wetering D, van Dissel J, van de Vosse E. IL-23 and IL-12 responses in activated human T cells retrovirally transduced with IL-23 receptor variants. Mol Immunol. 2008;45:3889–3895. doi: 10.1016/j.molimm.2008.06.029. [DOI] [PubMed] [Google Scholar]
- Di Cesare A, Di Meglio P, Nestle FO. The IL-23/Th17 axis in the immunopathogenesis of psoriasis. J Invest Dermatol. 2009;129:1339–1350. doi: 10.1038/jid.2009.59. [DOI] [PubMed] [Google Scholar]
- Di Meglio P, Di Cesare A, Laggner U, Chu C, Napolitano L, Villanova F, Tosi I, Capon F, Trembath RC, Peris K, et al. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23-induced Th17 effector response in humans. PLoS One. 2011;6:e17160. doi: 10.1371/journal.pone.0017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meglio P, Villanova F, Napolitano L, Tosi I, Terranova Barberio M, Mak RK, Nutland S, Smith CH, Barker JN, Todd JA, Nestle FO. The IL23R A/Gln381 allele promotes IL-23 unresponsiveness in human memory T-helper 17 cells and impairs Th17 responses in psoriasis patients. J Invest Dermatol. 2013;133:2381–2389. doi: 10.1038/jid.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Goldschmitt J, Peschel C, Brakenhoff J, Kallen K, Wollmer A, Grötzinger J, Rose-John S. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat Biotechnol. 1997;15:142–145. doi: 10.1038/nbt0297-142. [DOI] [PubMed] [Google Scholar]
- Floss DM, Mrotzek S, Klocker T, Schroder J, Grotzinger J, Rose-John S, Scheller J. Identification of canonical tyrosine-dependent and non-canonical tyrosine-independent STAT3 activation sites in the intracellular domain of the interleukin 23 receptor. J Biol Chem. 2013;288:19386–19400. doi: 10.1074/jbc.M112.432153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss DM, Schröder J, Franke M, Scheller J. Insights into IL-23 biology. From structure to function. Cytokine Growth Factor Rev. 2015;26:269–578. doi: 10.1016/j.cytogfr.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Ishizaka-Ikeda E, Pan CX, Seto Y, Nagata S. Functional domains of the granulocyte colony-stimulating factor receptor. EMBO J. 1991;10:2855–2865. doi: 10.1002/j.1460-2075.1991.tb07835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbers C, Hermanns H, Schaper F, Müller-Newen G, Grötzinger J, Rose-John S, Scheller J. Plasticity and cross-talk of interleukin 6-type cytokines. Plasticity and cross-talk of interleukin 6-type cytokines. Cytokine Growth Factor Rev. 2012;23:85–97. doi: 10.1016/j.cytogfr.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Gearing D, Ziegler S, Comeau M, Friend D, Thoma B, Cosman D, Park L, Mosley B. Proliferative responses and binding properties of hematopoietic cells transfected with low-affinity receptors for leukemia inhibitory factor, oncostatin M, and ciliary neurotrophic factor. Proc Natl Acad Sci USA. 1994;91:1119–1123. doi: 10.1073/pnas.91.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin D, Rogers N, Briscoe J, Witthuhn B, Watling D, Horn F, Pellegrini S, Yasukawa K, Heinrich P, Stark GR, et al. A major role for the protein tyrosine kinase JAK1 in the JAK/STAT signal transduction pathway in response to interleukin-6. EMBO J. 1995;14:1421–1429. doi: 10.1002/j.1460-2075.1995.tb07128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SJ, Lee CH, Lee SB, Kim CM, Jang KL, Shin HS, Sung YC. A novel function of IL-12p40 as a chemotactic molecule for macrophages. J Immunol. 1999;163:2902–2908. [PubMed] [Google Scholar]
- Haan C, Haan S. Co-immunoprecipitation protocol to investigate cytokine receptor-associated proteins, e.g., Janus kinases or other associated signaling proteins. Methods Mol Biol. 2013;967:21–38. doi: 10.1007/978-1-62703-242-1_2. [DOI] [PubMed] [Google Scholar]
- Ihle JN, Kerr IM. Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet. 1995;11:69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- Jacobson NG, Szabo SJ, Weber-Nordt RM, Zhong Z, Schreiber RD, Darnell JE, Jr, Murphy KM. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J Exp Med. 1995;181:1755–1762. doi: 10.1084/jem.181.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaghiosoff M, Neubauer H, Lassnig C, Kovarik P, Schindler H, Pircher H, McCoy B, Bogdan C, Decker T, Brem G, et al. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity. 2000;13:549–560. doi: 10.1016/s1074-7613(00)00054-6. [DOI] [PubMed] [Google Scholar]
- Ketteler R, Glaser S, Sandra O, Martens U, Klingmüller U. Enhanced transgene expression in primitive hematopoietic progenitor cells and embryonic stem cells efficiently transduced by optimized retroviral hybrid vectors. Gene Ther. 2002;9:477–487. doi: 10.1038/sj.gt.3301653. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Kotenko SV, Izotova LS, Pollack BP, Muthukumaran G, Paukku K, Silvennoinen O, Ihle JN, Pestka S. Other kinases can substitute for Jak2 in signal transduction by interferon-gamma. J Biol Chem. 1996;271:17174–17182. doi: 10.1074/jbc.271.29.17174. [DOI] [PubMed] [Google Scholar]
- Minegishi Y, Saito M, Morio T, Watanabe K, Agematsu K, Tsuchiya S, Takada H, Hara T, Kawamura N, Ariga T, et al. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25:745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Murakami M, Narazaki M, Hibi M, Yawata H, Yasukawa K, Hamaguchi M, Taga T, Kishimoto T. Critical cytoplasmic region of the interleukin 6 signal transducer gp130 is conserved in the cytokine receptor family. Proc Natl Acad Sci USA. 1991;88:11349–11353. doi: 10.1073/pnas.88.24.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeger L, McKinney J, Salvekar A, Hoey T. Identification of a STAT4 binding site in the interleukin-12 receptor required for signaling. J Biol Chem. 1999;274:1875–1878. doi: 10.1074/jbc.274.4.1875. [DOI] [PubMed] [Google Scholar]
- Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- Nishikomori R, Usui T, Wu CY, Morinobu A, O’Shea JJ, Strober W. Activated STAT4 has an essential role in Th1 differentiation and proliferation that is independent of its role in the maintenance of IL-12R beta 2 chain expression and signaling. J Immunol. 2002;169:4388–4398. doi: 10.4049/jimmunol.169.8.4388. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454–467. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, et al. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, Pflanz S, Zhang R, Singh KP, Vega F, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- Park H, Li Z, Yang XO, Hee Chang S, Nurieva R, Wang Y, Wang Y, Hood L, Zhu Z, Tian Q, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidasheva S, Trifari S, Phillips A, Hackney JA, Ma Y, Smith A, Sohn SJ, Spits H, Little RD, Behrens TW, et al. Functional studies on the IBD susceptibility gene IL23R implicate reduced receptor function in the protective genetic variant R381Q. PLoS One. 2011;6:e25038. doi: 10.1371/journal.pone.0025038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prchal-Murphy M, Semper C, Lassnig C, Wallner B, Gausterer C, Teppner-Klymiuk I, Kobolak J, Müller S, Kolbe T, Karaghiosoff M, et al. TYK2 kinase activity is required for functional type I interferon responses in vivo. PLoS One. 2012;7:e39141. doi: 10.1371/journal.pone.0039141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presky D, Yang H, Minetti L, Chua A, Nabavi N, Wu C, Gately M, Gubler U. A functional interleukin 12 receptor complex is composed of two beta-type cytokine receptor subunits. Proc Natl Acad Sci USA. 1996;93:14002–14007. doi: 10.1073/pnas.93.24.14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell TD, Yan Q, Fan G, Khalifah AP, Bishop DK, Brody SL, Walter MJ. IL-12 p40 homodimer-dependent macrophage chemotaxis and respiratory viral inflammation are mediated through IL-12 receptor beta 1. J Immunol. 2003;171:6866–6874. doi: 10.4049/jimmunol.171.12.6866. [DOI] [PubMed] [Google Scholar]
- Sakamaki K, Miyajima I, Kitamura T, Miyajima A. Critical cytoplasmic domains of the common beta subunit of the human GM-CSF, IL-3 and IL-5 receptors for growth signal transduction and tyrosine phosphorylation. EMBO J. 1992;11:3541–3549. doi: 10.1002/j.1460-2075.1992.tb05437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin R, Wu X, Abraham C. Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ and CD8+ human T-cell functional responses. Proc Natl Acad Sci USA. 2011;108:9560–9565. doi: 10.1073/pnas.1017854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda K, Kato K, Aoki K, Matsuda T, Miyamoto A, Shibamori M, Yamashita M, Numata A, Takase K, Kobayashi S, et al. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity. 2000;13:561–571. doi: 10.1016/s1074-7613(00)00055-8. [DOI] [PubMed] [Google Scholar]
- Suthaus J, Tillmann A, Lorenzen I, Bulanova E, Rose-John S, Scheller J. Forced homo- and heterodimerization of all gp130-type receptor complexes leads to constitutive ligand-independent signaling and cytokine-independent growth. Mol Biol Cell. 2010;21:2797–2807. doi: 10.1091/mbc.E10-03-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner JW, Chen W, Young RL, Longmore GD, Shaw AS. The conserved box 1 motif of cytokine receptors is required for association with JAK kinases. J Biol Chem. 1995;270:6523–6530. doi: 10.1074/jbc.270.12.6523. [DOI] [PubMed] [Google Scholar]
- Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DAA, Doherty PC, Grosveld GC, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- Trinchieri G, Pflanz S, Kastelein RA. The IL-12 family of heterodimeric cytokines: new players in the regulation of T cell responses. Immunity. 2003;19:641–644. doi: 10.1016/s1074-7613(03)00296-6. [DOI] [PubMed] [Google Scholar]
- Usacheva A, Sandoval R, Domanski P, Kotenko SV, Nelms K, Goldsmith MA, Colamonici OR. Contribution of the Box 1 and Box 2 motifs of cytokine receptors to Jak1 association and activation. J Biol Chem. 2002;277:48220–48226. doi: 10.1074/jbc.M205757200. [DOI] [PubMed] [Google Scholar]
- Vignali DA, Kuchroo V. IL-12 family cytokines: Immunological playmakers. Nat Immunol. 2012;13:722–728. doi: 10.1038/ni.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallweber HJ, Tam C, Franke Y, Starovasnik MA, Lupardus PJ. Structural basis of recognition of interferon-alpha receptor by tyrosine kinase 2. Nat Struct Mol Biol. 2014;21:443–448. doi: 10.1038/nsmb.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watford WT, Hissong BD, Bream JH, Kanno Y, Muul L, O’Shea JJ. Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4. Immunol Rev. 2004;202:139–156. doi: 10.1111/j.0105-2896.2004.00211.x. [DOI] [PubMed] [Google Scholar]
- Watford WT, Moriguchi M, Morinobu A, O’Shea JJ. The biology of IL-12: Coordinating innate and adaptive immune responses. Cytokine Growth Factor Rev. 2003;14:361–368. doi: 10.1016/s1359-6101(03)00043-1. [DOI] [PubMed] [Google Scholar]
- Wu C, Wang X, Gadina M, O’Shea J, Presky D, Magram J. IL-12 receptor beta 2 (IL-12R beta 2)-deficient mice are defective in IL-12-mediated signaling despite the presence of high affinity IL-12 binding sites. J Immunol. 2000;165:6221–6228. doi: 10.4049/jimmunol.165.11.6221. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Yamaguchi M, Miyasaka N, Miura O. SOCS-3 inhibits IL-12-induced STAT4 activation by binding through its SH2 domain to the STAT4 docking site in the IL-12 receptor beta2 subunit. Biochem Biophys Res Commun. 2003;310:1188–1193. doi: 10.1016/j.bbrc.2003.09.140. [DOI] [PubMed] [Google Scholar]
- Zou J, Presky DH, Wu CY, Gubler U. Differential associations between the cytoplasmic regions of the interleukin-12 receptor subunits beta1 and beta2 and JAK kinases. J Biol Chem. 1997;272:6073–6077. doi: 10.1074/jbc.272.9.6073. [DOI] [PubMed] [Google Scholar]