Abstract
Purpose
Ocular refraction is measured in spherical equivalent as the power of the external lens required to focus images on the retina. Myopia (nearsightedness) and hyperopia (farsightedness) are the most common refractive errors, and the leading causes of visual impairment and blindness in the world. The goal of this study is to identify rare and low-frequency variants that influence spherical equivalent.
Methods
We conducted variant-level and gene-level quantitative trait association analyses for mean spherical equivalent, using data from 1,560 individuals in the Beaver Dam Eye Study. Genotyping was conducted using the Illumina exome array. We analyzed 34,976 single nucleotide variants and 11,571 autosomal genes across the genome, using single-variant tests as well as gene-based tests.
Results
Spherical equivalent was significantly associated with five genes in gene-based analysis: PTCHD2 at 1p36.22 (p = 3.6 × 10−7), CRISP3 at 6p12.3 (p = 4.3 × 10−6), NAP1L4 at 11p15.5 (p = 3.6 × 10−6), FSCB at 14q21.2 (p = 1.5 × 10−7), and AP3B2 at 15q25.2 (p = 1.6 × 10−7). The variant-based tests identified evidence suggestive of association with two novel variants in linkage disequilibrium (pairwise r2 = 0.80) in the TCTE1 gene region at 6p21.1 (rs2297336, minor allele frequency (MAF) = 14.1%, β = –0.62 p = 3.7 × 10−6; rs324146, MAF = 16.9%, β = –0.55, p = 1.4 × 10−5). In addition to these novel findings, we successfully replicated a previously reported association with rs634990 near GJD2 at 15q14 (MAF = 47%, β = –0.29, p=1.8 × 10−3). We also found evidence of association with spherical equivalent on 2q37.1 in PRSS56 at rs1550094 (MAF = 31%, β = –0.33, p = 1.7 × 10−3), a region previously associated with myopia.
Conclusions
We identified several novel candidate genes that may play a role in the control of spherical equivalent. However, further studies are needed to replicate these findings. In addition, our results contribute to the increasing evidence that variation in the GJD2 and PRSS56 genes influence the development of refractive errors. Identifying that variation in these genes is associated with spherical equivalent may provide further insight into the etiology of myopia and consequent vision loss.
Introduction
Uncorrected refractive errors, the leading cause of visual impairment and blindness worldwide [1], are also associated with several ocular disorders, such as glaucoma, cataract, retinal detachment, and macular degeneration, further increasing the risk of vision loss in later life [2,3]. In the United States, myopia affects 1 in 3 individuals 20 years of age or older, and the prevalence of hyperopia increases with increasing adult age [4]. Although mild refractive errors can be easily managed with non-invasive optical means or refractive surgery, determining the underlying causes of refractive errors may lead to better management of their clinical complications.
The etiology of refractive errors is complex and not fully understood. Many environmental factors have been linked to increased myopia, including high intensity near-work [5], high intelligence [6-8], and low outdoor exposure [5,9-11]. These associations have been inconsistent across studies [9,12,13], and the causative relationship between these factors and spherical equivalent has not been established. In addition to environmental factors, genetic factors play an important role in control of spherical equivalent. The estimated heritability of spherical equivalent in populations of European descent ranges from 0.50 to 0.86 [14-21]. Genome-wide linkage studies have reported evidence of genetic linkage to more than 30 chromosomal regions for myopia and spherical equivalent [22]. Genome-wide association studies (GWASs) targeting more common variants have identified susceptibility loci associated with spherical equivalent or myopia in East Asian populations [23-30] and in populations of European ancestry [30-35]. However, in most cases the biologic basis of these associations have yet to be experimentally validated [36].
Evolutionary theory and empirical data suggest that rare and low-frequency variants could have relatively large effects on the risk of developing complex traits, and collectively rare variants may make significant contributions to disease susceptibility in a given population [37]. However, these variants were poorly characterized by earlier generations of GWAS genotyping arrays [38]. Sequencing of the exome or whole genome directly assays all variants, including rare and low-frequency variants, but it is currently costly to perform on thousands of study subjects. Exome arrays, with enhanced resolution of coding variants, provide a less costly alternative to sequencing to evaluate the role of rare (minor allele frequency (MAF) < 1.0%) and low-frequency (MAF = 1.0 – 5.0%) variants in phenotypes.
To examine the impact of rare and low-frequency coding variants on spherical equivalent, we conducted an exome array analysis on participants in the Beaver Dam Eye Study (BDES). Additionally, data permitting, we evaluated known regions of association reported in other populations for spherical equivalent and myopia.
Methods
Study participants
The BDES is a population-based cohort established in 1987 to study age-related eye disorders [39]. A total of 5,924 individuals aged from 43 to 84 years were identified from 3,715 households through a private census of Beaver Dam, Wisconsin. Between 1988 and 1990, 4,926 (83.14%) individuals participated in the baseline examination. Familial relationships of 2,783 participants were confirmed and the data was assembled into pedigrees. To improve the sampling efficiency, we genotyped individuals with intraocular pressure or spherical equivalent values at the baseline, resulting in a nested sub-cohort of 2,032 individuals with the full spectrum of spherical equivalent values. The study adhered to the ARVO statement on human subjects and was approved by the institutional review board at the University of Wisconsin. The study complied with the guidelines of the Declaration of Helsinki. Informed consent was obtained from all participants in the study.
Clinical evaluation
All participants in the BDES received a complete eye exam that included standardized evaluation of refraction using the Humphrey 530 refractor (San Leandro, CA) [39]. When the automated refraction yielded visual acuity of 20/40 or worse, a modification of the Early Treatment Diabetic Retinopathy Study (ETDRS) protocol [40] was followed to obtain the best-corrected visual acuity. The mean spherical equivalent (MSE) in diopters for each eye was calculated using the standard formula (MSE = sphere + 0.5 × cylinder). Nuclear lens opacity was determined by grading slit-lamp lens photographs using a standard protocol, resulting in a five-level scale [41]. Eyes without a lens, with an intraocular lens, or with best-corrected visual acuity of 20/200 or worse were excluded. Age, sex, and education were also obtained at the baseline visit.
Genotyping and quality control
DNA from 2,032 BDES participants, 22 blind duplicate samples, and 44 HapMap samples were genotyped on the Illumina Infinium HumanExome-12 v1.1 (Illumina, Inc., San Diego, CA) array at the Genetic Resources Core Facility (GRCF), the Johns Hopkins School of Medicine, the Institute of Genetic Medicine (Baltimore, MD). Of the 2,032 BDES participants, 1,908 (93.9%) were successfully genotyped and had a call rate >98%. After related individuals (n = 126) and individuals with unresolved gender inconsistencies (n = 15), missing spherical equivalent measurements of either eye (n=118), differences in spherical equivalent > ± 4 D between the eyes (n = 15), and missing values in age, sex, education, or nuclear sclerosis (n = 74) were excluded, 1,560 individuals remained in the final analytical cohort.
Of the 242,901 variants genotyped, we excluded the following: sex chromosomal variants (n = 5,465), insertion/deletion variants (n = 134), variants with call rates ≤98% (n = 6,027), monomorphic variants (n = 132,213), and duplicate variants (n = 314). The final data set included 98,748 polymorphic variants. We evaluated Hardy–Weinberg equilibrium (HWE) on these variants and reported the p value estimated from the HWE exact test for significant associations.
To evaluate genetic ancestry, we performed principal component analysis (PCA) using SMARTPCA in EIGENSTRAT (version 4.2) [42]. All BDES participants genotyped were self-reported non-Hispanic white. The first two principal components were included in the analysis to control for potential population stratification, resulting in a genomic inflation factor (λ) of 1.02.
Trait definitions
Age, education, and nuclear sclerosis have known effects on spherical equivalent and were significant covariates in our previous linkage analysis [43]. Using linear models, we regressed age (mean centered), education (mean centered), sex, nuclear sclerosis (sum of the nuclear lens opacity score of both eyes), and the first two principal components on the average of spherical equivalent in the right and left eyes. The residuals from this regression were used as the quantitative outcome in our association tests. For replications of loci associated with myopia in prior GWASs, we performed logistic regression analysis that compared myopia to hyperopia and myopia to non-myopia, adjusting for the covariates. We defined individuals with spherical equivalent < −1 D in either eye and < −0.5 D in the opposite eye as myopic and those with spherical equivalent > +1 D in either eye and > + 0.5 D in the opposite eye as hyperopic. Emmetropia was defined as spherical equivalent ranged from −1 D to +1 D in both eyes. The non-myopic group included emmetropes and hyperopes. Individuals who fell outside these categories were excluded from the logistic regression analysis.
Statistical analysis
We applied single-variant and gene-based tests to investigate the association of rare and low-frequency genetic variants with spherical equivalent as a quantitative trait. Single-variant association analysis was performed on variants with MAF ≥1.0% (n = 34,976) under an additive model using PLINK (version 1.07) [44]. To determine the number of independent tests, we tabulated the number of single nucleotide polymorphisms (SNPs) with pairwise linkage disequilibrium (LD) <0.2. There were 23,342 independent variants, which translated to a Bonferroni corrected significance threshold of p<2.1 × 10−6.
Gene-based analysis can be a powerful alternative approach to single-variant analysis in detecting effects of rare variants [45]. Rare variants in our analysis were defined as those with MAF of 1% or less. To examine the impact of rare variants on spherical equivalent, we used the following tests: (1) the burden test, where all variants were collapsed into a gene-based score [46], and (2) the sequence kernel association test (SKAT) [47]. Analysis was restricted to 11,571 genes with at least two rare variants. To optimize the statistical power of SKAT, we applied the Madsen-Browning weighting
which slightly up-weights the rare variants [48]. A p value of <4.3 × 10−6, corresponding to a Bonferroni correction for 11,571 independent gene tests, was used to determine significance.
We evaluated gene regions with GWAS significant associations in published studies or the region spanning two genes if the GWAS association was intergenic [23-34]. For each known GWAS locus, we performed a single-variant association analysis for quantitative refraction and myopia, targeting (1) the exact variant or (2) variants in strong LD (r2 >0.8) with the reported variants. Variants were considered as replicated at p<0.01 if the magnitude and direction of their effects in our study were consistent with those reported in previous studies.
Results
Characteristics of the Beaver Dam Eye Study and the genotyped sub-cohort included in these analyses are presented in Table 1. The baseline mean spherical equivalent in the subset was −0.20 D with a standard deviation of 2.9 D (Appendix 1). Compared to the entire BDES cohort, our subset included a higher proportion (38.6% versus 21.5%, respectively) of myopes and a similar proportion of hyperopes (37.0% versus 37.8%, respectively).
Table 1. The characteristics of the Beaver Dam Eye Study (BDES) participants.
| Characteristic | BDES Exome (n=1,560) | BDES Fulla (n=4,452) |
|---|---|---|
| Age (years), mean ± SD (range) |
59.5±10.6 (43 – 85) |
61.1±10.8 (43 – 86) |
| Female gender, N (%) |
884 (56.7) |
2,493 (56.0) |
| Education (years), mean ± SD |
12.7±3.1 |
12.0±2.8 |
| Nuclear Sclerosis b, mean ± SD |
4.7±1.6 |
4.8±1.6 |
| Spherical Equivalent (D)c, mean ± SD (range) |
−0.20±2.9 (−13.0 – 10.3) |
0.28±2.2 (−13.0 – 10.3) |
| Myopia, N (%) |
602 (38.6) |
958 (21.5) |
| Emmetropia, N (%) |
242 (15.5) |
1,217 (27.3) |
| Hyperopia, N (%) | 577 (37.0) | 1,681 (37.8) |
a All BDES individuals with reliable spherical equivalent measurements of both eyes at baseline visit. bThe sum of nuclear lens opacity grading of the right and left eyes at baseline. cThe average of spherical equivalent of the right and left eyes at baseline. Myopia was defined as < −1 D in either eye and < −0.5 D in the opposite eye; hyperopia was defined as > +1 D in either eye and > +0.5 D in the opposite eye; emmetropia was defined as between −1 D and +1 D in both eyes.
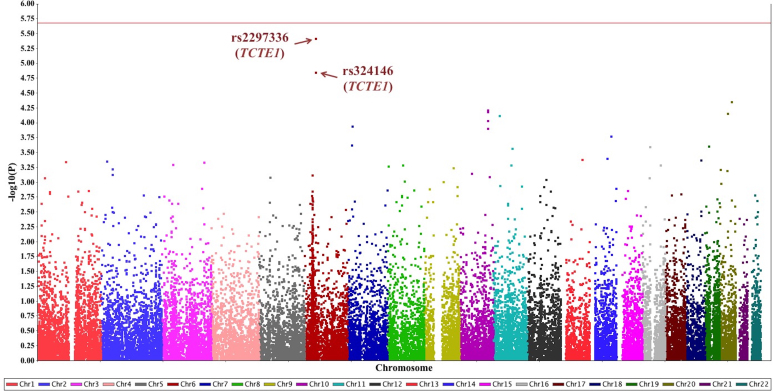
We tested 34,976 variants for their individual effect on spherical equivalent (Figure 1). No variant reached significance (p<2.1 × 10−6). However, suggestive associations were identified with two variants on chromosome 6p21.1 in the TCTE1 gene (OMIM 186975) region (Appendix 2). A missense SNP, rs2297336 (MAF = 14.1%), causes a phenylalanine to serine substitution (p.Phe261Ser). Each copy of the minor allele G of rs2297336 was associated with a 0.62 D (95% CI = [–0.88, −0.36], p = 3.7 × 10−6) decrease in spherical equivalent. Functional prediction programs predict this variant to be “deleterious” (SIFT [49]) and “probably damaging” (PolyPhen [50]) indicating this variant is likely to have a functional impact. This variant is within a transcription factor (TF) binding site [51]. In addition, a missense SNP rs324146 (p.Pro35Leu, MAF = 16.9%) in LD with rs2297336 (r2 = 0.80) in this study sample also had suggestive evidence of association. Each additional copy of the minor allele G of rs324146 was associated with a 0.55 D lower spherical equivalent (95% CI = [–0.79, −0.30], p = 1.4 × 10−5).
Figure 1.
Manhattan plot for the single-variant association results. A total of 34,976 variants with minor allele frequency (MAF) ≥1.0% were tested for single-variant associations. The significance threshold across the exome was set to p<2.1×10−6 (red line). Suggestive associations were discovered on two variants in the TCTE1 gene region at 6p21.1: rs2297336 (p = 3.7 ×10−6) and rs324146 (p = 1.4 ×10−5).
A total of 11,571 gene regions were tested in the gene-based analysis. In these analyses, five gene regions showed significant (p<4.3 × 10−6) evidence of association (Table 2, Appendix 3) with spherical equivalent. The most significant association was with the FSCB gene (OMIM 611779) on chromosome 14q21.2 (p = 1.5 × 10−7). Six protein-altering (one stop-gain and five missense) variants contributed to the gene-based finding.
Table 2. Significant gene-based association results (p<4.3 × 10−6).
| Gene | Locationa | Best Test | P value | No. of variants | Amino Acid Alteration (N)b |
|---|---|---|---|---|---|
|
PTCHD2 |
1p36.22 |
SKAT |
3.6 × 10−7 |
4 |
p.Arg260Pro(2), p.Arg542His(16),
p.Pro1034=(1), p.Val1178Ile(2) |
|
CRISP3 |
6p12.3 |
SKAT |
4.3 × 10−6 |
3 |
p.Pro197Thr(3), p.Gly141Ala(2),
p.Arg103Gln(1) |
|
NAP1L4 |
11p15.5 |
SKAT |
3.6 × 10−6 |
2 |
p.Ala369Val(1),
p.Thr111Ile(2) |
|
FSCB |
14q21.2 |
SKAT |
1.5 × 10−7 |
6 |
p.Val714Ile(2), p.Ala392Thr(8), p.Ala310Val(1),
p.Ala281Val(19), p.Lys251Arg(1), p.Gln74Ter(8) |
| AP3B2 | 15q25.2 | Burden | 1.6 × 10−7 | 3 | p.Ser946Thr(3), p.Lys880Arg(1), p.Lys262Thr(2) |
aGene locations reported in GRCh37. bAmino acid alteration reported in dbSNP. n=no. of minor allele copies in the analyzed cohort.
We identified four additional gene regions with significant association with spherical equivalent: the AP3B2 gene (OMIM 602166) on chromosome 15q25.2 (p = 1.6 × 10−7), the PTCHD2 gene (OMIM 611251) on chromosome 1p36.22 (p = 3.6 × 10−7), the NAP1L4 gene (OMIM 601651) on chromosome 11p15.5 (p = 3.6 × 10−6), and the CRISP3 gene on chromosome 6p12.3 (p = 4.3 × 10−6). Although the SKAT and the burden test supported a significant association between rare variants in the AP3B2 gene and quantitative refraction, the p value was more significant in the burden test, possibly suggesting that the variants included in the test influence spherical equivalent in the same direction and with similar magnitudes. Although these findings are intriguing, due to the low frequency of rare variants in these four genes (Table 2, Appendix 3), the observed associations were driven by a small number of observations and should be interpreted with caution.
The exome array contained either the exact variant or at least one variant in strong LD for eight previously reported GWAS loci: SIX6 (OMIM 606326) [30], GJD2 (OMIM 607058) [30,31,34], RASGRF1 (OMIM 606600) [30,32,34], CNDP2 (OMIM 169800) [30], PDE11A (OMIM 604961) [34], PRSS56 (OMIM 613858) [30,34], 4q25 [24], and MIPEP (OMIM 602241) [26]. Variants of GJD2, RASGRF1, and PRSS56 had been previously associated with spherical equivalent and myopia. In our analysis, we had evidence of replication for two of these eight regions (Table 3). We replicated the association of rs634990 located 37.2 kb upstream of the GJD2 gene on chromosome 15q14 with spherical equivalent (Appendix 4). In the Beaver Dam Eye study, the minor allele C of rs634990 was associated with a lower spherical equivalent (MAF = 47%, β = –0.29, 95% CI = [–0.48, −0.11], p = 1.8 × 10−3). In the comparison between myopic and hyperopic individuals, the odds ratio (OR) of having myopia was 1.57 greater (95% confidence interval (CI) = [1.15, 2.15], p = 4.5 × 10−3) for those carrying one copy of the C allele of rs634990 and 1.85 greater (95% CI = [1.27, 2.70], p = 1.4 × 10−3) for the homozygous C carriers. As extreme phenotype samples have been shown to increase power in genetic association studies [52,53], when emmetropic individuals were included in the control group, the association of rs634990 with myopia became smaller and marginally significant (heterozygous carriers: OR = 1.36 [1.03, 1.78], p = 0.03; homozygous carriers: OR = 1.48 [1.07, 2.04], p = 0.02). In addition, rs1550094 on chromosome 2q37.1 in the PRSS56 gene region was significantly associated with spherical equivalent (Appendix 4). Each copy of the minor allele G of rs1550094 on average reduced spherical equivalent by 0.33 D (MAF = 31%, 95% CI = [–0.53, −0.12], p = 1.7 × 10−3). Although rs1550094 was associated with myopia in the analysis of myopia versus hyperopia, this association was not statistically significant for the homozygous carriers (OR = 1.28 [0.80, 2.05], p = 0.30), only for the heterozygous carriers (OR = 1.44, 95% CI = [1.09, 1.91], p = 0.01). A similar trend was observed in the comparison of the myopia versus non-myopia (heterozygous carriers: OR = 1.41 [1.10, 1.79], p = 5.8 × 10−3; homozygous carriers: OR = 1.33 [0.88, 2.00], p = 0.17).
Table 3. Association results for genetic variants in published GWAS loci for spherical equivalent or myopia.
| Loci |
Gene |
GWAS variantsb |
GWAS effect size |
Tested variant (r2)c |
MAF |
β (SE) |
P value |
|---|---|---|---|---|---|---|---|
| Spherical equivalent | |||||||
| 14q23.1 |
SIX6 [30] |
rs1254319 |
−0.09 |
rs1254319 (1.00) |
29% [A] |
−0.17 (0.10) |
>0.01 |
| 15q14 |
GJD2 [31] |
rs634990 |
−0.23 |
rs634990 (1.00) |
47% [C] |
−0.29 (0.09) |
1.8 × 10−3 |
| 15q24.2 |
RASGRF1 [32] |
rs939658 |
−0.15 |
rs939658 (1.00) |
50% [A] |
−0.04 (0.10) |
>0.01 |
| 18q22.3 | CNDP2 [30] | rs12971120 | 0.11 | rs2278161 (1.00) | 22% [C] | 0.14 (0.12) | >0.01 |
| Myopia | |||||||
|---|---|---|---|---|---|---|---|
| 2q31.2 |
PDE11A [34] |
rs17400325 |
1.14d |
rs17400325 (1.00) |
3% [C] |
−0.15 (0.28) |
>0.01 |
| 2q37.1 |
PRSS56 [34] |
rs1550094 |
1.09d |
rs1550094 (1.00) |
31% [G] |
−0.33 (0.10) |
1.7 × 10−3 |
| 4q25 |
4q25 [24] |
rs10034228 |
0.81e |
rs10034228 (1.00) |
31% [C] |
−0.02 (0.10) |
>0.01 |
| 13q12 | MIPEP [26] | rs9318086 | 1.64/1.32f | rs9318086 (1.00) | 44% [A] | −0.03 (0.10) | >0.01 |
aLocations are reported in GRCh37 coordinates. bThe index variant reported in published GWAS. cExact variant or top variant in strong LD with the reported GWAS variant (r2 >0.80 in 1000 Genome Pilot 1 CEU population). dHazard ratio. eAllelic odds ratio. fHomozygous/Heterozygous odds ratio.
Discussion
In this study, we performed an exome array analysis on 1,560 BDES participants to identify rare and low-frequency variants associated with spherical equivalent. We discovered novel associations with rare variants in five gene regions, PTCHD2, CRISP3, NAP1L4, FSCB, and AP3B2, and suggestive associations with common variants in TCTE1. In addition, we replicated the previously reported association of GJD2 with spherical equivalent and myopia and discovered an association of spherical equivalent on PRSS56, a locus that had been previously identified for myopia.
The rare variants in the identified genes may not explain much of the phenotypic variation in quantitative refraction in the general population due to their low frequencies. However, the novel loci associated with spherical equivalent may not only be of great influence in a small number of families that harbor these rare variants but also provide valuable insights into novel genes and pathways involved in the etiology of refractive errors. The AP3B2 gene is highly expressed in adult RPE cells [54,55]. The AP3B2 protein is a subunit of the neuron-specific AP3 complex involved in neuronal membrane trafficking via mediating the direct transport of lysosomal proteins from the trans-Golgi network to the lysosomes and lysosome-related organelles [56,57]. A recent study found binding activity between the AP3B2 protein and a membrane scaffold protein, 4.1G, whose deficiency can lead to mislocalization of photoreceptor terminals and visual acuity impairments in mice [58]. The AP3B2 gene has been implicated in the lysosome pathway [59-62]. At the significance level of 0.01, we also found evidence of association with two other genes in the lysosome pathway in the gene-based analysis: SLC17A5 (OMIM 604322) on chromosome 6q13 (p = 2.0 × 10−3) and MFSD8 (OMIM 611124) on chromosome 4q28.2 (p = 2.7 × 10−3; Appendix 5). These findings may suggest the potential involvement of one or more genes of the lysosome pathway in the development of refractive errors.
The gene PTCHD2 on chromosome 1p36.22 is located in the myopia locus 14 (MYP14, OMIM 610320), a known linkage region for myopia and spherical equivalent [63,64]. PTCHD2 is also overlapped with a linkage region for the Volkmann type of cataract (CTRCT8, OMIM 115655) and a locus for primary infantile glaucoma (GLC3B, OMIM 600975), suggesting its potential role in the development of several ocular conditions. This gene is highly expressed in retinal ganglion cells [54]. The PTCHD2 protein interacts with several other proteins in the Hedgehog signaling pathway and may be involved in regulation of lipid transport and cholesterol homeostasis [65,66].
The genes FSCB, NAP1L4, and CRISP3 are all expressed in the retina [54] but do not have a clear mechanism by which they may influence spherical equivalent. The product of the FSCB gene is colocalized with a calcium-binding protein, CABYR, in the mouse testis, and was postulated to be involved in the later stages of fibrous sheath assembly [67]. Alterations of the FSCB gene have also been reported in patients with osteosarcoma [68] and in families with cerebral palsy [69]. The NAP1L4 gene encodes a member of the nucleosome assembly protein (NAP) family. This protein is a highly conserved histone chaperone protein that regulates the histone H2A-H2B heterodimer and mediates nucleosome formation [70,71]. This gene is located near the imprinted gene domain of 11p15.5, an important tumor-suppressor gene region that has been linked to growth-related disorders such as Beckwith-Wiedemann syndrome and various human cancers [72]. The CRISP3 gene is widely distributed in tissues with an exocrine function [73] and is suggested to play an role in the pathogenesis of Sjögren’s syndrome [74,75] and prostate cancer [76-79].
To further infer the potential mechanisms of PTCHD2, CRISP3, NAP1L4, FSCB, and AP3B2 gene underlying refractive errors, we identified genes that were predicted with strong evidence (confidence score >0.70) to be functional partners of these five genes from the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database [80]. Due to the limits of the exon array, not all pathway members were included among the 11,571 genes in our gene-based analysis. For those tested, we found no association at the significance level of 0.01 (Appendix 6), suggesting that these five genes may contribute to the development of refractive error either independently or through other functional partners that have yet to be identified. As refractive errors often develop due to incorrect morphology (i.e., incorrect axial length), further studies are necessary to understand how variation in these genes may influence ocular growth and development.
Our results contribute to an increasing body of evidence that variants in the GJD2 gene are associated with spherical equivalent. Multiple SNPs in vicinity of the GJD2 gene on chromosome 15q14 have been reported to be associated with refractive error and myopia. rs634990 was first discovered as a susceptibility locus for spherical equivalent in a Dutch population-based study (MAF = 47%, β = –0.25, p = 1.03 × 10−7) [31]. This association was later replicated by the Blue Mountain Eye Study (BMES) [81] and the Consortium for Refractive Error and Myopia (CREAM) [82], as well as in this analysis of the BDES cohort. This variant, rs634990, is also associated with myopia. When we compared the myopes to hyperopes, as was reported by Verhoeven et al. (OR = 1.88 [1.64, 2.16], p<0.001 for homozygous carriers; OR = 1.33 [1.19. 1.49], p<0.001 for the heterozygous carriers) [82], our findings were consistent with this previous study, confirming the association of rs634990 with myopia and indicated a protective effect of its C allele on hyperopia. We did not have data on rs524952, a locus that is strongly correlated with rs634990 (r2 = 0.90) and associated with spherical equivalent [30,81,82] and myopia [34,83] in European populations. Associations of GJD2 variants with refractive errors and myopia have also been replicated in studies conducted in Asian populations [84,85]. GJD2 encodes a member of the connexin family of proteins (hCx36) that form hexameric intercellular gap junction channels. Expression of the GJD2 gene has been found in the inner and outer plexiform layers of the retina in mice, rats, and humans [86]. Although the function of GJD2 in the human retina remains unknown, animal studies suggest an essential role for gap junction channels containing Cx36 (counterpart protein of hCx36 in mice) in the transmission of visual signals through photoreceptor cells, particularly the rod-cone pathways [87]. Disruption of Cx36 expression in mice can cause visual transmission deficits [88]. However, further investigation is necessary to understand how variation in the GJD2 gene influences spherical equivalent.
Our results support the potential involvement of PRSS56 in spherical equivalent. Associations with spherical equivalent or myopia have been found for two SNPs (rs1656404 and rs1550094) in the PRSS56 gene region. We did not have data on rs1656404, a locus that was first reported for spherical equivalent (MAF = 21.0%, β = –0.151, p = 2.38 × 10−9) in a multiancestry cohort [30] and later for myopia [83]. However, rs1550094 was discovered for myopia development (i.e., age of onset) in the GWAS of 23andMe participants of European ancestry (HR = 1.09 [1.07–1.11], MAF = 30.5%, p = 1.3 × 10−15) [34] and is associated with spherical equivalent and myopia in the present study. Due to the low correlation (r2<0.50) between these two SNPs, rs1550094 and rs1656404 were independently associated with a decreased spherical equivalent. Experimental evidence also supports a potential role for PRSS56 in controlling spherical equivalent. The PRSS56 gene encodes a trypsin-like serine peptidase expressed in the human retina, cornea, sclera, and optic nerve [89]. Mutations of the PRSS56 gene cause autosomal recessive posterior microphthalmos and familial nanophthalmos, two rare ocular disorders characterized by abnormally small eyes and severe hyperopia due to reduced axial length [89,90]. In animal studies, mutations of the Mfrp gene caused a progressive accumulation of Prss56 throughout postnatal development and together led to shortening of photoreceptor outer segments and retinal degeneration [91].
We employed an extreme value sampling strategy to maximize the phenotypic differences among individuals. This could lead to the oversampling of variants that are more common among individuals with extreme trait values and thus increase the statistical power of our study. In this study, we directly examined coding variants for association with spherical equivalent. We applied single-variant analysis and gene-based analysis to optimize the power to detect associations for low-frequency and rare variants. However, the exome array was designed using exome sequencing data from ethnically diverse populations. Given that our study population is ethnically homogenous, more than 50% of the genotyped variants were monomorphic, and exclusion of these variants resulted in the mean coverage of six variants in each gene region. This low coverage can be particularly influential when variants in a gene region are mostly singletons or doubletons, resulting in an inflated association driven by a limited number of extreme observations. Therefore, we are cautious with the interpretation of such genes in our gene-based analysis. In addition, variants in sequencing data have to reach certain frequencies to be selected into the exome array. Thus, there is a lack of coverage in the exome array of extremely rare variants, which prohibited us from examining these variants for association with quantitative refraction.
Additional studies focused on assessing the role of low-frequency coding variants across diverse populations are necessary to further validate the reported findings. Such studies would complement the existing GWAS that highlight common variations, and together, these approaches may begin to elucidate the causal alleles and the underlying mechanisms for refractive errors. A better understating of the genetics of spherical equivalent, in combination with the interaction of environmental exposure, may be useful in determining individual risks for myopia and hyperopia and lead to novel or improved measures of prevention, management, and treatment.
Acknowledgments
Grant Information: Research reported in this publication was supported by the National Eye Institute of the National Institutes of Health under award number R01EY021531 and U10006594, and the Burroughs-Wellcome Fund Maryland: Genetics, Epidemiology and Medicine Training Grant.
Appendix 1. Spherical equivalent measurements in the Beaver Dam Eye Study samples
(A) The average of spherical equivalent in left and right eyes at baseline visit was distributed with a mean of -0.20 D and a standard deviation of 2.9 D (B) The residual spherical equivalent after adjusting for age, sex, years of education, nuclear sclerosis and the first two principal components was distributed with a mean of 0 D and a standard deviation of 2.7 D. (C) Compared to the full BDES cohort, the distribution of spherical equivalent in the analyzed cohort was more heavy-tailed. To access the data, click or select the words “Appendix 1.”
Appendix 2. Box plots of spherical equivalent by genotypes at rs2297336 and rs324146 of TCTE1
The distributions of spherical equivalent residuals were plotted for each observed genotype at rs2297336 (left) and rs324146 (right) of TCTE1 on chromosome 6p21.1. Genotypes are showed on x-axis. The mean spherical equivalent residuals for each genotype is marked by a dark red diamond with the value listed above. The three horizontal bars from the bottom to the top indicate the 25th, 50th and 75th percentile of the spherical equivalent residuals. Observations beyond the 25th and 75th percentile range are represented by black dots. For both SNPs, each copy of its minor allele is associated with decreased spherical equivalent. To access the data, click or select the words “Appendix 2.”
Appendix 3. Distributions of spherical equivalent for five identified genes
(Continue) Five gene regions reached the significant threshold in the gene-based analysis (p < 4.3 × 10-6). Spherical equivalent residuals were plotted by the presence of the minor allele of each variant in the gene region. Majority of study samples are homozygotes for major alleles (Red). The remaining groups are individuals with one copy of the minor allele at the variant indicated on the X-axis (Black). The dash line represents the group mean of spherical equivalent residuals. No homozygote or compound heterozygous of the minor allele was seen for these genes. To access the data, click or select the words “Appendix 3.”
Appendix 4. Box plots of spherical equivalent by genotypes at rs634990 of GJD2 and rs1550094 of PRSS56
The distributions of spherical equivalent residuals were plotted for each observed genotype at rs634990 of GJD2 (left) and rs150094 of PRSS56 (right). After adjusting for age, sex, years of education, nuclear sclerosis and the first two principal components, (A) each copy of the minor allele C of rs634990 is associated with a 0.29 D decrease in spherical equivalent (p = 1.8 × 10-3), and (B) the minor allele G of rs150094 is associated with a 0.33 D decrease in spherical equivalent (p = 1.7 × 10-3). To access the data, click or select the words “Appendix 4.”
Appendix 5. Gene-based association results for genes in lysosome pathways
a Genes in Lysosome pathway were identified from KEGG (the Kyoto Encyclopedia of Genes and Genomes) database. bGenes reached significant threshold at p < 0.01 were highlighted in bold. To access the data, click or select the words “Appendix 5.”
Appendix 6. Gene-based association results for predicted functional partners of PTCHD2, CRISP3, NAP1L4, FSCB, and AP3B2
aGenes in the STRING database with strong evidence (confidence score > 0.70) of protein-protein interactions with reported genes. The number in the parentheses indicates the proportion of genes tested in our gene-based analysis. bThe number in parentheses represents the number of genes out of the total number of genes identified as functional partners that were tested in our gene-based analysis. To access the data, click or select the words “Appendix 6.”
References
- 1.Wong TY, Zheng Y, Jonas JB, Flaxman SR, Keeffe J, Leasher J, Naidoo K, Pesudovs K, Price H. White R a, Resnikoff S, Taylor HR, Bourne RR a. Prevalence and causes of vision loss in East Asia: 1990–2010. Br J Ophthalmol. 2014;98:599–604. doi: 10.1136/bjophthalmol-2013-304047. [DOI] [PubMed] [Google Scholar]
- 2.Saw S-M, Gazzard G, Shih-Yen EC, Chua W-H. Myopia and associated pathological complications. Ophthalmic Physiol Opt. 2005;25:381–91. doi: 10.1111/j.1475-1313.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 3.Verhoeven VJ, Wong KT, Buitendijk GH, Hofman A, Vingerling JR, Klaver CC. Visual consequences of refractive errors in the general population. Ophthalmology. 2015;122:101–9. doi: 10.1016/j.ophtha.2014.07.030. [DOI] [PubMed] [Google Scholar]
- 4.Vitale S, Ellwein L, Cotch MF, Ferris FL, Sperduto R. Prevalence of refractive error in the United States, 1999–2004. Arch Ophthalmol. 2008;126:1111–9. doi: 10.1001/archopht.126.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.French AN, Morgan IG, Mitchell P, Rose KA. Risk factors for incident myopia in Australian schoolchildren: the Sydney adolescent vascular and eye study. Ophthalmology. 2013;120:2100–8. doi: 10.1016/j.ophtha.2013.02.035. [DOI] [PubMed] [Google Scholar]
- 6.Williams C, Miller LL, Gazzard G, Saw SM. A comparison of measures of reading and intelligence as risk factors for the development of myopia in a UK cohort of children. Br J Ophthalmol. 2008;92:1117–21. doi: 10.1136/bjo.2007.128256. [DOI] [PubMed] [Google Scholar]
- 7.Saw S-M, Shankar A, Tan S-B, Taylor H, Tan DTH, Stone RA, Wong T-Y. A cohort study of incident myopia in Singaporean children. Invest Ophthalmol Vis Sci. 2006;47:1839–44. doi: 10.1167/iovs.05-1081. [DOI] [PubMed] [Google Scholar]
- 8.Saw S-M, Tan S-B, Fung D, Chia K-S, Koh D, Tan DTH, Stone RA. IQ and the association with myopia in children. Invest Ophthalmol Vis Sci. 2004;45:2943–8. doi: 10.1167/iovs.03-1296. [DOI] [PubMed] [Google Scholar]
- 9.Jones-Jordan LA, Mitchell GL, Cotter SA, Kleinstein RN, Manny RE, Mutti DO, Twelker JD, Sims JR, Zadnik K. Visual activity before and after the onset of juvenile myopia. Invest Ophthalmol Vis Sci. 2011;52:1841–50. doi: 10.1167/iovs.09-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guggenheim JA, Northstone K, McMahon G, Ness AR, Deere K, Mattocks C, Pourcain BS, Williams C. Time outdoors and physical activity as predictors of incident myopia in childhood: a prospective cohort study. Invest Ophthalmol Vis Sci. 2012;53:2856–65. doi: 10.1167/iovs.11-9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirani M, Tong L, Gazzard G, Zhang X, Chia A, Young TL, Rose KA, Mitchell P, Saw S-M. Outdoor activity and myopia in Singapore teenage children. Br J Ophthalmol. 2009;93:997–1000. doi: 10.1136/bjo.2008.150979. [DOI] [PubMed] [Google Scholar]
- 12.Zadnik K, Sinnott LT, Cotter SA, Jones-Jordan LA, Kleinstein RN, Manny RE, Twelker JD, Mutti DO. Prediction of Juvenile-Onset Myopia. JAMA Ophthalmol. 2015;133:683–9. doi: 10.1001/jamaophthalmol.2015.0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramamurthy D, Lin Chua SY, Saw S-M. A review of environmental risk factors for myopia during early life, childhood and adolescence. Clin Exp Optom. 2015 doi: 10.1111/cxo.12346. [DOI] [PubMed] [Google Scholar]
- 14.Teikari JM, Kaprio J, Koskenvuo MK, Vannas A. Heritability estimate for refractive errors–a population-based sample of adult twins. Genet Epidemiol. 1988;5:171–81. doi: 10.1002/gepi.1370050304. [DOI] [PubMed] [Google Scholar]
- 15.Wojciechowski R, Congdon N, Bowie H, Munoz B, Gilbert D, West SK. Heritability of refractive error and familial aggregation of myopia in an elderly American population. Invest Ophthalmol Vis Sci. 2005;46:1588–92. doi: 10.1167/iovs.04-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dirani M, Chamberlain M, Shekar SN, Islam AFM, Garoufalis P, Chen CY, Guymer RH, Baird PN. Heritability of refractive error and ocular biometrics: the Genes in Myopia (GEM) twin study. Invest Ophthalmol Vis Sci. 2006;47:4756–61. doi: 10.1167/iovs.06-0270. [DOI] [PubMed] [Google Scholar]
- 17.Peet JA Cotch M-F, Wojciechowski R, Bailey-Wilson JE, Stambolian D. Heritability and familial aggregation of refractive error in the Old Order Amish. Invest Ophthalmol Vis Sci. 2007;48:4002–6. doi: 10.1167/iovs.06-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CY-C, Scurrah KJ, Stankovich J, Garoufalis P, Dirani M, Pertile KK, Richardson AJ, Mitchell P, Baird PN. Heritability and shared environment estimates for myopia and associated ocular biometric traits: the Genes in Myopia (GEM) family study. Hum Genet. 2007;121:511–20. doi: 10.1007/s00439-006-0312-0. [DOI] [PubMed] [Google Scholar]
- 19.Paget S, Vitezica ZG, Malecaze F, Calvas P. Heritability of refractive value and ocular biometrics. Exp Eye Res. 2008;86:290–5. doi: 10.1016/j.exer.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Lopes MC, Andrew T, Carbonaro F, Spector TD, Hammond CJ. Estimating heritability and shared environmental effects for refractive error in twin and family studies. Invest Ophthalmol Vis Sci. 2009;50:126–31. doi: 10.1167/iovs.08-2385. [DOI] [PubMed] [Google Scholar]
- 21.Klein A, Suktitipat B. Heritability analysis of spherical equivalent, axial length, corneal curvature, and anterior chamber depth in the Beaver Dam Eye Study. Arch. 2009;127:649–55. doi: 10.1001/archophthalmol.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hawthorne F a Young TL. Genetic contributions to myopic refractive error: Insights from human studies and supporting evidence from animal models. Exp Eye Res. Elsevier Ltd. 2013;114:141–9. doi: 10.1016/j.exer.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 23.Nakanishi H, Yamada R, Gotoh N, Hayashi H, Yamashiro K, Shimada N, Ohno-Matsui K, Mochizuki M, Saito M, Iida T, Matsuo K, Tajima K, Yoshimura N, Matsuda F. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;5:e1000660. doi: 10.1371/journal.pgen.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Qu J, Xu X, Zhou X, Zou H, Wang N, Li T, Hu X, Zhao Q, Chen P, Li W, Huang K, Yang J, He Z, Ji J, Wang T, Li J, Li Y, Liu J, Zeng Z, Feng G, He L, Shi Y. A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum Mol Genet. 2011;20:2861–8. doi: 10.1093/hmg/ddr169. [DOI] [PubMed] [Google Scholar]
- 25.Li Y-J, Goh L, Khor C-C, Fan Q, Yu M, Han S, Sim X, Ong RT-H, Wong T-Y, Vithana EN, Yap E, Nakanishi H, Matsuda F, Ohno-Matsui K, Yoshimura N, Seielstad M, Tai E-S, Young TL, Saw S-M. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011;118:368–75. doi: 10.1016/j.ophtha.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Qu J, Zhang D, Zhao P, Zhang Q, Tam POS, Sun L, Zuo X, Zhou X, Xiao X, Hu J, Li Y, Cai L, Liu X, Lu F, Liao S, Chen B, He F, Gong B, Lin H, Ma S, Cheng J, Zhang J, Chen Y, Zhao F, Yang X, Chen Y, Yang C, Lam DSC, Li X, Shi F, Wu Z, Lin Y, Yang J, Li S, Ren Y, Xue A, Fan Y, Li D, Pang CP, Zhang X, Yang Z. Genetic variants at 13q12.12 are associated with high myopia in the Han Chinese population. Am J Hum Genet. 2011;88:805–13. doi: 10.1016/j.ajhg.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan Q, Barathi VA, Cheng C-Y, Zhou X, Meguro A, Nakata I, Khor C-C, Goh L-K, Li Y-J, Lim W, Ho CEH, Hawthorne F, Zheng Y, Chua D, Inoko H, Yamashiro K, Ohno-Matsui K, Matsuo K, Matsuda F, Vithana E, Seielstad M, Mizuki N, Beuerman RW, Tai E-S, Yoshimura N, Aung T, Young TL, Wong T-Y, Teo Y-Y, Saw S-M. Genetic variants on chromosome 1q41 influence ocular axial length and high myopia. PLoS Genet. 2012;8:e1002753. doi: 10.1371/journal.pgen.1002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Gong B, Chen L, Zuo X, Liu X, Tam POS, Zhou X, Zhao P, Lu F, Qu J, Sun L, Zhao F, Chen H, Zhang Y, Zhang D, Lin Y, Lin H, Ma S, Cheng J, Yang J, Huang L, Zhang M, Zhang X, Pang CP, Yang Z. A genome-wide meta-analysis identifies two novel loci associated with high myopia in the Han Chinese population. Hum Mol Genet. 2013;22:2325–33. doi: 10.1093/hmg/ddt066. [DOI] [PubMed] [Google Scholar]
- 29.Khor CC, Miyake M, Chen LJ, Shi Y, Barathi VA, Qiao F, Nakata I, Yamashiro K, Zhou X, Tam POS, Cheng C-Y, Tai ES, Vithana EN, Aung T, Teo Y-Y, Wong T-Y, Moriyama M, Ohno-Matsui K, Mochizuki M, Matsuda F, Yong RYY, Yap EPH, Yang Z, Pang CP, Saw S-M, Yoshimura N. Genome-wide association study identifies ZFHX1B as a susceptibility locus for severe myopia. Hum Mol Genet. 2013;22:5288–94. doi: 10.1093/hmg/ddt385. [DOI] [PubMed] [Google Scholar]
- 30.Verhoeven VJM, Hysi PG, Wojciechowski R, Fan Q, Guggenheim JA, Höhn R, MacGregor S, Hewitt AW, Nag A, Cheng C-Y, Yonova-Doing E, Zhou X, Ikram MK, Buitendijk GHS, McMahon G, Kemp JP, Pourcain BS, Simpson CL, Mäkelä K-M, Lehtimäki T, Kähönen M, Paterson AD, Hosseini SM, Wong HS, Xu L, Jonas JB, Pärssinen O, Wedenoja J, Yip SP, Ho DWH, Pang CP, Chen LJ, Burdon KP, Craig JE, Klein BEK, Klein R, Haller T, Metspalu A, Khor C-C, Tai E-S, Aung T, Vithana E, Tay W-T, Barathi VA, Chen P, Li R, Liao J, Zheng Y, Ong RT, Döring A, Evans DM, Timpson NJ, Verkerk AJMH, Meitinger T, Raitakari O, Hawthorne F, Spector TD, Karssen LC, Pirastu M, Murgia F, Ang W, Mishra A, Montgomery GW, Pennell CE, Cumberland PM, Cotlarciuc I, Mitchell P, Wang JJ, Schache M, Janmahasatian S, Janmahasathian S, Igo RP, Lass JH, Chew E, Iyengar SK, Gorgels TGMF, Rudan I, Hayward C, Wright AF, Polasek O, Vatavuk Z, Wilson JF, Fleck B, Zeller T, Mirshahi A, Müller C, Uitterlinden AG, Rivadeneira F, Vingerling JR, Hofman A, Oostra BA, Amin N, Bergen AAB, Teo Y-Y, Rahi JS, Vitart V, Williams C, Baird PN, Wong T-Y, Oexle K, Pfeiffer N, Mackey DA, Young TL, van Duijn CM, Saw S-M, Bailey-Wilson JE, Stambolian D, Klaver CC, Hammond CJ. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet. 2013;45:314–8. doi: 10.1038/ng.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solouki AM, Verhoeven VJM, van Duijn CM, Verkerk AJMH, Ikram MK, Hysi PG, Despriet DDG, van Koolwijk LM, Ho L, Ramdas WD, Czudowska M. Kuijpers RW a M, Amin N, Struchalin M, Aulchenko YS, van Rij G, Riemslag FCC, Young TL, Mackey D a, Spector TD, Gorgels TGMF, Willemse-Assink JJM, Isaacs A, Kramer R, Swagemakers SM a, Bergen A a B, van Oosterhout A a LJ, Oostra B a, Rivadeneira F, Uitterlinden AG, Hofman A, de Jong PTVM, Hammond CJ, Vingerling JR, Klaver CCW. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. Nature Publishing Group. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hysi PG, Young TL. Mackey D a, Andrew T, Fernández-Medarde A, Solouki AM, Hewitt AW, Macgregor S, Vingerling JR, Li Y-J, Ikram MK, Fai LY, Sham PC, Manyes L, Porteros A, Lopes MC, Carbonaro F, Fahy SJ, Martin NG, van Duijn CM, Spector TD, Rahi JS, Santos E, Klaver CCW, Hammond CJ. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42:902–5. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng W, Butterworth J, Bradley DT, Hughes AE, Soler V, Calvas P, Malecaze F. A genome-wide association study provides evidence for association of chromosome 8p23 (MYP10) and 10q21.1 (MYP15) with high myopia in the French Population. Invest Ophthalmol Vis Sci. 2012;53:7983–8. doi: 10.1167/iovs.12-10409. [DOI] [PubMed] [Google Scholar]
- 34.Kiefer AK, Tung JY, Do CB. Hinds D a, Mountain JL, Francke U, Eriksson N. Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS Genet. 2013;9:e1003299. doi: 10.1371/journal.pgen.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stambolian D, Wojciechowski R, Oexle K, Pirastu M, Li X, Raffel LJ, Cotch MF, Chew EY, Klein B, Klein R, Wong TY, Simpson CL, Klaver CCW, van Duijn CM, Verhoeven VJM, Baird PN, Vitart V, Paterson AD, Mitchell P, Saw SM, Fossarello M, Kazmierkiewicz K, Murgia F, Portas L, Schache M, Richardson A, Xie J, Wang JJ, Rochtchina E, Viswanathan AC, Hayward C, Wright AF, Polasek O, Campbell H, Rudan I, Oostra BA, Uitterlinden AG, Hofman A, Rivadeneira F, Amin N, Karssen LC, Vingerling JR, Hosseini SM, Döring A, Bettecken T, Vatavuk Z, Gieger C, Wichmann H-E, Wilson JF, Fleck B, Foster PJ, Topouzis F, McGuffin P, Sim X, Inouye M, Holliday EG, Attia J, Scott RJ, Rotter JI, Meitinger T, Bailey-Wilson JE. Meta-analysis of genome-wide association studies in five cohorts reveals common variants in RBFOX1, a regulator of tissue-specific splicing, associated with refractive error. Hum Mol Genet. 2013;22:2754–64. doi: 10.1093/hmg/ddt116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stambolian D. Genetic susceptibility and mechanisms for refractive error. Clin Genet. 2013;84:102–8. doi: 10.1111/cge.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. Nature Publishing Group. 2011;13:135–45. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, McLaren PJ, Gupta N, Sklar P, Sullivan PF, Moran JL, Hultman CM, Lichtenstein P, Magnusson P, Lehner T, Shugart YY, Price AL, de Bakker PIW, Purcell SM, Sunyaev SR. Exome sequencing and the genetic basis of complex traits. Nat Genet. Nature Publishing Group. 2012;44:623–30. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linton K, Klein B, Klein R. The validity of self-reported and surrogate-reported cataract and age-related macular degeneration in the Beaver Dam Eye Study. Am J. 1991;33:2224–8. doi: 10.1093/oxfordjournals.aje.a116049. [DOI] [PubMed] [Google Scholar]
- Early Treatment Diabetic Retinopathy Study Coordinating Center. Manual of Operations. Baltimore: Center, Diabetic Retinopathy Coordinating; 1980. [Google Scholar]
- 41.Klein BE, Klein R, Linton KL. Prevalence of age-related lens opacities in a population. The Beaver Dam Eye Study. Ophthalmology. 1992;99:546–52. doi: 10.1016/s0161-6420(92)31934-7. [DOI] [PubMed] [Google Scholar]
- 42.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 43.Klein AP, Duggal P, Lee KE, Cheng C-Y, Klein R, Bailey-Wilson JE, Klein BEK. Linkage analysis of quantitative refraction and refractive errors in the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 2011;52:5220–5. doi: 10.1167/iovs.10-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asimit J, Zeggini E. Rare variant association analysis methods for complex traits. Annu Rev Genet. 2010;44:293–308. doi: 10.1146/annurev-genet-102209-163421. [DOI] [PubMed] [Google Scholar]
- 46.Li B, Leal S. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–21. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. The American Society of Human Genetics. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kähäri AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SMJ, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. Ensembl 2015. Nucleic Acids Res. 2014;43(D1):D662–9. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schork NJ, Nath SK, Fallin D, Chakravarti A. Linkage disequilibrium analysis of biallelic DNA markers, human quantitative trait loci, and threshold-defined case and control subjects. Am J Hum Genet. 2000;67:1208–18. doi: 10.1086/321201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li D, Lewinger JP, Gauderman WJ, Murcray CE, Conti D. Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genet Epidemiol. 2011;35:790–9. doi: 10.1002/gepi.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar R, Mazor Y, Rinon A, Blumenthal J, Golan Y, Buzhor E, Livnat I, Ben-Ari S, Lieder I, Shitrit A, Gilboa Y, Ben-Yehudah A, Edri O, Shraga N, Bogoch Y, Leshansky L, Aharoni S, West MD, Warshawsky D, Shtrichman R. LifeMap DiscoveryTM: the embryonic development, stem cells, and regenerative medicine research portal. PLoS One. 2013;8:e66629. doi: 10.1371/journal.pone.0066629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faúndez V, Horng JT, Kelly RB. A function for the AP3 coat complex in synaptic vesicle formation from endosomes. Cell. 1998;93:423–32. doi: 10.1016/s0092-8674(00)81170-8. [DOI] [PubMed] [Google Scholar]
- 57.Grabner CP, Price SD, Lysakowski A, Cahill AL, Fox AP. Regulation of large dense-core vesicle volume and neurotransmitter content mediated by adaptor protein 3. Proc Natl Acad Sci USA. 2006;103:10035–40. doi: 10.1073/pnas.0509844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanuki R, Watanabe S, Sugita Y, Irie S, Kozuka T, Shimada M, Ueno S, Usukura J, Furukawa T. Protein-4.1G-Mediated Membrane Trafficking Is Essential for Correct Rod Synaptic Location in the Retina and for Normal Visual Function. Cell Rep. The Authors. 2015;10:796–808. doi: 10.1016/j.celrep.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 59.Dell’Angelica EC, Ohno H, Ooi CE, Rabinovich E, Roche KW, Bonifacino JS. AP-3: an adaptor-like protein complex with ubiquitous expression. EMBO J. 1997;16:917–28. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dell’Angelica EC, Klumperman J, Stoorvogel W, Bonifacino JS. Association of the AP-3 adaptor complex with clathrin. Science. 1998;280:431–4. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- 61.Faundez VV, Kelly RB. The AP-3 complex required for endosomal synaptic vesicle biogenesis is associated with a casein kinase Ialpha-like isoform. Mol Biol Cell. 2000;11:2591–604. doi: 10.1091/mbc.11.8.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivan V, Martinez-Sanchez E, Sima LE, Oorschot V, Klumperman J, Petrescu SM, van der Sluijs P. AP-3 and Rabip4′ coordinately regulate spatial distribution of lysosomes. PLoS One. 2012;7:e48142. doi: 10.1371/journal.pone.0048142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wojciechowski R, Moy C, Ciner E, Ibay G, Reider L, Bailey-Wilson JE, Stambolian D. Genomewide scan in Ashkenazi Jewish families demonstrates evidence of linkage of ocular refraction to a QTL on chromosome 1p36. Hum Genet. 2006;119:389–99. doi: 10.1007/s00439-006-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li YJ. Guggenheim J a., Bulusu A, Metlapally R, Abbott D, Malecaze F, Calvas P, Rosenberg T, Paget S, Creer RC, Kirov G, Owen MJ, Zhao B, White T, MacKey D a., Young TL. An international collaborative family-based whole-genome linkage scan for high-grade Myopia. Invest Ophthalmol Vis Sci. 2009;50:3116–27. doi: 10.1167/iovs.08-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zikova M, Corlett A, Bendova Z, Pajer P, Bartunek P. DISP3, a sterol-sensing domain-containing protein that links thyroid hormone action and cholesterol metabolism. Mol Endocrinol. 2009;23:520–8. doi: 10.1210/me.2008-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katoh Y, Katoh M. Identification and characterization of DISP3 gene in silico. Int J Oncol. 2005;26:551–6. [PubMed] [Google Scholar]
- 67.Li Y-F, He W, Jha KN, Klotz K, Kim Y-H, Mandal A, Pulido S, Digilio L, Flickinger CJ, Herr JC. FSCB, a novel protein kinase A-phosphorylated calcium-binding protein, is a CABYR-binding partner involved in late steps of fibrous sheath biogenesis. J Biol Chem. 2007;282:34104–19. doi: 10.1074/jbc.M702238200. [DOI] [PubMed] [Google Scholar]
- 68.Choy E, Hornicek F, MacConaill L, Harmon D, Tariq Z, Garraway L, Duan Z. High-throughput genotyping in osteosarcoma identifies multiple mutations in phosphoinositide-3-kinase and other oncogenes. Cancer. 2012;118:2905–14. doi: 10.1002/cncr.26617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McMichael G, Girirajan S, Moreno-De-Luca A, Gecz J, Shard C, Nguyen LS, Nicholl J, Gibson C, Haan E, Eichler E, Martin CL, MacLennan A. Rare copy number variation in cerebral palsy. Eur J Hum Genet. Nature Publishing Group. 2014;22:40–5. doi: 10.1038/ejhg.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Okuwaki M, Kato K, Nagata K. Functional characterization of human nucleosome assembly protein 1-like proteins as histone chaperones. Genes Cells. 2010;15:13–27. doi: 10.1111/j.1365-2443.2009.01361.x. [DOI] [PubMed] [Google Scholar]
- 71.Rodriguez P, Munroe D, Prawitt D, Chu LL, Bric E, Kim J, Reid LH, Davies C, Nakagama H, Loebbert R, Winterpacht A, Petruzzi MJ, Higgins MJ, Nowak N, Evans G, Shows T, Weissman BE, Zabel B, Housman DE, Pelletier J. Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics. 1997;44:253–65. doi: 10.1006/geno.1997.4868. [DOI] [PubMed] [Google Scholar]
- 72.Crider-Miller SJ, Reid LH, Higgins MJ, Nowak NJ, Shows TB, Futreal PA, Weissman BE. Novel transcribed sequences within the BWS/WT2 region in 11p15.5: tissue-specific expression correlates with cancer type. Genomics. 1997;46:355–63. doi: 10.1006/geno.1997.5061. [DOI] [PubMed] [Google Scholar]
- 73.Krätzschmar J, Haendler B, Eberspaecher U, Roosterman D, Donner P, Schleuning WD. The human cysteine-rich secretory protein (CRISP) family. Primary structure and tissue distribution of CRISP-1, CRISP-2 and CRISP-3. Eur J Biochem. 1996;236:827–36. doi: 10.1111/j.1432-1033.1996.t01-1-00827.x. [DOI] [PubMed] [Google Scholar]
- 74.Laine M, Porola P, Udby L, Kjeldsen L, Cowland JB, Borregaard N, Hietanen J, Ståhle M, Pihakari A, Konttinen YT. Low salivary dehydroepiandrosterone and androgen-regulated cysteine-rich secretory protein 3 levels in Sjögren’s syndrome. Arthritis Rheum. 2007;56:2575–84. doi: 10.1002/art.22828. [DOI] [PubMed] [Google Scholar]
- 75.Porola P, Virkki L, Przybyla BD, Laine M, Patterson TA, Pihakari A, Konttinen YT. Androgen deficiency and defective intracrine processing of dehydroepiandrosterone in salivary glands in Sjögren’s syndrome. J Rheumatol. 2008;35:2229–35. doi: 10.3899/jrheum.080220. [DOI] [PubMed] [Google Scholar]
- 76.Asmann YW, Kosari F, Wang K, Cheville JC, Vasmatzis G. Identification of differentially expressed genes in normal and malignant prostate by electronic profiling of expressed sequence tags. Cancer Res. 2002;62:3308–14. [PubMed] [Google Scholar]
- 77.Kosari F, Asmann YW, Cheville JC, Vasmatzis G. Cysteine-rich secretory protein-3: a potential biomarker for prostate cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:1419–26. [PubMed] [Google Scholar]
- 78.Bjartell A, Johansson R, Björk T, Gadaleanu V, Lundwall A, Lilja H, Kjeldsen L, Udby L. Immunohistochemical detection of cysteine-rich secretory protein 3 in tissue and in serum from men with cancer or benign enlargement of the prostate gland. Prostate. 2006;66:591–603. doi: 10.1002/pros.20342. [DOI] [PubMed] [Google Scholar]
- 79.Ribeiro FR, Paulo P, Costa VL, Barros-Silva JD, Ramalho-Carvalho J, Jerónimo C, Henrique R, Lind GE, Skotheim RI, Lothe RA, Teixeira MR. Cysteine-rich secretory protein-3 (CRISP3) is strongly up-regulated in prostate carcinomas with the TMPRSS2-ERG fusion gene. PLoS One. 2011;6:e22317. doi: 10.1371/journal.pone.0022317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schache M, Richardson AJ, Mitchell P, Wang JJ, Rochtchina E, Viswanathan AC, Wong TY, Saw SM, Topouzis F, Xie J, Sim X, Holliday EG, Attia J, Scott RJ, Baird PN. Genetic association of refractive error and axial length with 15q14 but not 15q25 in the Blue Mountains Eye Study cohort. Ophthalmology. 2013;120:292–7. doi: 10.1016/j.ophtha.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 82.Verhoeven VJM, Hysi PG, Saw S-M, Vitart V, Mirshahi A, Guggenheim JA, Cotch MF, Yamashiro K, Baird PN, Mackey DA, Wojciechowski R, Ikram MK, Hewitt AW, Duggal P, Janmahasatian S, Khor C-C, Fan Q, Zhou X, Young TL, Tai E-S, Goh L-K, Li Y-J, Aung T, Vithana E, Teo Y-Y, Tay W, Sim X, Rudan I, Hayward C, Wright AF, Polasek O, Campbell H, Wilson JF, Fleck BW, Nakata I, Yoshimura N, Yamada R, Matsuda F, Ohno-Matsui K, Nag A, McMahon G, St Pourcain B, Lu Y, Rahi JS, Cumberland PM, Bhattacharya S, Simpson CL, Atwood LD, Li X, Raffel LJ, Murgia F, Portas L, Despriet DDG, van Koolwijk LME, Wolfram C, Lackner KJ, Tönjes A, Mägi R, Lehtimäki T, Kähönen M, Esko T, Metspalu A, Rantanen T, Pärssinen O, Klein BE, Meitinger T, Spector TD, Oostra BA, Smith AV. de Jong PTVM, Hofman A, Amin N, Karssen LC, Rivadeneira F, Vingerling JR, Eiríksdóttir G, Gudnason V, Döring A, Bettecken T, Uitterlinden AG, Williams C, Zeller T, Castagné R, Oexle K, van Duijn CM, Iyengar SK, Mitchell P, Wang JJ, Höhn R, Pfeiffer N, Bailey-Wilson JE, Stambolian D, Wong T-Y, Hammond CJ, Klaver CCW. Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Hum Genet. 2012;131:1467–80. doi: 10.1007/s00439-012-1176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simpson CL, Wojciechowski R, Oexle K, Murgia F, Portas L, Li X, Verhoeven VJM, Vitart V, Schache M, Hosseini SM, Hysi PG, Raffel LJ, Cotch MF, Chew E, Klein BEK, Klein R, Wong TY, van Duijn CM, Mitchell P, Saw SM, Fossarello M, Wang JJ, Polašek O, Campbell H, Rudan I. Oostra B a, Uitterlinden AG, Hofman A, Rivadeneira F, Amin N, Karssen LC, Vingerling JR, Döring A, Bettecken T, Bencic G, Gieger C, Wichmann H-E, Wilson JF, Venturini C, Fleck B, Cumberland PM, Rahi JS, Hammond CJ, Hayward C, Wright AF, Paterson AD, Baird PN, Klaver CCW, Rotter JI, Pirastu M, Meitinger T, Bailey-Wilson JE, Stambolian D. Genome-wide meta-analysis of myopia and hyperopia provides evidence for replication of 11 loci. PLoS One. 2014;9:e107110. doi: 10.1371/journal.pone.0107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yamashiro K, Yoshikawa M, Miyake M, Oishi M, Akagi-Kurashige Y, Kumagai K, Nakata I, Nakanishi H, Oishi A, Gotoh N, Yamada R, Matsuda F, Yoshimura N. Comprehensive replication of the relationship between myopia-related genes and refractive errors in a large Japanese cohort. Invest Ophthalmol Vis Sci. 2014;55:7343–54. doi: 10.1167/iovs.14-15105. [DOI] [PubMed] [Google Scholar]
- 85.Miyake M, Yamashiro K, Tabara Y, Suda K, Morooka S, Nakanishi H, Khor C-C, Chen P, Qiao F, Nakata I, Akagi-Kurashige Y, Gotoh N, Tsujikawa A, Meguro A, Kusuhara S, Polasek O, Hayward C, Wright AF, Campbell H, Richardson AJ, Schache M, Takeuchi M, Mackey DA, Hewitt AW, Cuellar G, Shi Y, Huang L, Yang Z, Leung KH, Kao PYP, Yap MKH, Yip SP, Moriyama M, Ohno-Matsui K, Mizuki N, MacGregor S, Vitart V, Aung T, Saw S-M, Tai E-S, Wong TY, Cheng C-Y, Baird PN, Yamada R, Matsuda F, Yoshimura N. Identification of myopia-associated WNT7B polymorphisms provides insights into the mechanism underlying the development of myopia. Nat Commun. 2015;6:6689. doi: 10.1038/ncomms7689. [DOI] [PubMed] [Google Scholar]
- 86.Söhl G, Joussen A, Kociok N, Willecke K. Expression of connexin genes in the human retina. BMC Ophthalmol. BioMed Central Ltd. 2010;10:27. doi: 10.1186/1471-2415-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seeliger MW, Brombas A, Weiler R, Humphries P, Knop G, Tanimoto N, Müller F. Modulation of rod photoreceptor output by HCN1 channels is essential for regular mesopic cone vision. Nat Commun. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. 2011;2:532. doi: 10.1038/ncomms1540. [DOI] [PubMed] [Google Scholar]
- 88.Güldenagel M, Ammermüller J, Feigenspan A, Teubner B, Degen J, Söhl G, Willecke K, Weiler R. Visual transmission deficits in mice with targeted disruption of the gap junction gene connexin36. J Neurosci. 2001;21:6036–44. doi: 10.1523/JNEUROSCI.21-16-06036.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gal A, Rau I, El Matri L, Kreienkamp HJ, Fehr S, Baklouti K, Chouchane I, Li Y, Rehbein M, Fuchs J, Fledelius HC, Vilhelmsen K, Schorderet DF, Munier FL, Ostergaard E. Thompson D a., Rosenberg T. Autosomal-recessive posterior microphthalmos is caused by mutations in PRSS56, a gene encoding a trypsin-like serine protease. Am J Hum Genet. 2011;88:382–90. doi: 10.1016/j.ajhg.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Orr A, Dubé MP, Zenteno JC, Jiang H, Asselin G, Evans SC, Caqueret A, Lakosha H, Letourneau L, Marcadier J, Matsuoka M, Macgillivray C, Nightingale M, Papillon-Cavanagh S, Perry S, Provost S, Ludman M, Guernsey DL, Samuels ME. Mutations in a novel serine protease PRSS56 in families with nanophthalmos. Mol Vis. 2011;17:1850–61. [PMC free article] [PubMed] [Google Scholar]
- 91.Soundararajan R, Won J, Stearns TM, Charette JR, Hicks WL, Collin GB, Naggert JK, Krebs MP, Nishina PM. Gene Profiling of Postnatal Mfrprd6 Mutant Eyes Reveals Differential Accumulation of Prss56, Visual Cycle and Phototransduction mRNAs. PLoS One. 2014;9:e110299. doi: 10.1371/journal.pone.0110299. [DOI] [PMC free article] [PubMed] [Google Scholar]