Abstract
The selectivity of metal sensors for a single metal ion is critical for cellular metal homeostasis. A suite of metal-responsive regulators is required to maintain a prescribed balance of metal ions ensuring that each apo-protein binds the correct metal. However, there are cases when non-essential metals ions disrupt proper metal sensing. An analysis of the Ni-responsive transcriptome of the green alga Chlamydomonas reinhardtii reveals that Ni artificially turns on the CRR1-dependent Cu-response regulon. Since this regulon also responds to hypoxia, a combinatorial transcriptome analysis was leveraged to gain insight into the mechanisms by which Ni interferes with the homeostatic regulation of Cu and oxygen status. Based on parallels with the effect of Ni on the hypoxic response in animals, we propose that a possible link between Cu, oxygen and Ni sensing is an as yet uncharacterized prolyl hydroxylase that regulates a co-activator of CRR1. This analysis also identified transcriptional responses to the pharmacological activation of the Cu-deficiency regulon. Although the Ni-responsive CRR1 regulon is composed of 56 genes (defined as the primary response), 259 transcripts responded to Ni treatment only when a copy of the wild-type CRR1 gene was present. The genome-wide impact of CRR1 target genes on the transcriptome was also evident from the 210 transcripts that were at least 2-fold higher in the crr1 strain, where the abundance of many CRR1 targets was suppressed. Additionally, we identified 120 transcripts that responded to Ni independent of CRR1 function. The putative functions of the proteins encoded by these transcripts suggest that high Ni results in protein damage.
Combinatorial genome-wide analyses of transcriptome changes in response to genetic mutation and environmental perturbations give insight into a network of oxygen, copper and nickel signaling.
Introduction
The promiscuous binding of transition metal ions to proteins can result in negative consequences for a cell. Normally, homeostatic pathways operate to confine metal ions to appropriate interactions in the cell, but under various circumstances, mechanisms for maintenance of homeostasis can be overwhelmed. Exposure to extreme levels of metals (high or low), abiotic stress (such as oxidative) and disease (such as a mutated metal transporter) can promote protein mismetallation and subsequent protein inactivation as a major source of damage.4-7 For example, in the bacterium Escherichia coli, zinc (Zn) overload (created with a Zn efflux pump knock-out and Zn supplementation) results in Zn-loading and inactivation of Fe-dependent threonine dehydrogenase,8 while in the yeast Saccharomyces cerevisiae, defects that increase the ratio of available iron (Fe) to manganese (Mn) in the mitochondrion, result in Fe-loading and inactivation of Mn-dependent superoxide dismutase.9 Therefore, regulated control of the metabolism of each transition metal in the cell is required to properly metallate the metalloproteome.
Non-essential metal ions are also found in the biosphere. These can enter cells using the pathways for essential metals and in so doing can disrupt metal homeostasis. In bacteria, cadmium (Cd) disrupts the homeostasis of several essential transition metals leading to symptoms reminiscent of deficiency for those essential metals.11-13 Recently, evidence from Streptococcus pneumonia suggests that dysregulation by Cd is due to mismetallation of metal-responsive transcriptional regulators and competition for essential metal ion uptake.12 While this study provides novel insight into the role of protein mismetallation in the toxicity of Cd, in general, knowledge gaps exists between metal toxins and their symptoms. For instance in animals, nickel (Ni) exposure can ultimately cause cancer with one possible source of carcinogenesis linked to inactivation of Fe-dependent proteins, but how those proteins are inactivated by Ni is debated. Either Ni directly substitutes for Fe causing protein inactivation,14, 15 or Ni depletes ascorbate an essential cofactor needed for Fe redox control.16, 17 This example highlights a common obstacle in understanding the mechanisms of metal ion toxicity: which physiological perturbations are directly caused by the metal ion and which are downstream consequences caused by inactivation of a molecular target?
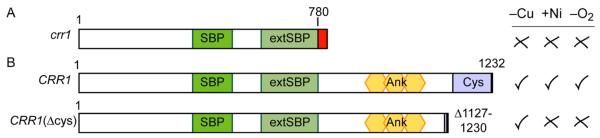
To gain foundational insight into how non-essential transition metals interfere with essential metal ion homeostasis, we leveraged a unique phenomenon in the unicellular experimental system Chlamydomonas reinhardtii. In C. reinhardtii, as with animals, there is no known catalytic role for Ni. Nevertheless, Ni acts as a pharmacological agent in activating hypoxic signalling in both animals and C. reinhardtii. In C. reinhardtii (but not in animals), Ni simultaneously activates copper (Cu)-deficiency signaling.18 Ni does not appear to inhibit Cu uptake or usage; thus the trivial explanation of Ni causing secondary Cu deficiency is ruled out.18 Evidence suggests that Ni interferes with regulation by the transcription factor CRR1, which is involved in inducing both the Cu-deficiency response and part of the response to hypoxia.2, 3, 10 Indeed, a cysteine-rich domain at the C-terminal end of CRR1 is required for activation of the CRR1 regulon by Ni and hypoxia but not for Cu-deficiency activation (Figure 1A and B).3
Fig 1. Schematic of CRR1 and effect of CRR1 mutations on CYC6 expression.
A, the crr1 strain used in this study has a single base-pair deletion in the CRR1 gene corresponding to codon 780 in the protein sequence. This mutation is predicted to cause a frameshift (indicated by a red box) and premature termination. B, the two other strains in this study were created by transforming the crr1 strain with either a WT copy of the CRR1 gene encoding the WT CRR1 protein (CRR1) or a deletion construct where the sequence encoding the cysteine-rich region was deleted (CRR1ΔCys).3 The response of the CYC6 gene in each strain to Cu deficiency (−Cu), Ni addition (+Ni) or hypoxia (−O2) is shown to the right3; an X indicates no response and a checkmark indicates a response was observed. The recombinantly produced SBP domain of CRR1 was shown to bind to the GTAC core of the Cu response element in the CYC6 and CPX1 promoters.10 Domain abbreviations: SBP, SQUAMOSA-promoter binding protein; extSBP, extended region with similarity to other SBPs; Ank, ankyrin repeats; Cys, cysteine-rich region.
It was thus concluded that Ni artificially induces the Cu and hypoxia responses through a mechanism that is independent of Cu sensing. Compared to the Cu and hypoxia responses, relatively little is understood concerning the interplay between Ni and CRR1 and the consequences this interaction has on the cell. While Ni is an essential cofactor for a few rare enzymes (such as Ni-Fe hydrogenase and urease), a Ni-dependent protein has yet to be identified in C. reinhardtii, suggesting that from a molecular point of view Ni is simply a pharmacological agent in the signalling pathway.
To understand the genome-wide impact of Ni addition on the C. reinhardtii transcriptome with specific focus on the activation of Cu-responsive gene expression, we performed high-throughput cDNA sequencing (RNA-Seq). A comparative transcriptomic analysis of three strains (crr1, crr1 transformed with a WT copy of CRR1, and crr1 transformed with a deletion construct of CRR1, where the region encoding the Ni-responsive domain was removed, Figure 1A and B) was used to delineate the CRR1-dependent Ni response. A comprehensive comparison with the transcriptomes of cells either deprived of Cu or oxygen was also leveraged to gain insight into the mechanisms by which Ni interferes with the homeostatic regulation of hypoxia and Cu deficiency.
Methods
C. reinhardtii strains, media, and culture conditions
Prior to RNA isolation C. reinhardtii mutant strains CC-3960 (crr1-219; referred here as crr1), CC-5073 (crr1-2-[CRR1(Δcys)]), PST353; referred here as CRR1(Δcys)), and CC-5071 (crr1-2-[CRR1], R423; referred here as CRR1) were grown in duplicate (two independent cultures of each strain) in Tris-acetate-phosphate (TAP) medium using Hutner’s trace element mix20 at 24°C with rotational shaking (180 rpm) under continuous light (~90 μmol/m2/s with a ratio of 1 warm fluorescent light bulb (3,000K) to 2 cool fluorescent light bulbs (4,100K)) in an Innova 44 incubator (Innova, New Brunswick Scientific, Edison, NJ). Cells were collected at a density of 3 × 106 cell ml−1 (steady-state) and the +Ni cells 6h hours after addition of NiCl2 to a final concentration of 50 μM in the medium.
Nucleic acid purification
Total RNA was prepared as described previously.21 RNA quality was assessed on an Agilent 2100 Bioanalyzer (electrophoresis and chromotographic separation on a microfluidics-based platform) and by hybridization as described previously.21 The probe used for detection, CβLP, is a 915-bp EcoRI fragment from the cDNA cloned in pcf8-13.22
RNA-Sequencing and data analysis
Genomic DNA was removed from the total RNA preparation by treatment with RQ1 DNase (Promega) according to the manufacturer’s instructions. cDNA libraries were prepared for single-end sequencing (75 nt reads) and sequenced by the Joint Genome Institute using the Illumina platform. The sequence reads were aligned to v5.5 of the reference genome23 (available at ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Creinhardtii/) using STAR24 with default parameters plus –alignIntronMax 10000. Transcript abundance estimates were calculated and normalized in terms of fragments per kb of exon per million fragments (FPKM) using default parameters in cuffdiff.25 Venny,26 eulerAPE27 or BioVenn28 were used for drawing Venn diagrams.
Except where noted otherwise, differentially abundant transcripts were defined as having at least a FPKM value of 10 and a 2.0-fold change. For re-analysis and comparisons with the previously published Cu1 and hypoxia2 datasets, the previously sequenced reads were re-aligned to v5.5 of the genome and transcript abundance estimates were calculated following the same parameters as for the Ni dataset. Hierarchal clustering was performed with Gene Cluster 3.0 and visualized with TreeView.29
Measurement of intracellular metal content
Total cellular metal content was measured by ICP-MS as previously described.30
Results
Ni activates a link between CRR1-dependent Cu and O2 signalling
Most of the target genes of the CRR1-dependent, nutritional Cu-signalling pathway respond also to hypoxia treatment.2 Nonetheless the individual responses of these targets to Cu or O2 are not identical, and there is variability to the extent with which the hypoxia response is attenuated in the crr1 and CRR1(Δcys) strains.31 In part, the differences can be explained by the presence of hypoxia-response elements in the upstream regions of some CRR1 target genes but not others.31, 32 In previous work, we also showed that the key sentinel genes of the nutritional Cu regulon, CYC6, CPX1 and CRD1, are expressed in response to Ni.18 As was shown for the CYC6 hypoxic response (but not the Cu response), the Ni response required a cysteine-rich domain at the C-terminus of CRR1.3 These results suggest that Ni treatment will activate induction of the entire CRR1 regulon and will do so in a manner similar to the hypoxic response.
To test this hypothesis we analysed the Ni transcriptomes from this work (Table S1) in the context of the older RNA-Seq1, 2 datasets from published work on nutritional Cu-deficiency responses and hypoxia responses. This analysis required re-aligning these published datasets to the latest iteration of gene models (v5.5) (Table S2) and redefining the Cu-responsive CRR1 regulon (Figure 2A; Table S3). The present reanalysis captured 48 of the original 63 CRR1 Cu targets1 and predicted 17 new targets. Another 6 of the original targets could be included in the set of new targets if the cutoffs are moved (1.6-fold change and <10 FPKM). This left 4 of the original targets whose gene models did not map forward to v5.5 and 4 transcripts whose abundance did not change between Cu status or between the crr1 and CRR1 strains in the reanalysis.
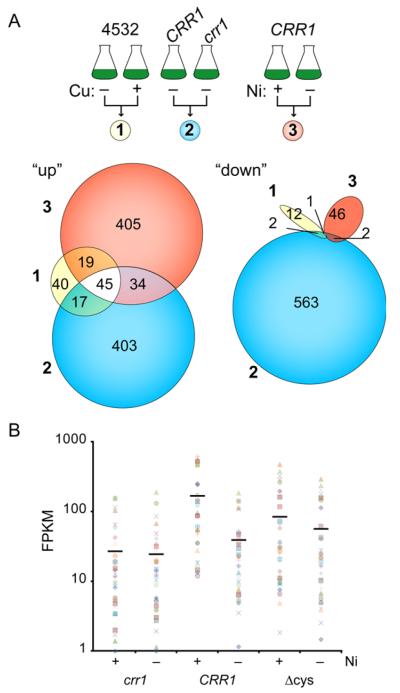
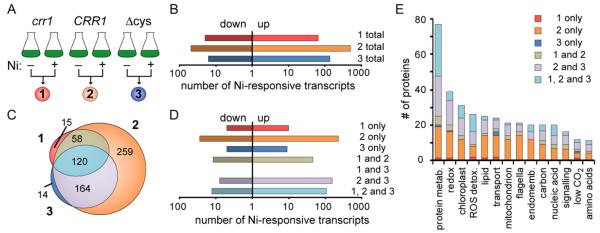
Fig 2. Identification of CRR1 Cu targets and effect of Ni on their expression.
A, schematic of strains (listed above flask cartoons) and culture conditions (listed below flask cartoons) that were compared to identify differentially accumulating transcripts. The numbers of transcripts resulting from these pairwise comparisons are shown as scaled Venn diagrams. “Up” corresponds to those transcripts with higher transcript abundance in absence of Cu (comparison 1), in CRR1 (comparison 2), or presence of Ni (comparison 3). “Down” corresponds to those transcripts with higher abundance in the presence Cu (comparison 1), in crr1 (comparison 2) and absence of Ni (comparison 3). B, transcript abundances of the 36 CRR1 targets that had at least a 2-fold increase in transcript abundance in the presence of Ni in the CRR1 strain but less than 2 fold increase in both the crr1 and CRR1(Δcys) strains. The solid black bars represent the average FPKM value for each strain and condition.
Ni affected the abundances of 46 transcripts out of the 65 CRR1 targets in the CRR1 strain (Figure 2A; Table S3). Another 10 targets can be included in this subset if the criteria are changed to at least a 1.5-fold change and >5 FPKM. Under standard growth conditions (without the addition of Ni), 11 (out of the 56 targets) had at least 2.0-fold lower transcript abundance in crr1 compared to the CRR1 strain (with at least 10 FPKM in CRR1) (Table S3), further supporting involvement of CRR1 in regulating their expression. A role for CRR1 in mediating at least part of the Ni response was evident for most of the target transcripts; the Ni effect was attenuated or abolished in the crr1 or CRR1(Δcys) strains for 52 and 49 out of the 56 targets, respectively. The Ni response of only half of these, however, was completely abolished in both strains, including transcripts encoding the Cu transporter CTR3 and the ferredoxin FDX6 and the new targets of unknown function CGL120 and FAP79. Only 1 CRR1 target, ERG3, responded to Ni treatment in all strains and therefore likely responds to Ni independently of CRR1. While most of the target genes in the CRR1-dependent, nutritional Cu-signalling pathway respond to Ni supplementation (Table S3), the extent to which expression is affected in the crr1 and CRR1(Δcys) strains varies (Figure 2B), as was found for the hypoxic response.2
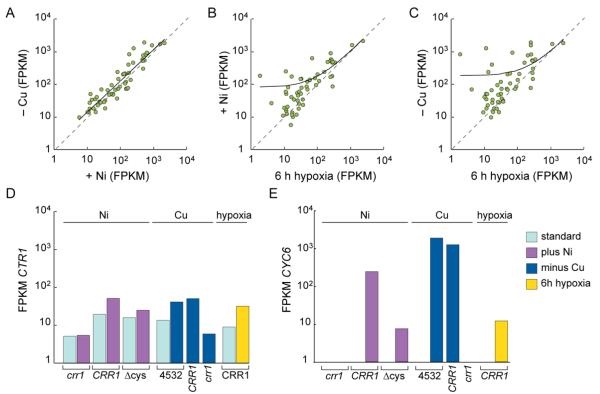
To tease apart the similarities and distinctions in the outputs of the three signaling pathways, we undertook pairwise comparisons of transcript abundances for the 56 Ni-responsive CRR1 targets upon each treatment (−Cu, +Ni and −O2). The transcript abundances for these 56 targets in +Ni were relatively accurate at predicting abundances in −Cu (power-law relationship with a R2 value of 0.85) (Figure 3A). Similar to the pairwise comparison with −Cu, transcript abundances from +Ni and −O2 had a strong correlation (R2 value of 0.83) but fit a linear relationship (Figure 3B). The correlation between −Cu and −O2 for these targets was poor; transcript abundances fit a linear relationship with a R2 value of 0.41 (Figure 3C).
Fig 3. Ni addition mimics the Cu-deficiency response of CRR1 target transcripts.
A, scatter plot of transcript abundances from cells grown in absence of Cu and cells grown in the presence of Ni. The power regression trend line (R2 value of 0.85) is shown as a solid black line. The grey dashed lines represent a 1-to-1 correlation. B, scatter plot of transcript abundances from cells grown in presence of Ni and cells grown under 6 h of anoxic conditions. The linear regression trend line (R2 value of 0.83) is shown as a solid black line. The grey dashed lines represent a 1-to-1 correlation. C, scatter plot of transcript abundances from cells grown in absence of Cu and cells grown under 6 h of anoxic conditions. The linear regression trend line (R2 value of 0.41) is shown as a solid black line. The grey dashed lines represent a 1-to-1 correlation. D, transcript abundances of CTR1 from Ni, Cu and hypoxia transcriptomes. E, transcript abundances of CYC6 from Ni, Cu and hypoxia transcriptomes. In the legend to panels D and E on the right, “standard” refers to untreated, standard growth condition.
Although most of the CRR1 target genes respond to both Cu deficiency and hypoxia, the extent of the response to each condition varies between these genes. The relatively poor correlation between transcript abundances is likely attributable to one or more additional regulatory factors involved in the expression of some CRR1 target genes during hypoxia.31, 32 Indeed, we had previously identified separate elements in the promoter of CPX1, but not of CYC6, which are required for either the Cu response or the hypoxic response.31 Distinct regulatory mechanisms could explain why many target genes respond differently to Cu and hypoxia. In contrast, there is a relatively strong correlation between the transcript abundances of these genes in the presence of Ni and absence of Cu, suggesting that Ni treatment and Cu deficiency make a relatively equal contribution to the expression of these CRR1 target genes.
The magnitude of the Ni response of these targets appears to be somewhere in between the Cu and hypoxia responses. For instance, CYC6 transcript abundance was estimated at ~0 FPKM in the absence of stimuli and increased to 1280, 250 and 11 (two orders of magnitude range in expression) after the depletion of Cu, addition of Ni or depletion of oxygen, respectively (Figure 3E). CTR1 (encoding a Cu transporter), conversely, responded to −Cu, +Ni and −O2 with a 2.7 to 3.9-fold increase (Figure 3D).
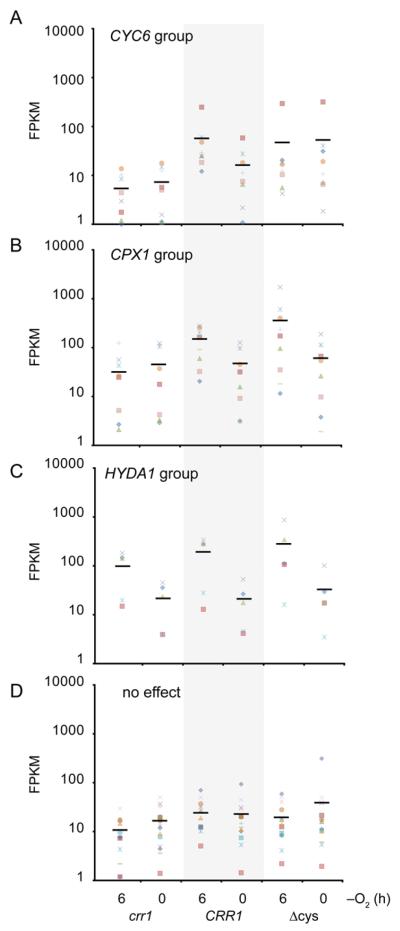
For these 56 targets, there was also a more consistent attenuation of the Ni response than of the hypoxia response in the CRR1(Δcys) and crr1 strains. We identified a core set of 37 CRR1 targets that respond to Ni in the CRR1 strain but not in either crr1 or CRR1(Δcys) (Table S3). Except for the CTH1 transcript, whose abundance decreases upon hypoxia (and Cu deficiency), the hypoxia response of these target genes can be further clustered into four general groups (Figure 4A-D). The hypoxic response of the CYC6 group is present only in CRR1. The hypoxic response of the CPX1 group is present in CRR1 and CRR1(Δcys). The hypoxic response of the HYDA1 group is present in all three strains, while the response in the fourth group did not meet our cutoffs in any strain. Interestingly, based on previous experiments31, 32, the CYC6 hypoxic response is mediated solely through CRR1, the CPX1 response is dependent on both CRR1 and an unknown factor, and the HYDA1 hypoxia response in partly due to CRR1 and partly due to another factor. The patterns of expression for the three groups in the three strains mirror these experimental results. As noted previously2, neither the CPX1 nor HYDA1 hypoxic response requires the cysteine-rich C-terminus of CRR1. Also in contrast to CYC6, the hypoxic response of CPX1 involves cis-acting elements that are distinct from the Cu-response elements, and the hypoxic response of HYDA1 is retained in the crr1 mutant.31, 32 Therefore, the cysteine-rich region may only be required for those CRR1 target genes (such as CYC6) whose hypoxic response is mediated solely through the Cu-response element. Additional experiments are needed to determine if this reasoning holds true as predicted for other members of the CYC6, CPX1 and HYDA1 groups. Since the cysteine-rich region of CRR1 is required for the Ni response of these 36 targets, Ni may be activating the CRR1-dependent portion of the hypoxia response.
Fig 4. Ni-responsive CRR1 target genes can be group into four hypoxia-response sets.
Transcript abundances of the 36 CRR1 targets from Figure 1C prior to (0 h) and 6 hours (6 h) after the crr1, CRR1 and CRR1(Δcys) strains were transferred to dark anoxic conditions. FPKM values were calculated with realigned reads from the published anoxic RNA-Seq dataset of Hemschemeier, et al.2 The solid black bars represent the average FPKM value for each strain and condition. A, transcript abundances of CRR1 target genes with a similar expression pattern to CYC6. B, transcript abundances of CRR1 target genes with a similar expression pattern to CPX1. C, transcript abundances of CRR1 target genes with a similar expression pattern to HYDA1. D, transcript abundances of CRR1 target genes that did not meet our cutoffs for differential accumulation in each pairwise comparison.
Pleiotropic effects of CRR1 mutation on Ni-induced changes to the transcriptome
Although the Ni-responsive CRR1 Cu regulon has only 56 members, we found a total of 259 transcripts that responded to Ni uniquely in CRR1 (Figure 5C). The response of another 164 transcripts to Ni was common between CRR1 and CRR1(Δcys), and only 120 transcripts were shared between the three strains (Figure 5C and D). All together we observed a strain-dependent Ni response for 510 transcripts (Table S1). This result suggests that CRR1 has direct or indirect target genes outside of the Cu-responsive regulon that respond to Ni treatment. Conversely, these transcript changes may result from a knock-on effect of CRR1 target gene expression during Ni treatment.
Fig 5. Identification of differentially accumulating transcripts in response to Ni treatment.
A total of 559 transcripts were at least 2-fold higher in abundance in the presence of Ni vs. the absence of Ni in any given strain and 71 transcripts that were at least 2-fold lower in abundance in the presence of Ni (Table S1). A, schematic of culture conditions and strains compared in panels B, C, D and E. B, using a filter of 2.0-fold change and 10 FPKM, the number of transcripts that are less abundant in the presence of Ni (“down”) and those that are more abundant in the presence of Ni (“up”) for each strain is shown. C, scaled Venn diagram representing the number of differentially accumulating transcripts identified in each pairwise comparison illustrated in panel A and the overlaps between the three sets. We used the q-values from CuffDiff for each pairwise comparison to determine if the abundances of a transcript changed in the same direction between strains. D, the number of transcripts that are less abundant in the presence of Ni (“down”) and those that are more abundant in the presence of Ni (“up”) for each area of the Venn diagram is given. E, manually annotated function enrichment for the “up” transcripts in panel D.
To address these possibilities, we performed hierarchal clustering of transcript abundances from the Ni and Cu datasets. We identified a connection between some of these strain-dependent transcripts and the identified CRR1 Cu targets. A cluster of 120 transcripts co-expressed with the majority of putative CRR1 targets (56 transcripts) (Figure 6A). Of the non-CRR1 targets in this cluster, we found several cytochrome oxidase assembly factors (COX11, COX17 and CMC1 involved in Cu insertion and COX15 involved in heme insertion). Indeed, of the eight assembly factors that have been identified in the delivery and insertion of Cu into cytochrome oxidase33, the abundances of transcripts for most are increased at least 2 fold by Ni addition in the CRR1 strain. In each case, induction was blocked in the crr1 and CRR1(Δcys) strains (Figure 6D). COX17, COX19 and CMC1 transcript abundances are also increased during Cu deficiency (2.0-, 2.4-, 1.9-fold, respectively), but unlike the Ni response, the abundances of these three transcripts appear to be the same in the CRR1 and crr1 strains in the Cu dataset, suggesting that the corresponding genes are not direct targets of CRR1.
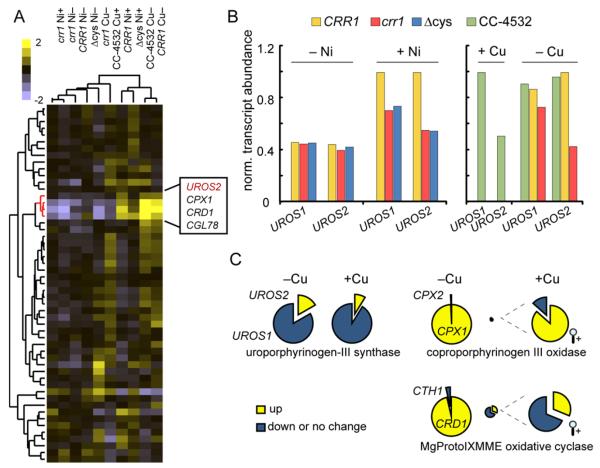
Fig 6. Ni addition and Cu deficiency affect expression of cytochrome oxidase maturation components.
A, hierarchical clustering of transcript abundances from crr1, CRR1, CRR1(Δcys), and CC-4532 in the presence or absence of Ni or Cu. The genes identified as CRR1 targets are marked with a red block to the right. The scale bar is on a log scale. B, transcript abundances normalized to the max (either in Ni experiments or Cu experiments) encoding Ni-responsive cytochrome oxidase Cu chaperones. Note that Castruita, et al.1 did not perform RNA-Seq on the CRR1 and crr1 strains grown in the presence of Cu (+Cu). C, transcript abundances normalized as in B of selected CRR1 target transcripts. D, overview of transcript abundance changes in the presence of Ni vs. absence of Ni for proteins involved in Cu delivery to plastocyanin in the chloroplast or cytochrome oxidase in the mitochondrion. Fold change is on a linear scale. TM, thylakoid membrane; IE inner envelope; OE, outer envelope; OM, outer membrane; IM, inner membrane.
In Cu-deficient C. reinhardtii, Cu allocation to cytochrome oxidase in the mitochondrion is prioritized over plastocyanin in the chloroplast.34 Indeed, we found that the transcripts encoding the two Cu transporting P-type ATPases that function in tandem to provide Cu to plastocyanin are reduced in the presence of Ni, while transcripts encoding proteins involved in Cu delivery to cytochrome oxidase are increased in abundance following Ni addition (Figure 6D). Although Cu does not become limiting after Ni addition18, there may be a CRR1-dependent process that mediates re-prioritization of Cu between organelles.
In this cluster, we also identified an isoform of uroporphyrinogen-III synthase, which is involved in tetrapyrrole biosynthesis. Tetrapyrroles are important cofactors, especially in photosynthetic organisms where the pathway branches to yield both heme and chlorophyll. Two oxygen-dependent enzymes that act at rate-limiting steps in the pathway, coproporphyrinogen III oxidase and the aerobic oxidative cyclase, are expressed at higher levels in copper-deficient or hypoxic cells.35-37 This results from increased expression of one of two isoforms that encode these activities, namely CPX1 and CRD1.1, 2 In this work, we identified another step in tetrapyrrole biosynthesis that may be a direct or indirect target of CRR1. UROS2 is one of 2 genes encoding uroporphyrinogen-III synthase. The corresponding transcript increases in abundance both upon Ni addition and in Cu-deficient cells. Unlike for the cytochrome oxidase chaperones above, the increase of UROS2 transcript abundance upon Ni treatment and Cu depletion is dependent on CRR1 function (Figure 7B). This gene was not included in the CRR1 target list (Table S3) because the change in its expression (1.9) did not meet the 2.0 fold cutoff. Uroporphyrinogen-III synthase may represent a third step in tetrapyrrole biosynthesis that is regulated by CRR1. Nevertheless, the increase is not as large as that observed for CPX1 and CRD1. Indeed, while CPX1 and CRD1 become the dominant isoforms during Cu deficiency, UROS2 does not (Figure 7C). Also, while the combined abundances for CPX1/CPX2 and CTH1/CRD1 increase (Figure 7C), combined abundances for UROS1 and UROS2 transcripts remain constant between Cu sufficiency and deficiency.
Fig 7. Transcript abundances for genes in the tetrapyrrole biosynthesis pathway.
A, hierarchal clustering and corresponding heatmap of transcripts involved in tetrapyrrole biosynthesis and catabolism. The experiment labels are the same as in Fig. 6. The CRR1-target cluster is highlighted. The target identified in this study, UROS2, is in red. The scale bar is on a log scale. B, transcript abundances normalized to the max (either in Ni experiments or Cu experiments) corresponding to the genes for uroporphyrinogen-III synthase. C, relative transcript abundant (FPKM) of isoforms corresponding to Cu-regulated steps of the tetrapyrrole pathway. Transcripts that are higher in Cu deficiency compared to Cu-replete growth medium are in yellow. The relative sizes of the pie charts for each step are normalized to total FPKM. Magnification of the CPX1/CPX2 and CTH1/CRD1 charts representing relative transcript abundances in the presence of Cu is shown on the right.
This CRR1-target cluster also contained 5 transcripts encoding enzymes that are documented or are candidates for function in ergosterol biosynthesis (Figure 6A). ERG3, ERG1, CYP710B1 and CYP51G1 made the cutoffs used to identify CRR1 targets, while the fold change of SMT1 between the Cu-deficient CRR1 and crr1 strains (1.8) did not meet the 2.0-fold cutoff. Ergosterol is a sterol found in membranes and has roles in maintaining membrane fluidity and permeability.38 Whether these transcript changes affect ergosterol membrane content and/or are related to the previously measured increase in unsaturated fatty acids in Cu-deficient relative to Cu-replete cells1 is yet to be determined.
Proteostasis among the most highly affected processes in response to Ni treatment
In addition to activating the CRR1-dependent regulon, Ni is a toxin that inhibits the growth of C. reinhardtii. 39 We manually annotated the proteins encoded by the 559 transcripts whose abundances increase in the presence of Ni (in any strain) and classified them by function (Table S1). In the wild-type CRR1 strain (Table S4), Ni addition caused reductions in the abundances of transcripts encoding cell-wall proteins and proteins involved in DNA replication, such as MCM (mini-chromosome maintenance) complex proteins, DNA topoisomerase II, and the eukaryotic δ subunit of DNA polymerase. At the same time, Ni caused the increased abundances of transcripts encoding proteins involved in protein folding/degradation and lipid biosynthesis, including 7 candidate enzymes of ergosterol biosynthesis. We also found transcripts encoding proteins involved or putatively involved in apoptosis and autophagy. These changes suggest a reprioritization of cellular resources from cell division to repair of protein and lipid damage.
Proteostasis was also among the most highly affected processes in the strain-independent response to Ni treatment (107 transcripts increased in abundance and 13 transcript decreased in abundance following Ni treatment in all strains; Table S5). The main functional categories include protein degradation (both proteases and proteins potentially involved in ubiquitination and the proteasome), molecular chaperones (such as HSP22C, HSP22E and HSP22F), ROS detoxification (specifically protein-oxidation repair, such as two methionine-sulfoxide reductases and thioredoxin-like proteins, three proteins with glutathione-S-transferase domains, and one protein involved in ascorbate biosynthesis), and proteins associated with the endoplasmic reticulum, Golgi apparatus and the secretory pathway (Figure 5E). The enrichment of transcripts corresponding to proteostasis and ROS detoxification suggests that Ni stress causes transcriptomic changes consistent with protein damage and perturbed redox balance.40 Many of these transcriptional changes could also be connected to the induction of autophagy (a self-degradation process that involves clearing of damaged cellular components ranging from mis-folded proteins to entire organelles41), as previously described.40
Among transcripts shared between the strains, we did not find a candidate transporter for Ni efflux. We did find a single transcript with similarity to the ATPase component of ABC-type transporters, but this protein and similar proteins have yet to be characterized. Of the 22 transcripts without a predicted domain, 3 had at least 2 putative transmembrane helixes, but it is unknown whether they could have a role in metal efflux. Alternatively, the large number of transcripts that fell into categories related to protein stress and the role of autophagy in responding to Ni stress40 could point to exocytosis of Ni through a general detoxification pathway.
Of the transcripts reduced in abundances in the presence of Ni, most of these (9 out of 13) are cell wall proteins, including 5 pherophorins. Pherophorins are hydroxyproline-rich glycoproteins that are thought to make up a large proportion of the cell wall and are involved in cell wall synthesis.42-44 These transcript changes could, therefore, be an indication of de-prioritization of cell growth in response to Ni stress.
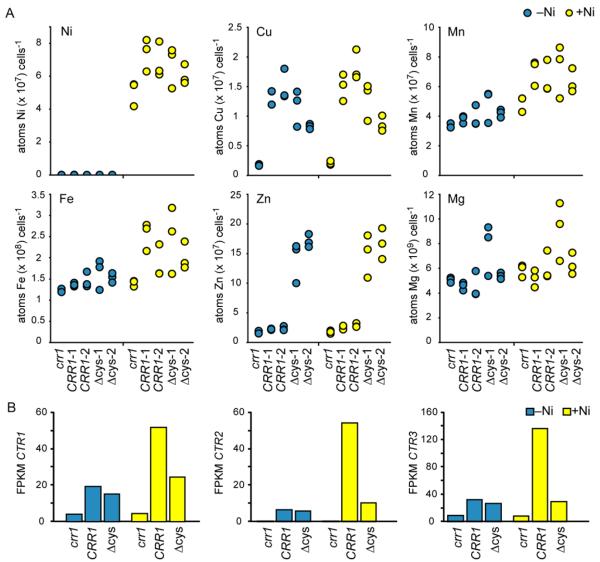
Effect of Ni addition on metal quota
The abundances of transcripts encoding Cu importers and a predicted Fe transporter are increased in Ni-treated cells, dependent on CRR1 function. To assess the impact of these increases, we measured the metal content of the crr1, CRR1 and CRR1(Δcys) strains grown in the absence or presence of Ni. Although the crr1 strain had significantly less Cu than the other strains in either the presence or absence of Ni (roughly 85% less than the CRR1 strains, pvalue <0.02), which correlated with the reduced abundances of the CTR1, CTR2 and CTR3 transcripts (encoding assimilatory Cu transporters), there was no significant effect of Ni addition (pvalue >0.2) on the Cu quota (Figure 8A). On the other hand, Ni addition did affect the Fe and Mn content of the strains (pvalue 0.001 and <0.001, respectively) (Figure 8A). Although the increase was also observed for the crr1 strain (10% increase in Fe content, pvalue <0.02, and 30% increase in Mn content, pvalue <0.008, upon Ni treatment), the crr1 strain had significantly less Mn and Fe compared to the other strains upon Ni treatment (pvalue <0.0002 for Fe and <0.02 for Mn). It is possible that these differences upon Ni treatment are attributable to differential abundance of the transcripts encoding the divalent metal cation transporter IRT2 and/or the Fe-assimilatory proteins FEA1 and FEA2 (Table S1). As noted previously3, the CRR1(Δcys) strain hyper-accumulated Zn, and we found that this phenotype was not affected by Ni addition.
Fig 8. Effect of Ni addition on the metal content.
A, The metal content of the crr1 strain, two independent transformants of crr1 transformed with the WT copy of CRR1 (CRR1-1 and CRR1-2) and two independent transformants of crr1 transformed with the CRR1(Δcys) construct (Δcys-1 and Δcys-2) was measured by ICP-MS. Each data point represents a biological replicate for each strain and condition (cells were collected independently from three separate cultures for each strain and condition combination). B, transcript abundances of the assimilatory Cu transporters that likely affect the measured Cu quotas.
In addition to the reduced Cu quota (resulting presumably from reduced expression of the assimilatory CTR transporters), we found that crr1 also accumulated less Ni (Figure 8A) (although, we note that the difference between the strains only reaches significance between crr1 and CRR1-1, pvalue 0.03). The Ni content of the strains may have an impact on the number of differentially expressed transcripts. There are 35% and 68% fewer differentially expressed transcripts from the crr1 strain than from the CRR1(Δcys) and CRR1 strains, respectively. However, at this point we cannot differentiate between a possible effect caused by unequal Ni accumulation and the contribution mis-regulation of CRR1 targets during Cu sufficiency has on the observed differences between the strains.
Discussion
Ni treatment leads to substantial transcriptome remodelling in C. reinhardtii. In part these changes are due to activation of the CRR1-dependent regulon. Transcript abundances for 85% of the putative CRR1 Cu targets were affected by Ni addition, and the Ni-responsiveness of 90% of these transcripts was abolished in the crr1 mutant. We were also able to determine that the effect of Ni on transcript abundances of these targets parallels the effect of Cu deficiency (Figure 3A), whereas the correlation between the Cu response and hypoxia response was not as strong. This observation suggests that Ni addition mimics Cu deficiency and activates CRR1 whereas additional factors are involved in the hypoxia-induced expression of several CRR1 targets.31
This analysis also provides genome-wide insight of Ni-stress on C. reinhardtii. As shown previously, Ni treatment induces autophagy40 and transcriptomic changes are consistent with protein and ROS stress. This response and the cellular perturbations that cause this response are particularly relevant to studies that employ the CYC6 promoter as a Ni-inducible system for the expression of transgenes.18, 45 Researchers employing the CYC6 promoter should be aware of the stresses highlighted here that are caused by Ni treatment. Correlating physiology and other parameters with transgene expression should be performed side-by-side with wild-type cells (not expressing the transgene) treated with Ni.
Parallels with the animal HIF system
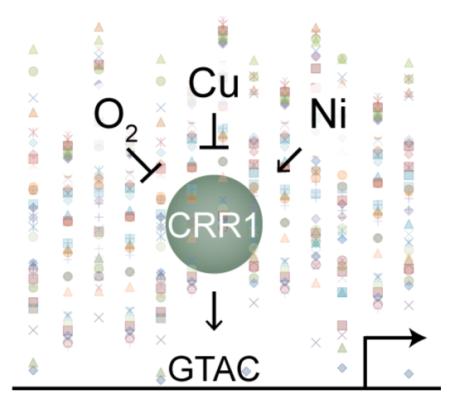
The ability of Ni to interfere with signaling is not unique to Cu sensing in C. reinhardtii. Interestingly, Ni mimics oxygen deficiency in animals46, 47, where both oxygen deficiency and Ni lead to inactivation of the prolyl hydroxylases that are involved in targeting the alpha subunit of the hypoxic response regulator, HIF-1, for degradation.15, 48 The result is that in the presence of Ni (or absence of O2) the HIF-1α subunit is not modified by the prolyl hydroxylase and is not degraded leading to activation of the hypoxic-response regulon. A similar mechanism involving prolyl-hydroxylase-mediated regulation of CRR1 could explain in part why both hypoxia and Ni lead to increased transcript abundances of CRR1 targets in C. reinhardtii.
Another parallel between the HIF system and our current data is that roughly half of the CRR1 targets are also affected by Fe limitation.49 Because Fe is an essential cofactor for the HIF-1α prolyl hydroxylases, Fe limitation also inhibits HIF-1α degradation and activates hypoxic signaling.50 In C. reinhardtii, a physiological link between Fe limitation and Cu homeostasis exists. A higher demand for Cu (and thus activation of CRR1) would be needed during Fe limitation to support the maturation of a multi-Cu oxidase that is homologous to ceruloplasmin in human and involved in high-affinity Fe uptake across the plasma membrane.51, 52 Therefore, a yet to be characterized prolyl hydroxylase could tie together the Cu-, Ni-, oxygen- and Fe-dependent regulation of CRR1 targets. Based on the involvement of the cysteine-rich region of CRR1 in the Ni and hypoxia responses, but not the Cu response, one possibility is that activity of this prolyl hydroxylase leads to degradation of a co-activator that interacts with CRR1 through this cysteine-rich domain. During hypoxia and Ni treatment, the prolyl hydroxylase is inactivated and the co-activator binds CRR1 leading to induction of the target genes.
The role of post-translational modification and co-activators in the regulation of CRR1 activity has yet to be explored, but there are several prolyl hydroxylases (Cre02.g084450, Cre09.g408464, Cre03.g179500, Cre02.g084400, and Cre10.g443050) among the putative CRR1 targets. Cre09.g408464 is a promising candidate in connecting CRR1 to Ni, hypoxia and Fe status. This uncharacterized protein is fungal-like (not found in land plants) and shares similarity with Ofd1, a prolyl hydroxylase that regulates the degradation of a hypoxic-response regulator in fission yeast.53
Consequences of CRR1 mis-regulation
While the mechanism responsible for CRR1-dependent regulation remains to be elucidated, this study has provided systems-level insight into the effect of Ni on CRR1-regulated transcripts and the consequence of mis-regulation during Cu sufficiency. Although the Ni-responsive CRR1 Cu regulon has only 56 members, there were a total of 259 transcripts whose abundance was affected by Ni in the CRR1 strain but not in either the crr1 or CRR1(Δcys) strains. This pattern suggests that genes for these transcripts are responding to increased (or decreased) abundance of the CRR1 targets during Cu sufficiency. While we found some evidence for chloroplast stress, such as increased abundance of PTOX2 transcript (encoding an oxidase involved in chlororespiration54), we were surprised to find that several transcripts encoding assembly factors of cytochrome c oxidase in the mictochondrion were increased in abundance (Figure 6A and B). These transcript changes could be in response to the mis-regulation of CRR1 targets in the presence of Ni.
Cytochrome oxidase in the second most abundant Cu-dependent protein in C. reinhardtii (during Fe sufficiency) and receives Cu with higher priority during Cu deficiency.34 While transcripts encoding Cu chaperones involved in Cu delivery to cytochrome oxidase are marginally increased in abundance during Cu deficiency, many of them are at least 2 fold higher in the presence of Ni. The effect of Ni was attenuated or absent in the crr1 or CRR1(Δcys) strains. In addition to Cu chaperones and subunits of cytochrome oxidase, transcripts encoding cytochrome c were also increased specifically in CRR1 as were several transcripts encoding enzymes in ubiquinone biosynthesis, suggesting that multiple steps along the electron transfer chain are affected by mis-regulation of CRR1 targets.
We were also surprised to find that the transcript abundances for some CRR1 targets were lower in the crr1 mutant than in the CRR1 strain under standard growth conditions. This difference between strains may account in part for the 210 transcripts that were at least 2-fold higher in crr1 compared to the CRR1 strain. These transcripts could be responding to the lower Cu content of crr1 (Figure 8A) (and thus responding independently of CRR1 to Cu deficiency) and/or to the consequence of lower transcript abundance of CRR1 targets. For instance, transcript encoding alternative oxidase is increased 2.3 fold in crr1 compared to the complemented strain suggesting that the activity of cytochrome c oxidase may be compromised. There are also several transcripts in this subset that respond also to high light and/or low CO2 (which are stress conditions in C. reinhardtii) suggesting that Ni treatment generates stress in the chloroplast. While there have not been any obvious phenotypes reported for the crr1 mutant grown under this condition, it is likely that either the reduced abundances of these CRR1 targets are physiologically subtle (except for the noticeable reduction of cellular Cu concentration) or these 210 transcripts are responsible for acclimating to their loss.
Conclusion
Genome-wide and comparative analyses of transcriptome changes in response to environmental perturbations provide the opportunity to gain insight into the mechanisms that link regulatory networks. Because of their central roles in catalysis and protein activity, the regulation of metal ion homeostasis is often linked to the physiological state of the cell. One such relationship exists between Cu homeostasis and oxygen deficiency in C. reinhardtii, and we have found that to a certain extent Ni artificially activates this link. It is clear from this analysis and from previous work that the connection between Cu and oxygen involves at least an additional yet to be identified factor. One promising candidate would be a prolyl hydroxylase such as the CRR1 target Cre09.g408464, which is homologous to a prolyl hydroxylase in yeast that is responsible for oxygen sensing. The transcriptome analyses presented here promote a model where the inactivation of a prolyl hydroxylase (by oxygen deficiency or Ni) leads to the interaction between a co-activator and CRR1. Additionally, the cysteine-rich region that is deleted in the CRR1(Δcys) mutant would be a likely interaction domain, since deletion of this domain abolishes or attenuates the ability of oxygen deficiency or Ni to activate CRR1-dependent expression.
Supplementary Material
Significance to Metallomics statement.
For most eukaryotic organisms, Ni does not have a catalytic role in the cell, and its presence can be detrimental. However, understanding how toxicity arises is challenging. This study provides transcriptome-level insight into the consequences of Ni treatment on the cell with specific attention on the pharmacological interaction between Ni and the regulation of Cu and oxygen homeostasis.
Acknowledgements
This work was supported by the National Institutes of Health (NIH) (GM092473 (for RNA-Seq analysis) and GM42143 to SSM). CB-H acknowledges support from an Individual Kirschstein National Research Service Award (GM100753) and the Office of Biological and Environmental Research of the Department of Energy.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
References
- 1.Castruita M, Casero D, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, Yan W, Cokus S, Loo JA, Benning C, Pellegrini M, Merchant SS. Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell. 2011;23:1273–1292. doi: 10.1105/tpc.111.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hemschemeier A, Casero D, Liu B, Benning C, Pellegrini M, Happe T, Merchant SS. Copper response regulator1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell. 2013;25:3186–3211. doi: 10.1105/tpc.113.115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sommer F, Kropat J, Malasarn D, Grossoehme NE, Chen X, Giedroc DP, Merchant SS. The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell. 2010;22:4098–4113. doi: 10.1105/tpc.110.080069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imlay JA. The mismetallation of enzymes during oxidative stress. J Biol Chem. 2014;289:28121–28128. doi: 10.1074/jbc.R114.588814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaby-Haas CE, Merchant SS. Lysosome-related organelles as mediators of metal homeostasis. J Biol Chem. 2014;289:28129–28136. doi: 10.1074/jbc.R114.592618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang M, Parker MJ, Stubbe J. Choosing the right metal: case studies of class I ribonucleotide reductases. J Biol Chem. 2014;289:28104–28111. doi: 10.1074/jbc.R114.596684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macomber L, Hausinger RP. Mechanisms of nickel toxicity in microorganisms. Metallomics. 2011;3:1153–1162. doi: 10.1039/c1mt00063b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu M, Imlay JA. Superoxide poisons mononuclear iron enzymes by causing mismetallation. Mol Microbiol. 2013;89:123–134. doi: 10.1111/mmi.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang M, Cobine PA, Molik S, Naranuntarat A, Lill R, Winge DR, Culotta VC. The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 2006;25:1775–1783. doi: 10.1038/sj.emboj.7601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kropat J, Tottey S, Birkenbihl RP, Depège N, Huijser P, Merchant S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci U S A. 2005;102:18730–18735. doi: 10.1073/pnas.0507693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Begg SL, Eijkelkamp BA, Luo Z, Couñago RM, Morey JR, Maher MJ, Ong CL, McEwan AG, Kobe B, O'Mara ML, Paton JC, McDevitt CA. Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae. Nat Commun. 2015;6:6418. doi: 10.1038/ncomms7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ammendola S, Cerasi M, Battistoni A. Deregulation of transition metals homeostasis is a key feature of cadmium toxicity in Salmonella. Biometals. 2014;27:703–714. doi: 10.1007/s10534-014-9763-2. [DOI] [PubMed] [Google Scholar]
- 13.Blaby-Haas CE, Flood JA, Crécy-Lagard V, Zamble DB. YeiR: a metal-binding GTPase from Escherichia coli involved in metal homeostasis. Metallomics. 2012;4:488–497. doi: 10.1039/c2mt20012k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Giri NC, Zhang R, Yamane K, Zhang Y, Maroney M, Costa M. Nickel ions inhibit histone demethylase JMJD1A and DNA repair enzyme ABH2 by replacing the ferrous iron in the catalytic centers. J Biol Chem. 2010;285:7374–7383. doi: 10.1074/jbc.M109.058503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson TL, Chen H, Di Toro DM, D'Angelo G, Costa M. Soluble nickel inhibits HIF-prolyl-hydroxylases creating persistent hypoxic signaling in A549 cells. Mol Carcinog. 2006;45:479–489. doi: 10.1002/mc.20176. [DOI] [PubMed] [Google Scholar]
- 16.Salnikow K, Donald SP, Bruick RK, Zhitkovich A, Phang JM, Kasprzak KS. Depletion of intracellular ascorbate by the carcinogenic metals nickel and cobalt results in the induction of hypoxic stress. J Biol Chem. 2004;279:40337–40344. doi: 10.1074/jbc.M403057200. [DOI] [PubMed] [Google Scholar]
- 17.Kaczmarek M, Cachau RE, Topol IA, Kasprzak KS, Ghio A, Salnikow K. Metal ions-stimulated iron oxidation in hydroxylases facilitates stabilization of HIF-1 alpha protein. Toxicol Sci. 2009;107:394–403. doi: 10.1093/toxsci/kfn251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinn JM, Kropat J, Merchant S. Copper response element and Crr1-dependent Ni2+-responsive promoter for induced, reversible gene expression in Chlamydomonas reinhardtii. Eukaryot Cell. 2003;2:995–1002. doi: 10.1128/EC.2.5.995-1002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eriksson M, Moseley JL, Tottey S, Del Campo JA, Quinn J, Kim Y, Merchant S. Genetic dissection of nutritional copper signaling in chlamydomonas distinguishes regulatory and target genes. Genetics. 2004;168:795–807. doi: 10.1534/genetics.104.030460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutner SH, Provasoli L, Schatz A, Haskins CP. Some approaches to the study of the role of metals in the metabolism of microorganisms. P Am Philos Soc. 1950;94:152–170. [Google Scholar]
- 21.Quinn JM, Merchant S. Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 1998;297:263–279. doi: 10.1016/s0076-6879(98)97020-3. [DOI] [PubMed] [Google Scholar]
- 22.Schloss JA. A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol Gen Genet. 1990;221:443–452. doi: 10.1007/BF00259410. [DOI] [PubMed] [Google Scholar]
- 23.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernández E, Fukuzawa H, González-Ballester D, González-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral JP, Riaño-Pachón DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen CJ, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martínez D, Ngau WC, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, Grossman AR. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveros JC. Venny. An interactive tool for comparing lists with Venn's diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- 27.Micallef L, Rodgers P. eulerAPE: drawing area-proportional 3-Venn diagrams using ellipses. PLoS One. 2014;9:e101717. doi: 10.1371/journal.pone.0101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kropat J, Hong-Hermesdorf A, Casero D, Ent P, Castruita M, Pellegrini M, Merchant SS, Malasarn D. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 2011;66:770–780. doi: 10.1111/j.1365-313X.2011.04537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinn JM, Eriksson M, Moseley JL, Merchant S. Oxygen deficiency responsive gene expression in Chlamydomonas reinhardtii through a copper-sensing signal transduction pathway. Plant Physiol. 2002;128:463–471. doi: 10.1104/pp.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pape M, Lambertz C, Happe T, Hemschemeier A. Differential expression of the Chlamydomonas [FeFe]-hydrogenase-encoding HYDA1 gene is regulated by the copper response regulator1. Plant Physiol. 2012;159:1700–1712. doi: 10.1104/pp.112.200162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soto IC, Fontanesi F, Liu J, Barrientos A. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim Biophys Acta. 2012;1817:883–897. doi: 10.1016/j.bbabio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kropat J, Gallaher SD, Urzica EI, Nakamoto SS, Strenkert D, Tottey S, Mason AZ, Merchant SS. Copper economy in Chlamydomonas: Prioritized allocation and reallocation of copper to respiration vs. photosynthesis. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1422492112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinn JM, Nakamoto SS, Merchant S. Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript. J Biol Chem. 1999;274:14444–14454. doi: 10.1074/jbc.274.20.14444. [DOI] [PubMed] [Google Scholar]
- 36.Tottey S, Block MA, Allen M, Westergren T, Albrieux C, Scheller HV, Merchant S, Jensen PE. Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc Natl Acad Sci U S A. 2003;100:16119–16124. doi: 10.1073/pnas.2136793100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moseley J, Quinn J, Eriksson M, Merchant S. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 2000;19:2139–2151. doi: 10.1093/emboj/19.10.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Róg T, Pasenkiewicz-Gierula M, Vattulainen I, Karttunen M. Ordering effects of cholesterol and its analogues. Biochim Biophys Acta. 2009;1788:97–121. doi: 10.1016/j.bbamem.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 39.Macfie SM, Welbourn PM. The cell wall as a barrier to uptake of metal ions in the unicellular green alga Chlamydomonas reinhardtii (Chlorophyceae) Arch Environ Contam Toxicol. 2000;39:413–419. doi: 10.1007/s002440010122. [DOI] [PubMed] [Google Scholar]
- 40.Pérez-Martín M, Blaby-Haas CE, Pérez-Pérez ME, Andrés-Garrido A, Blaby IK, Merchant SS, Crespo JL. Activation of Autophagy by Metals in Chlamydomonas reinhardtii. Eukaryot Cell. 2015;14:964–973. doi: 10.1128/EC.00081-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ender F, Godl K, Wenzl S, Sumper M. Evidence for autocatalytic cross-linking of hydroxyproline-rich glycoproteins during extracellular matrix assembly in Volvox. Plant Cell. 2002;14:1147–1160. doi: 10.1105/tpc.000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hallmann A. The pherophorins: common. Plant J. 2006;45:292–307. doi: 10.1111/j.1365-313X.2005.02627.x. [DOI] [PubMed] [Google Scholar]
- 44.Kubo T, Abe J, Oyamada T, Ohnishi M, Fukuzawa H, Matsuda Y, Saito T. Characterization of novel genes induced by sexual adhesion and gamete fusion and of their transcriptional regulation in Chlamydomonas reinhardtii. Plant Cell Physiol. 2008;49:981–993. doi: 10.1093/pcp/pcn076. [DOI] [PubMed] [Google Scholar]
- 45.Ferrante P, Catalanotti C, Bonente G, Giuliano G. An optimized, chemically regulated gene expression system for Chlamydomonas. PLoS One. 2008;3:e3200. doi: 10.1371/journal.pone.0003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salnikow K, An WG, Melillo G, Blagosklonny MV, Costa M. Nickel-induced transformation shifts the balance between HIF-1 and p53 transcription factors. Carcinogenesis. 1999;20:1819–1823. doi: 10.1093/carcin/20.9.1819. [DOI] [PubMed] [Google Scholar]
- 47.Salnikow K, Blagosklonny MV, Ryan H, Johnson R, Costa M. Carcinogenic nickel induces genes involved with hypoxic stress. Cancer Res. 2000;60:38–41. [PubMed] [Google Scholar]
- 48.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 49.Urzica EI, Casero D, Yamasaki H, Hsieh SI, Adler LN, Karpowicz SJ, Blaby-Haas CE, Clarke SG, Loo JA, Pellegrini M, Merchant SS. Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell. 2012;24:3921–3948. doi: 10.1105/tpc.112.102491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schofield CJ, Ratcliffe PJ. Signalling hypoxia by HIF hydroxylases. Biochem Biophys Res Commun. 2005;338:617–626. doi: 10.1016/j.bbrc.2005.08.111. [DOI] [PubMed] [Google Scholar]
- 51.La Fontaine S, Quinn JM, Nakamoto SS, Page MD, Göhre V, Moseley JL, Kropat J, Merchant S. Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot Cell. 2002;1:736–757. doi: 10.1128/EC.1.5.736-757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terzulli A, Kosman DJ. Analysis of the high-affinity iron uptake system at the Chlamydomonas reinhardtii plasma membrane. Eukaryot Cell. 2010;9:815–826. doi: 10.1128/EC.00310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes BT, Espenshade PJ. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 2008;27:1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houille-Vernes L, Rappaport F, Wollman FA, Alric J, Johnson X. Plastid terminal oxidase 2 (PTOX2) is the major oxidase involved in chlororespiration in Chlamydomonas. Proc Natl Acad Sci U S A. 2011;108:20820–20825. doi: 10.1073/pnas.1110518109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.