Abstract
Two urea transporters, UT-A1 and UT-A3, are expressed in the kidney terminal inner medullary collecting duct (IMCD) and are important for the production of concentrated urine. UT-A1, as the largest isoform of all UT-A urea transporters, has gained much attention and been extensively studied; however the role and the regulation of UT-A3 are less explored. In this study, we investigated UT-A3 regulation by glycosylation modification. A site-directed mutagenesis verified a single glycosylation site in UT-A3 at Asn279. Loss of the glycosylation reduced forskolin-stimulated UT-A3 cell membrane expression and urea transport activity. UT-A3 has two glycosylation forms, 45 kDa and 65 kDa. Using sugar specific-binding lectins, the UT-A3 glycosylation profile was examined. The 45 kDa form was pulled down by lectin Con A and GNL, indicating an immature glycan with a high amount of mannose (Man); whereas the 65 kDa form is a mature glycan composed of acetylglucosamine (GlcNAc), poly-N-acetyllactosame (poly-LacNAc) that was pulled down by WGA and tomato lectin, respectively. Interestingly, the mature form of UT-A3 glycan contains significant amounts of sialic acid. We explored the enzymes responsible for directing UT-A3 sialylation. Sialyltransferase ST6GalI, but not ST3GalIV, catabolizes UT-A3 α2, 6-sialylation. Activation of PKC by PDB treatment promoted UT-A3 glycan sialylation and membrane surface expression. PKC inhibitor chelerythrine blocks ST6GalI-induced UT-A3 sialylation. Increased sialylation by ST6GalI increased UT-A3 protein stability and urea transport activity. Collectively, our study reveals a novel mechanism of UT-A3 regulation by ST6GalI-mediated sialylation modification that may play an important in kidney urea reabsorption and the urinary concentrating mechanism.
Keywords: Glycosylation, Sialyltransferase, Urea transporter, Protein kinase C
Introduction
In mammals, there are two types of urea transporters, UT-A and UT-B, encoded by the SLC14A2 and SLC14A1 genes, respectively. These genes share a high degree of homology and are tandemly aligned at chromosome 18q12.3 in humans (15). The UT-A urea transporter yields 6 distinct isoforms, of which 3 are found chiefly in the kidney medulla. UT-A1 and UT-A3 are expressed in the inner medullary collecting duct (IMCD) epithelial cells, while UT-A2 is found in the thin descending limb. UT-B is located in the endothelial cells of descending vasa recta (11, 15, 31). Over the past decade, multiple urea transporter knockout mouse models have been generated and demonstrated that all of these transporters actively participate in intra-renal urea recycling and in the urinary concentrating mechanism (10, 32, 35).
UT-A1 is the longest form with an open reading frame of 929 amino acids in rat. Structurally, UT-A3 is the N-terminal half of UT-A1 and UT-A2 represents a C-terminal half of UT-A1. Both UT-A1 and UT-A3 are transcribed from the same promoter. By eliminating UT-A1 exon 10, Fenton et al generated a knockout mouse lacking both UT-A1 and UT-A3 (10). UT-A1/UT-A3 knockout mice have seriously impaired urea concentration ability (10), indicating that UT-A1 and UT-A3 play a crucial role in the overall capacity to concentrate urine. UT-A1, the largest urea transporter isoform in the kidney, has been extensively investigated. It is generally believed that the UT-A1 urea transporter, expressed in the IMCD epithelial cells, plays a major role in the urinary concentrating mechanism (10, 22).
Protein analysis of kidney inner medulla (IM) tissue demonstrated a large abundance of UT-A3 protein expressed in the IMCD (29, 30), suggesting that UT-A3 may also be important in handling renal urea. However, the function and regulation of UT-A3 in the kidney urinary concentrating mechanism are largely underexplored. Although UT-A1 and UT-A3 are both expressed in the kidney IMCD epithelial cells, these two transporters do not form a protein-protein interaction complex, suggesting that UT-A1 and UT-A3 function independently as separate transporters (3). To support it, microinjection of UT-A1 and UT-A3 alone shows that they function individually (26, 27). Since UT-A1 is expressed in the apical membrane of IMCD epithelial cells and mediates urea reabsorption from the tubular lumen, one possible function for UT-A3 in the IMCD is to serve as the basolateral urea transporter, permitting urea to exit from the cells. Indeed, studies by Smith et al. showed that UT-A3 is in the basolateral membrane in mouse kidney IMCD cells and basolateral urea flux is detected from mUT-A3-MDCK cells (27, 28). However, the study by Terris et al. showed that UT-A3 is located in the apical membrane in rat kidney IMCD cells. Double immunostaining revealed no overlap of UT-A3 with Na-K-ATPase at the basolateral surface, making it unlikely that UT-A3 represents the basolateral urea transporter, at least in rat (30). Recently, transgenic restoration of UT-A1 was shown to confer maximal urinary concentration in the absence of UT-A3 (16). Therefore, the role of UT-A3 in kidney urea handling is still unclear.
Glycosylation is a key post-translational modification that affects the structure and function of glycoproteins. Like many other glycoproteins at the plasma membrane, both UT-A1 and UT-A3 are heavily modified by glycosylation. Immunoblotting studies of rat IM demonstrate that the UT-A1 protein exhibits two different glycosylation forms that differ in size, 97 and 117 kDa (4, 14). In contrast, UT-A3 presents two bands at ~44- and ~65-kDa in IM (30). Removing N-glycans by PNGase F treatment, these two bands disappear and yield a single band at ~40 kDa. Our recent studies report that the mature glycosylation of UT-A3, containing a large amount of poly-N-acetyllactosamine (poly-LacNAc), is associated with increased urea transport activity of UT-A3 (29), showing that glycosylation modification plays an important regulatory role for UT-A3 function.
In this study, by sugar specific-binding lectins, we profiled UT-A3 glycan structure components and found that the mature 65 kDa UT-A3 glycan is modified by sialic acid. We further discovered that sialyltransferase ST6GalI, but not ST3GalIV, catalyzes UT-A3 α 2, 6-sialylation. Activation of PKC promotes UT-A3 glycan sialylation and membrane surface expression. Sialylation modification increases UT-A3 protein stability and activity. Taken together, ST6GalI-mediated sialylation may play an important role in kidney UT-A3 functional regulation.
Methods and Materials
Plasmid construction
Rat UT-A3 in p3xFLAG-CMV-10 and pGH19 vector were generated as previously described (29). The asparagine (N) residue at the position 279 in UT-A3 was substituted with glutamine (Q) by a site-directed mutagenesis using primers 5’-GCGTCTTCAGCGCCCCAGATCACCTGGTCAGA-3’ and 5’-CTCTGACCAGGTGATCTGGGGCGCTGAAGACGC-3’. To generate ST6GalI and ST3GalIV constructs, a 1212-bp coding region gene of rat ST6GalI and a 1002-bp coding region gene of rat ST3GalIV were PCR amplified from kidney cDNA and subcloned into pcDNA3 vector or pGH19 vector. Oligonucleotide primers were designed based on the rat ST6GalI (NM_207602) and ST3GalIV gene (NM_203337). For amplification of ST6GalI, the forward primer including Hind III site: 5’-CCAAGCTTATGATTCATACCAACTTGAAGAA A-3’ and the reverse primer including XbaI site: 5’-CTCTAGATCAACAACGAATGTTCCGG AAGCC -3’ were used. For ST3GalIV, the forward primer including Hind III site: 5’-CCAAGCTTATGACCAGCAAATCTCACTGG and the reverse primer including XbaI site: CTCTAGACATCCAAGTCAGAAGTATGTGAGGTT were used. All constructs were verified by nucleotide sequence analysis.
Animals and tissue collection
All animal protocols were approved by the Institutional Animal Care and Use Committee of Emory University. Male Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA) weighing 125–200 g were used in this study. Rats were euthanized by asphyxiation. Kidneys were removed and the IM was collected for lipid raft isolation.
Cell culture, transfection, and treatment
Human embryonic kidney (HEK)-293 cells were maintained in DMEM supplemented with 10% (v/v) FCS at 37 °C in 5% (v/v) CO2. Cells were grown in six-well plates to 80% confluency and transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen) for 48 h. Before cell collection, some cells were treated with 2 μM PDBu (Sigma) for 2 h, 10 μM chelerythrine (Sigma) for 2 h and/or 10 μM forskolin (FSK) for 30 min. For the protein degradation study, the cells were incubated with 100 μg/ml cycloheximide (Calbiochem) and chased for 12 h. Cells were then lysed in a modified RIPA buffer (150 mM NaCl, 10 mM Tris _HCl, pH 7.5, 1 mM EDTA, 1% (v/v) Triton X-100, 1% (m/v) sodium deoxycholate, 0.1% (m/v) SDS, and protease inhibitors) and processed for Western blot analysis.
Cell surface biotinylation
Cell-surface expression level of UT-A3 was examined using the membrane-impermeant biotinylation reagent NHS-SS-biotin (Pierce, 21331). Cell surface biotinylation assays were performed as described previously (7).
Lipid raft isolation
HEK-293 cells were grown in 10 cm plates and were transfected with pcDNA3-FLAG-UT-A3 or UT-A3-N279Q mutant. After 48 h, cells were homogenized in 0.5% (v/v) Brij 96V (Sigma)/TNEV buffer on ice for 30 min. Supernatant (500 μl) was mixed with an equal volume of 80% (v/v) sucrose in TNEV and transferred into a polyallomer centrifuge tube (13×51 mm; Beckman Coulter). Three milliliters of 35% (v/v) sucrose in TNEV was carefully layered on top of the mixture, followed by another 1-ml layer of 5% (v/v) sucrose. The sucrose gradient was then centrifuged in a SW 50.1 rotor (Beckman Coulter) at 34,000 rpm (110,000g) for 20 h at 4°C. After centrifugation, fractions were collected starting from the top to bottom of the tube. Thirteen fractions (~400 μl) were collected, and equal volumes of each fraction were analyzed by 4 −15% (v/v) gradient SDS-PAGE and immunoblotted with relevant antibodies.
Glycosidase PNGase F treatment
For removal of N-glycans from glycoproteins, samples from lipid raft membrane fractions were first denatured in 1.5% (m/v) SDS and 2.5% (m/v) β-mercaptothanol, heated at 100°C for 10 min, then incubated in 50 mM sodium phosphate buffer, 1% (v/v) Noniet P-40, and 1μl of peptide N-glycosidase F (PNGase F, New England Biolabs ) at 37°C for 2 h. The samples were then processed for Western blot.
Lectin pulldown assay
Equal amounts of lipid raft membrane fractions (fractions 2~5) were incubated with 30 μl agarose-conjugated lectins at 4°C overnight. UT-A3 proteins precipitated by different lectins were detected by Western blot. Agarose-bound concanavalin A (Con A), wheat germ agglutinin (WGA), Galant husnivalis lectin (GNL), Tomato lectin (lycopersicum esculentum lectin), datura stramonium lectin (DSL), phaseolus vulgaris leucoagglutinin (PHA-L), Sambucus nigra lectin (SNA), and Aleuria aurantia lectin (AAL) were purchased from Vector Laboratories (Burlingame, CA). Maackia amurensis agglutinin (MAA) was purchased from EY Laboratories.
Oocyte urea flux experiment
X.laevis oocytes were prepared and maintained in OR3 medium as described previously (8). Capped cRNAs were transcribed in vitro from linearized cDNAs with T7 polymerase using the mMESSAGE mMACHINE T7 Ultra Kit (Ambion); cRNAs (2 ng) of UT-A3 alone or with ST6GalI in total volume of 23 nl water were injected into each oocyte. Three days later, healthy oocytes were selected for functional study (8). Urea flux data are expressed as means ± SDs. Oocyte biotinylation (~15 cells/group) was performed as described (13). For tunicamycin treatment, at 2 h prior to cRNA injection, oocytes were pre-injected with 10 ng tunicamycin (Sigma).
Western blot analysis
Western blotting was performed as described previously (18). Blots were probed with primary antibody, followed by anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies (GE Healthcare) and developed by ECL (GE Healthcare). Antibodies used in this study were FLAG (Sigma), UT-A1 (8), GAPDH (Santa Cruz). NIH ImageJ software was used to quantify the band density. The two glycosylation bands of UT-A3 were calculated together and normalized to GAPDH. The change of UT-A3 was expressed as % of control groups.
Statistical analysis
All data are presented as means ± SD. Statistical analysis was determined by one-way analysis of variance (ANOVA) followed by Tukey HSD tests.
Results
1. Disruption of glycosylation site at N279 reduces FSK-stimulated UT-A3 membrane expression
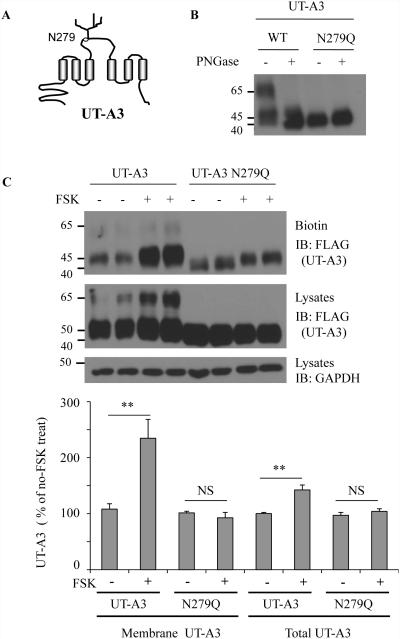
Immunoblotting of kidney IM tissue shows that UT-A3 is a highly glycosylated proteins with two different glycosylation forms (30) due to different extent of glycosylation modification. Amino acid analysis of UT-A3 sequence shows two consensus glycosylation sites in rat UT-A3 at N13 and N279. However, N13 is in the intracellular N-terminus which is unlikely to serve as a glycosylation conjugate site. Only N279 located in the large extracellular loop (Fig.1A) might be the potential glycosylation site. To prove it, we performed site-directed mutagenesis by substituting glutamine (Q) for asparagine (N) at N279 and generated a non-glycosylation site mutant of UT-A3. Wild type UT-A3 and N279Q UT-A3 mutant were transiently transfected into HEK 293 cells. After 48 h, cells were collected for lipid raft isolation. The cell membrane lipid raft fractions were processed for glycosidase digestion. As shown in Fig.1B, after PNGase treatment, the two bands of wild type UT-A3 collapsed to a single band at ~40kDa. In N279Q UT-A3, there is no further size change after glycosidase digestion and the size is the same as the deglycosylated form of UT-A3 treated by PNGase F.
Figure 1. A single glycosylation site at N279 in UT-A3.
A: Structure diagram showing a glycosylation site at N279 located in the large extracellular loop. B: PNGase F digestion. FLAG-UT-A3 WT and FLAG-UT-A3 N279 were transiently transfected into HEK293 cells. 2 days later, the cell membrane lipid raft fractions were collected and de-glycosylated by PNGase F at 37°C for 2h. UT-A3 was analyzed by Western blot with FLAG antibody. C: UT-A3 membrane expression. HEK293 cells were transiently transfected with UT-A3 WT or N279Q for 48h. Cells were pre-treated with 10 μM forskolin (FSK) for 20 min and processed for cell surface biotinylation. Membrane UT-A3 proteins were precipitated by steptavidin beads followed by Western blot with FLAG antibody. The bar graphs show the percent increase in UT-A3 protein levels resulting from FSK treatment (** p<0.01, NS: no significance, n=3).
Activation of the cAMP-PKA pathway stimulates UT-A3, as well as UT-A1, membrane trafficking (2, 3). We then tested whether the glycosylation modification at N279 site regulates UT-A3 trafficking to the cell membrane by cell surface biotinylation. A 30 min treatment with 10 μM FSK significantly increased cell surface UT-A3 expression by ~130 % and total UT-A3 by ~42 % in WT cells but not in N279Q cells (Fig.1C).
2. Mutation of glycosylation site results in reduced UT-A3 urea transport activity
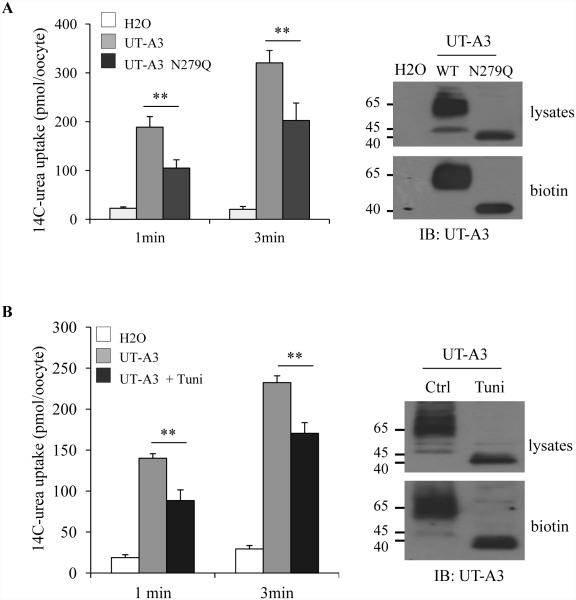
To investigate whether the glycosylation at N279 is important for UT-A3 urea transport activity, we used the Xenopus oocytes expression system. Equal amounts (2 ng/23 nl/oocyte) of UT-A3 WT or N279Q cRNAs were injected into oocytes. Three days later, urea transport activity was measured by 14C-urea uptake. Fig.2A shows that loss of the N-glycans results in reduced urea transport activity. UT-A3 protein expression level on the cell membrane was examined by surface biotinylation and followed by Western blot. The reduced urea transport activity in UT-A3 N279Q is consistent with a decrease of UT-A3 cell surface expression. The role of glycosylation on UT-A3 was further confirmed by pre-treatment of tunicamycin, which blocks protein glycosylation. Figure 2B showed inhibition of glycosylation reduces UT-A3 cell membrane expression and transport activity.
Figure 2. Un-glycosylated UT-A3 has reduced urea transport activity.
Oocytes were injected with UT-A3 or UT-A3 N279Q cRNAs (A) or pre-injected with 10 ng tunicamycin (Tuni) then injected with UT-A3 cRNA (B). After 3 days. Urea transport activity was measured by 14C-labeled urea flux (n=6 oocyte/time point; mean ± SD). UT-A3 protein expression on the cell membrane was examined by biotinylation and followed by Western blot with a UT-A1 NH2-terminal antibody. A representative blot was shown from 3 different experiments.
3. Sialylation modification of UT-A3 in kidney and HEK293 cells
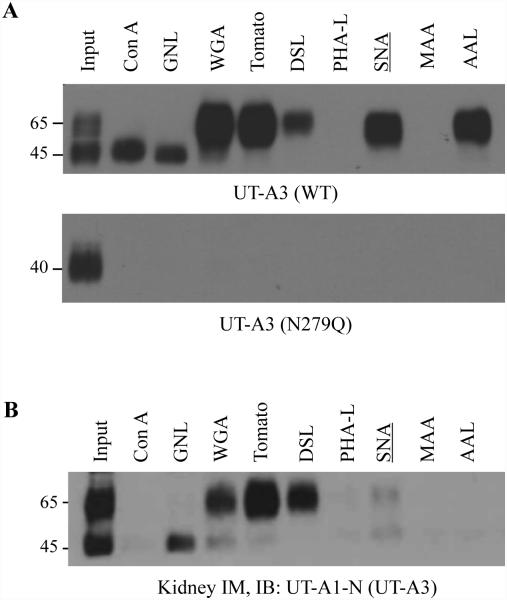
To further analyze UT-A3 modification by glycosylation, we took advantage of sugar-specific binding lectins and profiled UT-A1 glycan structures. Different lectins specifically recognize different structural determinants of glycan carbohydrates. We are particularly interested in UT-A3 on the cell surface. Since cell membrane UT-A3 (as well as UT-A1) is mainly located in the lipid raft microdomains (8, 29), we isolated cell membrane lipid raft fractions (fractions 2-5) from HEK293 cells transfected with UT-A3 or UT-A3 N279Q, as well as rat kidney inner medullary tissues for lectin pulldown assay. UT-A3 has two glycosylation forms, 45 kDa and 65 kDa. Figure 3A shows that the 45 kDa UT-A3 was pulled down by Con A and GNL indicating an immature glycan with high amount of mannose (Man); whereas the 65 kDa form is a mature glycan composed of acetylglucosamine (GlcNAc), poly-N-acetyllactosame (poly-LacNAc) and was pulled down by WGA and tomato lectin. Interestingly, the mature form of UT-A3 glycan also binds to SNA, which specifically binds α-2,6 linked sialic acid. As expected, unglycosylated N279Q UT-A3 was not pulled down by lectins. We also prepared lipid raft fraction samples from normal rat kidney IM and performed lectin pulldown experiments. The lectin pulldown results from IM tissues (Figure 3B) are similar to those for UT-A3 heterogeneously expressed in HEK 293 cells. A significant amounts of sialic acid in UT-A3 glycan indicated that sialic acid modification may play an important role in the regulation of UT-A3 urea transport activity.
Figure 3. Lectin affinity analysis of UT-A3 glycans.
HEK293 cells transfected with UT-A3 WT or N279Q (A) or rat kidney inner medulla (IM) tissues (B) were processed for lipid raft isolation by 5%-40% (v/v) sucrose gradient ultracentrifugation. Lipid raft fractions (2-5) were collected for lectin pull-down assay and followed by Western blot with antibodies to UT-A1 NH2-terminus. A representative blot was shown from >3 different experiments.
4. ST6GalI promotes UT-A3 sialylation modification
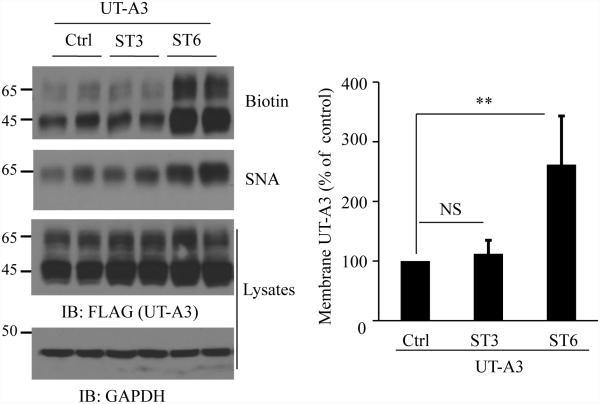
To explore the possible sialyltransferase that could modify UT-A3 sialylation and regulate UT-A3 membrane expression, we assessed sialylation enzymes ST3GalIV and ST6GalI by co-transfection with UT-A3 in HEK293 cells. ST6GalI, which catalyzes α2,6-sialylation, increased UT-A3 cell membrane accumulation as measured by membrane surface biotinylation. In contrast, ST3GalIV, which creates α2,3-sialic acid linkage, did not change UT-A3 plasma membrane expression (Figure 4). UT-A3 sialylation was assessed by agarose-conjugated SNA lectin pulldown assay. Overexpression of ST6GalI, but not ST3GalIV, promotes UT-A3 sialylation pulled down by SNA.
Figure 4. Sialytransferase ST6GalI directs UT-A3 sialylation and cell membrane expression.
UT-A3 membrane expression. HEK293 cells co-transfected UT-A3 without or with ST6GalI (ST6) or ST3GalIV (ST3) were processed for cell surface biotinylation. The total and the cell membrane UT-A3 proteins were examined by Western blot with FLAG antibody. UT-A3 protein sialylation was examined by SNA pull-down assay. Bar graph showed the cell membrane UT-A3 expression as a percent of control (** P<0.01, n=4).
5. ST6GalI promotes UT-A3 glycan sialylation via a PKC pathway.
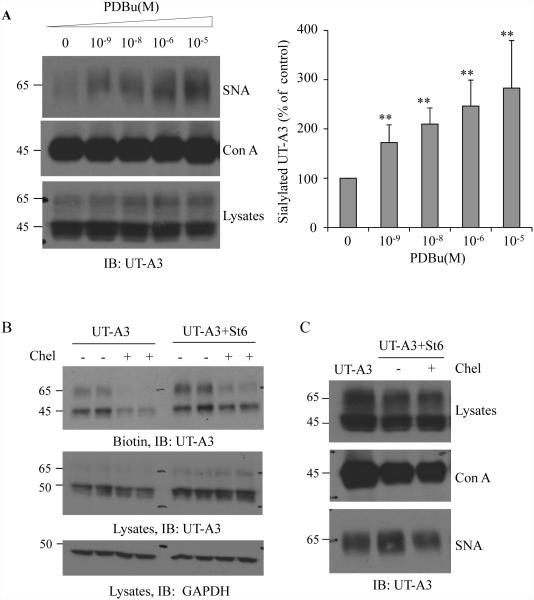
We recently reported that activation of PKC promotes UT-A1 glycan sialylation (18). Next, we examined whether activation of PKC also increases UT-A3 sialylation. UT-A3 transfected HEK293 cells were treated with different doses of PDBu for 4 h. Sialylated UT-A3 was evaluated by SNA pulldown assay. PKC stimulation promoted SNA-precipitated UT-A3 in a dose-dependent manner (Fig. 5A).
Figure 5. Activation of PKC pathway promotes UT-A3 sialylation.
A: UT-A3 sialylation by PDB. HEK 293 cells were transiently transfected with UT-A3 for 48h followed by PDBu treatment for 4h. Sialylated UT-A3 was pulled down by SNA and then examined by western blot. Con A lectin was used as a control. Bar graph showed the band densities of sialylated UT-A3 as a percent of control (0 PDBu) (** P<0.01, n=3). B: UT-A3 membrane expression. HEK293 cells were transiently transfected with UT-A3 alone or together with ST6GalI for 48h, and then treated with or without 2 μM chelerythrine (chel) for 2h. Cell surface proteins were biotinylated and analyzed by Western blot with FLAG antibody. C: Inhibition of ST6GalI-induced sialylation by PKC inhibitor. HEK293 cells transfected with UT-A3 alone or together with ST6GalI for 48h, then treated with chelerythrine for 2 h, sialylated UT-A3 was pulled down by SNA and then examined by western blot. B, C showed the representative western blots from three independent experiments.
We then investigated whether overexpression of ST6GalI increases UT-A3 cell membrane expression. The amount of UT-A3 at the cell membrane was increased when the cells overexpressed ST6Gal1; however this effect was largely prevented by PKC inhibition, indicating an important role of PKC in ST6GalI-mediated UT-A3 membrane expression. To prove that PKC is involved in ST6GalI-catalylzed UT-A3 sialylation, UT-A3 glycan sialylation was examined by SNA lectin pulldown. Inhibition of the PKC pathway by 4 h chelerythrine treatment decreased ST6GalI-induced sialylated UT-A3 (Figure 5C).
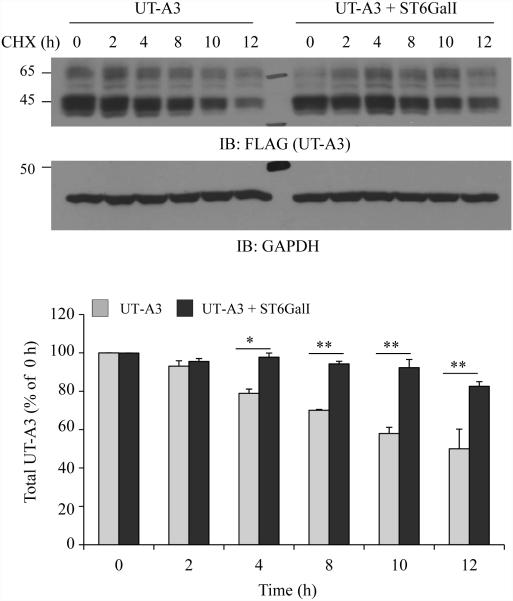
6. UT-A3 sialylation mediated by ST6GalI increases UT-A3 protein stability
One of the important roles of N-glycosylation is to increase protein stability (7). We next investigated whether sialylation modification will increase UT-A3 protein stability. UT-A3 was cotransfected with or without ST6GalI into HEK293 cells. After 48h, cells were treated with cycloheximide (CHX) to block new protein synthesis. Protein degradation was chased for 12 h. As seen in Figure 6, ST6GalI increased total UT-A3 protein stability.
Figure 6. Effect of ST6GalI on UT-A3 protein degradation.
HEK293 cells transfected with UT-A3 alone or together with ST6GalI were treated with 100 μg/ml cycloheximide (CHX) for the indicated time. The cells were lysed with RIPA buffer and total UT-A3 protein levels were evaluated by Western blot with FLAG antibody. Bar graph showed the band densities of UT-A3 (including both 65- and 45-kDa) as a percent of control (0 h) (* P<0.05, ** P<0.01, n=3).
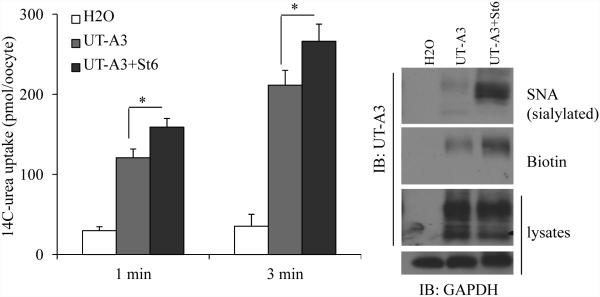
7. Increased sialylation induced by ST6GalI increases UT-A3 activity
As shown in Figure 4, 5 and 6, ST6GalI catalyzed UT-A3 sialylation and increased UT-A3 membrane expression and protein stability, we then asked whether these changes result in an increase of UT-A3 activity by oocyte expression system. Co-injection of ST6GalI cRNA promoted UT-A3 sialylation as pulled down by lectin SNA and increased UT-A3 cell membrane expression as judged by cell surface biotinylation (Figure 7). UT-A3 activity was measured by 14C-urea flux. Sialylation of UT-A3 by ST6GalI increased UT-A3 urea transport activity.
Figure 7. Effect of ST6GalI on UT-A3 activity.
Oocytes were injected with cRNAs of UT-A3 (2 ng/cell) alone or plus ST6GalI (2 ng/cell). After 3 days, rea transport activity was measured by 14C-labeled urea flux (n=6 oocyte/time point; mean ± SD). UT-A3 protein expression on the cell membrane was examined by biotinylation and sialylated UT-A3 was pulled down by SNA, followed by Western blot with a UT-A1 NH2-terminal antibody (experiments n=3).
Discussion
We previously reported that glycosylation is an important post-translational modification for both kidney urea transporter UT-A1 (7, 8, 18) and UT-A3 (29). In the current study, we extended our studies on UT-A3 regulation by N-glycans modification. The major findings are that: 1) UT-A3 has a single glycosylation site at N279 and mutation of N279 reduces UT-A3 urea transporter membrane expression and urea transport activity; 2) the mature form of 65 kDa UT-A3 is highly modified by sialic acid; 3) ST6GalI, but not ST3GalIV, catalyzes UT-A3 sialylation and enhances urea transport membrane trafficking and activity; 4) activation of PKC promoted UT-A3 glycan sialylation and inhibition of PKC activity blocks ST6GalI-induced UT-A3 sialylation and membrane surface expression; and 5) sialylation modification increases UT-A3 protein stability; 6) increased UT-A3 sialylation by ST6GalI is associated with increased urea transport activity.
Protein glycosylation is the most sophisticated post-translational modification because of the large number of enzymatic steps involved in glycan biosynthesis, glycan extension, and modification (20, 25). It starts with the addition of a 14 residue core oligosaccharide chain (Glc3Man9GlcNAc2) to the nascent polypeptide in the ER. After transit to the Golgi complex, these oligosaccharides are trimmed and modified by addition of N-acetylglucosamine (GlcNAc), galactose (Gal), fucose and sialic acid to complete the complex oligosaccharides. Different glycosylation is mainly due to different processing after α-mannosidase digestion by the sequential action of specific glycosyltransferases (17, 36). As seen in many transporter proteins (1, 9), UT-A3 is highly glycosylated with two forms due to different extent of glycosylation. We for the first time verified that the 65 kDa UT-A3 is the mature glycosylation form contained poly-LacNAc..
Interestingly, the glycan of 65 kDa UT-A3 is highly sialylated by α-2, 6-linked sialic acid. Sialic acid (N-acetylneuraminic acid) is a large and negatively charged sugar with a nine-carbon backbone. Sialylation occurring in the trans-Golgi apparatus is one of the major glycan maturation processes (23). Sialic acids can be linked to N-glycan via α2, 3, α-2, 6, and α2, 8, corresponding to three types of enzymes. The most common linkages on N-glycans are to the 3- or 6-position of galactose residues, termed α2,3 and α2,6 linkages, respectively. We recently performed RNA-seq analysis of gene expression profiles from kidney IM tissues and found that kidney IM expresses α2,3 sialyltransferase (ST3GalII, ST3GalIV, ST3GalVI) and α 2,6 sialyltransferase (ST6GalI, ST6GalnacII, ST6GalnacIII), but not α2,8-sialyltransferase (21). ST3GalIV is the most significantly increased sialyltransferase gene in IM from streptozotocin-induced diabetic rats (21). However, we found that ST6GalI increases UT-A3 α 2,6 sialylation. ST3GalIV may contribute to other glycoprotein sialylation in kidney IM.
Sialic acids typically are attached to the terminus of N-glycans and cap the terminal galactose in carbohydrate chains on the cell surface (6, 37). Glycoprotein sialylation modification is involved in a broad range of biological and pathological processes such as immune regulation, and bacterial and viral infections (34). A specific role for sialylation in the apical delivery of glycoproteins has been reported in Caco-2 (33) and MDCK cells (19, 24). Mo et al. found that sialylation of glycans is required for endolyn apical delivery (19), underlining the crucial role of sialylation in protein membrane trafficking. Our results demonstrated that sialylation modification of UT-A3 by ST6GalI increases UT-A3 activity and this effect is mainly through increased UT-A3membrane expression and protein stability.
Activation of PKC stimulates UT-A1 glycan sialylation both in vivo and in vitro, and this effect is blocked by PKC inhibition (18). Similarly, in this study we found that activation of PKC by PDBu increased UT-A3 sialylation. Furthermore, inhibition of PKC activity by chelerythrine blocks ST6GalI-induced UT-A3 sialylation and membrane surface expression. Therefore, activation of PKC pathway can promote both UT-A1 and UT-A3 sialylation. This is consistent with a study by Breen et al. showing that stimulation of PKC resulted in an increase in ST6GalI catalytic activity (5). Ma et al. reported the presence of phosphorylated serine and threonine sites on α2, 6-sialyltransferase (12), suggesting a functional involvement of ST6GalI-mediated sialylation by PKC activation.
In summary, UT-A3 is abundantly expressed in kidney IMCD epithelial cells. Compared with another urea transporter UT-A1, the knowledge of UT-A3 currently is very limited. This study provides evidence that N-linked glycosylation is functionally essential for regulating the activity of urea transporter UT-A3 and a pivotal role for α 2, 6-sialylation in the regulation of UT-A3 urea transport activity. Moreover, activation of PKC enhances ST6GalI-mediated UT-A3 glycan sialylation. We presume that at least one mechanism for PKC activation to increase kidney urea transport activity is through increasing UT-A3 sialylation, a novel mechanism of kidney UT-A3 regulation. In addition, sodium and urea are the two major solutes that set up the kidney medullary osmolarity gradient. However, there are presently no drugs that target the urea transporter. Future work by targeting ST6GalI that affects UT-A3 function in vivo may provide a promising pharmaceutical intervention for the water-overloaded diseases such as hypertension, congestive heart failure, cirrhosis, and nephrotic syndrome.
Acknowledgments
This work was supported by NIH grants R01-DK087838 (to G. Chen), R01-DK89828 (to J. Sands), and by Chinese National Natural Science Foundation Project 81570358, 81300248 (to X. Song), Heilongjiang Science and Technology funding LBH-Q13113 (to X. Song).
Reference
- 1.Alli AA, Bao HF, Liu BC, Yu L, Aldrugh S, Montgomery DS, Ma HP, Eaton DC. Calmodulin and CaMKII modulate ENaC activity by regulating the association of MARCKS and the cytoskeleton with the apical membrane. Am J Physiol Renal Physiol. 2015;309:F456–463. doi: 10.1152/ajprenal.00631.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blessing NW, Blount MA, Sands JM, Martin CF, Klein JD. Urea transporters UT-A1 and UT-A3 accumulate in the plasma membrane in response to increased hypertonicity. Am J Physiol Renal Physiol. 2008;295:F1336–1341. doi: 10.1152/ajprenal.90228.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blount MA, Klein JD, Martin CF, Tchapyjnikov D, Sands JM. Forskolin stimulates phosphorylation and membrane accumulation of UT-A3. Am J Physiol Renal Physiol. 2007;293:F1308–1313. doi: 10.1152/ajprenal.00197.2007. [DOI] [PubMed] [Google Scholar]
- 4.Bradford AD, Terris JM, Ecelbarger CA, Klein JD, Sands JM, Chou CL, Knepper MA. 97- and 117-kDa forms of collecting duct urea transporter UT-A1 are due to different states of glycosylation. Am J Physiol Renal Physiol. 2001;281:F133–143. doi: 10.1152/ajprenal.2001.281.1.F133. [DOI] [PubMed] [Google Scholar]
- 5.Breen KC, Georgopoulou N. The role of protein phosphorylation in alpha2,6(N)-sialyltransferase activity. Biochem Biophys Res Commun. 2003;309:32–35. doi: 10.1016/s0006-291x(03)01529-8. [DOI] [PubMed] [Google Scholar]
- 6.Cha SK, Ortega B, Kurosu H, Rosenblatt KP, Kuro OM, Huang CL. Removal of sialic acid involving Klotho causes cell-surface retention of TRPV5 channel via binding to galectin-1; Proceedings of the National Academy of Sciences of the United States of America; 2008. pp. 9805–9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G, Frohlich O, Yang Y, Klein JD, Sands JM. Loss of N-linked glycosylation reduces urea transporter UT-A1 response to vasopressin. J Biol Chem. 2006;281:27436–27442. doi: 10.1074/jbc.M605525200. [DOI] [PubMed] [Google Scholar]
- 8.Chen G, Howe AG, Xu G, Frohlich O, Klein JD, Sands JM. Mature N-linked glycans facilitate UT-A1 urea transporter lipid raft compartmentalization. FASEB J. 2011;25:4531–4539. doi: 10.1096/fj.11-185991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen M, Cai H, Klein JD, Laur O, Chen G. Dexamethasone increases aquaporin-2 protein expression in ex vivo inner medullary collecting duct suspensions. Front Physiol. 2015;6:310. doi: 10.3389/fphys.2015.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenton RA, Chou CL, Stewart GS, Smith CP, Knepper MA. Urinary concentrating defect in mice with selective deletion of phloretin-sensitive urea transporters in the renal collecting duct. Proc Natl Acad Sci U S A. 2004;101:7469–7474. doi: 10.1073/pnas.0401704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenton RA, Stewart GS, Carpenter B, Howorth A, Potter EA, Cooper GJ, Smith CP. Characterization of mouse urea transporters UT-A1 and UT-A2. Am J Physiol Renal Physiol. 2002;283:F817–825. doi: 10.1152/ajprenal.00263.2001. [DOI] [PubMed] [Google Scholar]
- 12.Gallagher HC, Murphy KJ, Foley AG, Regan CM. Protein kinase C delta regulates neural cell adhesion molecule polysialylation state in the rat brain. J Neurochem. 2001;77:425–434. doi: 10.1046/j.1471-4159.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- 13.Huang H, Yang Y, Eaton DC, Sands JM, Chen G. The N-terminal 81-aa fragment is critical for UT-A1 urea transporter bioactivity. J Epithel Biol Pharmacol. 2010;3:34–39. doi: 10.2174/1875044301003010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D, Sands JM, Klein JD. Changes in renal medullary transport proteins during uncontrolled diabetes mellitus in rats. Am J Physiol Renal Physiol. 2003;285:F303–309. doi: 10.1152/ajprenal.00438.2002. [DOI] [PubMed] [Google Scholar]
- 15.Klein JD, Blount MA, Sands JM. Urea transport in the kidney. Compr Physiol. 2011;1:699–729. doi: 10.1002/cphy.c100030. [DOI] [PubMed] [Google Scholar]
- 16.Klein JD, Wang Y, Mistry A, LaRocque LM, Molina PA, Rogers RT, Blount MA, Sands JM. Transgenic Restoration of Urea Transporter A1 Confers Maximal Urinary Concentration in the Absence of Urea Transporter A3. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2014121267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Yang B, Chen M, Klein JD, Sands JM, Chen G. Activation of protein kinase C-alpha and Src kinase increases urea transporter A1 alpha-2, 6 sialylation. J Am Soc Nephrol. 2015;26:926–934. doi: 10.1681/ASN.2014010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mo D, Costa SA, Ihrke G, Youker RT, Pastor-Soler N, Hughey RP, Weisz OA. Sialylation of N-linked glycans mediates apical delivery of endolyn in MDCK cells via a galectin-9-dependent mechanism. Mol Biol Cell. 2012;23:3636–3646. doi: 10.1091/mbc.E12-04-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nairn AV, York WS, Harris K, Hall EM, Pierce JM, Moremen KW. Regulation of glycan structures in animal tissues: transcript profiling of glycan-related genes. The Journal of biological chemistry. 2008;283:17298–17313. doi: 10.1074/jbc.M801964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian X, Li X, Ilori TO, Klein JD, Hughey RP, Li CJ, Alli AA, Guo Z, Yu P, Song X, Chen G. RNA-seq analysis of glycosylation related gene expression in STZ-induced diabetic rat kidney inner medulla. Front Physiol. 2015;6:274. doi: 10.3389/fphys.2015.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sands JM, Blount MA, Klein JD. Regulation of renal urea transport by vasopressin. Trans Am Clin Climatol Assoc. 2011;122:82–92. [PMC free article] [PubMed] [Google Scholar]
- 23.Schauer R. Achievements and challenges of sialic acid research. Glycoconjugate journal. 2000;17:485–499. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slimane TA, Lenoir C, Sapin C, Maurice M, Trugnan G. Apical secretion and sialylation of soluble dipeptidyl peptidase IV are two related events. Exp Cell Res. 2000;258:184–194. doi: 10.1006/excr.2000.4894. [DOI] [PubMed] [Google Scholar]
- 25.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 26.Stewart GS, Fenton RA, Wang W, Kwon TH, White SJ, Collins VM, Cooper G, Nielsen S, Smith CP. The basolateral expression of mUT-A3 in the mouse kidney. Am J Physiol Renal Physiol. 2004;286:F979–987. doi: 10.1152/ajprenal.00334.2003. [DOI] [PubMed] [Google Scholar]
- 27.Stewart GS, King SL, Potter EA, Smith CP. Acute regulation of mUT-A3 urea transporter expressed in a MDCK cell line. Am J Physiol Renal Physiol. 2007;292:F1157–1163. doi: 10.1152/ajprenal.00183.2006. [DOI] [PubMed] [Google Scholar]
- 28.Stewart GS, Thistlethwaite A, Lees H, Cooper GJ, Smith C. Vasopressin regulation of the renal UT-A3 urea transporter. Am J Physiol Renal Physiol. 2009;296:F642–648. doi: 10.1152/ajprenal.90660.2008. [DOI] [PubMed] [Google Scholar]
- 29.Su H, Carter CB, Frohlich O, Cummings RD, Chen G. Glycoforms of UT-A3 urea transporter with poly-N-acetyllactosamine glycosylation have enhanced transport activity. Am J Physiol Renal Physiol. 2012;303:F201–208. doi: 10.1152/ajprenal.00140.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terris JM, Knepper MA, Wade JB. UT-A3: localization and characterization of an additional urea transporter isoform in the IMCD. Am J Physiol Renal Physiol. 2001;280:F325–332. doi: 10.1152/ajprenal.2001.280.2.F325. [DOI] [PubMed] [Google Scholar]
- 31.Timmer RT, Klein JD, Bagnasco SM, Doran JJ, Verlander JW, Gunn RB, Sands JM. Localization of the urea transporter UT-B protein in human and rat erythrocytes and tissues. Am J Physiol Cell Physiol. 2001;281:C1318–1325. doi: 10.1152/ajpcell.2001.281.4.C1318. [DOI] [PubMed] [Google Scholar]
- 32.Uchida S, Sohara E, Rai T, Ikawa M, Okabe M, Sasaki S. Impaired urea accumulation in the inner medulla of mice lacking the urea transporter UT-A2. Mol Cell Biol. 2005;25:7357–7363. doi: 10.1128/MCB.25.16.7357-7363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulloa F, Franci C, Real FX. GalNAc-alpha -O-benzyl inhibits sialylation of de Novo synthesized apical but not basolateral sialoglycoproteins and blocks lysosomal enzyme processing in a post-trans-Golgi network compartment. The Journal of biological chemistry. 2000;275:18785–18793. doi: 10.1074/jbc.M000510200. [DOI] [PubMed] [Google Scholar]
- 34.Varki A, Gagneux P. Multifarious roles of sialic acids in immunity. Ann N Y Acad Sci. 2012;1253:16–36. doi: 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang B, Bankir L, Gillespie A, Epstein CJ, Verkman AS. Urea-selective concentrating defect in transgenic mice lacking urea transporter UT-B. J Biol Chem. 2002;277:10633–10637. doi: 10.1074/jbc.M200207200. [DOI] [PubMed] [Google Scholar]
- 36.Yousefi S, Higgins E, Daoling Z, Pollex-Kruger A, Hindsgaul O, Dennis JW. Increased UDP-GlcNAc:Gal beta 1-3GaLNAc-R (GlcNAc to GaLNAc) beta-1, 6-N-acetylglucosaminyltransferase activity in metastatic murine tumor cell lines. Control of polylactosamine synthesis. The Journal of biological chemistry. 1991;266:1772–1782. [PubMed] [Google Scholar]
- 37.Zhuo Y, Bellis SL. Emerging role of alpha2,6-sialic acid as a negative regulator of galectin binding and function. The Journal of biological chemistry. 2011;286:5935–5941. doi: 10.1074/jbc.R110.191429. [DOI] [PMC free article] [PubMed] [Google Scholar]