Abstract
DNA origami is a DNA-based nanotechnology that utilizes programmed combinations of short complementary oligonucleotides to fold a large single strand of DNA into precise 2-D and 3-D shapes. The exquisite nanoscale shape control of this inherently biocompatible material is combined with the potential to spatially address the origami structures with diverse cargos including drugs, antibodies, nucleic acid sequences, small molecules and inorganic particles. This programmable flexibility enables the fabrication of precise nanoscale devices that have already shown great potential for biomedical applications such as: drug delivery, biosensing and synthetic nanopore formation. In this Progress Report, we will review the advances in the DNA origami field since its inception several years ago and then focus on how these DNA-nanodevices can be designed to interact with cells to direct or probe their behavior.
Keywords: DNA origami, drug delivery, nanopores, biosensing, nanoparticles
1. Introduction
DNA has long been studied due to its ubiquity in biological function and as a fingerprint material for biological systems past and present. More recently it has received a surge in interest by engineers and scientists due to its ability to carry encoded information along its backbone that can be used as blueprints for self-assembly with nanometer spatial resolution due to its natural structural properties. This provides for precise control over the shape of an inherently biocompatible material on the nanoscale and the ability to spatially position cargos on/in these shapes with nanometer resolution. DNA-based nanostructures, therefore, have excellent potential for biomedical applications where nanoscale features are desirable for cell interaction (e.g., drug delivery or synthetic nanopore formation) or cell behaviour studies (i.e., biosensing applications). We begin by providing the reader with a background on the rapid evolution of the DNA-origami field. In Box 1, readers new to the origami field can learn the basic design principles of DNA-origami. We then focus on the progress of the field as it relates to applications in drug delivery, nanopore formation and biosensing before offering our perspective on the current challenges and future directions for the field.
Box 1: A Scaffold to Build DNA Origami Design Knowledge.
The great majority of DNA origami structures are based on the most natural DNA form, namely B-form DNA. B-form DNA is a right handed double helix, with a helical pitch (distance for one complete turn of DNA) of 10.5 base pairs (bp) or 3.4 – 3.6 nm.

The fundamental building blocks of structural DNA nanotechnology are DNA crossovers as initially described by Seeman[77]. A single crossover (immobile Holliday junction) keeps two DNA strands attached but cannot ascribe any relationship in terms of chain orientation. However, placing multiple crossovers between two double helices prescribes their relative orientation between the crossovers (assuming the length between crossovers is < the persistence length of DNA ≈ 50 nm). Of course, (1) a crossover can only occur at an integer number of bp increments, and (2) to design helices that are straight and parallel, the crossover must occur where the rotation of the two helices aligns them along their backbone following the B-form DNA structure. Alterations to the second point cause bundles to twist or bend[6, 17, 18].

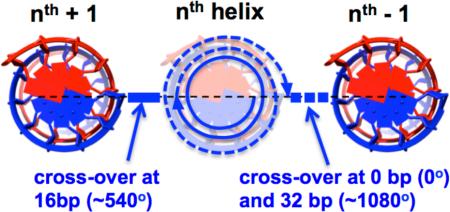
To create a planar (2-D) sheet of DNA, thus, the crossovers must generally align in a flat plane. That is, crossovers between the nth helix and its neighboring (nth-1) helix occur when the backbone of the nth DNA helix is orientead at 0° (or 360°, 720°,...), and crossovers to the (nth+1) helix occur when it has oriented at 180° (540°, 900°,...). To achieve this with an integer number of base pairs between crossovers, the helical pitch of DNA is adjusted slightly to 10.67 bp, meaning 32 bp is equivalent to 3 full turns of DNA (1080°, i.e., back to the 0° orientation pointing towards the nth-1 helix), and 16 bp equals 1.5 turns (540°, i.e. at the 180° orientation pointing towards the nth+1 helix). To create planar sheets, thus, alternating direction crossovers are placed at 16 bp intervals (or odd integer multiples thereof) between a helix and its two neighbouring helices. This allows for creation of 2-D structures with a resolution of 16 bp (~6 nm) in the helical axis direction and 2 neighbouring double helices of 6 – 7 nm in the lateral direction[5]. The violation of the natural pitch of B-form DNA, however, results in a torque that may cause 2-D sheets (or square lattice structures) to have a slight twist.
The initial move from 2-D to 3-D exploited the natural angles of B-form DNA[11,12,15,16]. At 2 turns = 21 bp = 720°, 7 bp = 240°. This can be used to have each DNA helix crossover with 3 neighbors at 120° intervals to create a honeycomb lattice. In this case, a DNA helix can cross over with one of three neighbouring helices at 7 bp intervals and with the same helix every 21 bp. This is often visualised by drawing planes through the DNA at 7 bp increments (all possible crossover sites to one neighbouring helix) and designing staple crossovers at those locations.
To create close-packed structures and to develop rectangular objects, a square lattice is required[17]. The square lattice is essentially an extension of the 2-D lattice design principles. Again we fix the helical pitch at 10.67 bp, so that 32 bp is equivalent to 1040°. We can now crossover in 8 bp increments, which equates to crossovers at 0°, 270°, 540°(180°) and 810° (90°) rotations. Thus, a helix can crossover with one of four neighbouring strands at 90° intervals every 8 bp and with the same strand every 32 bp.
This idea can, in fact, be extrapolated to access many angles between neighboring helices. Since each bp corresponds to 34.29°, crossovers can be placed at 1 bp increments to give multiples of 34.29°. Consider three neighbouring helices: the first two helices define the plane. Assuming the third helixlocation is not impeded by the first two helices, it can be positioned at increments of 34.29°. Indeed the flexibility of B-form DNA allows for slightly less than or slightly greater than 10.5 bp per turn. This principle can be exploited to access even more angles between neighbouring helices (described in detail in Ref. [20])
Finally, curvature in the plane parallel to the helices can be achieved by increasing/decreasing the number of base pairs between neighbouring helices[18]. For example if the crossovers between the nth and nth+1 helix are every 42 bp along the nth helix and only every 21 bp on the nth helix, the structure curves towards the nth helix. Note, that if the change in the number of bp between crossovers is a multiple of the number of potential crossover sites between bp (i.e., 21, 42, 63,..., for B-form DNA) no torque will be introduced into the system. If there is not an integer multiple of the potential crossover sites deleted between neighbouring helices, a compensatory torque will be introduced into the structure and this can be used to create twisted objects or can be balanced out between different helices to give pure bending (see Dietz et al. for full details[18]).
Combining these various design principles gives a huge amount of flexibility in generating 3-D structures. Although the latter two design principles involve complex evaluation of staples/crossovers, an open source software, caDNAno (www.cadnano.org) is available for researchers to design honeycomb and square lattice structures, and structure prediction software [78][12] has been developed as a tool to allow virtual prototyping and design refinement prior to fabrication. This software is readily accessible after watching short tutorials. Once designed, the scaffold strand and oligonucleotide staples are typically mixed in a 1:10 ratio and passed through thermal annealing cycles on PCR machines for a period of minutes to hours to days depending on complexity to yield the desired origami structures (full protocols and structures described by Castro et al. [12]).
2. From a 4-base Coding System to Artful Origami
Each DNA base can take on one of four nucleotides, which consist of a nitrogen-containing nucleobase – adenine (A), thymine (T), cytosine (C), guanine (G) — as well as a monosaccharide sugar (deoxyribose) and a phosphate group. Complementary strands in a double helix run antiparallel and binding occurs between the complementary nucleotides of the two strands: A with T and C with G. The sequence of nucleotides in a DNA strand can program spatial details on the nanoscale based on the structural characteristics of DNA, which were first fully defined in the 1950s by Watson-Crick [1] based on the excellent contributions of Levene and Chargaff before them[2].
The base-pair coding principle alone was used to initiate the field of DNA nanotechnology: the sequence of DNA strands can be designed to be complementary in a manner that enables programming the self-assembled geometry. The next critical step occurred with the description of naturally occurring crossovers by Holliday in the mid 1960s[3]. These Holliday junctions make specific connections between two DNA double helices. This allows for the creation of DNA lattices; however, given DNA's flexibility and the mobility of natural Holliday junctions, these crossovers between strands do little in terms of spatially defining DNA nanostructures. The crucial development, first conceived by Seeman in 1982[4], was to design DNA sequences that encoded immobile Holliday junctions. Immobilizing connection points between DNA double helices provided a foundation for the rational design of DNA nanostructures. This led to DNA crystals or tiles, which were experimentally demonstrated by Seeman's group in 1998[5]. Furthermore, these tiles can be joined together to make planar and periodic DNA structures using single stranded extensions (‘sticky ends’) that are complementary to single strands on other tiles. These structures have demonstrated immense potential and laid the foundation for a new type of DNA nanotechnology, namely, DNA Origami – named after the Japanese art of folding paper.
The DNA origami technique helps overcome some of the challenges associated with DNA tile method, specifically, that the DNA sequences must be fully designed from scratch and that the use of oligonucleotides throughout tile structures requires careful control of the stoichiometry[6]. By dedicating one of the strands as a scaffold strand that runs throughout the structure and either folds on itself[7] or is folded by complementary staple strands[6], the sequence design is simplified; the requirement to carefully control the stoichiometry is reduced; and structures with higher yields and enhanced geometric complexity can be formed. Interestingly, recent efforts by Yin and co-workers demonstrated that similarly complex structures can be achieved through the exclusive use of oligonucleotide strands by adopting a scaffold-like approach[8].
We will first describe the rapid evolution of DNA origami structures from the original 2-D and 3-D objects with defined spatial patterning to the vast array of objects that are implemented today. In box 1, the reader can get a brief guide to the design basics of DNA origami design and fabrication, as well as references for more in depth study. We will then describe the application of DNA origami particularly as it pertains to cellular applications that include: DNA origami based drug delivery, DNA origami based synthetic cell-membrane pores and biosensing applications. We conclude the article with a perspective of the future directions of this dynamic and promising field. Of note, we will almost exclusively focus on DNA origami in this review and only mention non-origami based techniques as they pertain to the development or advances in the origami field; readers are referred to the following recent excellent reviews that include non-origami DNA nanotechnology[9-11].
3. DNA Origami: A Technique for 2-D Objects That Leads to Endless 3-D Possibilities
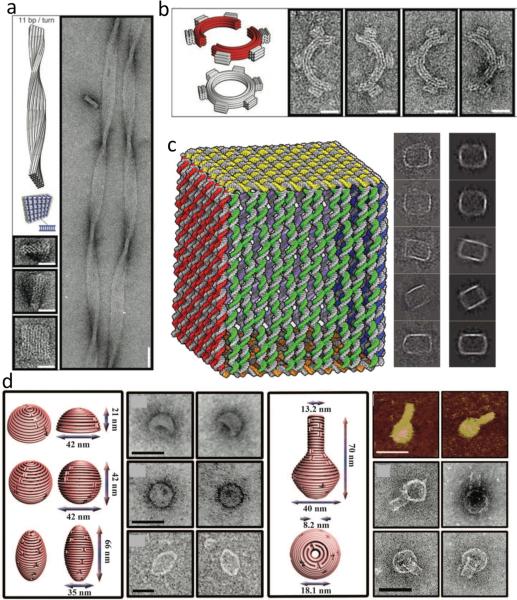
The phrase DNA origami was first used by Rothemund in 2006, who described the design principles for folding a single long strand of DNA (specifically the genomic DNA of M13mp18, which is still commonly used today), using a complementary set of short oligonucleotide staple strands[6]. The design principles for Rothemund's original 2-D DNA origami are more fully described in the boxed inset below and full protocols were described by Castro et al.[12]. Given the scaffold strand length and DNA origami shape characteristics, Rothemund essentially had a 100nm2 surface with 6 nm pixels[13] with which to design a vast array of DNA shapes. This was used to develop beautiful DNA origami shapes that could be imaged using atomic force microscopy (AFM) (Figure 1). Furthermore, complementary sticky ends on different objects could be hybridized together to create larger 1D ribbon or 2-D lattice type arrays of structures[6].
Figure 1.
The original DNA origami design technique and shapes. (a) Rothemund[6] used staple oligonucleotides (colored strands) to raster a single continuous scaffold strand (black line) into defined stapes. (b) DNA helix representation of the staples (colored strands) and scaffold (black strands) for the origami technique. (c) Designs for (rows 1 and 2) and AFM images of fabricated DNA origami shapes. Reproduced from [6].
Inspired by a non-origami hexagonal DNA tube described by Seeman's group in 2005[14], Shih and colleagues designed a 3-D DNA origami nanotube that is based on a honeycomb lattice[15]. In this nanotube, six DNA helices were stapled together at consecutive 240° angles (i.e. every 21 base-pairs) to form a hexagonal tube that could be used to enhance alignment in NMR techniques (further details below). Shih and colleagues established this honeycomb lattice approach more universally in 2009, describing a honeycomb shaped cross-sectional lattice in the X-Y plane looking end-on at DNA helices with the helical axis extending in the Z-direction as the basis for an array of structures including rods, square nuts, crosses and stacked crosses (Figure 2)[16]. Although the honeycomb lattice is versatile enough to design a huge range of structures, more close packing or flat cross-section edges may be desirable.
Figure 2.
The original 3-D origami folding designs, schematics and sample structures. (a) folding schematic for honeycomb arranged DNA origami lattices[16]. (b) Design and TEM imaging of honeycomb lattice shapes including a box, square nut, railed bridge, slotted cross and stracked cross (scale bar = 20nm) [16]. (c) Folding schematic for square lattice structures [17]. (d) 3- and 6-layer DNA origami cuboid designs and TEM imaging (scale bar 20nm) [17]. Reprinted with permission from[16] and [17] was reprinted with permission © 2009 American Chemical Society.
A second, close-packed lattice strategy was described in 2009 also by Shih and colleagues, which was based on a square lattice that allows for structures with more rectangular cross-sections (see Figure 2b, c)[17]. In this first paper, the authors described the design of various cuboids with consecutively lower aspect ratios (helices wide × layers of helices). However, while the honeycomb lattice follows the natural helical geometry of double-stranded DNA (1 full turn for 10.5 bp, or two turns in 21 bp), the square lattice instead forces 1 full turn every 10.67 bp (or three turns in 32 bp). This forced underwinding results in a global right handed twist on the origami structures, which can be exploited for design of select origami shapes. The same study described significant twisting in helices with 3 layers or less (note that in 2-D, these twists would also be observed). This twisting diminished as the number of layers increased to 6 or more[17], and could be reduced by locally deleting base-pairs to counteract the twist mismatch.
DNA origami shapes can be intentionally designed to twist and bend by forcing crossovers into the structures at non-natural intervals, such that compensatory deformation is introduced[18]. When crossovers in hexagonally packed DNA origami structure (typically 7 bp) are forced to be < 7 bp that section is stretched and over-wound; therefore, a local internal stress is induced that is relieved by a global shortening and a left-handed torque. Conversely, a full turn that is forced to be >7 bp is shortened and underwound leading to a global lengthening and right-handed torque. These principles were applied to a 10 × 6 helix bundle and, using sticky ends, ribbons of DNA with periodic twists of up to 3.5° per DNA helix turn were generated (Figure 3a). These under- and overwound segments can be carefully designed into the origami structures so that the global twists are cancelled out, leaving only the global lengthening/shortening deformations[18]. Dietz et al. designed a midregion in DNA origami beams such that the upper section forced shortening (and a left-handed torque) and the lower section forced lengthening (and a right-handed torque)[18]. Beams were designed such that they could bend from 0 – 180° in 5° increments and the global torques cancelled each other out. These origami structures were further augmented to generate curved structures such as gears and spirals (Figure 3b).
Figure 3.
Twisted, curved and folded DNA origami sheets. (a) DNA nanoribbon based on crossovers every 11 bp/turn of DNA (scale bar 20nm, Lower Left Images; 50nm ribbon image) [18]. (b) Six-tooth gear design developed by carefully bend DNA tubes in the plane (scale bar 20nm) [18]. (c) Hollow DNA origami box design (42 × 36 × 36 nm3), developed by folding a 2-D sheet and stapling along its edges [19]. (D) Strategies for sophisticated in- and out-of-plane curving can lead to intricate DNA origami shapes such as hemispheres, spheres, ellipsoids and nanoflasks (scale bar 50nm) [20]. Reproduced from [18-20].
An alternative approach to building 3-D objects is to fold up 2-D planes using sticky end technology to form a 3-D structure, more directly following the Japanese paper origami techniques. A particularly elegant example of this is a DNA origami box with a open/closeable lid (Figure 3c)[19]. The box was based on the techniques for 2-D structures described by Rothemund and the edges of the boxes were sealed together by staple strands that fold the box by bridging the edges, resulting in a ‘cuboid’ structure of external size 42 × 36 × 36 nm3 [19]. Due to the assumption of 10.67 bp forced into the design of 2-D structures as described above, there were concavities/convexities in the box walls that likely resulted from the underwinding of the 2-D planes.
This ability to selectively adjust the number of bp per turn for DNA was also exploited in 2011 by Han et al.[20] to develop a versatile approach for DNA origami design that allowed for in- and out-of-plane curvatures. Curvature was introduced in the plane by forcing crossover connections between neighboring helices each with a different number of bp between crossovers causing the bundle to bend in the direction of the shorter helix. The authors then positioned helices out-of-plane at a variety of angles by moving away from the restrictions set for the square and honeycomb lattices and allowing crossovers at any increments of bp. Using this flexible approach the authors designed and fabricated a variety of shapes including a sphere, an ellipsoid and a nanoflask[20] (Figure 3d).
The combination of these transformative efforts, along with the ability to grow structures by stapling them together or using sticky ends has empowered scientists and engineers with the ability to fabricate an essentially endless variety of origami structures. In the next section, we will describe the applications of these origami structures to medicine and, in particular, describe how they can be used to direct cell behavior or study biological systems.
4. DNA for Drug Delivery
Nanoparticles have long been recognized and studied for their use in drug delivery due to their ability to enhance the physicochemical properties of therapeutics; given DNA origami's structural and physicochemical flexibility/adaptability, it offers exciting potential for drug delivery. The earliest nanoparticles – dating back to the 1960s[21] –were liposomes that had the capability to prolong circulation time and to enhance delivery of hydrophobic drugs and other challenging therapeutics[22]. Now, many organic and inorganic nanoparticle structures are used to enhance drug delivery (see [23] for a recent review). Macroscale drug delivery systems[24] are designed to deliver drugs locally at the desired target site, to protect labile drugs and temporally control the release profile of their therapeutic cargo over a defined period of time. Nanoscale systems, by contrast, are typically introduced systemically and are designed to survive and persist in the circulatory system, protect their therapeutic cargo, target the diseased site and facilitate cell entry of their cargo. Although the potential to deliver drugs intravenously in these nanoparticle systems is attractive, the ability to remain in the circulatory system and for a high percentage of injected particles to collect at the target site remain significant challenges. The majority of approaches to overcoming these challenges focus on adapting the particle's size, shape and surface properties – all features that can be controlled using DNA origami.
Among the key advantages that DNA origami can exploit are its precise, programmable, and repeatable shape definition; its potential for cell uptake; its spatially addressable patterning of multiple drugs and/or targeting molecules at precisely defined locations; its potential for high density of drug loading; its ability to allow for tunable drug release; and its ability to be used as designed as an adaptable structure that can respond to environmental changes[9]. We address each of these advantages and provide salient examples in the following sections.
4.1 Precise Size and Shape Definition
It is a well-established phenomenon that the size and charge of particles affect their circulation time, renal clearance, uptake by the reticulo-endothelial system, and cell internalization potential; furthermore, many studies have explored the effect of size and charge on biodistribution for a wide spectra of nanoparticles[23].
DNA origami can be used to develop a wide range of particle shapes and sizes as described above and – from the perspective of defining optimum shapes for drug delivery – correctly folded and purified structures provide an essentially monodisperse size distribution. DNA origami is thus an excellent candidate for the study of, and application in, drug delivery that is also inherently biocompatible. The affect of shape and dimensions on DNA origami's ability to reach intracellular compartments is immediately apparent by comparing the enhanced effectiveness of DNA origami structures vs. unfolded control dsDNA in almost every study where this is presented (e.g. [25, 26]). When Jiang et al.[25] compared a 2-D DNA origami triangle with a 3-D DNA origami tube structure, for example, they found that both structures were equally effective in delivering doxorubicin (discussed in more detail below) intracellularly in a breast cancer cell line; both shapes were significantly more cytotoxic relative to unfolded controls. A similar 2-D DNA origami triangle was fabricated and compared to a 2-D rectangle and a 3-D origami tube by Zhang et al.[26] for in vivo breast tumor studies. These studies showed enhanced tumor uptake of all origami structures when compared to unfolded dsDNA, with triangle structures outperforming the tube and rectangle. These studies illustrate the potential for DNA origami – with its precisely defined structure – to inform the design of other nanoparticle system size and shape profiles for optimum efficacy. As additional tests and studies emerge, in vitro and in vivo comparative studies on the size and shape affects of DNA origami structures on both biodistribution and cell uptake will be of critical importance.
4.2 Spatially Addressing DNA Origami Structures Through Tagging
As DNA origami increasingly moves towards application in the biomedical field, techniques to label and identify DNA origami structures are essential. The original DNA origami article by Rothemund included a strategy for labeling of structures[6], and this basic principle has been extended by many authors for incorporation of a variety of cargos (e.g., small molecules, nucleic acids, fluorescent labels, inorganic particles and proteins) that can be precisely spatially addressed. Rothemund functionalized selected staples within the origami structures to include dumbbell hairpins, which were additional nucleotides within a staple that did not bind to the scaffold strand but instead formed a double stranded secondary structure piece of DNA above (or below, depending on the location and orientation) the scaffolded structure. This dumbbell hairpin provided height contrast to the origami structures – allowing for patterns/information to be drawn on the origami structures and read by AFM.
This basic principle has been extended for incorporating other molecules and is additionally informed by non-origami based DNA nanostructures for tailored drug delivery (for recent excellent reviews that cover non-origami DNA nanostructures for drug delivery, see for example[10, 11, 27]). Non-origami DNA nanostructures such as self-assembled tubes and tetrahedra have yielded promising success by functionalizing the structures through extended/intercalated domains that are tagged with therapeutics such as siRNA[28], fluorophores[29] and vaccine complexes[30]; these techniques are also applicable to origami based structures. DNA-origami based structures have been tagged with many therapeutic and biosensing molecules, including proteins[31], RNA[32], inorganic (gold) nanoparticles[33] and quantum dots[34].
The most popular techniques for conjugation of DNA origami include biotinylation of staple strands[31, 34, 35], addition of chemical groups such as amine groups to the ends of staples [31] and extension of staple strands with non scaffold-conjugating sequences for hybridization with molecules[32] and particles[33].
Perhaps the simplest solution for conjugating molecules is to extend the staple strands with non-scaffold binding tethers. Non scaffold-binding tethered extensions off the DNA staple oligonucleotides can be sequenced to attach to select complementary strands for direct bioactivity or to bind molecules conjugated to the complementary strands. For example, Ke et al. extended staple strands with complementary RNA sequences to Rag-1, C-myc and β-actin[17]. These staple strands were flexible and non-detectable by AFM until they hybridized with their complementary RNA sequence and became rigid and AFM-readable. The staple tether extensions have also been used to tether molecules and particles that are bound to the tether strand, including virus capsids[36]. In addition, several previous studies have employed non-scaffold binding tethers to attach large proteins bearing the binding tether complementary sequence including Fab’ antibody fragments[37], protein ligands[38], and antibodies[39]. This strategy was utilized by Douglas et al. and Shaw et al. to study the influence of functionalized DNA origami nanostructures on intracellular signaling. [37, 38]. Similarly, conjugation of gold nanoparticles is typically accomplished by complementary hybridization to strands that are bonded to gold nanoparticles through thiol-terminated groups[33]. The same thiol processing can be used on staple strands to bind them to other molecules. For example, thiol groups on staple strands were used to bind Meleimido-C3-NTA; the nitrilotriacetic acid (NTA) group is subsequently able to bind Histidine-tagged proteins in the presence of metal cations[40]. The versatility in the location of His-tagging on proteins makes this an attractive method for patterning proteins on DNA surfaces with orientation of the protein optimized for activity.
Inspired by some of the techniques developed for biomolecule conjugation of non-origami DNA nanostructures, a versatile set of reactions has been explored. Voigt et al.'s work is one of the earliest studies that incorporated principles from non-origami DNA nanotechnology into DNA origami structures[31]. They demonstrated a broad set of conjugation strategies and exploited the spatial addressability of nanoscale structures to analyze single-molecule chemical reactions[31]. For example, staple strands tethered with amine groups were subsequently tagged with azide, biotin or alkyne group through NHS (N-Hydroxysuccinimide) Ester reactions[31]. This work also utilized staples tethered with biotin for subsequent conjugation with streptavidin. Therein, the streptavidin provided structural information – its height profile could identify its location on AFM vs non-conjugated molecules; however, streptavidin can be directly conjugated to a protein to bind it to biotin or the remainder of its four binding sites can be used to conjugate a biotinylated protein. Pedersen et al. combined this capability and the careful spatial addressability to attach a cluster of TGF-β-receptor binding peptides to origami rectangles and enhance cell sensitivity to TGF-β ligand[35].
Although these techniques are extremely useful, to interrogate the effects or cellular localization of DNA origami structures, approaches that use unmodified and untagged structures remove potential confounding effects. One very elegant study on the intracellular localization of DNA origami by Shen et al.[41] exploits a label-free carbazole-based biscyanine molecule. These probes are weakly fluorescent in solution, yet become strongly fluorescent when they bind to DNA molecules; these probes are introduced just prior to the imaging study and thus should not interfere with the origami structure during the localization phase of the study. Using 3-D origami tubes as a model particle, the study demonstrated the lysosomal localization of DNA origami structures. Although the fluorescent intensity in these systems could potentially be used to qualitatively compare the uptake of various origami structures, it is best to combine these studies with a robust quantitative method, such as the technique demonstrated by Okholm et al.[42] for 2-D rectangles in a cervical carcinoma cell line. This study identified suitable primers for the M13mp18 scaffold (that did not amplify staple strands), which were able to detect the quantity of origami in solution with a linear range from 103 – 108 origami. In addition, samples recovered from cell lysates demonstrated a similar detectability; the recovery was lower than in the pure solution and, though this did not affect the linearity, it could be improved by adding glycogen to lysed cell solution to enhance DNA precipitation.
4.3 Loading Drug on DNA Origami
The densely packed nature of the origami structures and the flexibility in cargo loading confer the ability to load drugs that directly bind to DNA (e.g., the anthracyclines Doxorubicin and Daunorubicin) or to attach drugs to origami structures with high nanoscale density.
4.3.1 Anthracycline Loading Of Origami Structures
Precise tagging and locating of bioactive agents to DNA-origami structures is exciting; however, direct binding of a bioagent offers a simpler approach that, if successful, could exploit the size and shape characteristics of DNA origami structures directly. Obvious candidates for this strategy are drugs that intercalate in DNA. Doxorubicin (Dox), an anthracycline chemotherapeutic, is a promising drug for proof-of-concept as it has already been shown that delivering Dox via nanocarriers reduces toxic side effects and enhances tumor uptake[43].
Given the high number of intercalation sites per DNA-origami nanostructure, it could be used to deliver an especially high local dose of Dox. Several authors have presented invitro[25, 44] and in-vivo[26] studies of DNA-origami based structures for delivery of Dox. Jiang et al., loaded Dox into 2-D origami triangles and 3-D origami tubes, which both enhanced drug loading compared to unstructured dsDNA and DNA origami tube slightly outperforming the triangle in terms of loading characteristics[25]. They subsequently tested the effectiveness of these structures in a breast cancer cell lines: regular MCF-7 cells (reg-MCF7) and doxorubicin resistant MCF-7 cells (res-MCF7). When compared to free Dox at equimolar Dox concentrations, origami structures had equivalent cytotoxicity to free Dox in reg-MCF7. Strikingly, however, origami structures demonstrated significantly enhanced cytotoxicity in the res-MCF7 cells. The authors attributed this enhanced efficacy to the Dox-origami's ability to enhance uptake and retention of the drug with slow lysosomal acidification of the origami leading to redistribution of Dox to active target sites[25].
Zhao et al. studied the effects on encapsulation efficiency and release of Dox in DNA origami tubes as a function of global twist, as global twist results in different amounts of local deformation in DNA helices, which may affect drug intercalation or release[44]. A straight tube, S-Nano, and a twisted tube, T-Nano, were loaded with increasing concentrations of Dox during the folding process[44]. When these structures were subsequently imaged, the S-Nano tube demonstrated decreased folding quality relative to unloaded controls; by contrast, the T-Nano structures had an increased folding quality with the addition of Dox, as well as a higher Dox encapsulation efficiency than the S-Nano. When the release rate of Dox from these structures was examined, the S-Nano structure released Dox at an equivalent rate to an unfolded control dsDNA. The T-Nano structure, however, demonstrated a more sustained release profile over the course of several hours. The cytotoxicity of the Dox-loaded T-Nano structures was then tested in three breast cancer cell lines: MDA-MB-231, MDA-MB-468, and MCF-7)[44]. The cytotoxicity of the Dox/T-Nano structures was significantly higher in these cell lines than the equivalent dose of free Dox.
As briefly described above, the tumor targeting efficiency of three different origami structures – 2-D triangle, 2-D rectangle, and a 3-D tube – revealed the enhanced biodistribution of the 2-D triangle in vivo; all groups outperformed unfolded DNA controls[26]. These triangle origami structures were then loaded with Dox post-folding and demonstrated a significantly enhanced chemotherapeutic effect in orthotopic mice breast cancer tumors (Figure 4a). Imaging studies attributed this enhanced efficacy to an increased localization of Dox-origami at the tumor site. Furthermore, their data support reduced side effects (e.g., hepatotoxicity, nephrotoxicity, weight loss) of Dox-origami compared to free Dox. The authors also noted the affect of pH on drug release rate, with a reduced pH – such as the pH found in acidic tumor regions and subcellular organelles – accelerating release of Dox from the nanostructures. This comprehensive in vivo study demonstrates the exciting clinical potential for DNA origami structures as it begins to address several concerns and aspects relating to translation by testing a full preclinical model.
Figure 4.
DNA origami strategies for drug delivery. (a) Doxorubicin can be directly intercalated in DNA-origami shapes and shows enhanced localization over controls to tumor regions following injection due to its carefully controlled size and shape that lead to an enhanced permeability and retention (EPR) effect. Once localized at the tumor, it undergoes digestion at the higher pH and releases the active drug. Reprinted with permission from [26]. © 2014 American Chemical Society. (b) Schematic of a DNA nanorobot. The blue and orange tethers on the barrel ends form a lock that is unlocked by aptamers (red globular proteins in the schematic). Once opened, the cargo (pink, which is tethered to the nanorobot via yellow strands) is revealed to the target cell (Reproduced with permission from [37]). (c) A virus-inspired approach to enhancing the circulation time and stability of origami. A DNA NanoOctahedron is encapsulated with a lipid bilayer membrane and when injected in mice shows excellent biodistribution and little bladder accumulations, indicative of it avoiding renal clearance. Reprinted with permission from[57]. © 2014 American Chemical Society.
In addition to Dox, another anthracycline, Daunorubicin (Dauno) is a strong drug candidate for proof-of-concept DNA origami drug delivery studies in the context of leukemia due to its widespread clinical use [45] and recent findings by Halley et al. that highlighted the ability of Dauno-loaded DNA origami nanostructures to circumvent drug-resistance in HL-60/ADR multidrug resistant acute promyelocytic leukemia cells[46]. The authors designed a rod-shaped DNA nanostructure ~100nm in length that could be self-assembled within 10-15 minutes and exhibited good stability in cell culture media over 24 hours. The Dauno-loaded DNA nanostructures showed effective entry via the endolysomal pathway yielding larger amounts of drug persisting in cells and ultimately improving drug efficacy. Taken together, these findings suggest that DNA origami nanostructures loaded with anthracyclines are an exciting novel nanotherapeutic approach that holds tremendous therapeutic potential for both solid and liquid tumor model systems.
4.3.2 Cytosine-Phosphate-Guanine/Guanosine (CpG) Oligonucleotides
CpG motifs are found with far greater abundance in microbial DNA than in vertebrate genomes and are therefore recognized by the immune system as a sign of pathogen invasion. When unmethylated, these sequences are recognized by Toll-like receptor 9 (TLR9) and both the innate and adaptive immune system can be strongly activated. This makes CpG oligodeoxynucleotides a good adjuvant candidate for immunotherapy vaccines. One challenge with their use, however, is that natural CpG dinucleotides are very susceptible to nuclease degradation and, therefore, stabilizing modifications, such as phosphorothioate (PTO) modified backbones, are explored. Unfortunately, these PTO-CpGs are not as effective an adjuvant and can cause lymphoid or organ damage, which motivates the testing of DNA origami as a potential nanocarrier of CpG sequences for immunotherapies.
Schüller et al. explored the use of DNA origami as a CpG nanocarrier by hybridizing up to 62 individual CpG sequences to staple tethers on the inner or outer surface of a 30-helix DNA origami tube[47]. The CpG sequences tested were either unmodified, PTO-backbone modified, or partly PTO-backbone modified. When incubated with freshly isolated spleen cells, these CpG-sequence-coated DNA origami tubes caused a potent immune response via the TLR9 pathway, outperforming standard carrier systems (e.g., Lipofectamine)[47]. In contrast to Lipofectamine, the origami carriers did not affect cell viability and no detectable cytotoxicity was observed. Interestingly, the effectiveness of the origami nanocarriers was independent of whether the CpG sequences coated the inner or outer part of the origami tube, consistent with the idea that the tubes disassemble intra-endosomaly. Of the CpG sequences tagged to the DNA-origami based tubes, the partly PTO-backbone modified yielded the strongest response; it is reported that this group outperformed the fully PTO-backbone modified group due to better hybridization efficiency[47]. The authors did highlight the fact that in the absence of the immunostimulatory CpG oligonucleotides, the origami structures could still activate the innate immunity using non-TLR9 pathways; this is an important consideration when exploring in vivo DNA origami applications[47]. These results are consistent with prior studies on DNA nanotechnologies with non-origami based DNA nanocarriers[48, 49]. Interestingly, partially folded structures loaded with CpG sequences were nearly as effective as the correctly folded origami tubes with CpG sequences. The authors suggest that this demonstrates that the compactness, size and stability was more critical than the exact 3-D shape for cell uptake and immune activation; however, when incubated with FBS the origami structures demonstrated enhanced stability, which suggests folded nanostructures may be more suitable for in vivo applications.
4.4 Switchable Drug Delivery from Origami Robots
The potential for dynamic origami structures that can receive, process, and respond to input signals, for example by adapting its structure to release a drug is an especially promising possibility for DNA origami. Andersen et al.[19] first demonstrated a DNA-origami box with a dual lock key system that functioned liked an AND gate requiring two inputs to open. Temporary closing strands on the lid were shorter than the complementary strands on the box's front face leaving a toehold binding site for the “key” strands; the key strands could then replace the closing strands via toehold-mediated DNA strand displacement[50] to open the box. Kjems’ research group further advanced this box by reducing its dimensions and inserting a reversible locking system that would facilitate multiple opening and closing cycles of the boxes lid[51]. This was accomplished by design of locks where a single strand attaches to both the box face and the lid; these strands form a hairpin to close the box leaving a loop region that functions as a toehold for key strands. In the absence of keys, these locks keep the lid closed. Keys that bind the toehold region and unzip the locks are then used to open the lid. These keys can subsequently be displaced from the lid via their own toehold binding region using second input strand that is fully complementary to the key strands. Once the key strands are displaced, the original lock strands re-hybridize to close the lid. This process was repeated up to three times[51]. This offers the exciting potential for loading these boxes with a therapeutic that can be selectively released on instruction.
Several of the above concepts were elegantly demonstrated with a sophisticated logic-gated nanorobot for targeted drug delivery[37]. A barrel-type design was fabricated, consisting of two halves with ssDNA entropic springs on one end and two locks, similar to the box design above, on the other end (Figure 4b). In this design, however, the keys used to open locks were aptamers. The cargo – gold nanoparticles or Fab’ antibody fragments – was loaded inside the barrel by complementary hybridization of staple tethers facing the inside of the barrel. The authors first showed the ability of these nanorobots to selectively open depending on the lock sequence used (combinations of: the 4lt receptor which binds PDGF, TE17 and sgc8c) and tested their opening when cultured with a variety of cancer cell lines. Strikingly, these nanorobots were able to selectively detect cells from a mixed population down to a single cell level[37]. Finally, the authors demonstrated two potential therapeutic applications of the nanorobots. First, they showed the ability to bind to natural killer (NKL) cells using antibodies to CD33 and CDw328 Fab’ and demonstrated that the nanorobots induced growth arrest in these cells. Second, they incorporated antibodies to CD3ε and flagellin as the cargo in the nanorobots. These nanobots could ‘scavenge’ flagellin from their environment and then were subsequently opened by T-cells and activated the T-cells through the CD3ε and flagellin binding. This diverse tool shows an exciting potential for precise and selective cell delivery of cargos and the scavenging of molecules that could subsequently be delivered to cells with revealing of the cargo occurring only in the presence of defined inputs.
4.5 Addressing the Challenges With In vivo Translation
Despite the great promise of DNA origami nanoparticles for delivery and the in vivo demonstrations of their potential[26] there are concerns and challenges remaining regarding their in vivo stability due to native DNases. While more advanced progress has been made in implementing smaller strand based DNA nanodevices in vivo, for example the implementation of DNA-based pH sensors in c. elegans[52], characterization and optimization of DNA origami for in vivo applications is still in early stages. Castro et al. initially demonstrated that the in vitro enzymatic degradation of DNA origami bundles was significantly slower than bare double-stranded DNA by incubating a panel of DNA origami bundles with T7 endonuclease 1 or DNase 1[12]. Mei et al. further addressed some of these concerns by analyzing the affects on several origami structures (a 2-D rectangle, a 2-D triangle, and a 3-D multilayer parallelpiped structure) incubated with cell lysate from normal and cancerous cell lines[53]. The incubation was performed at room temperature or 4°C for up to 12 hrs. After 1hr, the unfolded single and double stranded controls already demonstrated degradation; however, all of the origami structures tested were recoverable and demonstrated no degradation on electrophoresis gels, TEM or AFM imaging. Indeed, following incubation with cell lysate and recovery of the structures, a functionalized origami structure – designed to bind β-actin mRNA – was successfully able to bind the mRNA. Findings for non-origami DNA nanostructures have shown their stability in various cell lysis buffers[54] and for up to 48hrs in live cells[48, 55]. Furthermore, DNA origami structures were demonstrated to be stable for at least 24hrs in multicellular organisms; however, these C elegans organisms lack innate immunity, which would be expected to increase the observed stability[56].
Despite these encouraging in vitro data, concerns regarding the longer-term stability and residence/circulatory time for DNA-origami structures motivate studies to enhance these characteristics. Perrault et al. recently reported on an elegant solution to enhance the stability and reduce immune activation of origami structures by taking inspiration from viruses (Figure 4c)[57]. A DNA NanoOctahedron (DNO) has staple tethers that can bind to lipidconjugated oligonucleotides; these lipid-conjugated oligonucleotides subsequently direct the assembly of a lipid bilayer that encapsulates the DNO. PEG inclusion in the bilayer structure facilitated tighter wrapping of the bilayer around the DNOs, perhaps related to its role in controlling micelle fusion[57]. The lipid membrane encapsulated-DNOs (E-DNOs) and the non-encapsulated DNOs (N-DNOs) were subjected to DNase I digestion for 24h at 37 °C and the lipid-protected structures were dramatically more stable (84% remaining vs. 30%). The encapsulated and non-encapsulated cells were next incubated with immune cells from mouse spleens. Although the N-DNO caused a potent inflammatory cytokine response, the response to E-DNOs was barely distinguishable from inactivated controls[57]. In vivo monitoring of fluorescently tagged E- and N-DNOs revealed a prominent difference. N-DNOs demonstrated a half-life of 49.5mins and they rapidly accumulated in the bladder. Since accumulation in the bladder necessitates sizes of <6nm this indicates the rapid structural degradation of the N-DNOs. By contrast, the E-DNOs had a half-life of 370 mins (comparable to PEGylated liposomes) and little bladder accumulation, which is illustrative of their stability.
Virus-like modification of DNA-origami structures has also been tested by Mikkilä et al.[58]. They coated DNA origami rectangles with virus capsid proteins (CPs) to enhance intracellular delivery[58]. The CPs have a positively charged N-terminus that facilitates binding to the origami structure with high yields; the result was a 13-fold increase in the transfection efficiency of DNA origami-CP complexes vs the naked DNA origami structure. These strategies for enhanced stability and efficacy are an active area of research and will strongly contribute to the translation efforts in the field.
5. DNA Origami Based Nanopores
Often in drug delivery of DNA-origami, the goal is to get sufficient origami structures across the lipid membrane; however, synthetic channels that can span lipid membranes or mimic natural membrane channels can be used to study membrane translocation or to direct molecules across the membrane. Bell et al. and Wei et al. demonstrated this potential by trapping DNA origami nanopores inside solid-state nanopores and demonstrating their ability to detect molecules translocating through the pore [59]. As the molecules translocate across the pores, a current blockade event is observed and these events can be studied to interrogate channel-molecule interactions that can enhance our understanding of cell membrane pore function. A hybrid channel was developed by combining a flat, two helix-thick origami structure that had a central pore with glass nanocapillaries (Figure 5a)[60]. Fluorescent labeling around the pore confirmed that the functional pore observations were in synchronicity with the trapping of the nanopores on the glass capillary. These origami pores were able to be repeatedly trapped and released from the glass[60]. Translocation of λ-DNA across the pores as a function of pore size was next examined. Unsurprisingly, the 5nm pores resulted in fewer λ-DNA folding states (i.e., it was more difficult for it to translocate through these smaller pores), demonstrating the sensing potential of these structures. The versatility of these pores was extended by chemically modifying the pores; ssDNA tethers on staple strands at the pore periphery were coded to attach to short sequences in the oligonucleotides used in subsequent translocation experiments[60]. These pore designs acted to delay oligonucleotides translocation and the length of the complementary sequences could control the delay. These detailed DNA nanopore systems can essentially imitate natural pores, selectively permitting cargo through; however, if these could be incorporated into membranes they could offer an exciting synthetic tool to affect cell behavior.
Figure 5.
DNA-origami based synthetic lipid membrane channels. (a) Schematic overview of a synthetic DNA-origami membrane channel that can be used to mimic lipid membrane channel behavior. By adjusting channel size, physical control over the passage of molecules (e.g., dsDNA) can be controlled. Enhanced control can be achieved by extending staple tethers into the channel that can trap ssDNA as it passes. Reprinted with permission from [60]. © 2013 American Chemical Society. (b) Schematic of a DNA origami based synthetic nanopore (Left) and eleven of these synthetic nanopores inserted in a lipid membrane vesicle Reproduced with permission [61].
Langecker and colleagues developed a synthetic DNA-origami based nanopore channel that can insert in lipid bilayers (Figure 5b)[61]. In this work, a DNA-origami based structure, consisting of an inner stem structure that crossed the membrane and an outer barrel structure that existed external to the membrane was created. To facilitate insertion in the lipid bilayer, 26 cholesterol moieties are attached to the membrane-facing surface of the outer barrel via hybridization of cholesterol-modified oligonucleotides with adaptor strands at the barrel face. Following fabrication, these channels successfully inserted in lipid bilayer membranes and functional testing revealed membrane channels with similar characteristics to natural ion channels. Conductance on the order of 1 nanosiemen was recorded and natural channel gating was seen. The natural channel gating was attributed to thermal fluctuations of the structure that caused stochastic unzipping/rezipping of the origami material within the channel[61]. This hypothesis was supported by tests on mutant channels designed to be less stable and undergo more frequent zipping/rezipping resulting in more pronounced gating in the channels[61]. Finally, the synthetic channels were used for experiments on DNA hairpin unzipping and guanine quadruplex unfolding[61]. For both molecules, application of positive voltages across the membranes resulted in transient current blockades that could be observed and demonstrates the potential of these synthetic channels as sensing devices to discriminate analyte molecules by interrogating their translocation characteristics. Given the myriad number of protein/peptide channels that span lipid bilayer membranes to control transport, the ability to insert synthetic channels in cell membranes may offer a new approach to controlling cellular delivery of therapeutics.
Although it is a non-origami technology, the potential for functional synthetic DNA nanopores has been demonstrated by Burns et al. [62], who designed synthetic pores that insert in cell membranes. These control the flux of molecules across the cell membrane and induce cytoxicity[62]. The membrane insertion strategy here was to include a hydrophobic belt of charge neutral ethyl phosphorotioate groups. Once incubated with cervical cancer cells, these synthetic pores insert in the lipid membranes facilitating transport across the membrane and inducing cell cytotoxicity. The combination of these studies illustrates the potential for the selective transport of drugs and molecules or even origami structures themselves across cell membrane that can induce bioactive effects (readers are referred to a recent review focusing on DNA nanopores by Hernández-Ainsa and Keyser[63])
6. DNA Origami for Biosensing and Studying Cell Behavior
Instructing cells in vivo and in vitro is a key biomedical technology but huge amounts of efforts also focus on technologies to probe and understand cell behavior. For similar reasons to applicability in cell instruction – i.e., nanoscale shaping and spatial addressability and flexible cargo loading – DNA origami is an exciting tool for biosensing and cell study. Furthermore, recent efforts implementing, and even multiplexing, smaller DNA nanodevices inside cells[64] demonstrate the immense potential of structural DNA nanotechnology for cellular and molecular applications.
One of the earliest examples of DNA-origami applications detailed the use of single-helix thick DNA origami tubes as a liquid crystal for alignment of membrane proteins to facilitate their nuclear magnetic resonance (NMR) structure determination[15]. NMR spectroscopy can be used to determine the structural properties of solubilized proteins; however, membrane proteins are particularly challenging to work with in NMR. Alignment media that can facilitate accurate measurements of residual dipolar coupling would facilitate the application of NMR techniques to their structure determination. Unfortunately, the majority of liquid crystals designed are unsuitable for use with the membrane-solubilizing detergents. Inspired by the liquid crystals of Pf1 phage – which are rigid, negatively charged rod-like particles that are commonly used for alignment of soluble proteins – a similar DNA-origami rod was designed[15]. This DNA origami crystal was used to measure the structural characteristics of the T-cell receptor. This new DNA-origami based liquid crystal could have widespread potential in measuring membrane proteins, which had previously been a major challenge in the NMR field.
DNA origami systems can be addressed with biosensing molecules and subsequently read by imaging techniques. One sophisticated example of a DNA origami logic gate for diagnostic purposes is based on the idea of staple tethering and strand displacement (Figure 6)[65]. When the correct miRNA signals are detected in solution, they bind staple tethers and release a biotin tagged output strand. This output strand in turn diffuses to the Output board and binds to staple tethers on it. When streptavidin is added to the output board prior to AFM imaging, it provides height resolution so that the biosensor can be read[65].
Figure 6.
Biosensing DNA-origami logic board for miRNA detection [65]. (a) When the correct miRNA staple strands are detected by the computation section of the board, biotinylated oliognucleotides are unzipped by these miRNA and diffuse to bind to the output section of the board. Following binding to the output strands, streptavidin is introduced and (b) the origami chips read by AFM. Reprinted with permission from [65]. © 2014 American Chemical Society.
An emerging approach for biosensing is through the use of so-called DNA-PAINT[66]. In this approach, overhangs on a DNA origami nanostructure are designed to be weakly complementary to target fluorescent strands in solution. The fluorescent strands bind transiently to the origami structure allowing localization of that overhang strand. This approach allows for super-resolution imaging since only a fraction of spots are labeled at any given time and single molecules can be localized with sub-diffraction limit accuracy as in other super-resolution methods. This approach has also recently been adapted by modifying antibodies with similar oligonucleotide strands that transiently bind fluorescent target strands[67].
For several biosensing applications, higher order origami structure positioning may be required. To this end, Gerdon et al. used a top down approach to position gold islands that could be functionalized with salt bridges to attract DNA origami(s)[68]. Following lithography of a silicon dioxide surface with the requisite pattern, gold was deposited on the surface using electron-beam evaporation and these regions could selectively bind single origami structures.
This sampling of origami-based biosensors demonstrates the potential and versatility of DNA origami as a tool for the interrogation of biological processes.
7. DNA Origami Technologies as Jigsaw Pieces for Larger Structures
Although many of the origami technologies are effective using a single unit of DNA origami (e.g., in drug delivery), from the very outset DNA origami techniques have included strategies to extend or polymerize identical or diverse DNA structure[6, 15]. The basic concept typically extends from conjugation strategies for any molecules to origami structures. Staple strands can extend between two origami pieces to give sticky ends that can be used to stitch them together[6]. Sugiyama's group demonstrated several beautiful examples of DNA jigsaw tiles that used the basic principle of single stranded overhangs and hierarchically shaped their origami structures to stitch together[69]. Another interesting concept is “superorigami” or “origami of origami”[70]. In this technique, individual origami tiles (can be visualized as typical origami structures) are fabricated with sticky ends. A second scaffold strand is partially folded by staple strands; however, gaps in this scaffold are filled with the DNA tethers from the origami tiles. When these structures are combined and hybridize, the tiles act as staples to completely fold the second scaffold strand and thereby a superstructural pattern is achieved.
Several attempts have been made to move away from the standard origami scaffold strands and by the use of superstructural patterning, shorter strands could be used. As previously mentioned, Yin and co-workers have developed frameworks for the modular design of 2-D and 3-D shapes using a similar architecture to scaffolded DNA origami but with structures made purely from staples[8]. They also adapted this method to form large crystals by extending their origami like structures in a planar fashion[71]. Another elegant idea is to design a shorter scaffold strand that has repeating sequences, which can be folded by the same set of staples in a repeating pattern. These shorter scaffold strands were produced by rolling circle amplification[72] and mixed with a small number of different staple strands to form 2-D origami rectangles and 3-D origami cuboids. Furthermore, the ends of the structures were designed in such a way that a single origami structure could polymerize with the next structure via open staple/scaffold sites. The length of the polymerized structures could be carefully controlled by the amplification processing time and the number of staple strands. Of course, the staple strands can be functionalized to incorporate molecules for imaging (i.e., via biotinylation of tethers) and for cell activation (i.e., via hybridization of unmethylated CpG strands to staple tethers). These techniques for building up from a single origami structure offer further functional and structural diversity to the DNA origami toolbox.
8. Summary and Perspectives
The structural DNA nanotechnology field has evolved dramatically since it was first described[4] and, within a decade of its inception [6], DNA origami has demonstrated great promise for clinical, biomedical, and research applications. From the first proposed 2-D shapes, which themselves have shown potential for biosensing[6, 65] and drug delivery[25, 26, 46] applications, there are now a plethora of examples of intricate 3-D shapes typically based on the folding of 2-D sheets[19], honeycomb lattices[16], square lattices[17] or they can be twisted or curved shapes[18] or objects with even more complex 3-D curvatures[20]. This provides an endless design space to the DNA origami field in terms of structure geometry, a number of which have been explored for a variety of applications including: drug delivery[25, 26, 37, 47], nanopore formation[59, 60, 61] and biosensing[6, 65].
Although the fabrication of detailed origami structures and shapes can be utilized as end products as is (e.g., the DNA nanopores can be used without addition of other molecules), often these structures serve as a templates for the incorporation of additional functional molecules that give enhanced functionality, including: small molecules, nucleic acids, fluorescent labels, inorganic particles and proteins. One particularly interesting clinical application is the incorporation of drug molecules that have affinity for DNA (e.g., the anthracyclines doxorubicin and daunorubicin). In these cases, the origami acts as a drug carrier that controls biodistribution systemically and enhances intracellular delivery of the drug[25, 26, 44, 46]. Inspired in part from the advances in the non-origami DNA nanotechnology field[10, 27], there are now several techniques for tagging cargo onto DNA-origami structures. These techniques all center around the idea of functionalizing the staple strands to have reactive end groups (e.g., biotinylation of staple strands[31, 34, 35] or addition of amine groups to staples[31]) or tethered staple extensions that do not react with the scaffold strand but instead can conjugate to other complementary strands that carry molecules[32], bioactive DNA sequences (e.g., CpG[47]) and particles[33]. These techniques have proven robust and readily accessible, simply by ensuring at the design stage that the correct staple strands are ordered from manufacturers with the appropriate chemical modifications. Indeed, since the modifications can be designed on a staple-by-staple basis, the DNA-to-modification ratio can be carefully controlled, as can the location of the modifications within the origami at nanoscale resolution[6].
A similar approach of hybridization of staple tethers can be used to build up multi-origami structures. This may be necessary for development of larger origami structures for select applications (e.g., creation of a liquid crystal media for NMR[15]) or for building up patterns at larger lengthscales, such as what would be need for molecular computing [69, 70] or for making DNA hydrogels for multicellular interaction.
Despite this diverse toolbox for DNA origami shape design, functionalization and multi-origami polymerization, there are still several challenges and obstacles that need to be addressed prior to the widespread uptake of DNA origami outside of academic labs. The first set of challenges relate to the ability to scale up the fabrication and facilitate production. Currently, origami structures are typically made in 100μL batches at 10-20nM and carefully controlled temperature ramps that typically last from several hours to several days[12]. Recently, a DNA origami nanopore was successfully folded in under one minute[73] and DNA origami tubes for drug delivery in 10 – 15 mins[46], which is a dramatic improvement in processing time and illustrates the remaining potential to enhance fabrication. This can still lead to low percentages of folding efficiencies for certain designs. Establishing robust techniques or modifications to these fabrication protocols to enable larger or more concentrated batch formation with higher yield percentages will be a critical step forward in translating these systems. In addition, the current cost of the oligonucleotide staples is relatively high for these systems. If a select object was being produced en masse, however, it is hopeful that other techniques (e.g., PCR amplification[72]) could be used to produce large numbers of staples at lower cost. PCR approaches have also been demonstrated for the development of diverse scaffold lengths up to 26 kb[74]. This capability will allow us to move away from the M13mp18 scaffold that is widely utilized and achieve shapes of different sizes and improve the flexibility in design.
For in vitro applications, specifically biosensing applications, similar challenges in terms of fabrication exist; however, two central challenges are (1) identifying systems where nanoscale resolution is essential, and (2) simplifying the readouts. As the DNA origami techniques are further explored and their capacity becomes more widespread and accessible, the most favourable applications will become more apparent. Some of these have already been elucidated: studying single molecule interactions[31], nanopore functional studies[59, 60], and studying receptor clustering[35] and we believe more applications will be forthcoming. The second challenge relates to the readouts. A lot of the DNA origami biosensors are currently read by AFM, using height contrast as the reading. Although AFM techniques are routine in many labs that work on DNA origami, it is difficult to imagine these systems being routinely used in the clinic or by non-DNA origami based research labs that simply want to purchase and apply the biosensor. Utilizing simpler readouts (e.g., based on fluorescence) or establishing a technology that can quickly read the height contrast on DNA origamis will be a key translational step. Finally, there is the exciting potential for Biocomputing – which DNA technology has helped enable – and to interface with these to help facilitate translation of DNA origami biosensors in the future.
DNA origami has shown exciting early potential in drug delivery applications and though there is already some data alleviating concerns regarding its translational potential, there are still several aspects that need to be addressed. As mentioned above, the ability to produce large volumes of the requisite origami structure at a competitive price is one central challenge. In addition to the fabrication limitations, a key remaining barrier is to fully establish the in vivo stability and biodistribution of origami structures and their ability to translocate across the cell membrane once at the correct site. Studies to date show the promising potential for the origami stability in vitro [12, 53] for timescales on the order of 24 hours and, in vivo testing of Dox loaded structures demonstrated favorable biodistribution, as well as therapeutic efficacy[26]. In vitro studies also demonstrated the potential for origami structures to enter cells and demonstrate enhanced therapeutic efficacy, most notably in drug resistant cancer cells[25, 44]. It remains to be fully elucidated for various DNA origami structures what their long-term stability is and whether additional strategies (e.g., encapsulation of the origami structures in transport vehicles[57]) are required to enhance stability and targeting. Although these additional strategies may ultimately be deemed necessary for select DNA-origami applications, especially in vivo, further studies are necessary to consider maintaining function (e.g. drug release) of DNA origami modified for improved stability. These modifications likely impact cellular interactions and complicate the fabrication process; hence, for some cellular applications it may be desirable to exploit DNA origami in its more natural state. Given the myriad techniques for tagging origami structures with proteins, it is easily conceivable that these origami structures can be functionalized with antibodies for targeting of specific cell populations[37] or for targeted refilling of local drug delivery depots[75]. The potential for this targeting to be combined with logic-gated control of therapeutic release holds exciting potential and these nanorobots certainly warrant in vivo exploration[37]. These drug delivery systems could be further activated by introducing programmable motion or mechanisms into the origami carriers that controls their structure and alters their interaction with select cells[76]. Localized delivery of origami structures in controlled release systems may also offer the opportunity to overcome the challenges of systemic administration and targeting by placing the origami structures local to the target site in a protective controlled release system that can maintain and deliver the origami structures and their cargo; these techniques are already well established for many cargos and other nucleic acid therapeutics[24]. Exploiting the potential for membrane inserted synthetic nanopores[61] that selectively allow drug translocation (DNA origami based or non-origami based) is a further avenue for drug delivery exploration. Ultimately, the challenge in translation relates to the identification of a first flagship application for these origami structures that drives the technology through the translational challenges. Those translational efforts will also identify and address the regulatory challenges that may arise in relation to the clinical translation of devices and pave the way for other DNA origami structures and applications.
The DNA origami field has witnessed explosive development in the last decade and increasing numbers of labs and researchers are now exploring the technology, with many potential exciting applications being described. We believe that the next phase of work will focus on overcoming the challenges associated with the translation of these technologies to the clinic and widespread biotechnology testing. Given the progress and excitement to date we are encouraged that these hurdles can be overcome and that the full potential of DNA origami realized.
Supplementary Material
Acknowledgements
The authors acknowledge Harumi Ramanayake and Jack Donohue for discussions and comments relating to the manuscript. For funding, CJK and FJO'B acknowledge RCSI's Office of Research and Innovation Summer Research School Programme and Seed Fund Award (Grant Number GR 14-0963); Science Foundation Ireland (SFI) under Grant Number SFI/12/RC/2278; and the European Union for a Marie Curie European Reintegration Grant under H2020 (Project Reference 659715). CRL is a recipient of a National Institutes of Health T32 Award in Oncology Training Fellowship at The Ohio State University Comprehensive Cancer Center, 5T32CA009338-37. CEC acknowledges the National Science Foundation (NSF) under Grant CMMI-1235060 and the Center for Emergent Materials at The Ohio State University, an NSF Materials Research Science and Engineering Center (Award DMR-0820414)
Contributor Information
Dr. Cathal J. Kearney, Department of Anatomy, Tissue Engineering Research Group and Advanced Materials and Bioengineering Research Center, Royal College of Surgeons in Ireland, 123 St. Stephen's Green, Dublin, Ireland.
Dr. Christopher R. Lucas, Department of Mechanical and Aerospace Engineering, The Ohio State University, Columbus, OH 43210, USA
Prof. Fergal J. O'Brien, Department of Anatomy, Tissue Engineering Research Group and Advanced Materials and Bioengineering Research Center, Royal College of Surgeons in Ireland, 123 St. Stephen's Green, Dublin, Ireland
Prof. Carlos E. Castro, Department of Mechanical and Aerospace Engineering, The Ohio State University, Columbus, OH 43210, USA
References
- 1.Watson JD, Crick FH. Nature. 1953;171:737. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Pray L. Nature Education. 2008;1:100. [Google Scholar]
- 3.Holliday R. Genetics Research. 1964;5:282. [Google Scholar]
- 4.Seeman NC. Journal of theoretical biology. 1982;99:237. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 5.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 6.Rothemund PWK. Nature. 2006;440:297. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 7.Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 8.Ke Y, Ong LL, Shih WM, Yin P. Science. 2012;338:1177. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wei B, Dai M, Yin P. Nature. 2012;485:623. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Fan C, Pei H, Shi J, Huang Q. Adv. Mater. 2013;25:4386. doi: 10.1002/adma.201300875. [DOI] [PubMed] [Google Scholar]
- 10.Chao J, Liu H, Su S, Wang L, Huang W, Fan C. Small. 2014;10:4626. doi: 10.1002/smll.201401309. [DOI] [PubMed] [Google Scholar]
- 11.Krishnan Y, Bathe M. Trends in cell biology. 2012;22:624. doi: 10.1016/j.tcb.2012.10.001. [DOI] [PubMed] [Google Scholar]; Surana S, Shenoy AR, Krishnan Y. Nat Nano. 2015;10:741. doi: 10.1038/nnano.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castro CE, Kilchherr F, Kim D-N, Shiao EL, Wauer T, Wortmann P, Bathe M, Dietz H. Nat Methods. 2011;8:221. doi: 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- 13.An individual double helix corresponds to 2 nm, thus an idealized double helix should correspond to 4 nm. However, an inter-helix gap, which is attributed to electrsotatic repulsion between the coils, of 1 – 1.5 nm is observed yielding the 6 – 7 nm.
- 14.Mathieu F, Liao S, Kopatsch J, Wang T, Mao C, Seeman NC. Nano Lett. 2005;5:661. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douglas SM, Chou JJ, Shih WM. Proc Nat Acad of Sci. 2007;104:6644. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Nature. 2009;459:414. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ke Y, Douglas SM, Liu M, Sharma J, Cheng A, Leung A, Liu Y, Shih WM, Yan H. J Am Chem Soc. 2009;131:15903. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dietz H, Douglas SM, Shih WM. Science. 2009;325:725. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J. Nature. 2009;459:73. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 20.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. Science. 2011;332:342. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 21.Bangham AD, Standish MM, Watkins JC. J. Mol. Biol. 13:238. doi: 10.1016/s0022-2836(65)80093-6. [DOI] [PubMed] [Google Scholar]
- 22.Farokhzad OC, Langer R. ACS Nano. 2009 doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 23.Singh R, Lillard JJW. Experimental and Molecular Pathology. 2009;86:215. doi: 10.1016/j.yexmp.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brannon-Peppas L, Blanchette JO. Adv Drug Deliv Rev. 2012;64:206. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]; Mura S, Nicolas J, Couvreur P. Nat Mat. 2013;12:991. doi: 10.1038/nmat3776. [DOI] [PubMed] [Google Scholar]
- 24.Kearney CJ, Mooney DJ. Nat Mat. 2013;12:1004. doi: 10.1038/nmat3758. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Q, Song C, Nangreave J, Liu X, Lin L, Qiu D, Wang Z-G, Zou G, Liang X, Yan H, Ding B. J Am Chem Soc. 2012;134:13396. doi: 10.1021/ja304263n. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q, Jiang Q, Li N, Dai L, Liu Q, Song L, Wang J, Li Y, Tian J, Ding B, Du Y. ACS Nano. 2014;8:6633. doi: 10.1021/nn502058j. [DOI] [PubMed] [Google Scholar]
- 27.Mohri K, Nishikawa M, Takahashi Y, Takakura Y. Eur J Pharm Sci. 2014;58:26. doi: 10.1016/j.ejps.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Lee H, Lytton-Jean AKR, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG. Nat Nanotechnol. 2012;7:389. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko S, Liu H, Chen Y, Mao C. Biomacromolecules. 2008;9:3039. doi: 10.1021/bm800479e. [DOI] [PMC free article] [PubMed] [Google Scholar]; Özhalıcı-Ünal H, Armitage BA. ACS Nano. 2009;3:425. doi: 10.1021/nn800727x. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Xu Y, Yu T, Clifford C, Liu Y, Yan H, Chang Y. Nano Lett. 2012;12:4254. doi: 10.1021/nl301877k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voigt NV, Tørring T, Rotaru A, Jacobsen MF, Ravnsbæk JB, Subramani R, Mamdouh W, Kjems J, Mokhir A, Besenbacher F, Gothelf KV. Nat Nanotechnol. 2010;5:200. doi: 10.1038/nnano.2010.5. [DOI] [PubMed] [Google Scholar]
- 32.Ke Y, Lindsay S, Chang Y, Liu Y, Yan H. Science. 2008;319:180. doi: 10.1126/science.1150082. [DOI] [PubMed] [Google Scholar]
- 33.Ding B, Deng Z, Yan H, Cabrini S, Zuckermann RN, Bokor J. J Am Chem Soc. 2010;132:3248. doi: 10.1021/ja9101198. [DOI] [PubMed] [Google Scholar]; Kuzyk A, Schreiber R, Fan Z, Pardatscher G, Roller E-M, Högele A, Simmel FC, Govorov AO, Liedl T. Nature. 2012;483:311. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]; Shen X, Song C, Wang J, Shi D, Wang Z, Liu N, Ding B. J Am Chem Soc. 2012;134:146. doi: 10.1021/ja209861x. [DOI] [PubMed] [Google Scholar]
- 34.Bui H, Onodera C, Kidwell C, Tan Y, Graugnard E, Kuang W, Lee J, Knowlton WB, Yurke B, Hughes WL. Nano Lett. 2010;10:3367. doi: 10.1021/nl101079u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedersen RO, Loboa EG, LaBean TH. Biomacromolecules. 2013;14:4157. doi: 10.1021/bm4011722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephanopoulos N, Liu M, Tong GJ, Li Z, Liu Y, Yan H, Francis MB. Nano Lett. 2010;10:2714. doi: 10.1021/nl1018468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Douglas SM, Bachelet I, Church GM. Science. 2012;335:831. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 38.Shaw A, Lundin V, Petrova E, Fördős F, Benson E, Al-Amin A, Herland A, Blokzijl A, Högberg B, Teixeira AI. Nat Methods. 2014;11:841. doi: 10.1038/nmeth.3025. [DOI] [PubMed] [Google Scholar]
- 39.Shaw A, Benson E, Högberg B. ACS Nano. 2015;9:4968. doi: 10.1021/nn507035g. [DOI] [PubMed] [Google Scholar]
- 40.Shen W, Zhong H, Neff D, Norton ML. J Am Chem Soc. 2009;131:6660. doi: 10.1021/ja901407j. [DOI] [PubMed] [Google Scholar]
- 41.Shen X, Jiang Q, Wang J, Dai L, Zou G, Wang Z-G, Chen W-Q, Jiang W, Ding B. Chem. Commun. 2012;48:11301. doi: 10.1039/c2cc36185j. [DOI] [PubMed] [Google Scholar]
- 42.Okholm AH, Nielsen JS, Vinther M, Sørensen RS, Schaffert D, Kjems J. Methods. 2014;67:193. doi: 10.1016/j.ymeth.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Barenholz Y. J Control Release. 2012;160:117. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 44.Zhao Y-X, Shaw A, Zeng X, Benson E, Nyström AM, Högberg B. ACS Nano. 2012;6:8684. doi: 10.1021/nn3022662. [DOI] [PubMed] [Google Scholar]
- 45.Lowenberg B, Ossenkoppele GJ, van Putten W, Schouten HC, Graux C, Ferrant A, Sonneveld P, Maertens J, Jongen-Lavrencic M, von Lilienfeld-Toal M, Biemond BJ, Vellenga E, van Marwijk Kooy M, Verdonck LF, Beck J, Dohner H, Gratwohl A, Pabst T, Verhoef G. The New England journal of medicine. 2009;361:1235. doi: 10.1056/NEJMoa0901409. [DOI] [PubMed] [Google Scholar]; Lancet JE, Cortes JE, Hogge DE, Tallman MS, Kovacsovics TJ, Damon LE, Komrokji R, Solomon SR, Kolitz JE, Cooper M, Yeager AM, Louie AC, Feldman EJ. Blood. 2014;123:3239. doi: 10.1182/blood-2013-12-540971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halley PD, Lucas CR, McWilliams EM, Webber MJ, Kural C, Lucas DM, Boyd JC, Castro CE. Small. 2015 doi: 10.1002/smll.201502118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schüller VJ, Heidegger S, Sandholzer N, Nickels PC, Suhartha NA, Endres S, Bourquin C, Liedl T. ACS Nano. 2011;5:9696. doi: 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Pei H, Zhu B, Liang L, Wei M, He Y, Chen N, Li D, Huang Q, Fan C. ACS Nano. 2011;5:8783. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 49.Nishikawa M, Matono M, Rattanakiat S, Matsuoka N, Takakura Y. Immunology. 2008;124:247. doi: 10.1111/j.1365-2567.2007.02762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rattanakiat S, Nishikawa M, Funabashi H, Luo D, Takakura Y. Biomaterials. 2009;30:5701. doi: 10.1016/j.biomaterials.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 50.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. Nature. 2000;406:605. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]; Zhang DY, Seelig G. Nat Chem. 2011;3:103. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 51.Zadegan RM, Jepsen MDE, Thomsen KE, Okholm AH, Schaffert DH, Andersen ES, Birkedal V, Kjems J. ACS Nano. 2012;6:10050. doi: 10.1021/nn303767b. [DOI] [PubMed] [Google Scholar]
- 52.Modi S, Goswami GSM,D, Gupta GD, Mayor S, Krishnan Y. Nat Nanotechnol. 2009;4:325. doi: 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- 53.Mei Q, Wei X, Su F, Liu Y, Youngbull C, Johnson R, Lindsay S, Yan H, Meldrum D. Nano Lett. 2011;11:1477. doi: 10.1021/nl1040836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keum J-W, Bermudez H. Chem. Commun. 2009:7036. doi: 10.1039/b917661f. [DOI] [PubMed] [Google Scholar]
- 55.Walsh AS, Yin H, Erben CM, Wood MJA, Turberfield AJ. ACS Nano. 2011;5:5427. doi: 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- 56.Surana S, Bhatia D, Krishnan Y. Methods. 2013;64:94. doi: 10.1016/j.ymeth.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perrault SD, Shih WM. ACS Nano. 2014;8:5132. doi: 10.1021/nn5011914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mikkilä J, Eskelinen A-P, Niemelä EH, Linko V, Frilander MJ, Törmä P, Kostiainen MA. Nano Lett. 2014;14:2196. doi: 10.1021/nl500677j. [DOI] [PubMed] [Google Scholar]
- 59.Bell NAW, Engst CR, Ablay M, Divitini G, Ducati C, Liedl T, Keyser UF. Nano Lett. 2012;12:512. doi: 10.1021/nl204098n. [DOI] [PubMed] [Google Scholar]; Wei R, Martin TG, Rant U, Dietz H. Angew. Chem. Int. Ed. 2012;51:4864. doi: 10.1002/anie.201200688. [DOI] [PubMed] [Google Scholar]
- 60.Hernández-Ainsa S, Bell NAW, Thacker VV, Göpfrich K, Misiunas K, Fuentes-Perez ME, Moreno-Herrero F, Keyser UF. ACS Nano. 2013;7:6024. doi: 10.1021/nn401759r. [DOI] [PubMed] [Google Scholar]
- 61.Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC. Science. 2012;338:932. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burns JR, Al-Juffali N, Janes SM, Howorka S. Angew. Chem. Int. Ed. 2014 doi: 10.1002/anie.201405719. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hernández-Ainsa S, Keyser UF. Nanoscale. 2014;6:14121. doi: 10.1039/c4nr04094e. [DOI] [PubMed] [Google Scholar]
- 64.Modi S, Nizak C, Surana S, Halder S, Krishnan Y. Nat Nanotechnol. 2013;8:459. doi: 10.1038/nnano.2013.92. [DOI] [PubMed] [Google Scholar]
- 65.Wang D, Fu Y, Yan J, Zhao B, Dai B, Chao J, Liu H, He D, Zhang Y, Fan C, Song S. Anal. Chem. 2014;86:1932. doi: 10.1021/ac403661z. [DOI] [PubMed] [Google Scholar]
- 66.Jungmann R, Steinhauer C, Scheible M, Kuzyk A, Tinnefeld P, Simmel FC. Nano Lett. 2010;10:4756. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]; Iinuma R, Ke Y, Jungmann R, Schlichthaerle T, Woehrstein JB, Yin P. Science. 2014;344:65. doi: 10.1126/science.1250944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jungmann R, Avendano MS, Woehrstein JB, Dai M, Shih WM, Yin P. Nat Meth. 2014;11:313. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gerdon AE, Oh SS, Hsieh K, Ke Y, Yan H, Soh HT. Small. 2009;5:1942. doi: 10.1002/smll.200900442. [DOI] [PubMed] [Google Scholar]
- 69.Endo M, Sugita T, Katsuda Y, Hidaka K, Sugiyama H. Chem. Eur. J. 2010;16:5362. doi: 10.1002/chem.200903057. [DOI] [PubMed] [Google Scholar]; Endo M, Sugita T, Rajendran A, Katsuda Y, Emura T, Hidaka K, Sugiyama H. Chem. Commun. 2011;47:3213. doi: 10.1039/c0cc05306f. [DOI] [PubMed] [Google Scholar]; Rajendran A, Endo M, Katsuda Y, Hidaka K, Sugiyama H. ACS Nano. 2011;5:665. doi: 10.1021/nn1031627. [DOI] [PubMed] [Google Scholar]
- 70.Zhao Z, Liu Y, Yan H. Nano Lett. 2011;11:2997. doi: 10.1021/nl201603a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ke Y, Ong LL, Sun W, Song J, Dong M, Shih WM, Yin P. Nat Chem. 2014;6:994. doi: 10.1038/nchem.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ouyang X, Li J, Liu H, Zhao B, Yan J, Ma Y, Xiao S, Song S, Huang Q, Chao J, Fan C. Small. 2013;9:3082. doi: 10.1002/smll.201300458. [DOI] [PubMed] [Google Scholar]
- 73.Göpfrich K, Zettl T, Meijering AEC, Hernández-Ainsa S, Kocabey S, Liedl T, Keyser UF. Nano Lett. 2015;15:3134. doi: 10.1021/acs.nanolett.5b00189. [DOI] [PubMed] [Google Scholar]
- 74.Pound E, Ashton JR, Becerril HA, Woolley AT. Nano Lett. 2009;9:4302. doi: 10.1021/nl902535q. [DOI] [PubMed] [Google Scholar]; Zhang H, Chao J, Pan D, Liu H, Huang Q, Fan C. Chem. Commun. 2012;48:6405. doi: 10.1039/c2cc32204h. [DOI] [PubMed] [Google Scholar]
- 75.Brudno Y, Silva EA, Kearney CJ, Lewin SA, Miller A, Martinick KD, Aizenberg M, Mooney DJ. Proceedings of the National Academy of Sciences. 2014;111:12722. doi: 10.1073/pnas.1413027111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marras AE, Zhou L, Su H-J, Castro CE. Proceedings of the National Academy of Sciences. 2015;112:713. doi: 10.1073/pnas.1408869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu TJ, Seeman NC. Biochemistry. 1993;32:3211. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 78. http://cando.dna-origami.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.