Abstract
Blood pressure (BP) is a complex trait that is the consequence of an interaction between genetic and environmental determinants. Previous studies have demonstrated increased blood pressure in mice with global deletion of TASK-1 channels contemporaneous with diverse dysregulation of aldosterone production. In humans, genome-wide association studies (GWAS) in ~100,000 individuals of European, East Asian and South Asian ancestry identified a single nucleotide polymorphism (SNP) in KCNK3 (the gene encoding TASK-1) associated with mean arterial pressure (MAP). The current study was motivated by the hypotheses that (1) association of KCNK3 SNPs with BP and related traits extends to African Americans and Hispanics, and (2) KCNK3 SNPs exhibit associations with plasma renin activity (PRA) and aldosterone levels. We examined baseline BP measurements for 7,840 participants from the Multi-Ethnic Study of Atherosclerosis (MESA), and aldosterone levels and PRA in a subset of 1,653 MESA participants. We identified statistically significant association of the previously reported KCNK3 SNP (rs1275988) with MAP in MESA African Americans (P=0.024) and a nearby SNP (rs13394970) in MESA Hispanics (P=0.031). We discovered additional KCNK3 SNP associations with systolic BP (SBP), MAP and hypertension. We also identified statistically significant association of KCNK3 rs2586886 with plasma aldosterone level in MESA and demonstrated that global deletion of TASK-1 channels in mice produces a mild-hyperaldosteronism, not associated with a decrease in renin. Our results suggest genetic variation in the KCNK3 gene may contribute to blood pressure variation and less severe hypertensive disorders in which aldosterone may be one of several causative factors.
Keywords: hypertension, hyperaldosteronism, genetic association, polymorphism, African American
Introduction
Blood pressure (BP) is a complex quantitative trait that is the consequence of an interaction between genetic and environmental determinants.1, 2 Genetic factors account for only 30% of intra-individual BP variability, with the most common forms of hypertension resulting from the cumulative burden of genetic variation at multiple loci with environmental determinants. Further confounding genetic discovery for this trait is the high variability of intra-individual BP measures.3 Thus, the detection of genes and variants contributing to variation in BP and hypertension has proven to be a significant challenge.
Two strategies that have been used effectively to detect genetic associations with modest size effects are to increase sample size, the approach of large-scale genome-wide association study (GWAS), and to perform long-term trait averaging to reduce variation. A combination of these approaches has led to the identification of a new genetic locus for hypertension, the KCNK3 gene, a two-pore domain leak potassium channel.3 of long-term average BP in over 40,000 individuals of European ancestry identified association between KCNK3 SNP rs1275988 with systolic BP (SBP) and mean arterial pressure (MAP), with replication in individuals of Chinese ancestry.3 Subsequently, a larger cross sectional GWAS of ~100,000 individuals of European, East Asian and South Asian ancestry confirmed association of this KCNK3 SNP with MAP.4
Our study was motivated by the hypotheses that KCNK3 SNPs are associated with plasma aldosterone and renin, and the association of KCNK3 SNPs with BP traits extends to African American and Hispanic race/ethnic groups. The first hypothesis is supported by several lines of evidence, (1) the unusually high expression of KCNK3 mRNA product, TASK-1, in the rodent and the human adrenal cortex5; (2) the generation of aldosterone-related hypertension in mice produced by the tandem global deletion of genes (Kcnk3 and Kcnk9) encoding leak/background TASK K+ channels6; and (3) the consensus that aldosterone acts as a causative and/or pathogenic factor in hypertension.7
To address the role of KCNK3 in the regulation of hypertension and aldosterone production in a multi-ethnic population, we performed a genetic association study of KCNK3 SNPs with diastolic blood pressure (DBP), systolic blood pressure (SBP), MAP, pulse pressure (PP), and prevalent hypertension (HTN) in the Multi-Ethnic Study of Atherosclerosis (MESA); we tested the same KCNK3 SNPs for associations with measures of aldosterone and renin in a subgroup of MESA and further examine replication of statistically significant SNPs in independent samples of the Atherosclerosis Risk in Communities (ARIC) study and the Framingham Heart Study (FHS) (Figure S1). We further examined aldosterone overproduction in mice in which only the Kcnk3 gene was deleted. Our studies extend the role of TASK-1 channels and variations in the coding gene (KCNK3) in hypertension and aldosterone production.
Methods
Study Design
The Multi-Ethnic Study of Atherosclerosis (MESA) is a longitudinal study of subclinical cardiovascular disease and risk factors that predict progression to clinically overt cardiovascular disease or subclinical disease. The first clinic visits occurred in 2000-2002 in which 6,814 participants were recruited from six field centers across the United States; all participants were free of CVD at the baseline exam. MESA and related ancillary studies were conducted under Institutional Review Board (IRB) approval of all participating studies sites including the University of Virginia with informed consent of all participants. The study was carried out in accordance with the principles of the Declaration of Helsinki. Additional details in the Online Supplement.
Phenotyping of MESA participants
At the baseline (Exam 1) and main exams for MESA Family (MESAFS) and Air, BP was measured three times; the average of the second and third reading was used for analysis. For the current analyses, measured BP was corrected for anti-hypertensive medication use by adding 10 mmHg (SBP) and 5 mmHg (DBP) to the observed values.8, 9 Using these medication-adjusted BP values, we determined MAP, (two-thirds DBP + one-third SBP), and PP (SBP -DBP). Hypertension status was defined as the presence of at least one of the following (1) measured SBP > 140 mmHg, (2) measured DBP > 90 mmHg, or (3) anti-hypertensive medication use.
At MESA Exams 2 and 3 (approximately 3 and 4.5 years after Exam 1, respectively), a random subsample of 1,960 MESA participants had aldosterone levels and plasma renin activity (PRA) measured as part of an ancillary study investigating renal artery disease.10
Genotype data
Participants recruited into the MESA cohort (6,814), MESAFS (2,128 from 528 families) and MESA Air (5,479 from MESA, 257 external cohort, and 490 from MESAFS) were genotyped in 2009 using the Affymetrix Human SNP Array 6.0. Combining these genome-wide genotypes with imputation to the 1,000 Genomes Project11, there were 85, 192, 113, and 66 KCNK3 region SNPs available for genetic association analysis in MESA White, African American, Hispanic, and Chinese, respectively. Additional details in the Online Supplement.
Measurement of aldosterone in TASK-1 KO mice
TASK-1 KO mice were generated and backcrossed to a C57Bl/6J background as described previously.6, 14 Male WT and T1KO mice habituated in metabolic cages were given free access to normal (NS, 0.3% Na+), low (LS, 0.02% Na+) and high (HS, 2.0% Na+) Na+ diets for one week. Twenty-four hour urine samples were collected on the last 4 days of each diet and analyzed for aldosterone (radioimmunoassay [RIA], Siemens) and creatinine (colorimetric assay, Cayman Chemicals) concentrations. At the end of each dietary condition, tail vein samples were collected and plasma renin concentration was measured (RIA, Diasorin). All animal studies were approved by the University of Virginia IRB and conducted according to the standards of the National Institutes of Health Guide for Care and Use of Laboratory Animals.6, 14, 15
Statistical analysis
Genetic association analysis in MESA
We performed genetic association analysis of DBP, SBP, MAP, PP, HTN, aldosterone levels, and PRA. Primary analyses examined association combined across race/ethnic groups in MESA, with secondary analyses stratified by MESA race/ethnic groups. Analysis of aldosterone levels and PRA was conducted with adjustment for age, sex, study site, principal components (PCs) of ancestry, while analysis of the BP traits included additional adjustment for age-squared and body mass index (BMI). Our approach to the genetic analyses is schematized in Figure S1 and detailed in the Online Supplement.
Replication analysis in ARIC and FHS
For those SNPs demonstrating association with BP traits and hypertension in MESA, we conducted replication analysis in the Atherosclerosis Risk in Communities (ARIC) Study and the Framingham Heart Study (FHS). Following the selection criteria for MESA, we examined only those FHS participants age 45 years or more and restricted both ARIC and FHS to those free of coronary heart disease (CHD) events at the time of examination. Aldosterone measures in the Framingham Heart Study (FHS) Offspring12 and Generation 3 participants13 were examined for replication of one aldosterone-associated SNP identified in MESA. Statistical analyses were conducted using regression models mirroring those used in MESA. Additional details are provided in the Online Supplement.
Analysis of aldosterone in TASK-1 KO mice
Aldosterone/creatinine ratio and PRC were log transformed due to unequal variance among dietary conditions, and tested for significance (P<0.05) using 2-way ANOVA and Bonferroni post hoc analysis.
Results
Characteristics of the study sample
At baseline (2000-2002), MESA participants were a relatively healthy group across all race/ethnicities (Table 1). The median age at baseline (Exam 1) was 60-63 years, with prevalence of diabetes ranging from 6.1%-18.7%, and percentage who currently smoke ranging from 5.5%-19.4%. African Americans had higher SBP (median 133.5, mmHg), DBP (median 76.5 mmHg), and MAP (median 95.5 mmHg) compared to the other race/ethnic groups (SBP: race/ethnic specific medians 124.0–126.0 mmHg; DBP: 71.5-72.5 mmHg; MAP: 89.5-90.8 mmHg) and a greater percentage were on anti-hypertensive medication (African Americans: 51.4%; other race/ethnicities: 28.1-35.2%). Aldosterone and PRA levels were lower in African Americans (aldosterone: median 11.8 ng/dL; PRA: 0.38 ng/mL/hr) compared to other race/ethnic groups (aldosterone: race/ethnic specific medians 12.9-13.9 ng/dL; PRA: 0.53-0.67 ng/mL/hr), as reported previously in MESA.10
Table 1.
Descriptives of the MESA participants included in genetic analyses.
| Variable | White | African American |
Hispanic | Chinese |
P-value for race/ethnicity‡ |
|---|---|---|---|---|---|
| Baseline participant characteristics * | |||||
| No. subjects | 2464 | 2556 | 2113 | 707 | |
| Women | 1274 (51.7) | 1436 (56.2) | 1140 (54.0) | 358 (50.6) | 0.005 |
| Age, years | 63 [54, 71] |
60 [53, 68] |
60 [53, 68] |
62 [53, 70] |
<0.0001 |
| BMI, kg/m2 | 27.1 [24.2, 30.4] |
29.4 [26.2, 33.8] |
28.6 [25.9, 32.1] |
23.8 [21.8, 26.0] |
<0.0001 |
| Diabetes | 151 (6.1) | 441 (17.3) | 394 (18.7) | 91 (12.9) | <0.0001 |
| Current smoke (yes/no) | 276 (11.5) | 492 (19.4) | 288 (13.8) | 39 (5.5) | <0.0001 |
| Ever smoke (yes/no) | 1368 (55.7) | 1348 (53.0) | 967 (45.8) | 176 (24.9) | <0.0001 |
| Blood pressure medication (yes/no) | 826 (33.5) | 1313 (51.4) | 743 (35.2) | 199 (28.1) | <0.0001 |
| Urinary sodium creatinine ratio (mmol/mmol) |
10.8 [7.0, 15.8] |
10.2 [6.6, 14.6] |
11.6 [7.4, 17.3] |
13.5 [8.5, 19.3] |
<0.0001 |
| Blood pressure traits | |||||
| No. subjects | 2464 | 2556 | 2113 | 707 | |
| Systolic blood pressure (mmHg)† | 124.0 [111.0, 140.0] |
133.5 [118.5, 150.5] |
126.0 [111.5, 143.0] |
124.2 [109.0, 142.5] |
<0.0001 |
| Diastolic blood pressure (mmHg)† | 71.5 [64.5, 78.9] |
76.5 [69.5, 83.5] |
72.5 [65.5, 79.5] |
72.5 [66.0, 80.0] |
<0.0001 |
| Mean arterial pressure (mmHg)† | 89.5 [81.0, 98.3] |
95.5 [87.2, 105.2] |
90.8 [82.2, 99.8] |
90.3 [81.3, 99.7] |
<0.0001 |
| Pulse pressure (mmHg)† | 51.5 [41.9, 64.5] |
56.0 [45.5, 70.0] |
52.5 [42.1, 66.5] |
50.5 [40.5, 64.5] |
<0.0001 |
| Prevalent hypertension (yes/no) | 1079 (43.8) | 1598 (62.5) | 955 (45.2) | 282 (39.9) | <0.0001 |
| Renin and aldosterone | |||||
| No. subjects | 697 | 321 | 422 | 213 | |
| Aldosterone (ng/dL) | 13.5 [9.8, 18.7] |
11.8 [7.9, 17.0] |
13.9 [10.0, 20.2] |
12.9 [9.8, 17.3] |
<0.0001 |
| Plasma renin activity (ng/mL/hour) | 0.61 [0.34, 1.26] |
0.38 [0.20, 0.89] |
0.67 [0.34, 1.40] |
0.53 [0.27, 0.98] |
<0.0001 |
| Aldosterone to renin ratio ( (ng/dL) / (ng/mL/hour)) |
22.1 [10.6, 39.0] |
28.9 [14.5, 57.8] |
19.7 [10.6, 38.5] |
23.7 [13.5, 41.9] |
0.003 |
Data are presented as n (%) for binary measures or median [IQR] for continuous measure.
Summary statistics are reported for the subset of individuals with data available for at least one of the phenotypes.
Blood pressure traits are shown after correction for medication use
Statistical significance of likelihood ratio test for race/ethnic differences in linear (or logistic) regression of continuous (binary) measures. Measures other than sex and age include covariate adjustment for sex and age in the regression models. Variable transformation applied before analyzing aldosterone (square-root), plasma renin activity (log) and aldosterone to renin ratio (log).
More than 80% of participants classified as using antihypertensive medication at baseline reported using no more than two classes of medication (Table S1). Among these participants, we observed expected differences in PRA with respect to medication exposure. Compared to non-hypertensives, renin levels were not statistically different in patients on alpha and calcium channel blockers (Table 2, P=0.34), were decreased in those on beta blockers (P=0.0001), and elevated markedly among patients on ARBs, ACE inhibitors and/or diuretics (P<0.0001).
Table 2.
Summary of aldosterone levels and plasma renin activity by medication class in MESA, compared to those without hypertension.
| Trait | Medication exposure | White | African American |
Hispanic | Chinese |
|---|---|---|---|---|---|
| Aldosterone (ng/dL) |
I. Beta-blockers only* | 12.6 [9.3, 16.0] (n=36) |
13.1 [9.8, 17.1] (n=11) |
12.4 [11.0, 15.3] (n=13) |
14.4 [10.8, 19.2] (n=12) |
| II. Alpha-blockers or calcium channel blockers only* |
16.3 [11.6, 21.4] (n=27) |
12.6 [8.7, 17.6] (n=24) |
18.7 [11.8, 25.4] (n=21) |
14.0 [11.2, 17.6] (n=10) |
|
| IIIA. Thiazide diuretics, Angiotensin receptor blockers, or ACE inhibitors* |
13.2 [9.5, 20.4] (n=131) |
12.3 [7.6, 16.9] (n=69) |
14.9 [10.0, 21.1] (n=57) |
12.4 [8.1, 15.5] (n=22) |
|
| IIIB. Angiotensin receptor blockers or ACE inhibitors without diuretics* |
11.5 [9.1, 15.6] (n=85) |
10.7 [6.3, 13.9] (n=35) |
13.0 [8.9, 19.1] (n=38) |
10.7 [6.7, 15.2] (n=16) |
|
| IV. No Hypertension | 12.8 [9.3, 17.3] (n=339) |
10.4 [7.3, 14.1] (n=96) |
13.0 [9.5, 18.0] (n=208) |
13.0 [9.7, 16.5] (n=123) |
|
| Plasma renin activity (ng/mL/hour) |
I. Beta-blockers only* | 0.36 [0.22, 0.66] (n=31) |
0.27 [0.20, 0.37] (n=10) |
0.34 [0.26, 0.71] (n=12) |
0.15 [0.03, 0.31] (n=12) |
| II. Alpha-blockers or calcium channel blockers only* |
0.74 [0.57, 1.27] (n=23) |
0.32 [0.11, 0.50] (n=21) |
0.45 [0.29, 1.06] (n=20) |
0.47 [0.24, 0.64] (n=9) |
|
| IIIA. Thiazide diuretics, Angiotensin receptor blockers, or ACE inhibitors* |
1.78 [0.87, 5.89] (n=129) |
0.91 [0.36, 2.16] (n=67) |
2.04 [0.61, 3.69] (n=56) |
1.42 [0.81, 3.50] (n=23) |
|
| IIIB. Angiotensin receptor blockers or ACE inhibitors without diuretics* |
2.04 [0.95, 5.72] (n=83) |
0.82 [0.28, 1.94] (n=35) |
2.06 [0.42, 3.53] (n=37) |
1.54 [0.92, 6.33] (n=18) |
|
| IV. No Hypertension | 0.55 [0.31, 0.89] (n=333) |
0.32 [0.20, 0.54] (n=94) |
0.66 [0.37, 1.10] (n=200) |
0.51 [0.30, 0.88] (n=120) |
|
| Aldosterone to renin ratio ( (ng/dL) / (ng/mL/hour)) |
I. Beta-blockers only* | 31.6 [21.4, 69.3] (n=31) |
34.3 [27.2, 61.0] (n=9) |
35.3 15.9, 47.9] (n=12) |
132.8 [34.3, 267.0] (n=11) |
| II. Alpha-blockers or calcium channel blockers only* |
21.9 [12.0, 31.0.] (n=22) |
49.0 [19.4, 99.3] (n=21) |
30.5 [19.1, 67.8] (n=20) |
34.2 [23.0, 64.7] (n=9) |
|
| IIIA. Thiazide diuretics, Angiotensin receptor blockers, or ACE inhibitors* |
7.7 [2.2, 18.3] (n=126) |
15.1 [4.6, 33.1] (n=65) |
9.1 [3.9, 25.6] (n=55) |
7.6 [3.9, 12.0] (n=21) |
|
| IIIB. Angiotensin receptor blockers or ACE inhibitors without diuretics* |
5.56 [1.97, 11.82] (n=81) |
11.0 [4.6, 40.0] (n=33) |
7.5 [3.6, 23.6] (n=36) |
6.5 [3.6, 12.0] (n=16) |
|
| IV. No Hypertension | 22.8 [14.5, 38.8] (n=330) |
30.7 [20.9, 53.4] (n=89) |
17.9 [11.9, 31.2] (n=198) |
23.4 [14.0, 39.0] (n=120) |
Data are presented median [IQR].
Medication exposure groups are constructed by grouping together those participants with exposure to any of the indicated classes of medications, and excluding all other classes of medications indicated in Table S1.
Likelihood ratio test performed for comparison of participants on Beta-blockers, alpha-blockers, or calcium channel blockers without diuretics compared to those on thiazide diuretics with or without K-sparing agents, angiotensin receptor blockers (with or without diuretics), or ACE inhibitors (with or without diuretics) for aldosterone levels (P=0.46) or plasma renin activity (P=3.0E-8) after covariate adjustment for sex, age and race/ethnicity.
KCNK3 SNPs across race/ethnic groups in MESA and association with BP traits
KCNK3 rs1275988, previously reported in GWAS of MAP3, 4, is located 1.3kb 5’ of KCNK3 suggesting a potential role in gene regulation. Although there is no evidence for this SNP as a cis-eQTL, examining the full range of tissues from GTEx (http://www.gtexportal.org/home/) or using the Blood eQTL browser16 (http://genenetwork.nl/bloodeqtlbrowser/). We examined race/ethnic-specific evidence of association between rs1275988 and MAP in MESA and found significant association of the C allele in stratified analyses of MESA White (Table S2, P=0.025) and African American (P=0.024) participants, with consistent direction of effect in MESA Chinese (not statistically significant). The rs1275988 genotype distribution in MESA Hispanic Americans deviated significantly from that expected under Hardy-Weinberg Equilibrium (HWE) assumptions and rs1275988 was not included in the analysis. Our finding of an association between rs1275988 and MAP in MESA African Americans represents a novel replication of the rs1275988 SNP association, as African Americans were not included in previous large-scale GWAS efforts.3, 4
In analysis of additional BP traits and prevalent hypertension, we identified associations in the full MESA cohort for KCNK3 rs1731243 with SBP (Table 3, P=1.9 × 10−4) and HTN (P=4.6 × 10−5), with consistent directions of effect in all race/ethnic groups. KCNK3 rs1731243 is in strong linkage disequilibrium (LD) (Figure S2, all 1000 Genomes11 Phase 1 v3 EUR, AFR, AMR and ASN r2 > 0.9) with the MAP-associated rs1275988. In addition, KCNK3 rs13394970 associated with MAP (P=1.9 × 10−4). While rs1339490 shows strong LD with rs1275988 among Europeans, Native Americans and Asians (all 1000G EUR, AMR and ASN r2 > 0.9), there is little LD between these two SNPs among Africans (1000G AFR r2=0.14). Both rs1731243 and rs13394970 are located in an intron of KCNK3 with predicted regulatory effects, particularly on enhancer histone marks and DNA binding motifs (HaploReg v4.1, http://www.broadinstitute.org/mammals/haploreg/haploreg.php). Conditional analysis including the most significantly associated SNP for SBP, MAP, and HTN (Table 3) as a covariate did not reveal any additional independently associated SNPs for these traits overall, nor in race/ethnic specific analyses.
Table 3.
Race/ethnic-specific association results for SNPs identified in association with blood pressure traits in MESA.
| Trait | SNP (Effect / other allele) |
Group | Effect allele frequency |
N | Beta | SE | P-value |
|---|---|---|---|---|---|---|---|
| SBP | rs1731243* (C/T) |
White | 0.38 | 2453 | 1.09 | 0.57 | 0.058 |
| African American | 0.76 | 2553 | 2.09 | 0.76 | 0.006 | ||
| Hispanic | 0.35 | 2106 | 1.20 | 0.71 | 0.093 | ||
| Chinese | 0.75 | 706 | 1.44 | 1.34 | 0.283 | ||
| Meta-analysis | 1.38 | 0.37 | 1.9E-04 | ||||
| MAP | rs13394970* (T/G) |
White | 0.38 | 2460 | 0.77 | 0.35 | 0.029 |
| African American | 0.45 | 2550 | 0.79 | 0.40 | 0.049 | ||
| Hispanic | 0.30 | 2107 | 0.97 | 0.45 | 0.031 | ||
| Chinese | 0.75 | 706 | 0.75 | 0.82 | 0.361 | ||
| Meta-analysis | 0.82 | 0.22 | 1.9E-04 | ||||
| HTN | rs1731243* (C/T) |
White | 0.38 | 2464 | 0.12 | 0.07 | 0.070 |
| African American | 0.76 | 2556 | 0.17 | 0.08 | 0.032 | ||
| Hispanic | 0.35 | 2113 | 0.20 | 0.08 | 0.008 | ||
| Chinese | 0.75 | 707 | 0.24 | 0.15 | 0.106 | ||
| Meta-analysis | 0.17 | 0.04 | 4.6E-05 |
Genetic association results for medication-corrected blood pressure traits reported under an additive 1 d.f. SNP dosage model. Analysis of blood pressure traits and prevalent hypertension conducted with adjustment for sex, age, age2, body mass index, study site and race/ethnic specific PCs of ancestry.
Most strongly associated SNP in meta-analysis for the indicated gene and trait.
To further characterize the main associations identified with BP traits, we performed sensitivity analyses using different statistical models, relevant covariates and stratification based on factors of additional interest. Notably, analyzing measured BP with covariate adjustment for antihypertensive medication exposure (Table S3), we observed results qualitatively similar to our main analysis in which phenotypic outcomes were corrected for antihypertensive medication exposure. Similarly, results for our main SNP associations remained qualitatively similar after additional covariate adjustment for urinary sodium/creatinine ratio (Table S4), and when long-term average BP traits (Table S5) were examined. In stratified analyses, we observed stronger evidence of association in participants over 60 years old vs. those 60 years and under (Table S6), in females vs. males (Table S7), and in post-menopausal vs. pre-menopausal women (Table S8), although overall the observed directions of effect were consistent across subgroups.
KCNK3 SNPs and variation in aldosterone levels and PRA in MESA
One KCNK3 SNP, rs2586886, demonstrated association with aldosterone levels in the MESA cohort (Table 4, P=3.0 × 10−4). The rs2586886 SNP is in an intron of KCNK3, affecting both promoter and enhancer histone marks (HaploReg v4.1), and is in high LD with rs1275988 in Europeans and Asians from the 1000 Genomes (Figure S2, 1000G EUR and ASN r2 > 0.9), with reduced LD in Native Americans (1000G AMR r2=0.72) and no LD in Africans (1000G AFR r2=0.12). The association showed consistent directional effects across the four race/ethnic groups, with stronger effects observed in African Americans and Chinese. In sensitivity analysis, we identified statistically significant evidence of heterogeneity for the association of rs2586886 with aldosterone based on antihypertensive medication exposure, with strong effects of the C allele on increased aldosterone levels observed only among individuals currently using antihypertensive medication (Table 4, Cochran’s Q heterogeneity P=0.004). To understand the significance of this finding, we examined the association of rs2586886 with aldosterone levels stratified by antihypertensive medication class (Table 2). Patients were subdivided into three major groups based on the established class effect of their medication on the renin-angiotensin system. Patients on medications that suppress the system were included in group I (beta-blockers only), those on drugs having little direct impact on the system, group II (alpha-blockers or calcium channel blockers) and those on medications that frankly stimulate renin group III (thiazide diuretics, angiotensin receptor blockers, or ACE inhibitors, alone or in combination). In a combined analysis across race/ethnic groups, we observed consistent directional effects in all medication subgroups. Nevertheless, the strongest (and significant) evidence of association of rs2586886 with elevated aldosterone was observed for those patients in group III (A&B) on agents that stimulate renin (Table S9). Notably, exclusion of participants taking diuretics from group IIIA, did not reduce the statistical significance of the association of aldosterone levels with rs2586886 (Table S9), even though diuretic-induced augmentation of aldosterone levels was absent from subgroup IIIB (Table 2). Analysis of rs2586886 with aldosterone levels in those without hypetension was not statistically significant (Table S9, P=0.283) but the direction of effect was consistent with that seen in those on medication. Sex-stratified analysis of rs2586886 for aldosterone levels demonstrated comparable effect sizes in males and females (Table S10, both nominal P<0.05). In addition, the C allele for the rs2586886 SNP also was associated with increases in measures of BP in the full MESA cohort (SBP, DBP, MAP, PP and HTN; Table S11, all nominal P<0.05), with consistent directions of effect in each of the four race/ethnic groups in MESA. There were no KCNK3 SNPs demonstrating statistical significance in the full analysis of renin across MESA race/ethnic groups. .
Table 4.
Race/ethnic-specific association results for KCNK3 SNP rs2586886 for aldosterone levels in MESA.
| Stratum | Group | Effect allele frequency |
N | Beta | SE | P-value |
|---|---|---|---|---|---|---|
| All | White | 0.37 | 683 | 0.068 | 0.059 | 0.252 |
| African American | 0.41 | 311 | 0.206 | 0.081 | 0.011 | |
| Hispanic | 0.30 | 415 | 0.158 | 0.081 | 0.052 | |
| Chinese | 0.75 | 211 | 0.205 | 0.103 | 0.047 | |
| Meta-analysis | 0.138 | 0.038 | 3.0E-04 | |||
| No hypertension medication |
White | 0.38 | 382 | −0.102 | 0.071 | 0.150 |
| African American | 0.35 | 118 | 0.033 | 0.120 | 0.783 | |
| Hispanic | 0.31 | 245 | 0.127 | 0.097 | 0.192 | |
| Chinese | 0.71 | 137 | 0.177 | 0.113 | 0.119 | |
| Meta-analysis | 0.020 | 0.047 | 0.664 | |||
| Hypertension medication |
White | 0.40 | 286 | 0.263 | 0.099 | 0.008 |
| African American | 0.42 | 174 | 0.210 | 0.113 | 0.065 | |
| Hispanic | 0.31 | 160 | 0.251 | 0.144 | 0.082 | |
| Chinese | 0.77 | 74 | 0.306 | 0.210 | 0.150 | |
| Meta-analysis | 0.248 | 0.063 | 8.4E-05 |
Genetic association results for square-root transformed aldosterone levels (in ng/dL) reported under an additive 1 d.f. SNP dosage model for rs2586886 effect allele C (other allele T). Analysis of aldosterone conducted with adjustment for sex, age, study site and race/ethnic specific PCs of ancestry.
Replication of SNP associations in ARIC and the Framingham Heart Study
Blood pressure traits
We examined SNPs across the KCNK3 gene in >10,000 participants from ARIC (Table S12) and >3,700 FHS (Table S12) participants for replication of association with BP traits for the most strongly associated SNPs identified in MESA (Tables 3 and 4). In meta-analysis that combined all replication samples from ARIC and FHS, each of the identified KCNK3 SNPs examined showed nominally significant association with the most strongly associated BP trait identified in MESA (Table S13, all nominal P<0.05). Among African American participants from ARIC, we did not observe statistically significant evidence of replication (nominal P>0.05), although the directions of effect were overall consistent in this group.
Aldosterone
Aldosterone levels in the MESA White and FHS cohorts, differed significantly (P<0.0001) with lower values observed in FHS Whites, whereas PRA was not different (P=0.07). We suggest that higher aldosterone values in MESA Whites may be attributable to greater disease progression as in aggregate MESA Whites were on more antihypertensive medications.
In the full set of FHS participants, we did not find statistical evidence of replication for the association of rs2586886 with aldosterone (Table S14, nominal P=0.559). However, in sensitivity analysis stratified by use of hypertension medication, we identified a suggestion of association in participants with (nominal P=0.094) and without (nominal P=0.095) medication use, with opposing directions of effect (beta=0.09, with medication use; beta=−0.04, without medication use) concordant with those in the same strata among the MESA participants (Table 4). Similar to MESA, we observed significant evidence of heterogeneity in the effects of KCNK3 rs2586886 on aldosterone levels based on medication use in FHS (Cochran’s Q heterogeneity P=0.026).
Hyperaldosteronism in TASK-1 KO mice
In a previous rodent study, the global deletion of TASK-1 channels produced a highly significant 15 mm Hg-increase in SBP in female mice, with a tendency toward elevated SBP in male mice.17 Canrenoate, a blocker of the mineralocorticoid receptor, normalized SBP between genotypes, suggesting that hypertension was aldosterone-dependent. Nevertheless, in that study hyperaldosteronism was not detected in males and that observed in females did not arise from adrenal zona glomerulosa cells, but rather from a sex-dependent zonation defect in which aldosterone is made in the zona fasciculata.
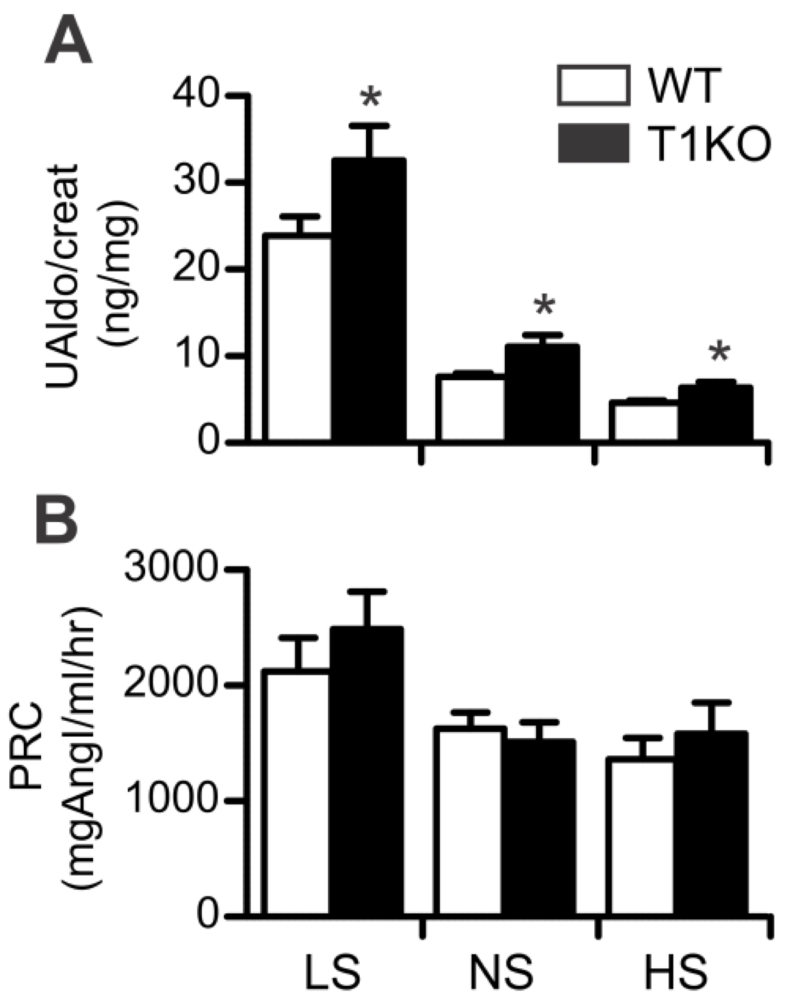
To determine if TASK-1 deletion causes hyperaldosteronism in animals in which adrenal zonation is normal and Cyp11b2 (aldosterone synthase) expression remains restricted to zona glomerulosa cells17, we studied the aldosterone/renin responses of male TASK-1 KO mice to changes in dietary sodium. Rather than assess plasma aldosterone concentration, which captures the adrenal response at a single point in time, we used an integrated measure of aldosterone status, 24 hour urinary aldosterone excretion (urinary aldosterone normalized to creatinine) that better reports modest changes in production.6, 15 Using this measure, we found that TASK-1 KO mice overproduce aldosterone on all sodium diets from 36 to 73 % (Figure 1A). Although enhanced, aldosterone production from TASK-1 KO mice remained under the control of the renin-angiotensin system. Thus, increasing dietary sodium lowered aldosterone production whereas decreasing dietary sodium raised aldosterone production in both WT and TASK-1 KO mice. TASK-1 KO mice maintained normal renin status (Figure 1B) on all sodium diets compared with WT mice (P=1.0, NS) in agreement with previous findings17, indicating that TASK-1 KO mice are not a model of low renin associated primary hyperaldostoneronism.7
Figure 1.
Effect of diet on urinary aldosterone (UAldo) and plasma renin concentration (PRC). A. 24-hour UAldo/creatinine (ng/mg) of wild type WT (white bars) and T1KO mice (black bars) on each of 3 salt diets: low Na (0.02% Na-LS; n=25/13), normal Na (0.3% (Na-NS; n=38/14); high Na (2.0% Na-HS; n=32/13). B. PRC (mg of angiotensin [Ang] 1 per ml/h) of WT and T1KO mice on salt diets: LS (n=21/12), NS (n=22/13), HS (n=18/11). Values represent mean ±SEM; * vs WT mice (P<0.05, two-way ANOVA)
Discussion
A substantial body of clinical and experimental data implicates aldosterone in the pathogenesis of hypertension, the prime example of which is primary aldosteronism, the most common secondary hypertensive disorder.18, 19 Moreover, additional evidence in support of a relationship between circulating aldosterone and hypertension incident is the reported 1.6-fold increase in hypertension risk among individuals with serum aldosterone concentrations in the highest quartile relative to those in the lowest quartile.20 Based on these studies and our own work characterizing the role of TASK channels in aldosterone-related hypertension, we hypothesized that previously reported genetic associations of KCNK3 with BP traits21 may relate to the dysregulation of renin and aldosterone. We therefore conducted a genetic association study of KCNK3 SNPs in relation to BP traits, hypertension, aldosterone and PRA in MESA.
In analysis of BP traits, we replicated association of the previously reported3, 4 KCNK3 SNP rs1275988 with MAP in MESA African Americans. We did not examine the association rs1275988 in MESA Hispanics because it failed the test for Hardy-Weinberg equilibrium. Nevertheless, we identified a nearby SNP with high LD (rs13394970) that demonstrated association with MAP in this group. We further identified another KCNK3 SNP, rs1731243, associated with SBP and hypertension status. Each of these SNP-trait associations replicated in the combined analysis of independent samples from ARIC and FHS, studies that have similar design and representative populations. The replication analysis did not achieve statistical significance in ARIC African Americans alone, although we observed overall consistent directions of effect in this group. In addition, we identified an aldosterone-associated SNP rs2586886 for which the C allele also demonstrated nominally significant evidence of association with increased SBP in MESA, an effect that replicated in the combined analysis of SBP in ARIC and FHS.
In examining replication of our selected SNPs from MESA in external cohorts, we recognize that European-ancestry samples from ARIC, MESA and FHS were included in previous large-scale GWAS efforts that identified the KCNK3 SNP rs1275988 in relation to SBP and MAP.3 As a consequence, we note that our replication sample is not entirely independent of previously published work. However, the effect size of the identified SNPs on BP traits in MESA is notably larger than those observed in the replication cohorts. This observation may reflect, in part, the Winner’s curse22 whereby effect estimates for SNPs are biased by the reporting of only those SNPs that reach a strict threshold for multiple comparisons. In addition, the larger effect sizes evident in MESA may reflect a better representation of the contemporary United States population in which environmental and dietary factors that promote hypertension are ample and could magnify the contribution of genetic variation to blood pressure and related traits.
Sensitivity analyses of the identified BP trait associated SNPs indicate the observed associations are strongest in females, subpopulations over the age of 60, and post-menopausal women. As BP itself rises with age, and in females post-menopause, it is noteworthy that the contribution of genetic variation in the KCNK3 gene to BP also becomes manifest at a later stage of life, as compensatory regulatory mechanisms decline.Thus, it is not surprising that our previous investigation that included teenagers and young adults did not identify KCNK3 SNPs demonstrating statistically significant associations with BP traits or aldosterone levels.23 Further, we note that this previous investigation used targeted genotyping of selected KCNK3 SNPs, and the SNPs reported in the current investigation were not included in that genotyping effort.
Interestingly, the rs2586886 SNP that associated with aldosterone levels in MESA showed significant evidence of heterogeneity with respect to antihypertensive medication status, with stronger effects of the C allele on increased aldosterone levels among participants on medication. Significance of this heterogeneity with respect to antihypertensive medication was also observed through replication in FHS. Although it is technically possible that this heterogeneity is caused directly by antihypertensive medication, we favor an alternate interpretation that medication suppressing the renin-angiotensin system unmasks the relationship between the C allele of rs2586886 and aldosterone to reveal dysfunction in the renin-angiotensin-aldosterone system. Plasma renin activity was significantly suppressed in the MESA White, Hispanic and Chinese participants taking beta-blockers alone compared with the non-hypertensive group. As expected, African Americans had low plasma renin activity whether or not they were taking beta-blockers24, 25 In the White, Hispanic and Chinese participants on beta-blockers alone, frank suppression of renin was not accompanied by suppression of aldosterone secretion; plasma aldosterone levels in group I were similar to those in the non-medicated, non-hypertensive group IV. Similarly, the response of group III participants to their antihypertensive medication is supported by very high levels of plasma renin activity, yet these participants, with presumptively low levels of AT1 receptor activation [a consequence of reduced Ang II generation (ACE inhibitors) or AT1 receptor inhibition (angiotensin receptor blockers)], maintained levels of aldosterone production that also were similar to those of group IV. Thus, the suppression of renin-angiotensin system by two classes of medication with divergent mechanisms of action provides substantial evidence for the association of the C allele with dysregulation of aldosterone production manifest as non-suppressibility of secretion.
While all of the KCNK3 SNPs analyzed were in strong LD with rs1275988 in EUR and ASN populations, LD was marginally reduced in AMR samples, and considerably reduced in AFR samples. Based on the patterns of LD across race/ethnic groups, we suggest that the aldosterone-associated KCNK3 rs2586886 represents a site that is distinct from the previously reported MAP-associated SNP rs1275988.21 The results in MESA and patterns of LD in the 1000 Genomes Project suggest that the previously reported MAP-associated SNP rs1275988 overlaps, in large part, the SNP most strongly associated with MAP (rs13394970) in the current investigation.
We have demonstrated that the global deletion of TASK-1 channels in mice produces a mild-hyperaldosteronism that is not associated with an increase in renin, suggesting either modest autonomous overproduction or an increased sensitivity to angiotensin II. Our studies build on the findings of Barhanin, Warth and colleagues who demonstrated that in TASK-1 KO male mice aldosterone synthase localizes normally in the ZG layer.17 However, in their animal cohort aldosterone production remained under strict control of the renin-angiotensin system, an outcome that could be explained by the reported age-dependent increase in TASK-3 channel mRNA in their cohort which would compensate for TASK-1 channel deletion.17 Also, these findings differ significantly from previous studies from our laboratory in which the deletion of TASK-3 channels alone (TASK-3−/−)15 or in combination with TASK-1 channels (TASK-1−/−::TASK-3−/−)6 produces variable, but higher, degrees of hyperaldosteronism, ranging from modest to exaggerated that were accompanied by a reduction in plasma renin. TASK-1 KO mice also differ from both TASK-3−/− and TASK-1−/−::TASK-3−/− mice in their aldosterone response to dietary sodium loading. The latter genotypes fail to suppress aldosterone output on a high salt diet. By contrast, TASK-1 KO mice reduce aldosterone output when salt intake increases and renin activity is decreased, similar to WT mice. Thus, TASK-1 deletion alone does not recapitulate the low renin phenotype that is characteristic of idiopathic primary hyperaldosteronism or low-renin essential hypertension, disorders that are associated with aldosterone production that is considered autonomous of the renin-angiotensin system.7
Our findings in mice are consistent with the failure of the aldosterone-associated SNP (rs2586886) to associate with renin. As rs2586886 is in the first intron containing putative regulatory elements, we hypothesize that attenuation of human KCNK3 gene transcription may recapitulate the mild dysfunction of aldosterone production from zona glomerulosa cells in mice with TASK-1 channel deletion that we report here. In rodents, Kcnk3 and Kcnk9 genes are highly expressed in the zona glomerulosa layer26; in contrast to the human adrenal cortex in which the expression of KCNK3 gene expression is dominant relative to other KCNK family members.5 Thus, although a reduction in the expression of any active hyperpolarizing membrane conductance from the plasma membrane of zona glomerulosa cells would be expected to increase intracellular calcium, the critical second messenger that drives aldosterone production,26 the extent of this increase, and thus the aldosterone size effect will depend upon the relative activity of other active conductances in the membrane. Disease causing variants in the exon-coding regions of several ion channels (KCNJ5, CACNA1D, CACNA1H) expressed in the zona glomerulosa of the adrenal cortex, have been discovered in patients with tumorigenic primary hyperaldosteronism that produce excessive levels of aldosterone or display marked and/or early onset-hypertension.27-30
Perspectives
We confirmed association of the previously reported KCNK3 rs1275988 with MAP in MESA African Americans, and identified additional association of KCNK3 SNPs with SBP, MAP and hypertension status in MESA that replicated in independent samples from FHS and ARIC. We further identified a novel association of KCNK3 rs2586886 with increased aldosterone levels in MESA, the association of which demonstrated evidence of heterogeneity with respect to hypertension medication status in both MESA and FHS. Interpreted in the context of our mouse studies, our results suggest that genetic variation in the KCNK3 gene may contribute to less severe polygenic hypertensive disorders in which aldosterone dysregulation may be one of several contributing factors.
Supplementary Material
Novelty and Significance.
What is New?
Our study validates the KCNK3 gene as a new genetic locus for hypertension.
Identified multiple new KCNK3 variants, within the same genomic region as a single previously reported allele, that associate with measures of blood pressure across race/ethnic groups.
Discovered a distinct KCNK3 variant associating with non-suppressible aldosterone and nominally with multiple BP traits.
Rodent studies demonstrate that genetic disruption of KCNK3 produces hyperaldosteronism in a hypertensive model without a reduction in renin.
What is Relevant?
The most common forms of hypertension result from the cumulative burden of genetic variation at multiple loci. Our study identifies KCNK3 gene variants as potential drivers of hypertension and aldosterone across multiple race/ethic groups.
Summary
Genetic variation in the KCNK3 gene across race/ethnic groups could contribute to less severe polygenic hypertensive disorders in which aldosterone elevation may be a contributing factor.
Acknowledgments
MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators.
Sources of Funding
Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, and UL1-TR-000040. MESA Family is supported by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071258, R01HL071259, by the National Center for Research Resources, Grant UL1RR033176, and the National Center for Advancing Translational Sciences, Grant UL1TR000124. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, California, USA) and the Broad Institute of Harvard and MIT (Boston, Massachusetts, USA) using the Affymetrix Genome-Wide Human SNP Array 6.0. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. This publication was developed under a STAR research assistance agreement, No. RD831697 (MESA Air), awarded by the U.S Environmental protection Agency. It has not been formally reviewed by the EPA. The views expressed in this document are solely those of the authors and the EPA does not endorse any products or commercial services mentioned in this publication. Support for the MESA Mineral Metabolite dataset was provided by grant R01HL096875 to MAA and by NIDDK grant R01 DK080015. This study also was supported by NHLBI grant R01 HL089717 to PQB and DAB.
Footnotes
Disclosures
All authors have read and agreed to the content within the article. All authors declare no financial or other conflict of interest relevant to the subject of this article.
References
- 1.Weder AB. Genetics and hypertension. J Clin Hypertens (Greenwich) 2007;9:217–223. doi: 10.1111/j.1524-6175.2007.06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper RS. Gene-environment interactions and the etiology of common complex disease. Ann Intern Med. 2003;139:437–440. doi: 10.7326/0003-4819-139-5_part_2-200309021-00011. [DOI] [PubMed] [Google Scholar]
- 3.Ganesh SK, Chasman DI, Larson MG, et al. Effects of long-term averaging of quantitative blood pressure traits on the detection of genetic associations. Am J Hum Genet. 2014;95:49–65. doi: 10.1016/j.ajhg.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato N, Loh M, Takeuchi F, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47:1282–1293. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nogueira EF, Gerry D, Mantero F, Mariniello B, Rainey WE. The role of task1 in aldosterone production and its expression in normal adrenal and aldosterone-producing adenomas. Clin Endocrinol (Oxf) 2010;73:22–29. doi: 10.1111/j.1365-2265.2009.03738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies LA, Hu C, Guagliardo NA, Sen N, Chen X, Talley EM, Carey RM, Bayliss DA, Barrett PQ. Task channel deletion in mice causes primary hyperaldosteronism. Proc Natl Acad Sci U S A. 2008;105:2203–2208. doi: 10.1073/pnas.0712000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young WF. Primary aldosteronism: Renaissance of a syndrome. Clin Endocrinol (Oxf) 2007;66:607–618. doi: 10.1111/j.1365-2265.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 8.Cui JS, Hopper JL, Harrap SB. Antihypertensive treatments obscure familial contributions to blood pressure variation. Hypertension. 2003;41:207–210. doi: 10.1161/01.hyp.0000044938.94050.e3. [DOI] [PubMed] [Google Scholar]
- 9.Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rifkin DE, Khaki AR, Jenny NS, McClelland RL, Budoff M, Watson K, Ix JH, Allison MA. Association of renin and aldosterone with ethnicity and blood pressure: The multi-ethnic study of atherosclerosis. Am J Hypertens. 2014;27:801–810. doi: 10.1093/ajh/hpt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newton-Cheh C, Guo CY, Gona P, Larson MG, Benjamin EJ, Wang TJ, Kathiresan S, O’Donnell CJ, Musone SL, Camargo AL, Drake JA, Levy D, Hirschhorn JN, Vasan RS. Clinical and genetic correlates of aldosterone-to-renin ratio and relations to blood pressure in a community sample. Hypertension. 2007;49:846–856. doi: 10.1161/01.HYP.0000258554.87444.91. [DOI] [PubMed] [Google Scholar]
- 13.O’Seaghdha CM, Hwang SJ, Vasan RS, Larson MG, Hoffmann U, Wang TJ, Fox CS. Correlation of renin angiotensin and aldosterone system activity with subcutaneous and visceral adiposity: The framingham heart study. BMC endocrine disorders. 2012;12:3. doi: 10.1186/1472-6823-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulkey DK, Talley EM, R.L S, Siegel A, West GH, Chen X, Sen N, Mistry AM, Guyenet PG, Bayliss DA. Task channels determine ph sensitivity in select respiratory neurons but do not contribute to central respiratory chemosensitivity. Journal of Neuroscience. 2007;27:14049–14058. doi: 10.1523/JNEUROSCI.4254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guagliardo NA, Yao J, Hu C, Schertz EM, Tyson DA, Carey RM, Bayliss DA, Barrett PQ. Task-3 channel deletion in mice recapitulates low-renin essential hypertension. Hypertension. 2012;59:999–1005. doi: 10.1161/HYPERTENSIONAHA.111.189662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eqtls as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heitzmann D, Derand R, Jungbauer S, et al. Invalidation of task1 potassium channels disrupts adrenal gland zonation and mineralocorticoid homeostasis. EMBO J. 2008;27:179–187. doi: 10.1038/sj.emboj.7601934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaisser F, Farman N. Emerging roles of the mineralocorticoid receptor in pathology: Toward new paradigms in clinical pharmacology. Pharmacol Rev. 2016;68:49–75. doi: 10.1124/pr.115.011106. [DOI] [PubMed] [Google Scholar]
- 19.Funder JW, Carey RM, Mantero F, Murad MH, Reincke M, Shibata H, Stowasser M, Young WF., Jr. The management of primary aldosteronism: Case detection, diagnosis, and treatment: An endocrine society clinical practice guideline. J Clin Endocrinol Metab. 2016:jc20154061. doi: 10.1210/jc.2015-4061. [DOI] [PubMed] [Google Scholar]
- 20.Vasan RS, Evans JC, Larson MG, Wilson PW, Meigs JB, Rifai N, Benjamin EJ, Levy D. Serum aldosterone and the incidence of hypertension in nonhypertensive persons. N Engl J Med. 2004;351:33–41. doi: 10.1056/NEJMoa033263. [DOI] [PubMed] [Google Scholar]
- 21.Ganesh SK, Zakai NA, van Rooij FJ, et al. Multiple loci influence erythrocyte phenotypes in the charge consortium. Nat Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang W, Yu W. Power estimation and sample size determination for replication studies of genome-wide association studies. BMC Genomics. 2016;17(Suppl 1):3. doi: 10.1186/s12864-015-2296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung J, Barrett PQ, Eckert GJ, Edenberg HJ, Xuei X, Tu W, Pratt JH. Variations in the potassium channel genes kcnk3 and kcnk9 in relation to blood pressure and aldosterone production: An exploratory study. J Clin Endocrinol Metab. 2012;97:E2160–2167. doi: 10.1210/jc.2012-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luft FC, Grim CE, Fineberg N, Weinberger MC. Effects of volume expansion and contraction in normotensive whites, blacks, and subjects of different ages. Circulation. 1979;59:643–650. doi: 10.1161/01.cir.59.4.643. [DOI] [PubMed] [Google Scholar]
- 25.Kotchen TA, Guthrie GP, Jr., Cottrill CM, McKean HE, Kotchen JM. Low renin-aldosterone in “prehypertensive” young adults. J Clin Endocrinol Metab. 1982;54:808–814. doi: 10.1210/jcem-54-4-808. [DOI] [PubMed] [Google Scholar]
- 26.Bandulik S, Tauber P, Lalli E, Barhanin J, Warth R. Two-pore domain potassium channels in the adrenal cortex. Pflugers Arch. 2015;467:1027–1042. doi: 10.1007/s00424-014-1628-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi M, Scholl UI, Yue P, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331:768–772. doi: 10.1126/science.1198785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholl UI, Stolting G, Nelson-Williams C, et al. Recurrent gain of function mutation in calcium channel cacna1h causes early-onset hypertension with primary aldosteronism. Elife. 2015;4:e06315. doi: 10.7554/eLife.06315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholl UI, Goh G, Stolting G, et al. Somatic and germline cacna1d calcium channel mutations in aldosterone-producing adenomas and primary aldosteronism. Nat Genet. 2013;45:1050–1054. doi: 10.1038/ng.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scholl UI, Healy JM, Thiel A, et al. Novel somatic mutations in primary hyperaldosteronism are related to the clinical, radiological and pathological phenotype. Clin Endocrinol (Oxf) 2015;83:779–789. doi: 10.1111/cen.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.