Summary
Pax7 is a nodal transcription factor that is essential for regulating the maintenance, expansion, and myogenic identity of satellite cells during both neonatal and adult myogenesis. Deletion of Pax7 results in loss of satellite cells and impaired muscle regeneration. Here we show that ectopic expression of the constitutively active intracellular domain of Notch1 (NICD1) rescues the loss of Pax7-deficient satellite cells and restores their proliferative potential. Strikingly NICD1-expressing satellite cells do not undergo myogenic differentiation and instead acquire a brown adipogenic fate both in vivo and in vitro. NICD-expressing Pax7-/- satellite cells fail to upregulate MyoD and instead express the brown adipogenic marker PRDM16. Overall these results show that Notch1 activation compensates for the loss of Pax7 in the quiescent state and acts as a molecular switch to promote brown adipogenesis in adult skeletal muscle.
Keywords: Pax7, Notch1, NICD1, satellite cells, myogenic, brown adipogenic
Introduction
Muscle regeneration is a process that involves the repair of damaged myofibers or the formation of new myofibers upon injury. The successful outcome of the regenerative process relies on the ability of satellite cells to self-renew as well as give rise to transient amplifying progenitors that are capable of differentiation (Collins et al., 2005; Kuang et al., 2007; Montarras et al., 2005; Sacco et al., 2008; Yin et al., 2013b). The central role of satellite cells in promoting and leading muscle regeneration is well documented by several genetic ablation studies (Lepper et al., 2011; McCarthy, 2012; Relaix and Zammit, 2012; Sambasivan et al., 2011).
Initially identified by their location underneath the basal lamina of muscle fibers (Mauro, 1961), satellite cells are also defined by the expression of the paired box transcription factor Pax7 (Seale et al., 2000). Pax7 is essential for satellite cell maintenance and function during the neonatal and post-natal period as shown by the absence of satellite cells and poor regenerative response of germ-line Pax7-deficient mice (Kuang et al., 2006; Relaix et al., 2006; Seale et al., 2000). Similarly, inactivation of Pax7 via tamoxifen-inducible Cre recombination of the Pax7 locus at all stages of adulthood results in a pronounced regeneration deficit and dramatic loss of satellite cells (Gunther et al., 2013; von Maltzahn et al., 2013).
The Notch pathway is intimately related to and required for the maintenance of satellite cells and it is down regulated during terminal differentiation (Bjornson et al., 2012; Brack et al., 2008; Buas et al., 2010; Conboy and Rando, 2002; Kuroda et al., 1999; Mourikis et al., 2012a; Mourikis et al., 2012b; Pisconti et al., 2010; Vasyutina et al., 2007). Signaling is activated by the physical interaction at the cell membrane between a Delta or Jagged ligands and one of the four Notch receptors. This in turn leads to the release of the Notch intracellular domain (NICD), which translocates into the nucleus where it binds to the transcription factor Rbp-j. The binding determines the release of transcriptional repressors and recruitment of co-activators of gene transcription. Canonical Notch target genes include the family of transcription factors Hes (1/5) and Hey (1/2) (Bray, 2006; Castel et al., 2013; Kopan and Ilagan, 2009).
Interestingly, deletion of Rbp-j during embryonic development results in loss of satellite cells and formation of small muscle fibers due to precocious terminal differentiation of Rbp-j-/- satellite cells (Vasyutina et al., 2007). In adult muscle, loss of Rbp-j leads to early satellite cell exit from quiescence and terminal differentiation, which closely resembles the Pax7-/- phenotype (Bjornson et al., 2012; Mourikis et al., 2012b). Importantly, Notch1 is expressed by satellite cells and is required for their proliferation (Conboy and Rando, 2002). More recently it was reported that over expression of the Notch1 intracellular domain (NICD1) promotes satellite cell self-renewal (Wen et al., 2012). These studies support the notion that the Notch pathway is an important regulator of satellite cell function and led us to investigate the effect of Notch signaling in Pax7-deficient satellite cells.
Results
NICD1 Rescues the Loss of Pax7-Deficient Satellite Cells
Previous studies have shown that germ line and conditional deletion of Pax7 results in satellite cell loss and impaired proliferation due in part to precocious differentiation. (Kuang et al., 2006; von Maltzahn et al., 2013). Gene expression and extensive in vivo studies imply that active Notch signaling is important for the maintenance of uncommitted satellite cells (Bentzinger et al., 2013; Fukada et al., 2007; Price et al., 2014). However, the extent to which Notch is essential for satellite cell function is currently unknown. Here, we over expressed a constitutively activated form of Notch1 (NICD1) in Pax7-deficient satellite cells to assess the role of Notch in satellite cell function.
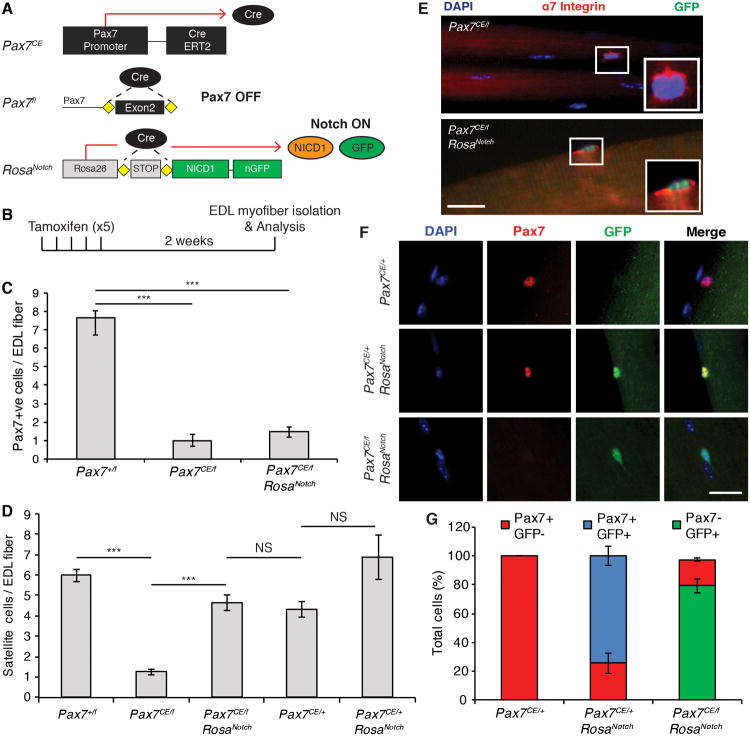
Cre-recombinase dependent conditional deletion of Pax7 in adult satellite cells was achieved by crossing Pax7CE/+ with Pax7f/f mice (Figure 1A and Figure S1) (Lepper et al., 2009). To conditionally activate Notch signaling in vivo, Pax7CE/+ or Pax7CE/f mice were crossed with RosaNotch mice in which the intracellular domain of Notch1 (NICD1) is driven from the Rosa26 locus (Murtaugh et al., 2003). Thus, in Pax7CE/f:RosaNotch mice, tamoxifen-induced CreER recombinase from the Pax7 locus results in the simultaneous inactivation of the Pax7 gene and the constitutive activation of NICD1 (Figure 1A). Expression of nuclear Green Fluorescent Protein (GFP) allowed us to distinguish satellite cells that have activated NICD1 (GFP+) from those that did not (GFP-). Efficient deletion of Pax7 expression was observed two weeks after the last tamoxifen injection (Figure 1B) and by enumerating the number of Pax7-expressing cells on isolated single EDL myofibers (Figure 1C).
Figure 1. NICD1 Rescues the Loss of Pax7CE/f Satellite Cells.
(A) Schematic describing the alleles used and the genetic approach used to activate NICD in Pax7CE satellite cells. CreERT2 driven from the Pax7 promoter simultaneously promotes the excision of exon 2 of Pax7 and the expression of the Notch intracellular domain (NICD1) from the Rosa 26 locus. NICD cells can be traced by the expression of iRES-GFP. See also Figure S1.
(B) Schematic of the tamoxifen regimen used in this study. 6-7 week old mice were given tamoxifen via IP injection for 5 consecutive days. Satellite cells were analyzed 2 weeks after the last tamoxifen injection.
(C) Graph shows the number of Pax7+ satellite cells per EDL fiber analyzed immediately after isolation. Data are shown as mean ± SEM. n=3; >50 fibers per genotype were counted. ***p≤0.001.
(D) Quantification of the number of satellite cells per EDL fiber. n=6; >50 fibers per genotype were counted. ***p≤0.001.
(E) Immunofluorescence staining of satellite cells on EDL myofibers stained for α7-Integrin (red) and GFP (green). Nuclei were counterstained with DAPI (blue). GFP is detected only in NICD+ satellite cells. Insets show cropped image of single satellite cells. Scale bar represents 100μm.
(F) Representative immunofluorescence staining showing lack of GFP expression in PaxCE/+:Rosa+/+ satellite cells (upper panel). Pax7 and GFP were detected in Pax7CE/+:RosaNotch satellite cells (middle panel). Pax7CE/f:RosaNotch satellite cells expressed GFP but not Pax7 (lower panel). Pax7 is shown in red; GFP is shown in green; and nuclei were counterstained with DAPI (blue). Scale bar represents 20μm.
(G) Graph shows the number of Pax7+ and GFP+ cells per EDL fiber isolated from mice of the indicated genotype. n=3; > 50 fibers per genotype were counted.
Myofibers isolated from EDL muscle from Pax7CE/f mice following Pax7 deletion exhibited a significant decrease in satellite cells as measured by counting the number of α7 integrin-expressing cells per myofiber relative to control Pax7+/f mice (1.26 ± 0.13 versus 5.98 ± 0.32 respectively) (Figure 1D). Myofibers isolated from heterozyogous Pax7+/- mice (Pax7CE/+), which express one functional Pax7 allele, also exhibit reduced numbers of satellite cells compared to Pax7+/f mice with 2 functional Pax7 alleles (4.31 ± 0.37 versus 5.98 ± 0.32 respectively) (Figure 1D).
Remarkably, the number of satellite cells on myofibers isolated from Pax7CE/f:RosaNotch mice was increased by 3.7-fold relative to Pax7CE/f mice (4.64 ± 0.37 versus 1.26 ± 0.13 respectively), and not significantly different from Pax7CE/+ control mice (Figure 1D). The increase in satellite cell number observed in Pax7CE/f:RosaNotch mice was not attributed to incomplete Pax7 excision as evidenced by the similar low level of Pax7-expressing cells on myofibers isolated from Pax7CE/f and Pax7CE/f:RosaNotch mice (Figure 1C). GFP expression was detected only in Pax7CE/f:RosaNotch satellite cells and not in Pax7CE/f cells (Figure 1E-F). Importantly, the expression of GFP and Pax7 was mutually exclusive in Pax7CE/f:RosaNotch mice clearly excluding the possibility that the rescue effect was dependent on residual Pax7 expression (Figure 1F-G). By contrast, the number of satellite cells on myofibers isolated from Pax7CE/+:RosaNotch mice compared to Pax7CE/+ littermate controls was only increased by about 1.6-fold (6.86 ± 1.07 versus 4.31 ± 0.37 respectively) (Figure 1D). Therefore, NICD1 expression prevents the loss of satellite cells that occurs following Pax7-deletion.
NICD1 Rescues the Proliferation of Pax7-Deficient Satellite Cells
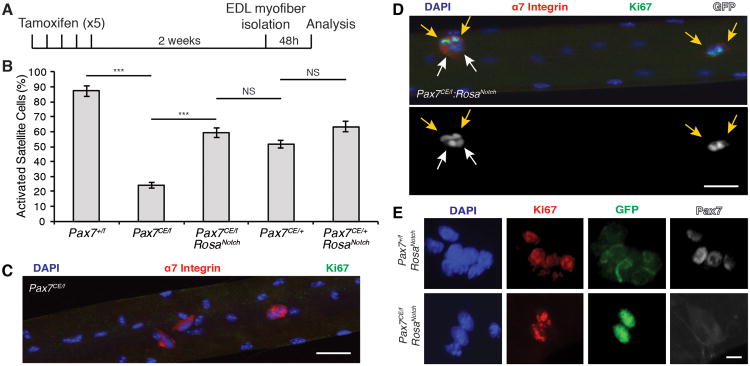
When cultured in high serum conditions, satellite cells exit quiescence, rapidly enter the cell cycle and divide. After 48h in culture (Figure 2A) most satellite cells on Pax7+/f myofibers expressed the proliferation marker Ki67 (87.33% ± 3.49) (Figure 2B). By contrast, only a minor fraction (23.9% ± 1.7) of Pax7CE/f satellite cells expressed Ki67 (Figure 2B-C).
Figure 2. NICD1 Rescues the Proliferation Deficit of Pax7CE/f Satellite Cells.
(A) Schematic detailing experimental procedures performed. EDL myofibers were isolated from mice 2 weeks after the last tamoxifen injection. Single fibers were cultured cultured for 48h in growth medium (GM) to allow satellite cell proliferation.
(B) Quantification of the number of activated satellite cells (α7-Integrin/Ki67/GFP+ cells) on EDL fibers. Data are shown as mean ± SEM. n=3 mice; >50 fibers per genotype were counted. ***p≤0.001.
(C) Immunofluorescence staining of Pax7CE/f satellite cells on EDL fibers. Note that most satellite cells are Ki67-. α7-Integrin is shown in red, Ki67 is shown in green, and nuclei were counterstained with DAPI (blue). Scale bar represents 100μm.
(D) Immunofluorescence staining of tamoxifen treated Pax7CE/f:RosaNotch satellite cells on EDL fibers. Note that NICD-GFP+ cells are Ki67+ (indicated with yellow arrows). White arrows indicate α7-Integrin+/NICD-GFP+ cells negative for Ki67. α7-Integrin is shown in red, Ki67 is shown in green, GFP is shown in grey, and nuclei were counterstained with DAPI (blue). Scale bar represents 100μm.
(E) Representative images of satellite cells on EDL fibers. NICD1-GFP+ cells from tamoxifen treated Pax7+/f:RosaNotch satellite cells are Pax7+ and GFP- (upper panel). Pax7CE/f:RosaNotch are KI67+ and Pax7− (lower panel). Ki67 is shown in red, GFP is shown in green, Pax7 is shown in grey, and nuclei were counterstained with DAPI (blue). Scale bar represents 10μm. See also Figure S2.
Strikingly, the number of proliferating satellite cells was significantly increased in myofibers isolated from tamoxifen-treated Pax7CE/f:RosaNotch mice compared to Pax7CE/f mice (59.21% ± 2.98 versus 23.9% ± 1.7 respectively) (Figure 2B). Immunofluorescence staining indicated that Pax7-deficient GFP-expressing cells on myofibers from Pax7CE/f:RosaNotch mice expressed both α7 Integrin and Ki67 (Figure 2D). Notably GFP+/Ki67+ cells were clearly Pax7 negative (Figure 2E). However, the presence of GFP+/Ki67- cells on myofibers isolated from Pax7CE/f:RosaNotch muscle suggested that some NICD1 expressing cells were not in the cell cycle (Figure 2D). No significant change in the proportion of Ki67-expressing satellite cells from Pax7CE/+:RosaNotch mice was observed compared to Pax7CE/+ littermate controls (Figure 2B and Figure S2). Together, our data indicates that ectopic expression of NICD1 is sufficient to rescue the proliferative disadvantage of Pax7-deficient satellite cells.
NICD1 inhibits MyoD expression in Pax7 null cells
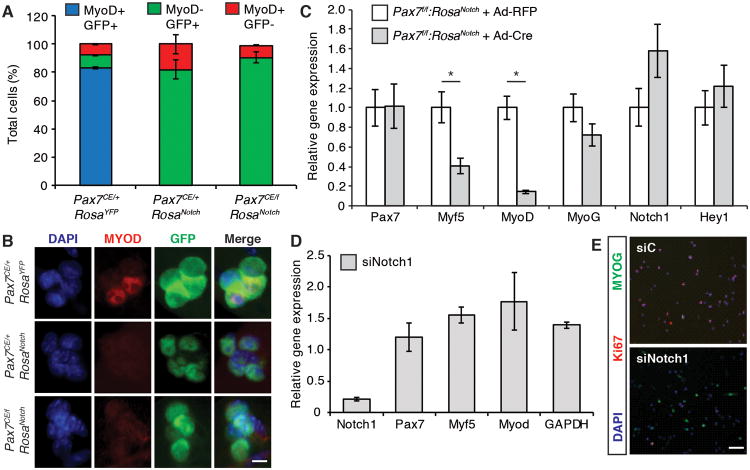
Notch signaling is a strong inhibitor of terminal myogenic differentiation (Buas et al., 2010; Conboy and Rando, 2002; Kuroda et al., 1999). Therefore, to investigate the myogenic status of NICD1-expressing satellite cells, EDL myofibers were isolated from tamoxifen-treated Pax7CE/+:RosaYFP, Pax7CE/f:RosaNotch, and Pax7CE/+:RosaNotch mice. Myofibers were cultured for 48h in standard myofiber culture media (Figure 2A) and MyoD expression assessed by immunostaining.
After 48 hours of culture, MyoD was detected in the majority of satellite cells on myofibers isolated from Pax7CE/+:RosaYFP EDL muscles (Figure 3A-B). Strikingly GFP expression was mutually exclusive with MyoD expression in Pax7CE/+:RosaNotch and Pax7CE/f:RosaNotch satellite cells (Figure 3A-B). These results suggest that ectopic NICD1 expression completely abrogates expression of MyoD in both the presence and absence of Pax7.
Figure 3. NICD1 Inhibits the Myogenic Program of Satellite Cells.
(A) Quantification of the number of MyoD+ and MyoD- satellite cells cultured on EDL myofibers cultured for 48h in standard myofiber media. Cell number was normalized to total GFP+ cells. Data are shown as mean ± SEM. n=3; >50 fibers were counted per genotype.
(B) MyoD expression in satellite cells from Pax7CE/+:RosaYFP (upper panel), Pax7CE/+:RosaNotch (middle panel) and Pax7CE/f:RosaNotch (lower panel) EDL fibers. MyoD is shown in red, GFP is shown in green, and nuclei were counterstained with DAPI (blue). Scale bar represents 10μm.
(C) qRT-PCR of Pax7, Myf5, MyoD, Myogenin (Myog), Notch1 and Hey1 from Pax7f/f RosaNotch primary myoblasts infected with an adenovirus expressing Cre recombinase (Ad-Cre) or a control virus expressing the Red Fluorescent Protein (Ad-RFP). Cell lysates were collected 48h post infection. Graph shows mean mRNA expression levels (± SEM) normalized to Gapdh. n=3. *p≤0.05. See also Figure S3.
(D) qRT-PCR of primary myoblasts transfected with a cocktail of siRNAs against Notch1 (siNotch1) or with a scrambled oligonucleotide control (siC). Graph shows mean relative mRNA expression level ± SEM normalized to Gapdh. n=3. See also Figure S4.
(E) Primary myoblasts transfected with siRNA against Notch1 (siNotch1) or with control siRNA (siC) were fixed 48h post transfection and stained for Myogenin (green) and Ki67 (red). Nuclei were counterstained with DAPI (blue). Scale bar represents 200μm.
Consistent with these results, over-expression of NICD1 in primary myoblasts resulted in down regulation of all three myogenic regulatory factors, MyoD, Myf5 and Myogenin (Figure 3C). Conversely, inhibition of Notch1 in primary myoblasts results in up-regulation of MyoD (Figure 3D) and Myogenin (Figure 3E). Of note MyoD and Myogenin were observed to be up regulated in Pax7-deficient satellite cells (germ line mutation Pax7LacZ/LacZ) (Figure S3). Moreover, RT-qPCR analysis revealed that multiple components of the Notch signaling pathway including Notch1, Notch2, Rbp-j, and Hey1 were down regulated in Pax7LacZ/LacZ cells (Figure S4A) and in siPax7 primary myoblasts relative to control (Figure S4B).
Together these results suggest that inactivation of Pax7 in adult satellite cells results in an increase in myogenic differentiation genes and a corresponding decrease in genes of the Notch pathway. In contrast, ectopic expression of NICD1 in Pax7-deficient cells abrogates expression of the MyoD-family of factors.
Notch Activation Promotes Brown Adipogenesis
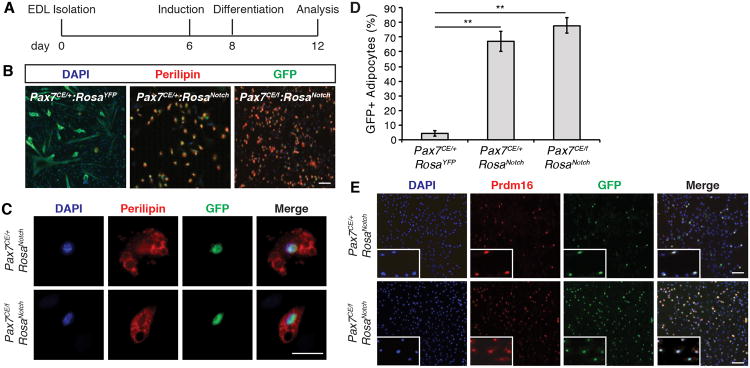
We previously demonstrated that satellite cells are bi-potential stem cells that have the ability to undergo both myogenic and brown adipogenic differentiation (Yin et al., 2013a). To investigate whether Notch signaling influences brown adipogenic differentiation of satellite cells, myofibers were cultured in proadipogenic media between 12 and 15 days (Figure 4A). Importantly, these culture conditions are permissive of brown adipogenesis but do not inhibit myogenic differentiation, as shown by the presence of fully differentiated myotubes in control Pax7CE/+:RosaYFP cultures (Figure 4B).
Figure 4. NICD1 Promotes Brown Adipogenesis.
(A) Timeline of the adipogenic differentiation protocol. See also Supplementary Information.
(B) Immunofluorescence of satellite cell-derived adipocytes from Pax7CE/+:RosaYFP (left); Pax7CE/+:RosaNotch (center) and Pax7CE/f:RosaNotch (right) fiber derived cultures respectively. Perilipin is shown in red, GFP is shown in green, and nuclei were counterstained with DAPI (blue). Scale bar represents 200μm.
(C) Cropped images of NICD-GFP+ cells from Pax7CE/+:RosaNotch and Pax7CE/f:RosaNotch fibers cultured in adipogenic medium. Perilipin is shown in red, GFP is shown in green, and nuclei were counterstained with DAPI (blue). Scale bar represents 100μm.
(D) Quantification of the number of GFP+/Perilipin+ cells in Pax7CE/+:RosaYFP, Pax7CE/+:RosaNotch, and Pax7CE/f:RosaNotch fiber derived cultures. Graph shows mean ± SEM; n=3; > 200 cells counted per genotype. **p≤0.01.
(E) Immunofluorescence images of NICD/GFP+ PRDM16+ cells after 15 days in culture under adipogenic conditions from Pax7CE/+:RosaNotch and Pax7CE/f:RosaNotch mice. Prdm16 is shown in red, GFP is shown in green, and nuclei were stained with DAPI (blue). Scale bar represents 200μm. Insets show cropped image of single GFP+/Prdm16+ cells.
Remarkably, expression of NICD1 in both Pax7CE/f and Pax7CE/+ cells resulted in the induction of very high numbers of adipocytes as shown by the co-expression of GFP and the adipose marker perilipin (Figure 4B-C). Consistent with this observation, quantification of the percentage of perilipin+ cells expressing GFP revealed that a significant number of adipocytes are derived from NICD1 (GFP+)-expressing cells (Figure 4D). Importantly all GFP+/perilipin+ cells express the brown adipogenic marker Prdm16 (Figure 4E).
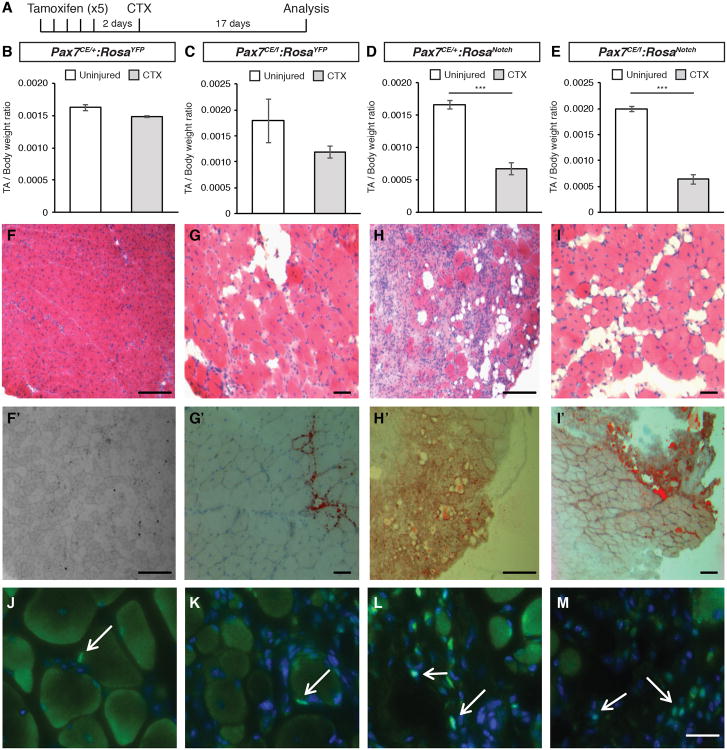
To assess the effect of NICD1 expression in satellite cells in vivo, the tibialis anterior (TA) muscles of tamoxifen-treated Pax7CE/f:RosaNotch and Pax7CE/+:RosaNotch mice were injected with cardiotoxin to induce acute injury (Figure 5A). Control (Pax7CE/+:RosaYFP) TA muscles underwent normal regeneration as evidenced by normal muscle weight (Figure 5B) and homogenous appearance of centrally nucleated myofibers, a hallmark of muscle regeneration (Figure 5F-F′). By contrast, Pax7-deficient (Pax7CE/f:RosaYFP) TA muscles following cardiotoxin-induced injury showed extensive fibrosis and widespread fatty infiltration (Figure 5G-G′). Notably, a significant loss of muscle mass (Figure 5D-E) and increased fat infiltration (Figure 5H-H′ and I-I′) was observed in both Pax7 heterozygous mutant (Pax7CE/+) and Pax7 homozygous mutant (Pax7CE/f) animals expressing NICD1. Examination of the uninjured contralateral muscles did not reveal any significant morphological differences across the genotypes (not shown).
Figure 5. NICD1 Promotes in vivo Brown Fat Formation.
(A) Schematic of the regeneration experiment. Mice were injected with tamoxifen daily for 5 days. Muscle injury was performed via cardiotoxin injection in the tibialis anterior (TA) muscle 2 days after the last tamoxifen injection. Mice were harvested and TA muscles were dissected and analyzed 17 days after injury.
(B-E) Ratio of TA muscle mass normalized to total body weight. Grey bars represent injured TA muscle mass, white bars represent contralateral (uninjured) TA. Data are shown as mean ± SEM. n≥4. ***p≤0.001.
(F-I) Representative Haematoxylin and Eosin (HE) staining of TA muscle sections. Scale bar repesents 200μm.
(F′-I′) Representative Oil red O staining of TA muscle sections. Scale bar represents 200μm.
(J-M) Detection of GFP expression in prefixed muscle sections from Pax7CE/+:RosaYFP (J); Pax7CE/f:RosaYFP (K); Pax7CE/+:RosaNotch (L) and Pax7CE/f:RosaNotch (M). GFP is shown in green. Nuclei were counterstained with DAPI (blue). Scale bar represents 100μm. See also Figure S5.
To address the contribution of NICD1 cells in the observed increase in muscle adiposity, regenerating TA muscles were prefixed and stained with GFP (Figure 5J-M). While Pax7CE/+ and Pax7CE/f YFP+ satellite cells were found in their anatomical position underneath the basal lamina of muscle fibers (Figure 5J-K), NICD1/GFP+ cells were clearly localized in the interstitial space of muscles in both Pax7CE/+:RosaNotch and Pax7CE/f:RosaNotch mice (Figure 5L-M, white arrows). Importantly NICD1 expressing muscles also showed an increase in the number of UCP-1 expressing interstitial brown adipocytes compared to control (Figure S5 C-C′; D-D′).
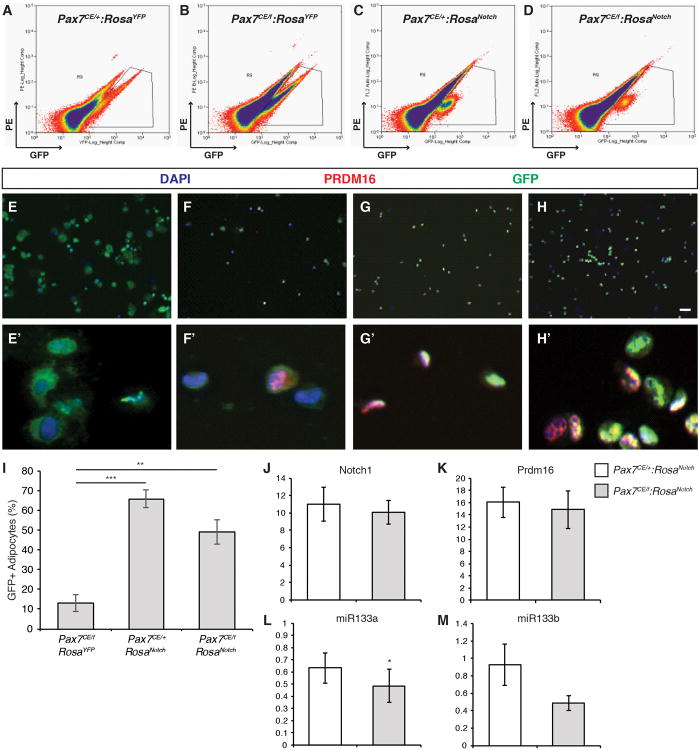
We next investigated the fate of NICD1/GFP+ over-expressing cells in vivo after regeneration by determining the proportion of Pax7-CreER marked cells that had activated the definitive brown adipose marker Prdm16. GFP-expressing cells were isolated 17 days after cardiotoxin injury by Fluorescence Activated Cell Sorting (FACS) (Figure 6A-D) (Pasut et al., 2012). Satellite cells isolated from Pax7CE/+:RosaYFP muscles did not express detectable levels of Prdm16 (Figure 6E-E′). A significant proportion (13.3% ± 4.4) of Pax7CE/f cells expressed Prdm16 (Figure 6I), suggesting that the regenerative deficit and loss of muscle mass observed in Pax7 deficient mice is due in part to brown adipogenic specification of presumptive satellite cells (Figure 6F-F′). Strikingly, NICD muscles exhibited a dramatic increase in the numbers of GFP+/Prdm16+ cells (Figure 6G-G′; H-H′; quantified in Figure 6I). In addition, qRT-PCR analysis of GFP+ sorted cells confirmed that Prdm16 mRNA was upregulated in satellite cells that express NICD1 (Figure 6J-K). Ectopic expression of NICD1 also resulted in down-regulation of miR-133a, which we previously demonstrated directly inhibits Prdm16 expression (Figure 6L-M) (Yin et al., 2013a).
Figure 6. NICD1 Cells Express Markers of Brown Adipocytes in vivo.
(A-D) FACS profiles of prospectively isolated satellite cells from injured muscles. Endogenous GFP expression was used to gate for satellite cells.
(E-H) A portion of FACS sorted satellite cells were cytospun and fixed immediately after sorting. GFP is shown in green, Prdm16 is shown in red, and nuclei were counterstained with DAPI (blue). Scale bar represents 100μm. E′-H′: cropped images of GFP+/Prdm16+ cells.
(I) Quantification of the number of GFP+/PRDM16+ cells from Pax7CE/f:RosaYFP, Pax7CE/+:RosaNotch, and Pax7CE/f:RosaNotch injured muscles expressed as the percentage of GFP+ cells that express PRDM16. Data are shown as mean ± SEM; >150 cells counted per genotype. **p≤0.01 and ***p≤0.001.
(J-M) q-PCR analysis of total Notch1 (J), Prmd16 (K), miR-133a (L) and miR-133b (M) in prospectively isolated NICD/GFP+ cells from Pax7CE/+:RosaNotch (white bars) and Pax7CE/f:RosaNotch (grey bars) injured muscles. Graphs show mean mRNA expression level ± SEM. Gene expression was normalized to ppia, and expressed relative to control (Pax7CE/+:RosaYFP). n≥3. *p≤0.05. See also Figure S6.
Over-expression of NICD1 results in markedly reduced MyoD expression (Figure 3A-B). Interestingly, chromatin immunoprecipitation (ChIP) sequencing experiments on cultured myoblasts identified putative binding sites for Pax7 (enhancer 2) and MyoD (enhancer 1 and 3) upstream of miR-133a (Figure S6A), which were confirmed to be functionally active by luciferase assays (Figure S6B). Altogether, these data support the notion that both Pax7 and MyoD enforce satellite cell myogenic identity and that this is achieved in part by maintaining high levels of miR-133a expression. In conclusion, our experiments suggest that NICD1 acts as a molecular switch to promote brown adipogenesis of satellite cells both in vitro and in vivo by down-regulating MyoD and miR-133a expression.
Discussion
Pax7 is absolutely required for normal satellite cell function in both neonatal and adult skeletal muscle (Gunther et al., 2013; Kuang et al., 2006; Relaix et al., 2006; Seale et al., 2000; von Maltzahn et al., 2013). By using a genetic approach, we found that over expression of the intracellular domain of Notch1 (NICD1) is sufficient to rescue the loss and proliferation of Pax7-deficient satellite cells. Moreover, we discovered that expression of NICD1 promoted a molecular lineage switch toward brown adipogenesis.
Notch signaling preserves satellite cell number during embryonic development and sustains their quiescent state in adult muscle (Bjornson et al., 2012; Brohl et al., 2012; Jiang et al., 2014; Mourikis et al., 2012b; Vasyutina et al., 2007). Quiescent satellite cells express high levels of Notch effector genes, which supports the notion that Notch signaling is active (Fukada et al., 2007). A potential mechanism has been proposed in which Notch signalling regulates genes involved in adhesion and cell-cell interaction (Vasyutina et al., 2007). The central role of Notch signaling in safeguarding stem cell number and quiescence has also been reported in other systems such as the adult brain suggesting that this is a general mechanism by which stem cell homeostasis is maintained during the life span of an organism (Androutsellis-Theotokis et al., 2006; Imayoshi et al., 2010).
Notch1 has been suggested to mediate satellite cell activation and expansion (Conboy and Rando, 2002; Sun et al., 2007). More recently, in vivo studies have shown that NICD myoblasts exhibit a slower growth rate compared to control cells (Wen et al., 2012) and Rbpj-/- cells directly differentiate without entering S-phase (Bjornson et al., 2012; Mourikis et al., 2012a). Notch signaling is initiated by ligand-receptor binding at the cell membrane. However, non-canonical ligand-independent activation of Notch signaling mediated by the ubiquitin ligase Deltex has recently been described (Hori et al., 2012). Importantly ligand-independent Notch signaling activation has been shown to act in a context dependent manner during blood cell development (Mukherjee et al., 2011). Our experiments indicate that NICD1 over-expression is sufficient to rescue the proliferative disadvantage of Pax7-deficient satellite cells (Figure 2B). Whether this rescue occurs via activation of canonical or non-canoical Notch signaling effectors remains to be determined.
Analysis of cell cycle kinetics indicates that NICD1 over-expressing satellite cells divide at a slower rate than wild type cells (Wen et al., 2012). We also found that Pax7CE/+:RosaNotch cells had a lower proportion of Ki67-expressing cells compared to Pax7+/f (51.72% ± 4.41 versus 87.33% ± 3.49 respectively) (Figure 2B). Activation of NICD1 may selectively favor the accumulation of slow cycling cells or result in an initially longer G1 phase. In this regard, an experiment in which cell divisions were traced by the use of an H2B-GFP reporter revealed that satellite cells are a heterogeneous population of slow and fast cycling cells and while slow cycling cells are responsible for self-renewal, fast cycling cells readily respond to injury and contribute to muscle repair (Chakkalakal et al., 2012). Interestingly, slow cycling cells can divide asymmetrically and generate both fast and slow cycling cells. It is tempting to speculate that this choice is regulated by Notch and that the level of Notch dictates the proliferative behavior of the daughter cells.
During embryonic development, brown adipocytes arise from Myf5-expressing myogenic cells (Seale et al., 2008). Moreover, lineage tracing has also shown that Pax7-expressing cells give rise to both brown adipocytes, and skeletal muscle (Lepper and Fan, 2010). In the adult, satellite stem cells are bipotential and are capable of differentiation into myogenic or brown adipogenic lineages. This lineage switch is regulated by microRNA-133 targeting the 3′UTR of Prdm16 (Yin et al., 2013a). Surprisingly, we found that NICD1 over-expression in Pax7-deficient satellite cells markedly stimulates their brown adipogenic determination (Figure 4D-E). Interestingly, Myf5-Cre activated NICD1 cells were found to express Pparᵧ (a marker of brown adipocytes) in the developing embryo (Mourikis et al., 2012a).
Our experiments suggest that high levels of NICD1 in adult satellite cells promote the up-regulation of Prdm16 and stimulate brown adipogenic specification by down-regulating miR-133a (Figure 6L-M) and MyoD expression (Figure 3C). Importantly, we now show that both Pax7 and MyoD can functionally bind to enhancer elements upstream of miR-133a and miR-133b, thereby providing mechanistic evidence of how miR-133 expression is regulated in satellite cells (Figure S6). As shown in Figure 6E-F and quantified in Figure 6I, the presence of brown adipocytes is more readily detectable in Pax7CE/f cells compared to Pax7CE/+ cells. The similar increase in BAT conversion observed in Pax7CE/f and Pax7CE/+ mice upon NICD over expression also suggests that MyoD inhibition is a critical and an important factor in promoting brown adipogenic lineage switching of satellite cells. In adult myogenesis, fibroadipogenic cells or other mesenchymal cells have been shown to contribute to ectopic fat infiltration within muscles (Joe et al., 2010; Uezumi et al., 2010). Our data show that satellite cells contribute at least in part to the formation of ectopic brown fat within regenerating muscles (Figure 4 and 6). However, not all interstitial cells were GFP+ and therefore we cannot exclude that over-expression of NICD1 may also have indirect effects on other cell types.
Gene expression studies of activated satellite cells from regenerating muscles show a transient decline in the expression of Notch pathway genes around 20h after injury. Notch activity is then up-regulated again with a peak observed around 4-5 days post injury (Mourikis and Tajbakhsh, 2014). NICD1 mice exhibit a profound regenerative deficiency characterized by an abnormal accumulation of mononucleated cells within the interstitial space that persists 15 days after injury (Figure 5). Over expression of NICD1 in the dystrophin/utrophin compound mutant mice correlates with an increased inflammation and impaired muscle regeneration (Mu et al., 2015). Thus an additional function of Notch signaling may be to regulate the initial recruitment of immune cells and that a transient down-regulation of the pathway allows for immune cell clearance. Notch activity is then up-regulated to allow for satellite cells expansion, differentiation and self-renewal.
In conclusion, our experiments suggest that Notch signaling partially compensates for the loss of Pax7. In normal muscle, high levels of Notch signaling are required to maintain the uncommitted state of satellite cells. Similarly, genetic over expression of NICD restores the proliferative potential of Pax7-deficient satellite cells. In addition, Notch signalling also plays a role in satellite cell fate as activation of Notch1 strongly promotes the lineage switch from myogenic towards brown adipogenic fate. These experiments provide important insights into the molecular circuits controlling lineage determination of adult satellite cells and the complex interplay of Notch signaling in satellite cell homeostasis and activation.
Experimental Procedures
Mice and Animal Care
Tamoxifen inducible Pax7 null mice (Pax7CE/f) were generated by crossing Pax7CE/+ mice with Pax7f/f mice (Lepper et al., 2009). To activate Notch in satellite cells, RosaNotch mice (Murtaugh et al., 2003) were crossed with Pax7CE/f mice. For satellite cell lineage tracing experiments, Pax7CE/+ and Pax7CE/f mice were crossed with Gt(Rosa)26Sortm9(CAG-YFP) mice (Jackson laboratories) to obtain Pax7CE/+:RosaYFP and Pax7CE/f:RosaYFP mice respectively. Tamoxifen (Sigma T5648) was pre-dissolved in corn oil at a concentration of 20mg/ml and injected intraperitoneally for 5 consecutive days. To induce muscle regeneration, 50μl of a cardiotoxin solution was administered intramuscularly to anesthetized animals. Cardiotoxin was prepared by dissolving Latoxan (Sigma) in physiological saline to a final concentration of 10 μM. Care of animals was in accordance with institutional guidelines as regulated by the Canadian Council of Animal Care (CCAC). Protocols were approved by Animal Research Ethics Board (AREB) at the University of Ottawa and are reviewed on an annual basis.
Satellite Cell Isolation and Culture
Satellite cells were isolated from hind limb muscles as previously described (Pasut et al., 2012). Briefly, muscles were dissected and minced to release cells by sequential incubation in collagenase-dispase. Antibody labeling was performed in PBS supplemented with 2% horse serum at 4°C. A complete list of antibodies can be found in Table S1 of the Supplementary Information. Hoechst 33342 (Sigma) was added at a final concentration of 1mg/ml. FACS was performed on a Moflo cytometer (Dako Cytomation) equipped with 5 lasers. The Summit v4.3 Suite was used for data acquisition and image processing. For gene expression analysis, RNA was isolated from freshly sorted satellite cells using the Arcturus® Picopure® RNA isolation kit as per manufacturer's instructions. Quantification of gene expression was performed using the REST 2009 software (Qiagen) or using the ∆∆Ct formula.
Myofiber Isolation and Analysis
Single EDL myofibers were isolated as previously described (Pasut et al., 2013). Briefly, single EDL muscles were dissected and digested at 37°C in 2% collagenase (Sigma). Single myofibers were either fixed right after the isolation or cultured in DMEM supplemented with Sodium Pyruvate (Invitrogen), 20% Fetal Bovine Serum (FBS, Wisent Inc), 1% Chicken Embryo Extract (CEE, New England Biolabs) and penicillin and streptomycin (P/S, Wisent Inc). For short-term culture, myofibers were cultured in suspension in horse serum-coated dishes. For long-term culture, myofibers were maintained in suspension for 24h and then transferred into Matrigel™ (BD Matrigel) coated dishes and cultured for up to 2 weeks. The medium was changed every two days. Adipogenic differentiation was induced as described in the Supplemental Experimental Procedures. For immunofluorescence, myofibers were fixed in 2% or 4% PFA (Sigma) followed by permeabilization in 0.05% Triton X100 (Sigma) in PBS and overnight blocking in 10% horse serum in PBS. Primary antibodies were diluted in blocking buffer and used as specified in Table S1 of the Supplementary Information. Respective secondary antibodies (Alexa, Invitrogen) were diluted in PBS and used at a 1:1000 dilution. DAPI 10 mg/ml (Sigma) was used to counter stain nuclei. Images were acquired using a Zeiss Axio Observer microscope or Zeiss Axioplan 2 microscope equipped with an AxioCam HR. Colors were added by using Photoshop Suite C5.
Cell Culture
Primary myoblasts were maintained in collagen-coated dishes and cultured in HAM's F10 (Invitrogen) supplemented with 2.5 ng/μl of bFGF (Cedarlane-Millipore), 20% fetal bovine serum (FBS, Wisent Inc), and penicillin and streptomycin (P/S, Wisent Inc). Virus infection was performed in HAM's F10 without antibiotics, supplemented with 10% fetal bovine serum (FBS, Wisent Inc). Adenoviruses containing either Cre (Ad-Cre) or Red Fluorescent Protein (Ad-RFP) were provided by Dr. Robin Parks. For gene expression analysis, cells were lysed in TRizol and RNA isolated using Nucleospin RNA Kit (Macherey Nachel) unless otherwise specified.
Histological Analysis
Tibialis anterior (TA) muscles were dissected and embedded into OCT-30% sucrose for sectioning. Primary antibodies were diluted in blocking solution (10% horse serum, 1% BSA in PBS) and used as specified in Table S1 of the Supplementary Information. Respective secondary antibodies (Alexa, Invitrogen) were diluted in PBS and used at a 1:1000 dilution. DAPI (Sigma) was used to counter stain nuclei.
Statistical Analysis
All quantitative data are expressed as mean +/- standard error of the mean (SEM), represented as error bars. Statistical analysis was performed on at least three biological replicates and significance determined by the Student t-test. Individual P-values are indicated in each figure legend.
Supplementary Material
Acknowledgments
Adenoviral vectors were a kind gift from Dr. Robin Parks, and the Prdm16 antibody was provided by Dr. Patrick Seale. We thank Jennifer Ritchie for mice breeding, colony maintenance and tamoxifen injections, and Nicolas Dumont for help with experiments. A.P. is supported by a CIHR-Training Program in Regenerative Medicine (TPRM) fellowship and holds an Excellence Scholarship from the University of Ottawa. These studies were carried out with support of grants to M.A.R. from the Canadian Institutes for Health Research (MOP-81288 and MOP-12080), the US National Institutes for Health (R01AR044031), the Stem Cell Network, and the Ontario Ministry of Economic Development and Innovation, and the Canada Research Chair Program. M.A.R. holds a Canada Research Chair in Molecular genetics.
Footnotes
Contributions of Authors: A.P. designed, conducted experiments, analyzed the data, and wrote the manuscript. N.C. conducted experiments, analyzed the data, and wrote the manuscript. U.G. and S.F. performed experiments shown in Figure 1C and G and Figure 2B. H.Y. performed and analyzed experiments shown in Figures 3C and S6. M.L. performed cardiotoxin injections and helped with analysis of muscle injury. M.A.R. designed experiments, analyzed the data, and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Wang YX, von Maltzahn J, Soleimani VD, Yin H, Rudnicki MA. Fibronectin regulates Wnt7a signaling and satellite cell expansion. Cell Stem Cell. 2013;12:75–87. doi: 10.1016/j.stem.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson CR, Cheung TH, Liu L, Tripathi PV, Steeper KM, Rando TA. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 2012;30:232–242. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Conboy IM, Conboy MJ, Shen J, Rando TA. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell. 2008;2:50–59. doi: 10.1016/j.stem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Brohl D, Vasyutina E, Czajkowski MT, Griger J, Rassek C, Rahn HP, Purfurst B, Wende H, Birchmeier C. Colonization of the satellite cell niche by skeletal muscle progenitor cells depends on Notch signals. Dev Cell. 2012;23:469–481. doi: 10.1016/j.devcel.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Buas MF, Kabak S, Kadesch T. The Notch effector Hey1 associates with myogenic target genes to repress myogenesis. J Biol Chem. 2010;285:1249–1258. doi: 10.1074/jbc.M109.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel D, Mourikis P, Bartels SJ, Brinkman AB, Tajbakhsh S, Stunnenberg HG. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013;27:1059–1071. doi: 10.1101/gad.211912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, Morgan JE. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- Fukada S, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Gunther S, Kim J, Kostin S, Lepper C, Fan CM, Braun T. Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell. 2013;13:590–601. doi: 10.1016/j.stem.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Regulation of ligand-independent Notch signal through intracellular trafficking. Commun Integr Biol. 2012;5:374–376. doi: 10.4161/cib.19995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Wen Y, Kuroda K, Hannon K, Rudnicki MA, Kuang S. Notch signaling deficiency underlies age-dependent depletion of satellite cells in muscular dystrophy. Dis Model Mech. 2014;7:997–1004. doi: 10.1242/dmm.015917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J Biol Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Fan CM. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis. 2010;48:424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy N. Cancer stem cells: Tracing clones. Nat Rev Cancer. 2012;12:579. doi: 10.1038/nrc3354. [DOI] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- Mourikis P, Gopalakrishnan S, Sambasivan R, Tajbakhsh S. Cell-autonomous Notch activity maintains the temporal specification potential of skeletal muscle stem cells. Development. 2012a;139:4536–4548. doi: 10.1242/dev.084756. [DOI] [PubMed] [Google Scholar]
- Mourikis P, Sambasivan R, Castel D, Rocheteau P, Bizzarro V, Tajbakhsh S. A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells. 2012b;30:243–252. doi: 10.1002/stem.775. [DOI] [PubMed] [Google Scholar]
- Mourikis P, Tajbakhsh S. Distinct contextual roles for Notch signalling in skeletal muscle stem cells. BMC Dev Biol. 2014;14:2. doi: 10.1186/1471-213X-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Tang Y, Lu A, Takayama K, Usas A, Wang B, Weiss K, Huard J. The role of Notch signaling in muscle progenitor cell depletion and the rapid onset of histopathology in muscular dystrophy. Hum Mol Genet. 2015;24:2923–2937. doi: 10.1093/hmg/ddv055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee T, Kim WS, Mandal L, Banerjee U. Interaction between Notch and Hif-alpha in development and survival of Drosophila blood cells. Science. 2011;332:1210–1213. doi: 10.1126/science.1199643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasut A, Jones AE, Rudnicki MA. Isolation and culture of individual myofibers and their satellite cells from adult skeletal muscle. J Vis Exp. 2013:e50074. doi: 10.3791/50074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasut A, Oleynik P, Rudnicki MA. Isolation of muscle stem cells by fluorescence activated cell sorting cytometry. Methods Mol Biol. 2012;798:53–64. doi: 10.1007/978-1-61779-343-1_3. [DOI] [PubMed] [Google Scholar]
- Pisconti A, Cornelison DD, Olguin HC, Antwine TL, Olwin BB. Syndecan-3 and Notch cooperate in regulating adult myogenesis. J Cell Biol. 2010;190:427–441. doi: 10.1083/jcb.201003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price FD, von Maltzahn J, Bentzinger CF, Dumont NA, Yin H, Chang NC, Wilson DH, Frenette J, Rudnicki MA. Inhibition of JAK-STAT signaling stimulates adult satellite cell function. Nat Med. 2014;20:1174–1181. doi: 10.1038/nm.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Zammit PS. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 2012;139:2845–2856. doi: 10.1242/dev.069088. [DOI] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R, Yao R, Kissenpfennig A, Van Wittenberghe L, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Sun H, Li L, Vercherat C, Gulbagci NT, Acharjee S, Li J, Chung TK, Thin TH, Taneja R. Stra13 regulates satellite cell activation by antagonizing Notch signaling. J Cell Biol. 2007;177:647–657. doi: 10.1083/jcb.200609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- Vasyutina E, Lenhard DC, Wende H, Erdmann B, Epstein JA, Birchmeier C. RBP-J (Rbpsuh) is essential to maintain muscle progenitor cells and to generate satellite cells. Proc Natl Acad Sci U S A. 2007;104:4443–4448. doi: 10.1073/pnas.0610647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Maltzahn J, Jones AE, Parks RJ, Rudnicki MA. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci U S A. 2013;110:16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Bi P, Liu W, Asakura A, Keller C, Kuang S. Constitutive Notch activation upregulates Pax7 and promotes the self-renewal of skeletal muscle satellite cells. Mol Cell Biol. 2012;32:2300–2311. doi: 10.1128/MCB.06753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van Ijcken W, Grosveld F, et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 2013a;17:210–224. doi: 10.1016/j.cmet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev. 2013b;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.