Abstract
Background
Genetic ancestry, sex and individual alleles have been associated with MS susceptibility.
Objective
To determine if established risk factors for disease onset are associated with relapse rate in pediatric MS.
Methods
Whole genome genotyping was performed for 181 MS or high-risk clinically isolated syndrome patients from two pediatric MS centers. Relapses and disease-modifying therapies were recorded as part of continued follow-up. Participants were characterized for 25-hydroxyvitamin D serum status. Ancestral estimates (STRUCTURE v2.3.1), HLA-DRB1*15 carrier status (direct sequencing), sex and a genetic risk score (GRS) of 110 non-HLA susceptibility SNPs were evaluated for association with relapse rate with Cox and negative binomial regression models.
Results
Over 622 patient-years, 408 relapses were captured. Girls had greater relapse rate than boys (IRR=1.40, 95% CI=1.04–1.87, p=0.026). Participants were genetically diverse; ~40%(N=75) had < 50% European ancestry. HLA-DRB1*15 status modified the association of vitamin D status (pixn=0.022) with relapse rate (per 10 ng/ml, in DRB1*15+ HR=0.72, 95% CI=0.58–0.88, p=0.002; in DRB1*15-HR=0.96, 95% CI=0.83–1.12, p=0.64). Neither European ancestry nor GRS was associated with relapse rate.
Conclusions
We demonstrate that HLA-DRB1*15 modifies the association of vitamin D status with relapse rate. Our findings emphasize the need to pursue disease-modifying effects of MS genes in the context of environmental factors.
Search terms: multiple sclerosis, genetics, vitamin D
Introduction
Multiple sclerosis (MS) is a demyelinating disease of the central nervous system triggered by genetic and environmental factors. Genetic ancestry has been linked to disease risk, but ancestry has also historically been tightly linked to geography with potential related environmental factors.1 To date the strongest known individual genetic risk factor for pediatric and adult MS is human leukocyte antigen (HLA)-DRB1*15:01, a class II allele encoded within the major histocompatibility complex on chromosome 6p21.2, 3 In individuals of African descent, HLA-DRB1*15:03, in addition to HLA-DRB1*15:01, is associated with MS risk.4 Over the last few years, many non-HLA single nucleotide polymorphisms (SNPs) have been associated with MS through genome wide association studies (GWAS).5, 6 Summary scores, such as the genetic risk score (GRS) and the MS genetic burden score, have been proposed to capture a single measure of overall genetic risk for MS.7, 8
While great progress has been made towards elucidating the genetic risk for MS, the role of genetic influences on the severity and course of MS is not yet clear. Consistent associations have not been observed between genetic risk variants and cross-sectional metrics of disability or MRI measures of brain atrophy or lesion load, but several SNPs have demonstrated association with MRI lesion distribution, clinical attack location and attack severity.9–13 Some genetic variants may also determine intrathecal antibody production.14 Most genetic studies of disease course have not included relapse rate as an outcome, despite this being a major disease measure in relapsing MS and the clinical indication for MS disease modifying therapies (DMTs). A recent study in adults suggests that a summary genetic burden score is associated with relapse rate.15 Most studies have not included information on environmental risk factors, although vitamin D status is associated strongly with relapse rate in both pediatric and adult patients.16, 17 Some genetic variants have recently been associated with 25-hydroxyvitamin D levels in adult MS patients, suggesting the potential for genetic modification of the vitamin D effects on the disease course.5, 18 Pediatric patients are ideal subjects for studying the effects of MS risk factors on the disease course, as increased genetic or environmental burden may result in early disease activity and, as previously reported, a higher relapse rate has been observed in children.19
The goal of this study was to determine whether established susceptibility factors including genetic ancestry, sex, HLA-DRB1*15, and a non-HLA GRS are associated with relapse rate in children with MS.
Materials and methods
Pediatric Subjects and Assessments
All children receiving clinical care through two Pediatric MS Centers of Excellence at Stony Brook University, New York and University of California, San Francisco were offered enrollment. The Institutional Review Boards from these centers and UC Berkeley approved this study. Informed consent was obtained from parents and assent from children. Participants had diagnoses of MS or clinically isolated syndrome suggestive of early MS based on available, published criteria with symptom onset at less than 18 years of age.20 All consecutive patients seen at the centers were offered enrollment from January 2006 to December 2011. While the clinics are tertiary referral centers they provide care for patients from wide geographic regions and provide free care for those children with limited access to health care. Blood samples were drawn upon enrollment and subjects were prospectively followed for clinical course.
Clinical, demographic and vitamin D serum assessments
Relapses were defined as new or significantly worsened neurological symptoms persisting greater than 24 hours in the absence of infection or fever (pseudo-exacerbation ruled out). Dates of DMT initiation and cessation were recorded for all medications used per patient and therapy use was treated as a time-varying covariate. Genetic ancestral estimates were obtained as below. Batched 25(OH) vitamin D levels on samples at study entry were determined by chemoluminescence assay as previously described (Heartlands, ARUP).21
Genotyping
Whole blood was collected, processed and extracted for DNA using standard procedures. HLA-DRB1 typing was performed by direct sequencing for each study participant. Both HLA-DRB1*1501 and 1503 were characterized (noted as HLA-DRB1*15). For whole genome SNP analysis, the Infinium 660K BeadChip or HumanOmniExpress BeadChip was used to genotype each study participant as previously described.5, 22 Briefly, overall sample and SNP call rates (<90%), sex discrepancies, reproducibility of replicates and checks for Mendelian inheritance using CEPH control trios, as well as performance of internal Illumina controls for each BeadChip were assessed using Illumina GenomeStudio software. Stringent quality control (QC) measures and comparison of sample genotypes across two Illumina platforms were performed using PLINK v.1.0723.
For Illumina 660K BeadChip, Mendelian consistency based on CEPH trio genotype results was >99.95%. SNPs that deviated from HWE (p<0.0001) (1.7%), had a MAF <0.01 (5.9%) or had a call-rate <90% (5.9%) were dropped from the data set. For OmniExpress BeadChip, Mendelian consistency based on CEPH trio genotype results was >99.97%. SNPs that deviated from HWE (p<0.0001) (0.4%), had a MAF <0.01 (7.3%) or call-rate <90% (0.25%) were dropped from the data set. Ten study replicates had >99.99% genotype matches among replicates of successfully typed SNPs. A total of 324,294 SNPs overlapped between the two Illumina BeadChips. As an additional measure of QC, a group of 21 samples were genotyped on both BeadChips with ~99% agreement. All case samples were successfully typed. No sex discrepancies were observed.
Phasing and imputation of genetic data
Additional genotypes were imputed using IMPUTE2 methodology24 with a pre-phasing step to produce estimated haplotypes using an accurate approach as implemented in ShapeIT24 and using the 1000 Genomes Phase I integrated variant set (February, 2012) that included reference data (approximately 37 million SNPs) for 1,092 individuals from Africa, Asia, Europe, and the Americas.25 A total of 110 SNP genotypes were extracted from the dataset for analyses described in the current manuscript.
Genetic ancestry estimates
Genetic ancestry estimates were determined using STRUCTURE Version 2.3.1, default parameters and k=5 ancestral populations for the current study (European, African, Central Asian, American, and East Asian).26 Briefly, data from the Human Genome Diversity Project (HGDP) were used as a reference (http://www.hagsc.org/hgdp/files.html).27 The reference panel has 1,043 samples from 7 populations: Africa, America, Central South Asia, East Asia, Europe, Middle East, and Oceania, and included 660,918 SNP genotypes. Our data were merged with the HGDP data. Following QC, a total of 75,545 SNPs were identified, common to both the pediatric MS patient and HGDP reference datasets. Pruning based on linkage disequilibrium produced genotypes for 37,215 independent SNPs. STRUCTURE analysis generated individual ancestry proportions for each patient in the current study.
Genetic Risk Score
A genetic risk score (GRS) for each patient was generated as previously described by combining the unweighted allele counts for each of 110 non-MHC MS susceptibility loci established through recent large GWAS of 14,498 White/European adult MS cases and 24,091 controls and replication.5–7 As weights used in risk scores for adult studies were determined using only White/European cases, these weights were not used in this diverse population of pediatric MS patients.
Statistical analysis
Repeated events hazard analyses and negative binomial regression models were used to evaluate the association between genetic risk factors and relapse rate. Cox regression models were employed, using time to relapse and with adjustment in standard errors to reflect multiple relapses per patient. The Schoenfeld residuals test was used to confirm the proportional hazards assumption in all presented models. As some predictors of interest violated this assumption analyses using negative binomial regression models were also performed. DMT use was treated as a time-varying covariate in the Cox models. As DMT did not contribute to the hazard for relapse in these analyses, it was not included in the negative binomial models. Predictors associated with relapse rate in univariate models were included in multivariable models with mutual adjustment. For examination of effect modification, genetic risk factor-vitamin D status interactions with p-values of p=0.2 or less were considered of interest. Annualized relapse rate (ARR), incident rate ratio (IRR) and hazard ratios (HR) and corresponding 95% confidence intervals (CI) were reported when appropriate. Analyses were performed using STATA v12.0 (Statacorp, TX).
Results
Genotyping and prospective clinical follow-up were performed on 181 subjects with pediatric MS or CIS. Characteristics of this cohort are similar to those previously reported in pediatric MS subjects (Table 1). Of the 131 of subjects (72% of total) exposed to DMT during the follow-up period, 105 (80%) were exposed to the interferon subclass of medications. A total of 408 relapses were captured over 622 patient-years of follow-up. Median ARR was 0.60 (IQR 0.17, 1.06). In an unadjusted analysis, females had higher relapse rate than males (IRR 1.41, 95% CI 1.04, 1.90, p=0.024), so sex was included in all multivariable models. Median ARR was lower in children with onset <11 years (n=42, median ARR=0.43, IQR 0.19, 1.03) vs. older (median ARR 0.61, IQR 0.14, 1.09) but in a model adjusting for sex, there was no association of younger onset with relapse rate (IRR=0.87, 95% CI 0.64, 1.18, p=0.47). Approximately 40% of pediatric MS patients were HLA-DRB1*15 positive.
Table 1.
Subject Characteristics
| MS or CIS cases (n=181) |
|
|---|---|
| Age of onset, mean yrs (± SD) | 13.1 (± 4.2) |
| Median relapses (range) | 1 (0–12) |
| Median follow-up time, months (range) | 34 (2–174) |
| White (self report, %) | 65.1 |
| Hispanic (self report, %) | 30.3 |
| Female (%) | 65.8 |
| Median baseline 25(OH) vitamin D ng/ml (range) | 23 (2–63) |
| HLA-DRB1*15:01/03 positive (%) | 41.4 |
| DMT exposure during follow-up period (%) | 72 |
Genetic ancestry
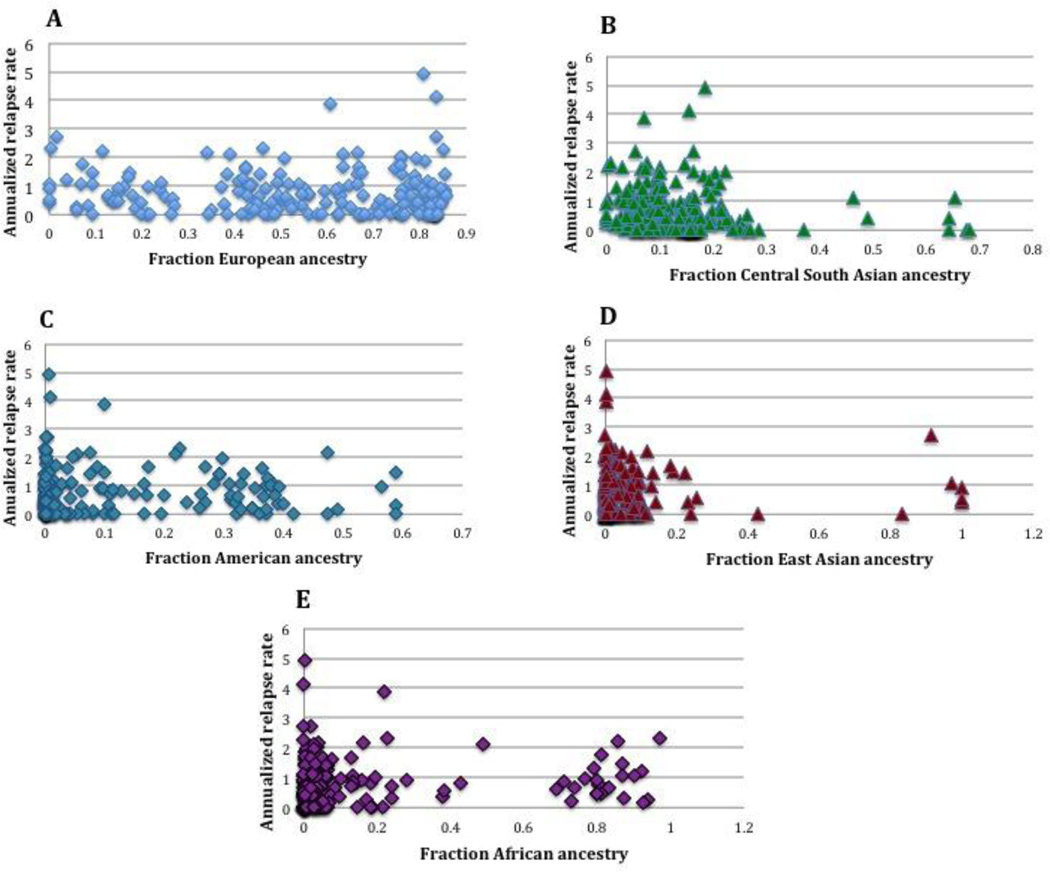
Genetic ancestry estimates, specifically percent European, African, Central South Asian, Native American, and East Asian, were determined for each subject and evaluated for association with relapse rate (Figure 2). Association with decreased relapse hazard was observed for Central South Asian and increased hazard for East Asian ancestry in Cox regression repeated events models (Table 2), but few subjects had estimates greater than 50% for these ancestries (Figure 2, Table 2). For two ancestral variables, European and American, the hazards were not proportional as determined by the Schoenfeld residuals test (Table 2). Addition of sex, DMT, and HLA-DRB1*15 status to the models did not change the results (data not shown). Given proportional hazards violations for two predictors of interest, we pursued two alternative models. First, we added an interaction term between the European estimate and time to the Cox regression model. For this model, there was no association between European ancestry and relapse rate (HR=0.84, 95% CI 0.43, 1.64, p=0.61, Schoenfeld p=0.15). An identical model for Native American ancestry also demonstrated no association with relapse hazard (HR=0.65, 95% CI 0.19, 2.24, p=0.50). For the second alternative model, we employed the negative binomial method to determine IRRs for relapses. Univariate models demonstrated association between Central South Asian ancestry and lower relapse rate (Table 3). Multivariable models did not demonstrate association between ancestral estimates and relapse rate (Table 3).
Figure 2. Plots of annualized relapse rate vs. ancestry.
Annualized relapse rate plotted vs. fractions of each of the 5 ancestral estimates: A) European B) Central South Asian C) Native American D) East Asian E) African.
Table 2.
Unadjusted association of ancestry estimates with relapse hazard
| HR | 95% CI | P value | Schoenfeld p value | |
|---|---|---|---|---|
| European | 0.62 | 0.37, 1.06 | 0.081 | 0.0414 |
| Central South Asian | 0.12 | 0.024, 0.59 | 0.009 | 0.18 |
| American | 1.19 | 0.55, 2.57 | 0.66 | 0.0009 |
| East Asian | 2.00 | 1.09, 3.70 | 0.026 | 0.28 |
| African | 1.31 | 0.89, 1.94 | 0.17 | 0.68 |
Cox regression model results for association of ancestral estimates (STRUCTURE Version 2.3.1) with the hazard to relapse. Schoenfeld residuals test was applied to determine violations of the proportional hazards assumption. P values <0.05 indicate violation of the assumption.
Table 3.
Association of ancestry estimates with relapse rate, univariate models and models adjusted for sex, 25-OH vitamin D level and HLA-DRB1*15
| Univariate Models | IRR | 95% CI | P value |
| European | 0.62 | 0.37, 1.05 | 0.076 |
| Central South Asian | 0.13 | 0.025, 0.64 | 0.013 |
| American | 1.27 | 0.53, 3.05 | 0.59 |
| East Asian | 1.66 | 0.80, 3.43 | 0.17 |
| African | 1.40 | 0.84, 2.34 | 0.19 |
| Adjusted Models | IRR | 95% CI | P value |
| European | 0.80 | 0.46, 1.42 | 0.45 |
| Central South Asian | 0.23 | 0.045, 1.14 | 0.072 |
| American | 1.05 | 0.44, 2.50 | 0.92 |
| East Asian | 1.39 | 0.68, 2.84 | 0.36 |
| African | 1.18 | 0.70, 1.98 | 0.54 |
Negative binomial regression model results for association of ancestral estimates (STRUCTURE Version 2.3.1) with the relapse rate.
Genetic risk score
The GRS comprised of 110 risk SNPs evaluated in these pediatric subjects showed no notable departure from a normal distribution (Supplemental Figure 1). No association between GRS and relapse hazard was observed (HR=0.99, 95% CI=0.98–1.01, p=0.34; Schoenfeld p=0.36). Adjustment for sex, DMT and HLA-DRB1* 15 status did not change these results (HR=0.99, 95% CI=0.97, 1.00, p=0.14). No evidence for interaction between GRS and vitamin D status in the association with relapse rate was observed (pixn=0.47). Analysis using negative binomial regression modeling gave similar results (IRR=0.99, 95% CI 0.97, 1.01, p=0.37). As the few subjects with low GRS scores had less than 50% European ancestry and non-HLA risk alleles have been identified in European populations, we repeated analysis in those of European descent (>50% for European ancestral estimate, n=106). No association was found, but as expected with smaller sample size the standard error was greater with wider confidence intervals (IRR 0.70, 95% CI 0.12, 4.21, p=0.70).
HLA-DRB1*15:01/15:03 and vitamin D
As previously reported in children17, higher vitamin D levels were associated with lower relapse rates (IRR=0.87, 95% 0.77, 0.98, p=0.027). HLA-DRB1* 15 allele carrier status in pediatric MS subjects did not demonstrate a marginal association with relapse rate (IRR= 0.97, 95% CI=0.73, 1.29, p=0.84); however, multivariable Cox regression modeling showed strong evidence for interaction between HLA-DRB1*15 status and vitamin D level, and association with relapse hazard (pixn=0.006), after adjustment for DMT and sex. Specifically, a 10 ng/ml higher vitamin D level was only associated with decreased hazard to relapse in subjects carrying at least one copy of 15:01 or 15:03 (HR 0.73, 95% CI 0.60–0.89, p=0.001), adjusting for DMT and sex. In subjects who were negative for HLA-DRB1*15, there was no association between vitamin D level with relapse rate (HR=1.00, 95% CI 0.89–1.13, p=0.95). There was also evidence for interaction based on the results from the negative binomial model adjusted for sex (p=0.022). Here, a ten ng/ml higher vitamin D level in DRB1*15 positive individuals resulted in a 28% lower relapse rate (IRR=0.72, 95% CI 0.58, 0.88. p=0.002) and in negative individuals there was no association (IRR=0.96, 95% CI 0.83,1.12, p=0.64). The interaction remained significant (p=0.018) for individuals with >0.50 European ancestry (n=106).
Discussion
Understanding the role of inherited factors and detectable environmental exposures in disease course is critical to developing a more personalized approach to MS treatment. In our clinically well-characterized cohort of pediatric MS subjects, we observed that the strongest known genetic risk factor modified the association of vitamin D level with relapse rate. Our results raise the possibility that vitamin D supplementation may be more effective in preventing relapses in those carrying this risk allele. Prior sequence analysis has demonstrated a putative vitamin D response element (VDRE) in the promoter region of HLA-DRB1*15 capable of binding a recombinant vitamin D receptor; increased cell surface expression of HLA-DRB1 upon stimulation with 1,25-dihydroxyvitamin D3 has also been reported.28 This VDRE motif has not been observed to contribute to MS susceptibility independent to the risk attributed to carrying the HLA-DRB1*15 haplotype29, but interestingly haplotypes HLA-DRB1* 04/*07/*09 (DR53) that express “nonresponsive” VDRE motifs have been associated with reduced risk of MS.29 Another study demonstrated that allelic variation in the vitamin D receptor is associated with reduced risk of MS in HLA-DRB1*15 haplotype carriers.30 The potential role of this motif in disease course is not known. Evaluation of the relationship between the HLA-DRB1*15 promoter region VDRE and relapse rate or formation of new lesions would help determine whether there is a clinically relevant biological interaction between vitamin D status and HLA-DRB1.
In a prior study of adults with MS, evidence for interaction between HLA-DRB1*15:01 and vitamin D status and association with new enhancing lesions was reported.31 Interestingly, the association between vitamin D level and enhancing lesions was stronger in individuals who did not carry HLA-DRB1*15 alleles. Our results do not support these findings. Differences may be due to the populations studied, inclusion of subjects with African ancestry and HLA-DRB1* 15:03 alleles in the current pediatric study, or other unmeasured genetic or environmental modifiers of the relationship between HLA-DRB1*15 and vitamin D status.
We found no evidence for association between European or African ancestry and relapse rate. In adults with MS, African ancestry has been associated with accelerated retinal injury32 and greater disability.33 The higher severity scores may be secondary to other features such as greater frequency of transverse myelitis33 or differences in recovery following neuronal injury rather than difference in relapse rate. The association between African ancestry and disability metrics requires further study in children. We did observe evidence in both Cox regression and negative binomial models for association between Central South Asian ancestry and relapse hazard or rate, respectively, but there were few subjects with ancestry estimates greater than fifty percent, which suggest only a few individuals may have contributed to this result. Nonetheless, the observed point estimate was of strong magnitude and these results should be investigated in a larger cohort. Prior work has suggested that MS in individuals of Central South Asian descent may have unique clinical or immunologic features.34
Despite recent report that a genetic burden score was associated with relapse rate in adults15, we found no evidence of association between a composite genetic risk score of 110 non-HLA susceptibility alleles and longitudinal risk of relapse. Results were similar based on both statistical methodologies and with 95% CI tightly centered around point estimates (HR and IRR) that included the null. These observations strongly argue that this metric of inherited risk burden is not associated with relapse rate in children.
DMT use was not found to be a significant covariate in the models studied, an expected finding since such an association would have implied that in a causal model, the genetic factors were associated with whether or not a patient was placed on DMT. This variable was included in the models to improve precision in the case that potential strong effects on the outcome would increase the standard error of the model. However, we did not observe this effect.
The diversity of racial and ethnic backgrounds in pediatric subjects is a strength of the current study. Other strengths include the well-characterized clinical phenotype of the patients, comprehensive longitudinal follow-up of relapse activity, and rigorous statistical modeling, including application of two well-established approaches for evaluating relapse activity as our outcome.
We acknowledge limitations of the current study. Our sample size was modest and constrained by the overall rarity of onset of MS in childhood, occurring in only 5% of MS cases.35 We did not have access to a similar dataset in children for replication of the observed HLA-DRB1*15-vitamin D status interaction. However, the length of follow-up and capture of 408 relapse events strengthened our study’s statistical power. Vitamin D status was captured only once at study entry from serum samples; the levels may only approximate their first event levels or pre-diagnosis levels. On the other hand, most subjects entered shortly after their first clinical events and further, levels are unlikely to have dramatically changed without supplementation, which was not routinely recommended for this group during the study period. Future studies in children with repeated serum measures of vitamin D will better address the effects of changes in vitamin D status over time.
The current study is the first to investigate the relationship between genetic factors and a clinical measure of disease activity in children with MS and the first to evaluate the role of ancestry estimates in relapse rate. Our results underscore the need for large, prospective studies of genetic effects on disease course in the context of environmental risk factors.
Supplementary Material
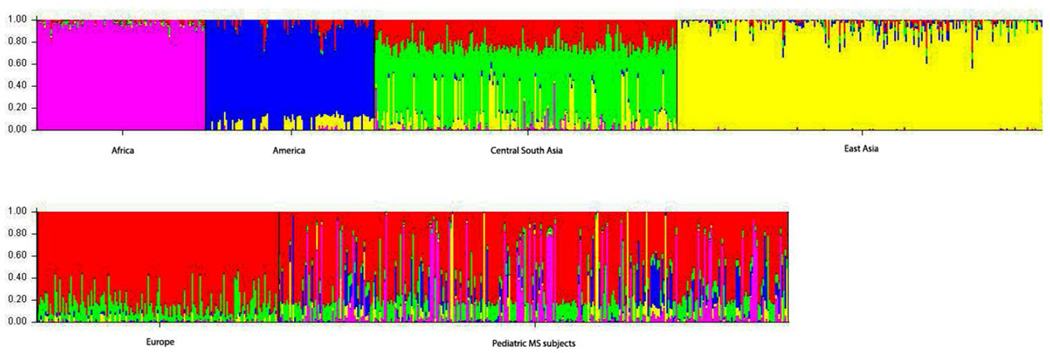
Figure 1. Diversity of ancestral components in pediatric MS subjects.
Bar plot of genetic ancestry of individuals sorted by population: 1) Africa 2) America 3) Central South Asia 4) East Asia 5) Europe and 6) Pediatric MS Subjects.
Contributor Information
Lisa F. Barcellos, Email: lbarcellos@genepi.berkeley.edu.
Xiaorong Shao, Email: xshao@genepi.berkeley.edu.
Janelle Noble, Email: jnoble@chori.org.
Ellen M. Mowry, Email: emowry1@jhmi.edu.
Hong Quach, Email: hquach@genepi.berkeley.edu.
Anita Belman, Email: abelman@stonybrook.edu.
T.C. Casper, Email: Charlie.Casper@hsc.utah.edu.
Lauren B. Krupp, Email: Lauren.Krupp@stonybrook.edu.
Emmanuelle Waubant, Email: Emmanuelle.Waubant@ucsf.edu.
REFERENCES
- 1.Page WF, Kurtzke JF, Murphy FM, Norman JE., Jr Epidemiology of multiple sclerosis in U.S. veterans: V. Ancestry and the risk of multiple sclerosis. Annals of neurology. 1993;33:632–639. doi: 10.1002/ana.410330612. [DOI] [PubMed] [Google Scholar]
- 2.Disanto G, Magalhaes S, Handel AE, et al. HLA-DRB1 confers increased risk of pediatric-onset MS in children with acquired demyelination. Neurology. 2011;76:781–786. doi: 10.1212/WNL.0b013e31820ee1cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patsopoulos NA, Barcellos LF, Hintzen RQ, et al. Fine-mapping the genetic association of the major histocompatibility complex in multiple sclerosis: HLA and non-HLA effects. PLoS genetics. 2013;9:e1003926. doi: 10.1371/journal.pgen.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oksenberg JR, Barcellos LF, Cree BA, et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. American journal of human genetics. 2004;74:160–167. doi: 10.1086/380997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawcer S, Hellenthal G, Pirinen M, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International Multiple Sclerosis Genetics C. Beecham AH, Patsopoulos NA, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nature genetics. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Jager PL, Chibnik LB, Cui J, et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8:1111–1119. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isobe N, Damotte V, Lo Re V, et al. Genetic burden in multiple sclerosis families. Genes Immun. 2013 doi: 10.1038/gene.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gourraud PA, Sdika M, Khankhanian P, et al. A genome-wide association study of brain lesion distribution in multiple sclerosis. Brain. 2013;136:1012–1024. doi: 10.1093/brain/aws363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalincik T, Guttmann CR, Krasensky J, et al. Multiple sclerosis susceptibility loci do not alter clinical and MRI outcomes in clinically isolated syndrome. Genes Immun. 2013;14:244–248. doi: 10.1038/gene.2013.17. [DOI] [PubMed] [Google Scholar]
- 11.Mowry EM, Carey RF, Blasco MR, et al. Multiple Sclerosis Susceptibility Genes Are Associated with Relapse Severity and Recovery. PLoS One. 2013;3:e75416. doi: 10.1371/journal.pone.0075416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mowry EM, Carey RF, Blasco MR, et al. Association of multiple sclerosis susceptibility variants and early attack location in the CNS. PLoS One. 2013;3:e75565. doi: 10.1371/journal.pone.0075565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genome-wide association study of severity in multiple sclerosis. Genes Immun. 2011;12:615–625. doi: 10.1038/gene.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goris A, Pauwels I, Gustavsen MW, et al. Genetic variants are major determinants of CSF antibody levels in multiple sclerosis. Brain : a journal of neurology. 2015;138:632–643. doi: 10.1093/brain/awu405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilven K, Patsopoulos NA, Dubois B, Goris A. Burden of risk variants correlates with phenotype of multiple sclerosis. Multiple sclerosis. 2015 doi: 10.1177/1352458514568174. [DOI] [PubMed] [Google Scholar]
- 16.Simpson S, Jr, Taylor B, Blizzard L, et al. Higher 25-hydroxyvitamin D is associated with lower relapse risk in multiple sclerosis. Ann Neurol. 2010;68:193–203. doi: 10.1002/ana.22043. [DOI] [PubMed] [Google Scholar]
- 17.Mowry EM, Krupp LB, Milazzo M, et al. Vitamin D status is associated with relapse rate in pediatric-onset multiple sclerosis. Annals of neurology. 2010;67:618–624. doi: 10.1002/ana.21972. [DOI] [PubMed] [Google Scholar]
- 18.Lin R, Taylor BV, Simpson S, Jr, et al. Association between multiple sclerosis risk-associated SNPs and relapse and disability - a prospective cohort study. Mult Scler. 2013 doi: 10.1177/1352458513496882. [DOI] [PubMed] [Google Scholar]
- 19.Gorman MP, Healy BC, Polgar-Turcsanyi M, Chitnis T. Increased relapse rate in pediatric-onset compared with adult-onset multiple sclerosis. Arch Neurol. 2009;66:54–59. doi: 10.1001/archneurol.2008.505. [DOI] [PubMed] [Google Scholar]
- 20.Krupp LB, Banwell B, Tenembaum S. Consensus definitions proposed for pediatric multiple sclerosis and related disorders. Neurology. 2007;68:S7–S12. doi: 10.1212/01.wnl.0000259422.44235.a8. [DOI] [PubMed] [Google Scholar]
- 21.Mowry EM, Krupp LB, Milazzo M, et al. Vitamin D status is associated with relapse rate in pediatric-onset multiple sclerosis. Ann Neurol. 2010;67:618–624. doi: 10.1002/ana.21972. [DOI] [PubMed] [Google Scholar]
- 22.Walsh KM, Chokkalingam AP, Hsu LI, et al. Associations between genome-wide Native American ancestry, known risk alleles and B-cell ALL risk in Hispanic children. Leukemia. 2013 doi: 10.1038/leu.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genomes Project C, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cann HM, de Toma C, Cazes L, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 28.Ramagopalan SV, Maugeri NJ, Handunnetthi L, et al. Expression of the multiple sclerosis-associated MHC class II Allele HLA-DRB1*1501 is regulated by vitamin D. PLoS genetics. 2009;5:e1000369. doi: 10.1371/journal.pgen.1000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nolan D, Castley A, Tschochner M, et al. Contributions of vitamin D response elements and HLA promoters to multiple sclerosis risk. Neurology. 2012;79:538–546. doi: 10.1212/WNL.0b013e318263c407. [DOI] [PubMed] [Google Scholar]
- 30.Agliardi C, Guerini FR, Saresella M, et al. Vitamin D receptor (VDR) gene SNPs influence VDR expression and modulate protection from multiple sclerosis in HLA-DRB1*15-positive individuals. Brain, behavior, and immunity. 2011;25:1460–1467. doi: 10.1016/j.bbi.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Mowry EM, Waubant E, McCulloch CE, et al. Vitamin D status predicts new brain magnetic resonance imaging activity in multiple sclerosis. Ann Neurol. 2012;72:234–240. doi: 10.1002/ana.23591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimbrough DJ, Sotirchos ES, Wilson JA, et al. Retinal damage and vision loss in African American multiple sclerosis patients. Annals of neurology. 2015;77:228–236. doi: 10.1002/ana.24308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cree BA, Khan O, Bourdette D, et al. Clinical characteristics of African Americans vs Caucasian Americans with multiple sclerosis. Neurology. 2004;63:2039–2045. doi: 10.1212/01.wnl.0000145762.60562.5d. [DOI] [PubMed] [Google Scholar]
- 34.Nicholas RS, Kostadima V, Hanspal M, et al. MS in South Asians in England: early disease onset and novel pattern of myelin autoimmunity. BMC neurology. 2015;15:72. doi: 10.1186/s12883-015-0324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh EA, Chitnis T, Krupp L, et al. Pediatric multiple sclerosis. Nature reviews Neurology. 2009;5:621–631. doi: 10.1038/nrneurol.2009.158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.