Abstract
Purpose
To develop a compressed sensing (CS) acceleration method with a high spectral bandwidth exploiting the spatial-spectral sparsity of MR spectroscopic imaging (MRSI).
Methods
Accelerations were achieved using blip gradients during the readout to perform non-overlapped and stochastically delayed random walks in kx-ky-t space, combined with block-Hankel matrix completion for efficient reconstruction. Both retrospective and prospective CS accelerations were applied to 13C MRSI experiments, including in vivo rodent brain and liver studies with administrations of hyperpolarized [1-13C] pyruvate at 7T and [2-13C] dihydroxyacetone at 3T, respectively.
Results
In retrospective undersampling experiments using in vivo 7T data, the proposed method preserved spectral, spatial and dynamic fidelities with R2 ≥ 0.96 and ≥ 0.87 for pyruvate and lactate signals, respectively, 750-Hz spectral separation and up to 6.6-fold accelerations. In prospective in vivo experiments, with 3.8-fold acceleration, the proposed method exhibited excellent spatial localization of metabolites and peak recovery for pyruvate and lactate at 7T as well as for dihydroxyacetone and its metabolic products with a 4.5-kHz spectral span (140 ppm at 3T).
Conclusion
This study demonstrated the feasibility of a new CS approach to accelerate high spectral bandwidth MRSI experiments.
Keywords: compressed sensing, random blip gradients, MR spectroscopic imaging, hyperpolarized carbon-13, Hankel matrix completion, calibrationless parallel imaging
Introduction
In hyperpolarized 13C experiments, MR spectroscopic imaging (MRSI) techniques provide real-time assessment of 13C labeling in multiple metabolites and their distribution in different organs or tissue types, leading to the measurement of enzyme kinetics, tissue perfusion, pH, redox state, and more (1–4). The major technical challenges facing hyperpolarized 13C MRSI with in vivo applications are the sub-minute lifetime of the hyperpolarized 13C signal and the relatively broad dispersion of 13C spectra. The short lifetime of the hyperpolarized 13C signal requires MRSI techniques that capture the dynamics of metabolites in a rapid manner and make efficient use of the 13C hyperpolarization. Therefore, it is desirable to reduce the number of phase encodings and excitations. Meanwhile, the broad spectral dispersion of many hyperpolarized 13C probes and their metabolic products make it necessary to develop high bandwidth methods. For example, monitoring [1-13C] lactate and 13C-bicarbonate production using hyperpolarized [1-13C] pyruvate requires a spectral dispersion of 20 ppm (5,6); hyperpolarized [2-13C] dihydroxyacetone and one of its metabolic products, [2-13C] glycerol-3-phosphate, have a spectral dispersion of 140 ppm, with several other products within this bandwidth (4); and even more challenging, a spectral dispersion of 180 ppm is necessary to follow [5-13C] glutamate and [2-13C] lactate production after injection of hyperpolarized [2-13C] pyruvate (6–8).
Previous hyperpolarized 13C studies have utilized several MRSI techniques, such as phase-encoded MRSI (2,5,9,10), echo planar spectroscopic imaging (EPSI) and compressed sensing (11–14), spiral spectroscopic imaging (spiral SI) (8,10,15), concentric rings spectroscopic imaging (16) and model-based spectral reconstruction (17–19). Although phase-encoded MRSI is slow, its intrinsically high spectral bandwidth is advantageous for many hyperpolarized 13C applications. The fast MRSI techniques, including EPSI, spiral SI, and concentric rings, utilize high-slew-rate gradient waveforms to cover k-t space repetitively (16,20,21). These techniques rapidly sample k-space, but have relatively low spectral bandwidths on clinical MRI systems (i.e. <1 kHz) due to limitations in gradient hardware performance. Model-based spectral reconstruction utilizes constraints based on the known chemical shifts of images that are acquired with multiple echo times (TEs) (17–19), but requires specific knowledge of the resonant frequencies and B0 inhomogeneity.
Compressed sensing (CS) is a promising tool for accelerating MRSI with two fundamental criteria: sparsity and incoherence (22). The capability of CS has been demonstrated in hyperpolarized 13C EPSI (11–14) as well as in 1H (23–29), 23Na (30), 31P (31) and 19F (32) MRSI applications. The spectral dimension of MRSI data is the sparsest, and several methods exploit this sparsity for substantial acceleration using incoherent spatial and spectral undersampling and CS reconstruction (11–14). Several reconstruction methods have also been successfully used in previous CS MRSI studies, e.g. L1-minimization (11–14,23,24), total variation minimization (25,27–29), and maximum entropy reconstruction (26), and group sparsity based reconstruction (33). Recently, Hankel or block-Hankel matrix completion has been used for recovering undersampled spectral data (34–36) as well as accelerating dynamic MRI (37,38) and calibrationless parallel imaging reconstruction (39–41).
Previously, random blip gradients have been utilized in hyperpolarized 13C studies to accelerate EPSI via randomizing the undersampling in the spatial (kx, ky), spectral (t), and dynamic (frame) dimensions (11–14). In the present study, four major improvements were made. First, the random blip scheme was optimized based on 2D random walks within the entire k-space sampling region. Second, random blip gradients were combined with stochastic time delays to preserve the inherent bandwidth of MRSI. Third, the CS reconstruction was formulated as a low rank matrix completion to efficiently exploit spatial-spectral sparsity and shared information along the dynamic dimension. Fourth, the proposed method was combined with calibrationless parallel imaging reconstruction (39–41). A retrospective study with varied acceleration factors was performed on an in vivo hyperpolarized 13C MRSI dataset. Then prospective accelerations were demonstrated in two hyperpolarized 13C MRSI experiments in vivo with hyperpolarized [1-13C] pyruvate at 7T and hyperpolarized [2-13C] dihydroxyacetone at 3T. We also evaluated the feasibility of CS MRSI with calibrationless parallel imaging in a phantom experiment.
Methods
Compressed Sensing MRSI
Random walk trajectory
The proposed method combined random blip gradients with random walk trajectories and stochastic time delays, allowing incoherent undersampling in k-t space. As shown in Figure 1a, triangular blip gradients were added to a MRSI sequence (FIDCSI, DV24 version at 3T, and DV23 at 7T, GE Healthcare, Waukesha, WI). In each repetition time (TR, Figure 1a), the modified MRSI sequence consisted of a slice selective RF excitation pulse followed by 20 pairs of Gx and Gy blip gradients (0.8 ms duration each) with random amplitudes and time delays. Each pair of Gx and Gy blip gradients moved the k-space trajectory in kx and ky, and a series of random blip gradients created a random walk trajectory in kx-ky-t space. The random walk trajectory was kept on Cartesian grid points when blip gradients were off, and MRSI data was continuously acquired during both blip gradient on and off periods. However, only the data sampled on the Cartesian grid during the blip off periods were used in reconstruction. Of note, time delays between blip gradients (in Fig. 1a) were randomized to create incoherent undersampling along the t dimension.
Figure 1.
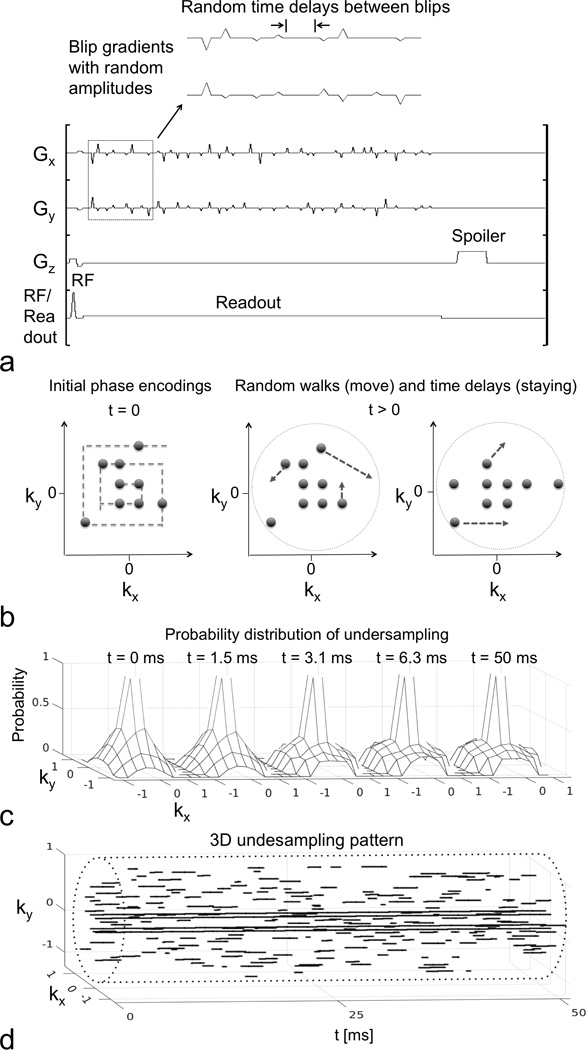
CS MRSI sequence, random walk and time delay scheme and undersampling pattern. (a) CS MRSI sequence for a single acquisition. Blip gradients with random amplitudes and time delays were applied on Gx and Gy during the readout period, which facilitated a random walk trajectory in the kx-ky plane. (b) The initial phase encoding locations followed a random spiral-out order. During the readout, some phase encodings were performing random walks, and some were not, due to the random time delays or the full sampling at the center. Random walks moved slower near the center, creating variable density sampling. Four phase encodings in the center were fully sampled along t dimension. (c) The probability distribution of undersampling at various time points. The central peak indicated full sampling. The sampling distribution due to the random walks in peripheral k-space became increasingly flat from t = 0 to 3.1 ms, and was relatively stationary with t > 3.1 ms. (d) The 3D kx-ky-t undersampling pattern (3.8-fold) for the first frame, with the cylindrical sampling boundary (dashed line).
Random walk trajectories with the blip gradients were designed with the following constraints (as illustrated in Fig. 1b): 1) initial phase encoding locations were randomly selected and followed a spiral-out order; 2) different random time delay and blip gradient schemes were used for each phase encoding; 3) trajectories in each frame were non-overlapped in kx-ky-t space; 4) trajectories were all confined within a cylinder boundary in kx-ky-t space; and 5) trajectories moved slower when they were closer to the center of the kx-ky plane to create variable density sampling. Probability distributions of this undersampling scheme were generated by Monte Carlo simulation with 1000 realizations, as shown in Figure 1c. The distribution became increasingly flat in the peripheral kx-ky plane from t = 0 to 3.1 ms and was relatively stationary for t > 3.1 ms.
Dynamic 3D undersampling pattern
The undersampling pattern was randomized along kx, ky, t and frame dimensions. Figure 1d shows the 3D undersampling pattern in kx-ky-t space for the first frame, and the undersampling factor was 3.8. For each frame, the undersampling scheme consisted of 14 phase encodings (i.e., 14 excitations). Among them, 10 phase encodings followed the random walk trajectories in kx-ky-t space with stochastic time delays as described above. The other 4 phase encodings fully sampled the t dimension for the center of the kx-ky plane. For different frames, the 3D undersampling patterns were all independently generated to create randomness along the frame dimension (as shown in Supporting Figure S1). These undersampling patterns were used in all prospective in vivo experiments.
Reconstruction
Block-Hankel reconstruction matrix
A concatenated Hankel matrix (or block-Hankel matrix) was used in this study to represent the dynamic MRSI data in a low-rank manner and to exploit the spatial-spectral sparsity and shared information along the dynamic dimension. As shown on the right side of Figure 2, a 3D sliding window with a size of 5 × 5 × 40 for retrospective experiment or 5 × 5 × 20 for prospective experiments was applied on kx, ky and t dimensions of MRSI data to form the block-Hankel matrix H.
Figure 2.
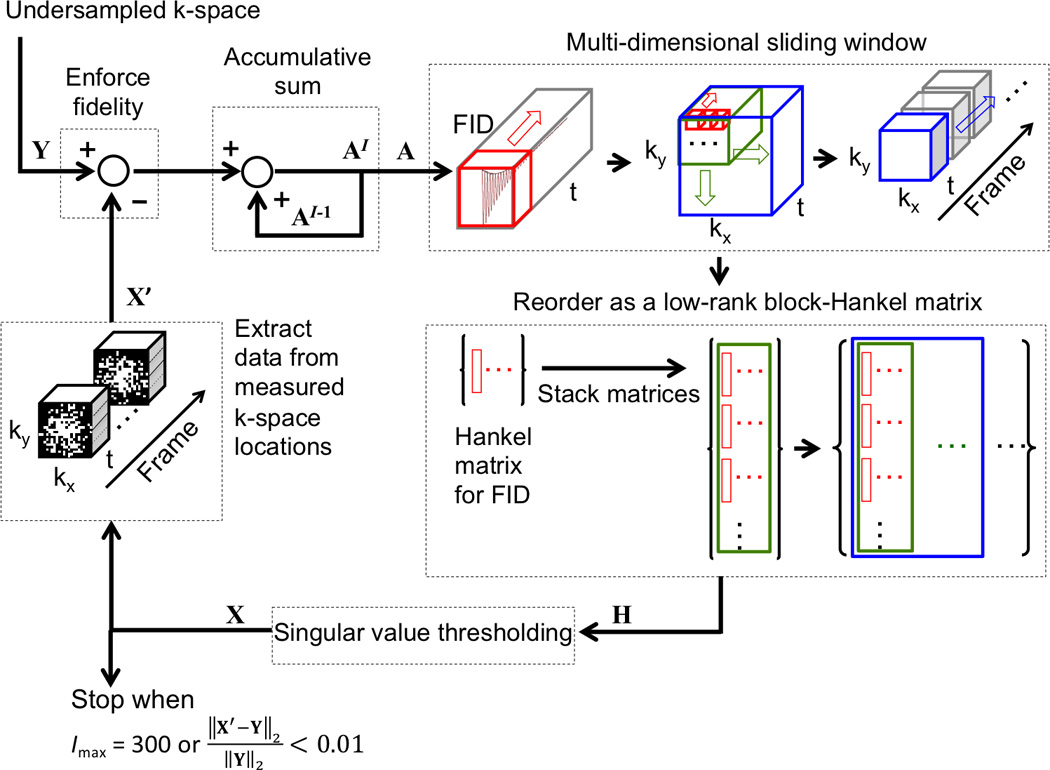
Illustration of the singular value thresholding method used to reconstruct CS datasets. Y is the undersampled k-space; X is the estimated or reconstructed k-space data; X’ the k-space data at measured locations extracted from X; A is the accumulative sum of the difference between Y and X’; H is the block-Hankel matrix of A and the input of the singular value thresholding operation; and Imax for maximum iteration number. Hankel matrices for FIDs (red windows) are first stacked in a column-wise order for all voxels within a kx-ky window (green window). Then, the column-wise block matrices for all kx-ky windows and frames (blue window indicates one frame) are concatenated along each row to form a large block-Hankel matrix, H.
To characterize the rank of this block-Hankel matrix, we modeled the MRSI data in x-y-frequency (f) space as a linear combination of spatial-spectral peaks that have Lorentzian line-shapes. In kx-ky-t space, signal contributed by a spatial-spectral peak at (xl, yl, fl) is the product of three separable exponential functions, that is
| Eq. 1 |
where l=1, 2, …, L is the index of peak, dl,u is the amplitude for frame u, Al and Bl are decay rates of the k-space signal along kx and ky (or line-broadening/blurring factors for spatial Lorentzian peak profiles), and R2* is the decay rate of the FID (or line-broadening/blurring factor for spectral Lorentzian peak profile). In this model, the block-Hankel matrix H for MRSI data is a linear combination of block-Hankel matrices for each spatial-spectral peak as given by
| Eq. 2 |
where L is the number of spatial-spectral peaks in the x-y-f space. Here rank(Hl) = 1 for l = 1, 2, …, L (see Appendix for details). Using subadditivity of rank, the rank of H has the following upper bound, that is
| Eq. 3 |
Therefore, the rank of block-Hankel matrix is not greater than the number of spatial-spectral peaks.
Block-Hankel matrix completion
The nuclear norm minimization for the block-Hankel matrix completion (34,36) can be written as
| Eq. 4 |
where M is a k-space undersampling operator, H a 3D sliding window and concatenation operator describing the conversion into a block-Hankel matrix, X the reconstructed MRSI dataset and Y the undersampled MRSI dataset. This nuclear norm minimization can be solved by a singular value thresholding method (42), as shown in Figure 2. The singular value thresholding S(H, μ) was defined as
| Eq. 5 |
where Sμ(Λ) was the soft thresholding on the diagonal entries of Λ with a threshold level μ, and UHΛHVH* was the singular value decomposition (SVD) of H. In this study, the threshold level μ was set to a large value to ensure accuracy (42).
All the SVD calculations were performed based on a randomized SVD algorithm for reducing the computation cost (43), using the source code that is available online (https://github.com/cvxr/TFOCS/randomizedSVD.m). Only 300 singular values were computed since the other singular values were normally below the threshold. All numerical calculations were carried out in MATLAB (Mathworks Inc., Natick, MA) on a desktop computer.
Broadband spatial-spectral (SPSP) selective RF excitation pulse
In this study, a broadband SPSP pulse was designed for the in vivo hyperpolarized [2-13C] dihydroxyacetone (DHAc) experiments. This pulse utilized aliased spectral bands to excite resonances that had a large spectral span (140-ppm, or 4.5-kHz at 3T). A method based on L2-regularization of an initial RF waveform using an echo-planar trajectory ensured the accuracy of spatial selectivity at several bands of interest across the large spectral span. The SPSP pulse was designed to apply small flip angles for [2-13C] DHAc and its hydrate, and large flip angles for its metabolic products: ≤ 0.3° at [2-13C] DHAc (213 ppm), 26° at phsophoenolpyruvate (151 ppm), 2.3° at DHAc hydrate (96 ppm) and 20° at glycerol 3-phosphate (G3P) (73 ppm). Additional details of this broadband SPSP pulse are presented in another paper (44).
MR experiments
In vivo mouse brain experiments
The in vivo mouse brain datasets were collected from a 7T whole-body MRI scanner (GE Healthcare, Waukesha, WI). A mouse head RF surface coil was used for both transmission and reception. Two in vivo 2D dynamic hyperpolarized 13C MRSI experiments with circular full k-space coverage (n = 1) or prospective 3.8-fold undersampling (n = 1) were performed in two nude mice. All animal experiments were carried out under a protocol approved by the Institutional Animal Care and Use Committee. Both the full k-space and prospectively accelerated MRSI scans were performed with slab thickness = 15 mm, matrix = 8 × 8, field of view (FOV) = 24 × 24 mm2, and spectral bandwidth = 5 kHz. For the full k-space MRSI, the other parameters were echo time (TE)/TR = 2.7/80 ms, flip angle = 5°, FID points = 256, readout time = 51.2 ms, excitations per frame = 54, number of frames = 10 and dynamic resolution = 4.1 s/frame. The phase encodings were sampled in a spiral-out order in k-space. The prospectively accelerated MRSI was performed with TE/TR = 2.6/140 ms, flip angle = 10°, FID points = 512, readout time = 102.4 ms, excitations per frame = 14, number of frames = 20, dynamic resolution = 3 s/frame, and undersampling factor = 3.8. The SNR efficiency for undersampling was 92% accounting for duty-cycle losses due to the blip gradients. All the dynamic MRSI data were acquired starting at the beginning of a bolus injection of approximately 0.35 mL hyperpolarized [1-13C] pyruvate solution (80 mM, pH = 7.0), produced using HyperSense polarizer (Oxford Instruments). T2-weighted 1H images were acquired with TE/TR = 17/4000 ms, FOV = 40 × 40 mm2, matrix size = 512 × 512, and slice thickness = 1 mm.
In vivo rat liver experiment
The in vivo rat liver dataset was collected from a 3T whole-body MRI scanner (GE Healthcare, Waukesha, WI). A quadrature volume RF coil was used for both transmission and reception. A Sprague-Dawley rat was examined after 24-hr food deprivation with ad libitum access to water. 3D balanced steady-state free precession anatomical 1H imaging was performed with TE/TR = 2.2/5.3 ms, FOV = 16 × 8 × 48 mm3 and matrix size = 256 × 256 × 80. Prospectively accelerated hyperpolarized 13C MRSI was performed with TE/TR = 10/150 ms, slice thickness = 20 mm, matrix = 8 × 8, FOV = 64 × 64 mm2, spectral bandwidth = 10 kHz, FID points = 512, excitations per frame = 14, number of frames = 20, and dynamic resolution = 3 s/frame. The readout time was 51.2 ms, undersampling factor = 3.8, and the SNR efficiency for undersampling was 83%. The data acquisition started at 15 s from the beginning of a bolus injection of approximately 3 mL hyperpolarized [2-13C] DHAc solution (80 mM, pH = 7.0), produced using HyperSense (Oxford Instruments).
Calibrationless parallel imaging experiment
A human-head-size thermal 13C phantom was filled with ethylene glycol (HOCH2CH2OH, anhydrous, 99.8%, Sigma-Aldrich, St. Louis, Missouri) and was scanned on the abovementioned 3T whole-body MRI. An eight channel 13C RF surface coil was used for signal reception (45). MRSI data has TE/TR = 2.8/1000 ms, matrix size = 12 × 12, voxel size = 16.6 × 16.6 × 50 mm3, spectral bandwidth = 5 kHz, no averages and no dynamics. The 4-fold undersampling pattern was generated according to the principles illustrated in Figure 1.
Data Analysis
For the phantom MRSI datasets, neither spectral nor spatial filters were used. Signal intensity was quantified by an integration method on absolute spectra. For all in vivo MRSI datasets, a 9 Hz line-broadening spectral filter was applied without spatial filtering. Signal intensities were obtained using the area integration method on the real part of complex spectra. All spectra were displayed in absolute form. In the prospective undersampling experiments, metabolite maps were linearly interpolated and were overlaid on anatomical references. In the retrospective undersampling experiment, metabolite maps were plotted without interpolation.
Results
Phantom experiments
We first performed thermal 13C phantom experiments where prospectively undersampled k-space with the proposed CS reconstruction and full k-space data showed a strong linear correlation with a slope = 1.02 and R2 = 0.94. Further details can be found in the Supporting Figure S2.
In vivo mouse brain experiments
In Figure 3, a full k-space in vivo dataset was reordered into the proposed block-Hankel matrix structure, which could be approximated by a few singular values (in Figure 3b). In Figure 3c, the different singular values and vectors of the block-Hankel matrix after low-rank approximation correspond to distinct spectral, dynamic and spatial features of this hyperpolarized [1-13C] pyruvate experiment. The first and second singular values and vectors were mostly [1-13C] pyruvate signal, with similar dynamics corresponding to the bolus injection and subsequent decay but slightly different spatial distributions. The sixth singular value was mostly [1-13C] lactate signal with dynamics shifted to later frames corresponding to the progressive build up of this metabolic product.
Figure 3.
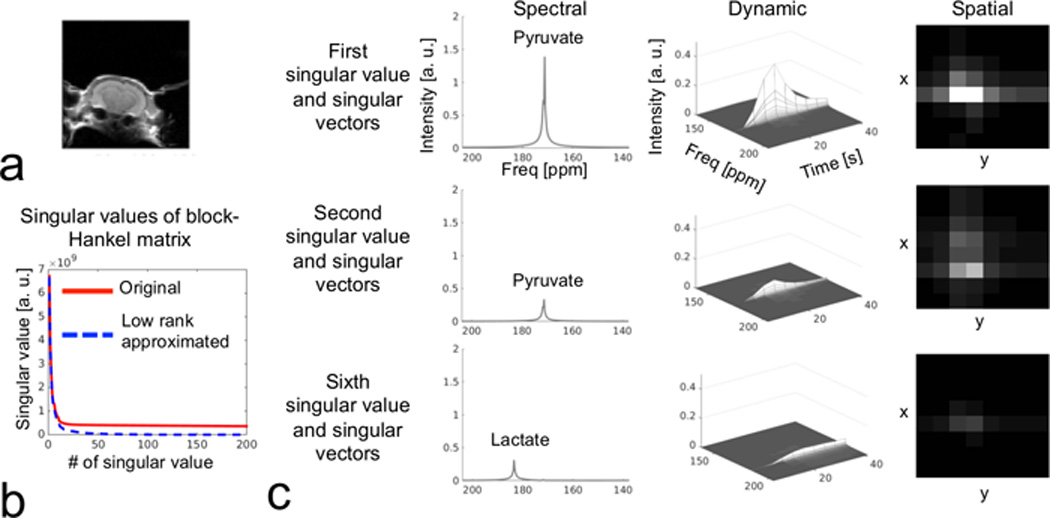
A typical full k-space in vivo hyperpolarized [1-13C] pyruvate MRSI dataset, reordered as a block-Hankel matrix with size of 1000 × 30000 (via sliding window and reordering as illustrated in Fig. 2). (a) The T2-weighted MRI depicts the field of view for the 2D 8 × 8 MRSI. (b) The first 200 singular values from the singular value decomposition of original block-Hankel matrix (red solid line) and its low-rank approximation (blue dash line) are plotted. Low-rank approximation (by Cadzow denoising on block-Hankel matrix) iteratively extracted the top 100 singular values and vectors, and was repeated 300 times. (c) Singular values and vectors from the block-Hankel matrix decomposition after low-rank approximation, showing distinct spectral, dynamic and spatial features.
The performance of the proposed method was examined by retrospective undersampling of the full k-space 13C MRSI dataset acquired on a normal mouse brain in vivo. In Figure 4, the reconstructed dynamic spectra with undersampling factors of 2, 2.7, 3.8 and 6.6 were compared with full k-space ground truth. The CS reconstruction preserved the spectral fidelity with acceleration factors up to 6.6-fold, even with the SNR losses introduced by retrospectively throwing away data. Spatial distributions of pyruvate and lactate were preserved in all undersampled datasets with CS reconstruction, resulting in similar lactate to pyruvate ratios within the studied brain region. Time courses of pyruvate and lactate signals in the undersampled datasets were linearly correlated with that in the full k-space dataset. For pyruvate, slopes of correlations were 0.96, 1.02, 0.98 and 0.96 for undersampling factors of 2, 2.7, 3.8 and 6.6, with R2 of 0.98, 0.98, 0.98 and 0.96, respectively. For lactate, slopes of correlations were 0.97, 0.85, 0.94 and 0.78 for undersampling factors of 2, 2.7, 3.8 and 6.6, with R2 of 0.96, 0.93, 0.95 and 0.87, respectively. The poorer lactate correlation at an undersampling factor of 6.6 led us to choose an undersampling factor of 3.8 for prospective experiments.
Figure 4.
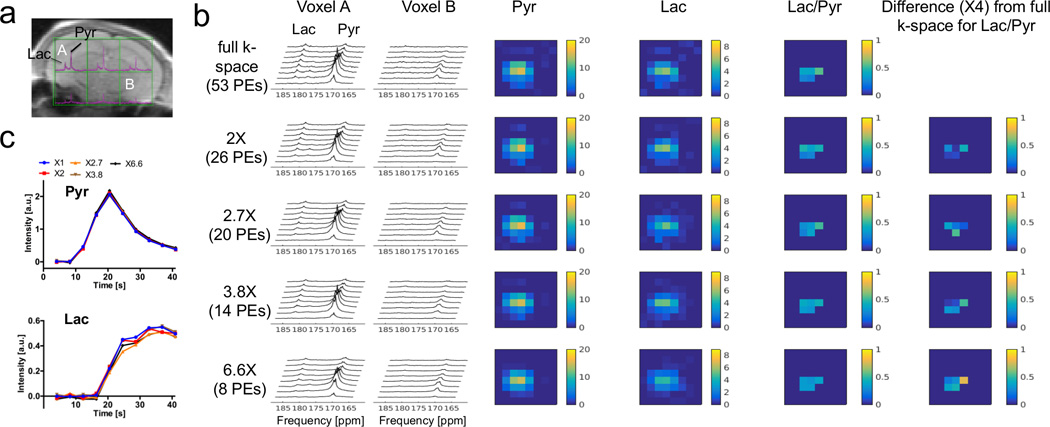
Retrospective CS experiment on a full k-space 13C mouse brain MRSI dataset following intravenous administration of hyperpolarized [1-13C] pyruvate (same as Fig. 3). The reconstructed dynamic spectra with undersampling factors of 2, 2.7, 3.8 and 6.6 were compared with full k-space ground truth. (a) Fully sampled k-space spectra are overlaid on top of a reference 1H image. Voxel A was selected as a high SNR voxel, and B was a low SNR voxel. (b) Dynamic spectra and metabolic maps with different undersampling factors. PEs stands for phase encodings. The pyruvate (Pyr) and lactate (Lac) peaks and their spatial features were well-preserved in all undersampled datasets. (c) The pyruvate and lactate time courses from the sum of six voxels on brain. CS largely preserved dynamic features as well.
Figure 5 shows the results of the prospective CS hyperpolarized [1-13C] pyruvate MRSI experiment with 3.8-fold undersampling on another normal mouse brain. Both pyruvate and lactate maps showed excellent agreement with the anatomical reference images, with metabolites primarily localized within the brain (5,9). The lactate and pyruvate also had dynamics typical of a bolus injection and metabolic conversion, and lactate was observed for over 54 s.
Figure 5.
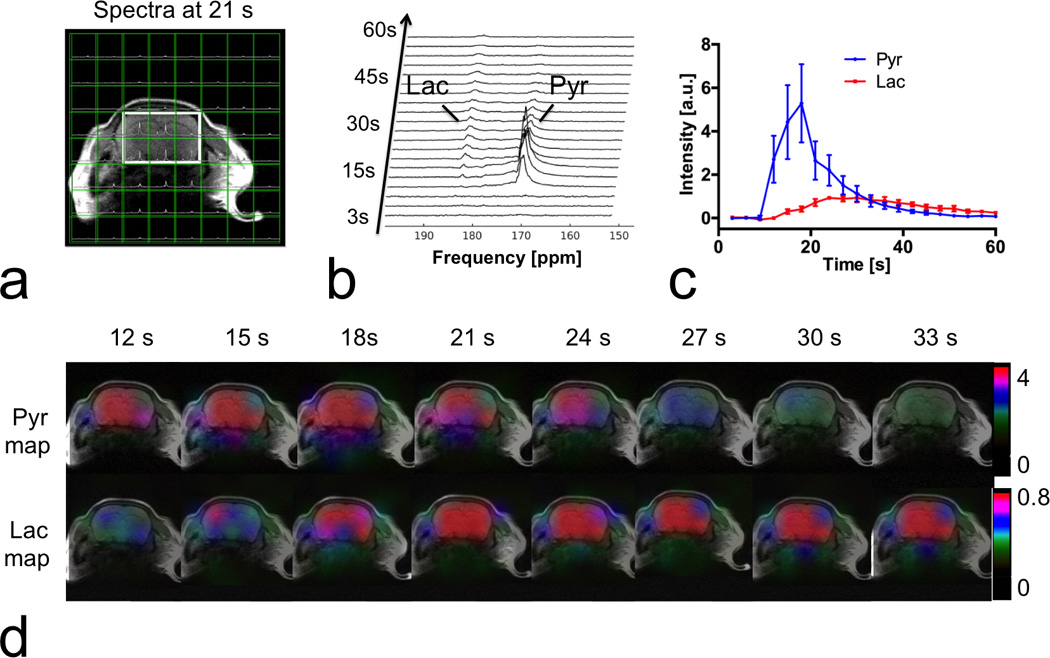
Prospectively 3.8-fold accelerated CS MRSI on mouse brain, following intravenous administration of hyperpolarized [1-13C] pyruvate. (a) A mosaic view of spectra and T2 weighted MRI. Spectra were spatially localized within the head area. (b) Dynamic spectra from the sum of six voxels marked by the solid box in (a). Lactate (Lac) and pyruvate (Pyr) peaks were clearly observed. (c) Time courses of pyruvate and lactate from the marked area. (d) The 13C metabolic maps showed pyruvate and lactate dynamics, with the majority of signal localized to the brain.
In vivo rat liver experiment
Figure 6 shows the results from the in vivo dynamic hyperpolarized [2-13C] DHAc MRSI experiment in a rat liver with a prospective acceleration factor of 3.8. Metabolites in the reconstructed spectra were spatially localized primarily within the liver. The spectra and time courses, summed among four voxels in the liver, revealed the metabolic conversion from hyperpolarized [2-13C] DHAc to G3P was primarily occurring in the rat liver. 13C resonances with a 140-ppm (or 4.5-kHz at 3T) span were effectively recovered by the proposed method.
Figure 6.
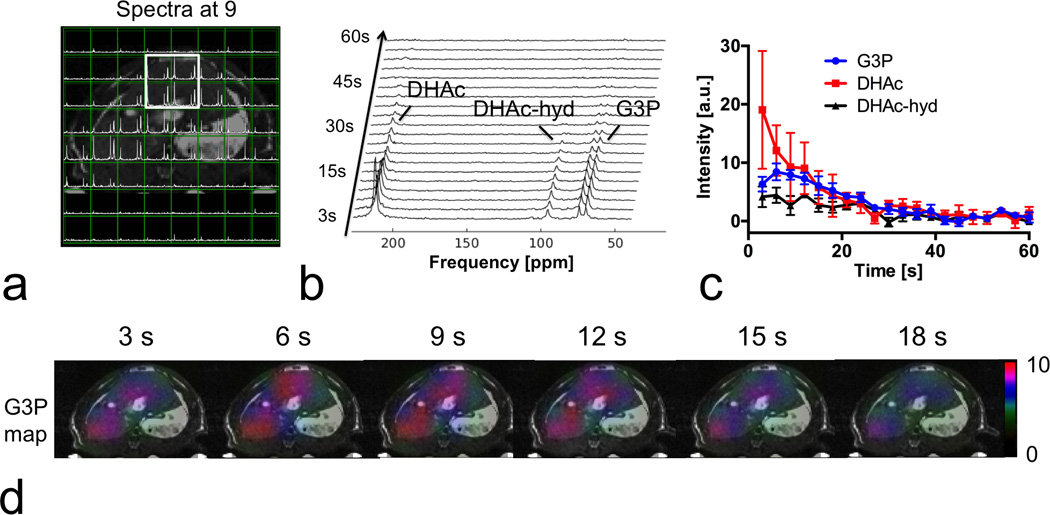
Prospectively 3.8-fold accelerated CS MRSI on rat liver, following intravenous administration of hyperpolarized [2-13C] dihydroxyacetone (DHAc), which results in a 140-ppm (4.5 kHz at 3T) range of metabolite chemical shifts. (a) A mosaic view of spectra and abdominal T2-weighted MRI. Spectra were spatially localized primarily within the liver as expected. (b) Dynamic spectra from the marked area in (a). The three peaks in the dynamic spectra are DHAc (≤0.3° flip, 213 ppm), DHAc hydrate (DHAc-hyd, 2.3° flip, 96 ppm) and glycerol 3-phosphate (G3P, 20° flip, 73 ppm), all of which were recovered by the proposed method. (c) Time courses of DHAc, DHA-hyd and G3P from the marked liver area. (d) The 13C metabolic map showed that G3P generated from DHAc was primarily distributed within the liver, indicating accurate reconstruction by our method.
Discussion
Compressed sensing MRSI with hyperpolarized 13C applications
The proposed undersampling scheme combined random blip gradients with random walk trajectories and stochastic time delays, allowing incoherent spatial and spectral undersampling for in vivo hyperpolarized 13C MRSI experiments. Meanwhile, the presented reconstruction method exploited the spatial-spectral sparsity and shared information along the dynamic dimension, where we extended the application of block-Hankel matrix completion to MRSI. The capability of the proposed method was demonstrated in the in vivo hyperpolarized 13C MRSI experiments with retrospective CS acceleration. Furthermore, in the prospective mouse brain experiment, CS was shown to preserve the spectral, spatial and dynamic characteristics of hyperpolarized [1-13C] pyruvate and lactate (which have 750-Hz spectral separation at 7T) with good sensitivity and high spectral bandwidth. Lactate and pyruvate signals were observed for over 54 s at 7T, a relatively long duration for in vivo experiments. In the prospective rat liver experiment, reconstructed spectra were spatially localized within the abdominal tissue area (G3P was mostly detected within the liver region). The recovered spectral peaks (with 4.5-kHz spectral separation at 3T) demonstrated the feasibility of CS acceleration for high spectral bandwidth MRSI in hyperpolarized 13C experiments in vivo.
The 15-mm slice thickness in the two mouse brain experiments and 20-mm slice thickness in the rat liver experiment likely caused partial volume effects, e.g. contaminations of DHAc and DHAc hydrate signals from the kidney in the rat liver experiment. In the retrospective experiment, the lactate had larger discrepancies with acceleration than pyruvate, likely due to the low SNR of [1-13C] lactate (typical in normal brain). The metabolite and ratio maps in Figure 4 also show some spatial smoothing with high acceleration factors. We used this experiment to choose the retrospective undersampling factor of 3.8, although we expect slightly better performance with prospective undersampling because throwing away data in the retrospective analysis results in SNR losses.
Calibrationless parallel imaging for compressed sensing MRSI
Figure 7 shows that the proposed CS MRSI method can be combined with the calibrationless parallel imaging reconstruction (39–41), since both are based on block-Hankel matrix completion. This only required changes to the construction of the block-Hankel matrix, where block-Hankel matrices for coils are concatenated as shown in Figure 7a. The combined CS and parallel imaging acceleration generally preserved the most of the spectral and spatial features of the 13C MRSI phantom dataset. This combination could be valuable for hyperpolarized 13C and other X-nuclear MRSI applications, where the coil calibration data and coil sensitivity maps are not available or technically difficult to acquire.
Figure 7.
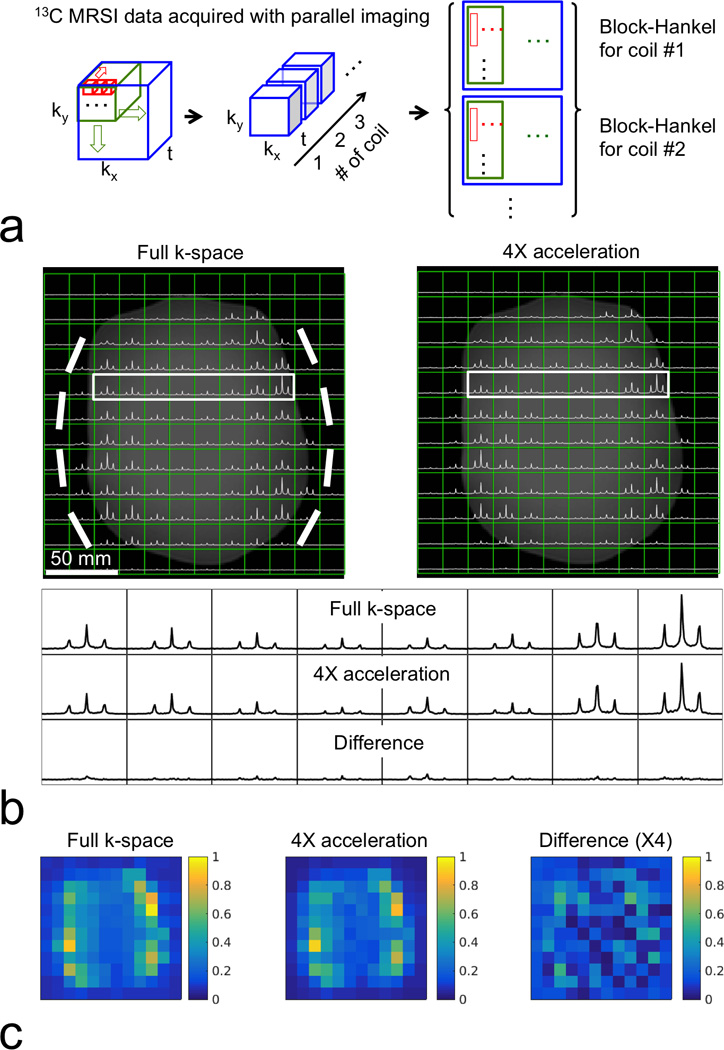
Retrospectively 4-fold accelerated 2D MRSI on a thermal 13C phantom (containing 99.8% ethylene glycol) including calibrationless parallel imaging. (a) For reconstruction incorporating calibrationless parallel imaging, Hankel matrices for FIDs (red windows) are stacked in a column-wise order, and kx-ky windows (green windows) are concatenated along each row, as done when not using parallel imaging (Fig. 2). Then, block-Hankel matrices for coils (blue windows) are concatenated along column. (b) Full k-space and CS accelerated spectra after the sum of squares coil combination. Bright bold lines indicate eight elements of the surface 13C RF receiver coil. The bottom panel shows the comparison of spectra from marked area (solid boxes). The triplet spectra of ethylene glycol (JC-H = 150 Hz) were well recovered by the combined CS and calibrationless parallel imaging, with most obvious differences occurring in the low SNR region in the middle of the phantom. (c) Integrated spectra maps comparison. The combined CS and parallel imaging acceleration generally preserved the most of spectral and spatial features of the 13C MRSI phantom dataset.
Conclusion
This study developed a CS method that uniquely combined random blip gradients with stochastic time delays, and exploited the spatial-spectral sparsity with a block-Hankel matrix completion reconstruction to accelerate high bandwidth MRSI. In retrospective hyperpolarized 13C experiments, CS preserved spectral, spatial and dynamic fidelities with up to 6.6-fold accelerations. In prospective experiments, 3.8-fold acceleration for dynamic MRSI was successfully performed in vivo in a mouse brain and a rat liver with hyperpolarized [1-13C] pyruvate and hyperpolarized [2-13C] dihydroxyacetone, respectively. These results demonstrated the feasibility of the proposed CS approach to accelerate high bandwidth MRSI in hyperpolarized 13C experiments in vivo.
Supplementary Material
Acknowledgments
The authors would like to thank Michael Lustig, John M. Pauly and Wenwen Jiang for their comments and suggestions. The authors also want to thank Lucas Carvajal for providing the raw data that was used in Figure 7. This work is supported by NIH grants R00EB012064, R01EB016741, P01CA118816, P41EB013598, R01CA154915 and R01CA172845.
Appendix
In kx-ky-t space, signal contributed by a Lorentzian spatial-spectral peak, l, at (xl, yl, fl) is given by the outer product (designated “◦”) of 4 vectors, that is
| Eq. A1 |
where d = [dl,1 ⋯ dl,u] a vector that contains the dynamic amplitudes for u frames, [−q, q] and [−a, a] the ranges of kx and ky dimensions, Al, Bl, and R2* are the decay rates (or line-broadening/blurring factors) and Δkx, Δky and Δt the minimal sampling intervals for kx, ky and t dimensions.
The 3D sliding window operation with a window size of p × b × n in kx, ky and t dimensions can apply to Sl, creating a block-Hankel matrix, Hl ∈ ℂ(2q−p+2)(2a−b+2)(m−n+1)×upbn. Hl can be written as the Kronecker product (designated “⊗”) of three Hankel matrices, i.e. Ht ∈ ℂ(m−n+1)×n, Hkx ∈ ℂ(2q−p+2)×p and Hky ∈ ℂ(2a−b+2)×b, and they are given respectively as
| Eq. A2 |
Notably in this model Hkx, Hky, and Ht are all rank-1 matrices, and rank(Hl) = rank(d)rank(Hkx)rank(Hky)rank(Ht) = 1.
References
- 1.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of >10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A. 2003;100(18):10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park I, Larson PE, Zierhut ML, Hu S, Bok R, Ozawa T, Kurhanewicz J, Vigneron DB, Vandenberg SR, James CD, Nelson SJ. Hyperpolarized 13C magnetic resonance metabolic imaging: application to brain tumors. Neuro Oncol. 2010;12(2):133–144. doi: 10.1093/neuonc/nop043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurhanewicz J, Vigneron DB, Brindle K, Chekmenev EY, Comment A, Cunningham CH, Deberardinis RJ, Green GG, Leach MO, Rajan SS, Rizi RR, Ross BD, Warren WS, Malloy CR. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia. 2011;13(2):81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno KX, Satapati S, DeBerardinis RJ, Burgess SC, Malloy CR, Merritt ME. Real-time detection of hepatic gluconeogenic and glycogenolytic states using hyperpolarized [2-13C]dihydroxyacetone. J Biol Chem. 2014;289(52):35859–35867. doi: 10.1074/jbc.M114.613265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurd RE, Yen YF, Mayer D, Chen A, Wilson D, Kohler S, Bok R, Vigneron D, Kurhanewicz J, Tropp J, Spielman D, Pfefferbaum A. Metabolic imaging in the anesthetized rat brain using hyperpolarized [1-13C] pyruvate and [1-13C] ethyl pyruvate. Magn Reson Med. 2010;63(5):1137–1143. doi: 10.1002/mrm.22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu S, Yoshihara HA, Bok R, Zhou J, Zhu M, Kurhanewicz J, Vigneron DB. Use of hyperpolarized [1-13C]pyruvate and [2-13C]pyruvate to probe the effects of the anticancer agent dichloroacetate on mitochondrial metabolism in vivo in the normal rat. Magn Reson Imaging. 2012;30(10):1367–1372. doi: 10.1016/j.mri.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JM, Josan S, Grafendorfer T, Yen YF, Hurd RE, Spielman DM, Mayer D. Measuring mitochondrial metabolism in rat brain in vivo using MR Spectroscopy of hyperpolarized [2-(1)(3)C]pyruvate. NMR Biomed. 2013;26(10):1197–1203. doi: 10.1002/nbm.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Josan S, Hurd R, Park JM, Yen YF, Watkins R, Pfefferbaum A, Spielman D, Mayer D. Dynamic metabolic imaging of hyperpolarized [2-(13) C]pyruvate using spiral chemical shift imaging with alternating spectral band excitation. Magn Reson Med. 2014;71(6):2051–2058. doi: 10.1002/mrm.24871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurd RE, Yen YF, Tropp J, Pfefferbaum A, Spielman DM, Mayer D. Cerebral dynamics and metabolism of hyperpolarized [1-(13)C]pyruvate using time-resolved MR spectroscopic imaging. J Cereb Blood Flow Metab. 2010;30(10):1734–1741. doi: 10.1038/jcbfm.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer D, Yen YF, Levin YS, Tropp J, Pfefferbaum A, Hurd RE, Spielman DM. In vivo application of sub-second spiral chemical shift imaging (CSI) to hyperpolarized 13C metabolic imaging: comparison with phase-encoded CSI. J Magn Reson. 2010;204(2):340–345. doi: 10.1016/j.jmr.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu S, Lustig M, Chen AP, Crane J, Kerr A, Kelley DA, Hurd R, Kurhanewicz J, Nelson SJ, Pauly JM, Vigneron DB. Compressed sensing for resolution enhancement of hyperpolarized 13C flyback 3D-MRSI. J Magn Reson. 2008;192(2):258–264. doi: 10.1016/j.jmr.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson PE, Kerr AB, Chen AP, Lustig MS, Zierhut ML, Hu S, Cunningham CH, Pauly JM, Kurhanewicz J, Vigneron DB. Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging. J Magn Reson. 2008;194(1):121–127. doi: 10.1016/j.jmr.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larson PE, Bok R, Kerr AB, Lustig M, Hu S, Chen AP, Nelson SJ, Pauly JM, Kurhanewicz J, Vigneron DB. Investigation of tumor hyperpolarized [1-13C]-pyruvate dynamics using time-resolved multiband RF excitation echo-planar MRSI. Magn Reson Med. 2010;63(3):582–591. doi: 10.1002/mrm.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larson PE, Hu S, Lustig M, Kerr AB, Nelson SJ, Kurhanewicz J, Pauly JM, Vigneron DB. Fast dynamic 3D MR spectroscopic imaging with compressed sensing and multiband excitation pulses for hyperpolarized 13C studies. Magn Reson Med. 2011;65(3):610–619. doi: 10.1002/mrm.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer D, Levin YS, Hurd RE, Glover GH, Spielman DM. Fast metabolic imaging of systems with sparse spectra: application for hyperpolarized 13C imaging. Magn Reson Med. 2006;56(4):932–937. doi: 10.1002/mrm.21025. [DOI] [PubMed] [Google Scholar]
- 16.Jiang W, Lustig M, Larson PE. Concentric rings K-space trajectory for hyperpolarized (13) C MR spectroscopic imaging. Magn Reson Med. 2016;75(1):19–31. doi: 10.1002/mrm.25577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon JW, Niles DJ, Fain SB, Johnson KM. Joint spatial-spectral reconstruction and k-t spirals for accelerated 2D spatial/1D spectral imaging of 13C dynamics. Magn Reson Med. 2014;71(4):1435–1445. doi: 10.1002/mrm.24796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiens CN, Friesen-Waldner LJ, Wade TP, Sinclair KJ, McKenzie CA. Chemical shift encoded imaging of hyperpolarized (13) C pyruvate. Magn Reson Med. 2015;74(6):1682–1689. doi: 10.1002/mrm.25532. [DOI] [PubMed] [Google Scholar]
- 19.Wiesinger F, Weidl E, Menzel MI, Janich MA, Khegai O, Glaser SJ, Haase A, Schwaiger M, Schulte RF. IDEAL spiral CSI for dynamic metabolic MR imaging of hyperpolarized [1-13C]pyruvate. Magn Reson Med. 2012;68(1):8–16. doi: 10.1002/mrm.23212. [DOI] [PubMed] [Google Scholar]
- 20.Posse S, Tedeschi G, Risinger R, Ogg R, Le Bihan D. High speed 1H spectroscopic imaging in human brain by echo planar spatial-spectral encoding. Magn Reson Med. 1995;33(1):34–40. doi: 10.1002/mrm.1910330106. [DOI] [PubMed] [Google Scholar]
- 21.Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, Spielman DM. Volumetric spectroscopic imaging with spiral-based k-space trajectories. Magn Reson Med. 1998;39(6):889–898. doi: 10.1002/mrm.1910390606. [DOI] [PubMed] [Google Scholar]
- 22.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 23.Geethanath S, Baek HM, Ganji SK, Ding Y, Maher EA, Sims RD, Choi C, Lewis MA, Kodibagkar VD. Compressive sensing could accelerate 1H MR metabolic imaging in the clinic. Radiology. 2012;262(3):985–994. doi: 10.1148/radiol.11111098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao P, Wu EX. Accelerating phase-encoded proton MR spectroscopic imaging by compressed sensing. J Magn Reson Imaging. 2015;41(2):487–495. doi: 10.1002/jmri.24553. [DOI] [PubMed] [Google Scholar]
- 25.Furuyama JK, Wilson NE, Burns BL, Nagarajan R, Margolis DJ, Thomas MA. Application of compressed sensing to multidimensional spectroscopic imaging in human prostate. Magn Reson Med. 2012;67(6):1499–1505. doi: 10.1002/mrm.24265. [DOI] [PubMed] [Google Scholar]
- 26.Nagarajan R, Iqbal Z, Burns B, Wilson NE, Sarma MK, Margolis DA, Reiter RE, Raman SS, Thomas MA. Accelerated echo planar J-resolved spectroscopic imaging in prostate cancer: a pilot validation of non-linear reconstruction using total variation and maximum entropy. NMR Biomed. 2015;28(11):1366–1373. doi: 10.1002/nbm.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarma MK, Nagarajan R, Macey PM, Kumar R, Villablanca JP, Furuyama J, Thomas MA. Accelerated echo-planar J-resolved spectroscopic imaging in the human brain using compressed sensing: a pilot validation in obstructive sleep apnea. AJNR Am J Neuroradiol. 2014;35(6 Suppl):S81–S89. doi: 10.3174/ajnr.A3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson NE, Iqbal Z, Burns BL, Keller M, Thomas MA. Accelerated five-dimensional echo planar J-resolved spectroscopic imaging: Implementation and pilot validation in human brain. Magn Reson Med. 2016;75(1):42–51. doi: 10.1002/mrm.25605. [DOI] [PubMed] [Google Scholar]
- 29.Chatnuntawech I, Gagoski B, Bilgic B, Cauley SF, Setsompop K, Adalsteinsson E. Accelerated H MRSI using randomly undersampled spiral-based k-space trajectories. Magn Reson Med. 2014 doi: 10.1002/mrm.25394. [DOI] [PubMed] [Google Scholar]
- 30.Maguire ML, Geethanath S, Lygate CA, Kodibagkar VD, Schneider JE. Compressed sensing to accelerate magnetic resonance spectroscopic imaging: evaluation and application to 23Na-imaging of mouse hearts. J Cardiovasc Magn Reson. 2015;17:45. doi: 10.1186/s12968-015-0149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Askin NC, Atis B, Ozturk-Isik E. Accelerated phosphorus magnetic resonance spectroscopic imaging using compressed sensing. Conf Proc IEEE Eng Med Biol Soc. 2012;2012:1106–1109. doi: 10.1109/EMBC.2012.6346128. [DOI] [PubMed] [Google Scholar]
- 32.Kampf T, Fischer A, Basse-Lusebrink TC, Ladewig G, Breuer F, Stoll G, Jakob PM, Bauer WR. Application of compressed sensing to in vivo 3D (1)(9)F CSI. J Magn Reson. 2010;207(2):262–273. doi: 10.1016/j.jmr.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Burns BL, Wilson NE, Thomas MA. Group sparse reconstruction of multi-dimensional spectroscopic imaging in human brain in vivo. Algorithms. 2014;7(3):276–294. [Google Scholar]
- 34.Cai J-F, Qu X, Xu W, Ye G-B. Robust recovery of complex exponential signals from random Gaussian projections via low rank Hankel matrix reconstruction. 2015 doi: 10.1016/j.acha.2016.02.003. arXiv preprint arXiv:150302893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu X, Mayzel M, Cai JF, Chen Z, Orekhov V. Accelerated NMR spectroscopy with low-rank reconstruction. Angew Chem Int Ed Engl. 2015;54(3):852–854. doi: 10.1002/anie.201409291. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Chi Y. Robust spectral compressed sensing via structured matrix completion. Information Theory, IEEE Transactions on. 2014;60(10):6576–6601. [Google Scholar]
- 37.Otazo R, Candes E, Sodickson DK. Low-rank plus sparse matrix decomposition for accelerated dynamic MRI with separation of background and dynamic components. Magn Reson Med. 2015;73(3):1125–1136. doi: 10.1002/mrm.25240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daňková M, Rajmic P, Jiřík R. Acceleration of Perfusion MRI Using Locally Low-Rank Plus Sparse Model. 12th International Conference, LVA/ICA 2015; Proceedings; August 25–28, 2015; Liberec, Czech Republic. 2015. pp. 514–521. [Google Scholar]
- 39.Shin PJ, Larson PE, Ohliger MA, Elad M, Pauly JM, Vigneron DB, Lustig M. Calibrationless parallel imaging reconstruction based on structured low-rank matrix completion. Magn Reson Med. 2014;72(4):959–970. doi: 10.1002/mrm.24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Y, Gordon JW, Shin PJ, von Morze C, Lustig M, Larson PE, Ohliger MA, Carvajal L, Tropp J, Pauly JM, Vigneron DB. Development and testing of hyperpolarized (13)C MR calibrationless parallel imaging. J Magn Reson. 2016;262:1–7. doi: 10.1016/j.jmr.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haldar JP, Zhuo J. P-LORAKS: Low-rank modeling of local k-space neighborhoods with parallel imaging data. Magn Reson Med. 2015 doi: 10.1002/mrm.25717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai JF, Candes EJ, Shen ZW. A Singular Value Thresholding Algorithm for Matrix Completion. Siam Journal on Optimization. 2010;20(4):1956–1982. [Google Scholar]
- 43.Halko N, Martinsson PG, Tropp JA. Finding Structure with Randomness: Probabilistic Algorithms for Constructing Approximate Matrix Decompositions. Siam Review. 2011;53(2):217–288. [Google Scholar]
- 44.Marco-Rius I, Cao P, von Morze C, Merrit M, Moreno KX, Chang G-Y, Ohliger MA, Pearce D, Kurhanewicz J, Larson PE, Vigneron D. Multiband Spectral-Spatial RF Excitation for Hyperpolarized [2 13C]Dihydroxyacetone 13C-MR Metabolism Studies. Magn Reson Med. 2016 doi: 10.1002/mrm.26226. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park I, Larson PE, Tropp JL, Carvajal L, Reed G, Bok R, Robb F, Bringas J, Kells A, Pivirotto P, Bankiewicz K, Vigneron DB, Nelson SJ. Dynamic hyperpolarized carbon-13 MR metabolic imaging of nonhuman primate brain. Magn Reson Med. 2014;71(1):19–25. doi: 10.1002/mrm.25003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


