Abstract
Rice contaminated with fungal species during storage is not only of poor quality and low economic value, but may also have harmful effects on human and animal health. The predominant fungal species isolated from rice grains during storage belong to the genera Aspergillus and Penicillium. Some of these fungal species produce mycotoxins; they are responsible for adverse health effects in humans and animals, particularly Aspergillus flavus, which produces the extremely carcinogenic aflatoxins. Not surprisingly, there have been numerous attempts to devise safety procedure for the control of such harmful fungi and production of mycotoxins, including aflatoxins. This review provides information about fungal and mycotoxin contamination of stored rice grains, and microbe-based (biological) strategies to control grain fungi and mycotoxins. The latter will include information regarding attempts undertaken for mycotoxin (especially aflatoxin) bio-detoxification and microbial interference with the aflatoxin-biosynthetic pathway in the toxin-producing fungi.
Keywords: Aflatoxin, Biodegradation, Biological control, Grain mold, Mycotoxin, Rice grain
Rice (Oryza sativa L.) is one of the most important food crops in the world. It is an intrinsic part of the Asian cultures, in which some people of Indonesia believed that it had a soul like humans. In Japan, rice was considered the food of gods and samurais, and individuals who squandered even a handful of rice were punished. Although these beliefs have changed over time, people still highly value rice, particularly in Asia [1]. Rice represents the primary food source for almost half of the world's population, providing 20% of the total human dietary energy supply (Food and Agriculture Organization [FAO] of the United Nations, http://faostat.fao.org/). In Korea, rice is the major cereal; the total area devoted to rice was 799,344 ha and total rice production was 4,327 thousand tons in 2015. Moreover, the annual consumption of rice is 62.9 kg/capita, higher than that of any other food crop. While the amount of harvested rice is increasing annually, consumer intake has gradually declined, decreasing by half between 1985 and 2015 (Statistics Korea, http://kostat.go.kr/portal/eng/). As a result, harvested rice is stored for increasingly longer periods, which alters its physical, chemical, and biological properties [2]. During storage, rice is prone to contamination by various fungal species that can reduce its quality and market value. Therefore, this review will cover the following topics: (1) previous studies on fungal genera and species frequently detected in stored rice grains, highlighting the adverse effects on the grains as a source of mycotoxins; (2) previously reported microbe-based (biological) methods to control fungal contamination during storage, focusing on mycotoxigenic fungi, especially Aspergillus flavus producing aflatoxins as the most serious mycotoxin; (3) possible microbial degradation of mycotoxins, especially aflatoxins, and suppression of aflatoxin biosynthesis in fungi via microbial interference; and (4) concluding remarks and suggestions on future studies.
FUNGAL SPECIES IN STORED RICE GRAINS
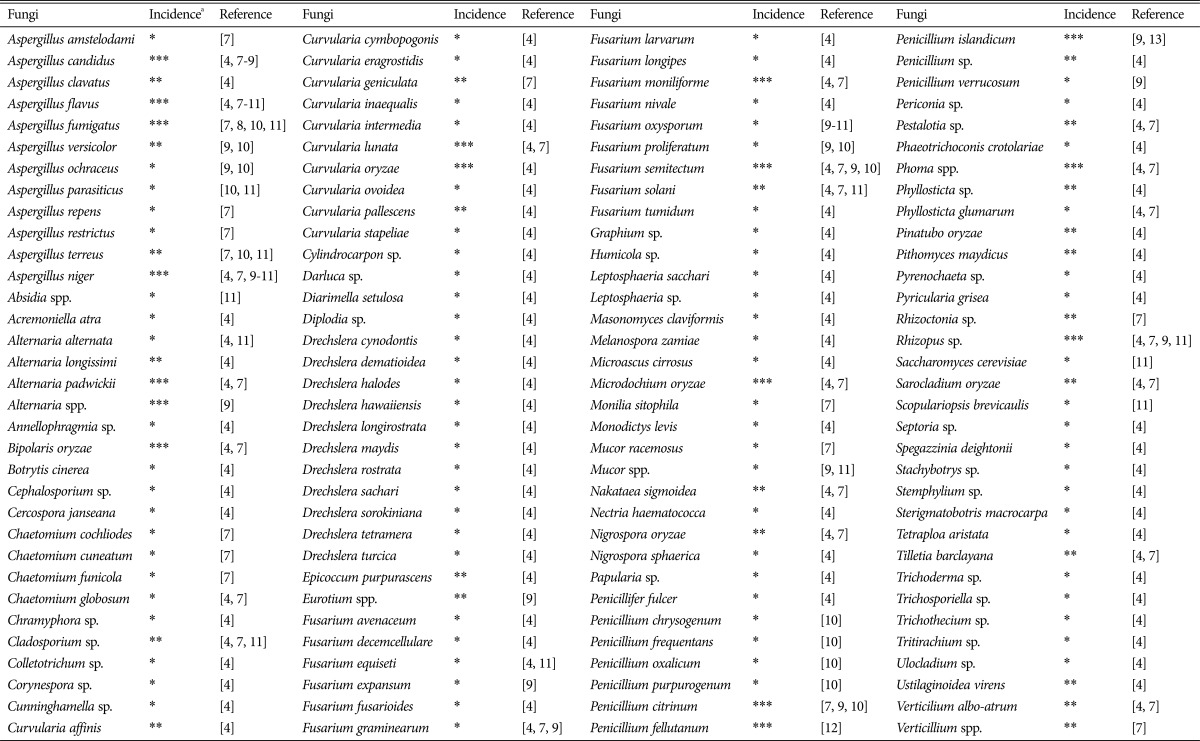
Fungal contamination of rice grains, a good substrate for storage fungi, starts in the field before harvest. The high relative humidity drops as the seeds enter the storage period, causing changes in the diversity and population of the fungi and the predominant fungal species [3]. In accordance with the rules of International Seed Testing Association, more than 500,000 seed lots have been tested by the International Rice Research Institute (IRRI) seed health unit and the fungal species detected were reported [4]. In general, rice is susceptible to a wide range of contaminating fungi, the growth of which is associated with grain spoilage, off-flavors, toxins, discoloration, and the production of harmful propagules [5]. The main reason for grain spoilage and tissue decomposition is the production of enzymes, such as lipases, proteases, carbohydrases, and volatile compounds (e.g., dimethyl disulfide, geosmin, and 2-methylisoborneol) by storage fungi [6]. As summarized from previous studies, fungal contamination and occurrence in rice grains [4,7,8,9,10,11,12,13] are listed in Table 1.
Table 1. A list of previously reported fungal species associated with rice grains.
a*, **, and *** indicate low, moderate, and frequent as concluded based on frequency of isolation in at least one of the mentioned report, respectively.
Predominant Aspergillus and Penicillium species
As mentioned above, low relative humidity during rice storage influences the structure of fungal communities, favoring dry condition-tolerant fungi. The most frequently detected fungal genera in stored rice are Aspergillus and Penicillium, both of which are relatively tolerant to storage conditions.
Previously, Oh et al. [2] evaluated the occurrence of fungi and bacteria on rice grains stored in the rice-processing complexes of the National Agricultural Cooperative Federation in Korea. They reported that Aspergillus and Penicillium were the predominant fungal genera, in spite of the high fungal diversity between regions. In a later assessment of microbial populations and aflatoxin contamination, they identified three predominant Aspergillus species, which were A. flavus, Aspergillus candidus, and Aspergillus fumigatus [8]. These findings were consistent with other studies, which identified Aspergillus, Penicillium, and Fusarium as the predominant genera [14], and A. candidus and A. flavus as the predominant species [9].
Aspergillus spp. are ubiquitous in stored food, particularly in cereals. Among the most frequent and important species of rice-infecting fungi is A. flavus, which is widespread across the world. This species normally exists as a saprophyte in the soil, where it associates with a variety of decaying organic matter [15]. A. flavus can tolerate a wide range of temperatures and a low level of relative humidity; this specific feature enables the fungus to efficiently colonize the surface of stored rice grains. Along with rice, it can infect several other crops, such as peanuts, maize, and tree nuts. Contamination with A. flavus is very serious because of the presence of aflatoxin, a secondary metabolite that was first isolated from the fungus [16]. Aflatoxin is the most harmful mycotoxin and causes various adverse health effects that will be discussed in the next section. A. candidus, belonging to the subgenus Circumdati, is characterized by slow growth and white conidia. This fungus is a common contaminant of stored grains and can cause respiratory problems in individuals exposed to dust from the contaminated grains [17]. In contrast, A. fumigatus belongs to the subgenus Fumigate and is known for its fast growing, turquoise or dark-green colonies [18]. Abad et al. [19] stated that though A. fumigatus does not infect humans, inhaled conidia can give rise to aspergillosis in immune-compromised individuals with altered or weakened immune responses. In the review [19], they also mentioned factors that made A. fumigatus a successful pathogen, as well as the genes and molecules involved in causing the disease.
Compared with the genus Aspergillus, Penicillium (the second most important fungal group in stored rice) is a more diverse genus in terms of both species number and habitat range [20]. Penicillium spp. usually have bluish or greenish gray-colored colonies and a closed texture [21]. Members of the genus Penicillium are known as serious grain contaminants owing to their production of mycotoxins (e.g., ochratoxin, patulin, and citrinin) and related fungal metabolites (e.g., alkaloids, amino acids, anthraquinone, and azaphilone) [22,23]. It is difficult to identify a specific Penicillium species due to their morphological similarities, which might have led to several misidentifications, particularly, of the ochratoxin-producing species. Recently, Penicillium verrucosum (the main source of ochratoxin contamination of cereals) and Penicillium nordicum have been accepted as ochratoxin-producing species of the genus Penicillium [24]. In Korea, two Penicillium species, Penicillium islandicum and Penicillium fellutanum, were found to be abundant in stored rice [12,13]. P. islandicum has been detected in imported and polished rice (15/45 samples) in Korea, harvested rice in Japan (3/100 samples), and paddy and milled rice in Argentina and Paraguay [9,13,25]. Besides, P. fellutanum is known to produce ergot alkaloids (agroclavine 1 and epoxyagroclavine 1) and diketopiperazine alkaloids (fellutanine A, B, C, and D) [22,26,27].
MYCOTOXINS ASSOCIATED WITH STORED RICE GRAINS
Mycotoxins are chemically diverse, toxic secondary metabolites produced by certain fungi, mostly those from the genera Aspergillus, Fusarium, and Penicillium. They contaminate food sources and have several toxicological effects on humans and animals when consumed directly or indirectly [28]. The earliest report on the impact of mycotoxins on human health was related to ergot-poisoning in Europe, which resulted in thousands of people suffering from severe symptoms, and also leading to death. The disease was named "Holy fire" or "St. Anthony's fire" but is known today as ergotism and is caused by the consumption of ergot-infected cereals [16]. The toxicity of the ergot-infected products is related to ergot alkaloids. These ergot alkaloids were originally named after their first known source, the sclerotia (ergot) of the fungus Claviceps purpurea. The ergot alkaloids have various toxic effects, such as painful spasms, diarrhea, nausea, headache, or gangrenous symptoms in the fingers and toes. However, the alkaloids have potential, biological functions, such as anti-herbivory defense and pharmaceutical applications, due to their structural similarities with three neurotransmitters, which can bind serotonin, dopamine, and adrenergic receptors [29,30]. Another famous early outbreak of mycotoxicosis was associated with the consumption of contaminated, discolored yellow rice in Japan in the early 19th century, which was later attributed to a toxigenic entity [31].
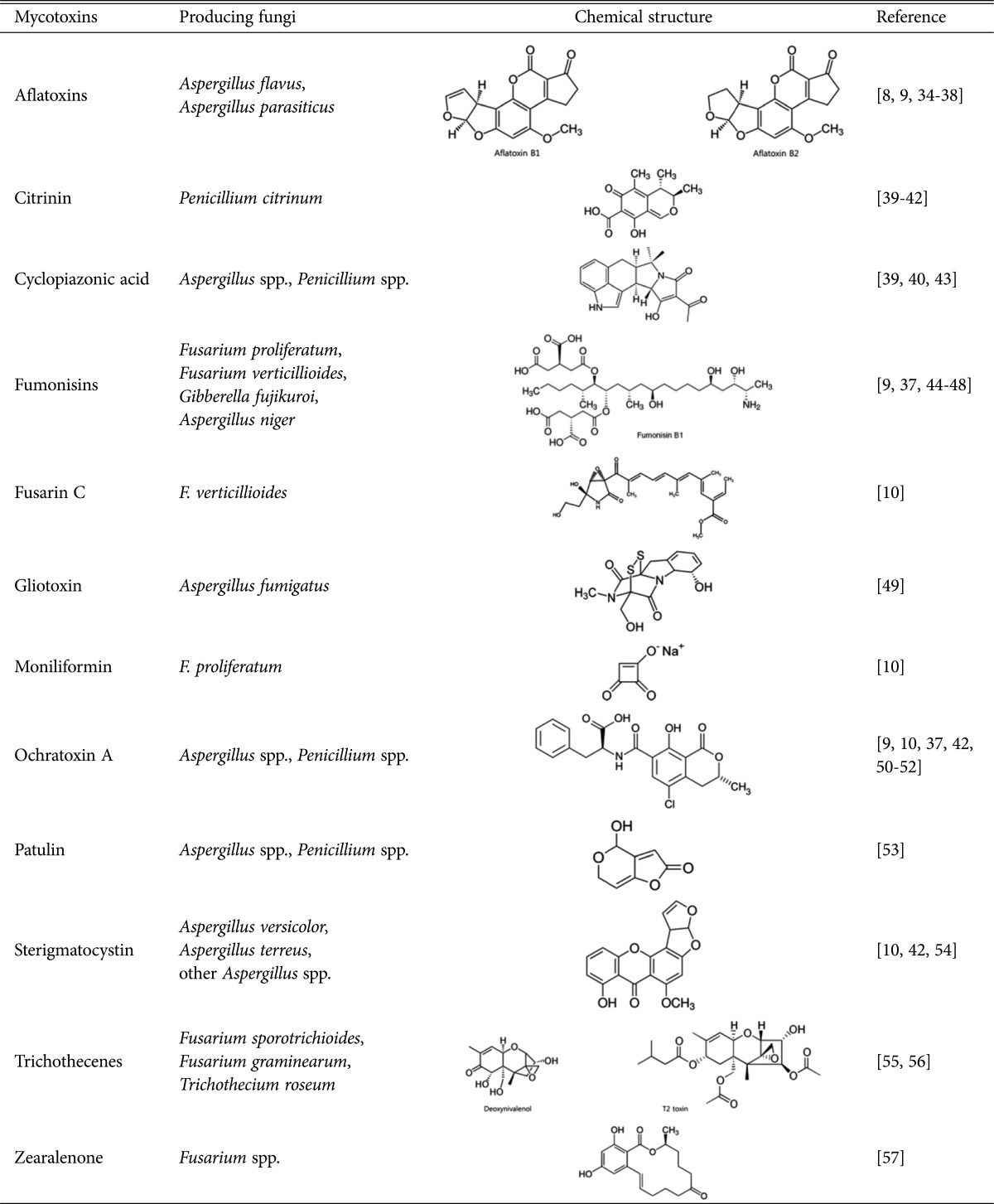
Rice, along with other cereal crops, is susceptible to contamination by a wide range of mycotoxin-producing filamentous fungi, which usually infect crops in the field and continue in storage facilities. In addition, rice is a good substrate for mycotoxin biosynthesis by grain fungi such as P. verrucosum [32] and Fusarium subglutinans [33]. For example, P. verrucosum was known to be related with ochratoxin A contamination in rice, which was reported as the most commonly detected mycotoxin with levels above the tolerated limits in Korean rice [9]. Several mycotoxins, including aflatoxins [8,9,34,35,36,37,38], citrinin [39,40,41,42], cyclopiazonic acid [39,40,43], fumonisins [9,37,44,45,46,47,48], fusarin C [10], gliotoxin [49], moniliformin [10], ochratoxin A [9,10,37,42,50,51,52], patulin [53], sterigmatocystin [10,42,54], trichothecenes [55,56], and zearalenone [57], have been found in rice grains. Rice-associated mycotoxins and their related fungi reported in previous studies are summarized in Table 2.
Table 2. Mycotoxins and their related fungi associated with rice grains.
Aflatoxins
Aflatoxins include 14 different types of fungal secondary metabolites with similar structures. In particular, among the naturally produced aflatoxins are B1, B2, G1, and G2; the names B and G are derived from the blue and green fluorescent colors, which are produced under UV light on thin layer chromatography plates [58]. Aflatoxins were first discovered after the mass deaths of turkeys due to liver disease in England in 1960, which was named turkey "X" disease since the cause was unknown. It was later revealed that the affected turkeys had been fed on aflatoxin-contaminated feed [59].
The most toxic aflatoxin B1 is exclusively produced by A. flavus and Aspergillus parasiticus. This toxin has been shown to have mutagenic, immunosuppressive, and teratogenic properties [60,61,62]. The relationship between aflatoxin B1 and liver cancer has been supported by epidemiological studies showing that regions with high aflatoxin dietary intake values had significantly higher incidence of liver cancer [63]. About 4.5 billion people are exposed to uncontrolled amounts of aflatoxins in developing countries, where poor cereal storage conditions promote aflatoxigenic fungi, leading to higher levels of aflatoxins in cereals. Due to concerns about human and animal health, regulatory limitations were issued on the quantity of aflatoxin allowed in food and animal feed in several countries. According to the FAO of the United Nations, the limits vary according to the commodity and the country. For example, in Europe, the limits range between 2~12 µg/kg for B1 and 4~15 µg/kg for total aflatoxins. In the United States, food safety regulations include a limit of 20 µg/kg for total aflatoxins in all food [64]. For these reasons, elimination of aflatoxin in food is crucial. Several attempts to eliminate or reduce aflatoxin contamination on food have been made; however, the most effective way to control this will be achieved by directly reducing or limiting exposure to the toxin-producing fungal species using traditional methods (e.g., physical separation, heat, or radiation treatments) and commercial fungicides.
CONTROL OF FUNGAL AND MYCOTOXIN CONTAMINATION IN STORED RICE GRAINS
Conventional control of grain fungi
The previous section introduced the predominant rice-storage fungal species and the hazards associated with their contamination of rice. This was analyzed in terms of the direct fungal effect on grain quality, and the impact on the environment and human health (e.g., due to mycotoxins). Several attempts have been made to control such important fungi and minimize the loss of quality and quantity of rice. Actions have included the use of host resistance, traditional practices, and chemical application to reduce the populations of fungal pathogens from field to storage [65]. According to the IRRI fact sheet on the management of storage fungi (available from http://irri.org/), several factors affect the populations of storage fungi on harvested rice grains. These include the temperature and moisture content of the stored grain, the condition of the grains before storage, storage period, and insect or mite activity in the grains. Contaminated grains are often treated by physical and chemical means, such as use of Dithane M-45 and Benlate (benomyl) fungicides. Benlate was reported to inhibit a broad spectrum of fungi, to be relatively non-toxic to plant cells and protoplasts, and to prevent fungal contamination [66]. However, increasing concerns about the risks of chemical control methods, limitations on their use against plant diseases, particularly at postharvest stage (unwanted health and environmental issues and the possible emergence of resistant strains) have sparked a search for safer alternatives to control and manage storage fungi. For example, waste fruit peels were investigated for their ability to inhibit A. flavus on rice; they were suggested as a safe method for reducing aflatoxin levels on rice grains for up to four months [67]. These control methods could be used for rice storage fungi; however, there is still a need to develop safe and environmentally sound measures to control rice-storage and aflatoxigenic fungi, possibly by using microbial antagonists as will be discussed in the following section.
Microbe-mediated control of grain fungi producing mycotoxins, including aflatoxins
The antifungal properties of beneficial microorganisms, including bacteria have been known since the 1930s. Extensive efforts have been made to use them for disease control as an alternative to chemical fungicides [68]. The best way for selecting potential biocontrol agents is to screen the diverse communities of non-pathogenic microorganisms that are adapted to the same environment. Such microorganisms are considered to be a large untapped source of protection against plant pathogens [69]. In rice, potential, antagonistic bacteria have been reported on seeds [70], which could be employed as biological control agents against plant pathogens [65]. Pseudomonas fluorescens and Pseudomonas aeruginosa have been described as effective bacterial agents against Rhizoctonia solani, which causes sheath blight [71]. These biocontrol studies generally focused on the field rice diseases; however, not many studies were conducted against storage fungal pathogens. In fact, research on the biocontrol of rice storage fungi should consider the microbial population colonizing the grains as a source of effective biocontrol agents. Adaptability to and survival on rice grains should be considered for the effectiveness of antagonistic bacteria from sources other than rice grains.
Among the bacteria with biocontrol activity reported to date, the genus Bacillus, one of the most studied and common biocontrol agents, is well characterized by its plant growth-promoting traits and its ability to protect against several plant pathogens by production of wide array of compounds, including antifungal peptides [72,73]. A marine strain of Bacillus megaterium was shown to have biocontrol activity against A. flavus on peanut kernels [74]. In that study, the bacterial treatment reduced the disease incidence on inoculated kernels, and also inhibited the biosynthesis of aflatoxins in a liquid medium, which was due to the inhibition of aflatoxin biosynthesis-regulatory genes (aflR and aflS) expression. This result was obtained by quantitative real-time polymerase chain reaction (qRT-PCR) analysis of the relative mRNA abundance of the A. flavus genes when co-cultured in the medium with the bacterium. Furthermore, the result was supported by a recent study [75], in which a mechanism of action involving the bacterial interference with aflatoxin biosynthesis was proposed; this will be discussed later. In another study, culture filtrates of biocontrol agents, such as Trichoderma spp., different strains of P. fluorescens, a strain of B. subtilis, and Rhodococcus erythropolis, showed considerable biocontrol activities against A. flavus and limited the production of aflatoxins. In particular, among the tested microbes, R. erythropolis completely inhibited the growth of A. flavus and decreased aflatoxin production [76].
On the other hand, for the biocontrol of Penicillium spp., a few studies showed biocontrol activity on stored rice grains. Among them, a preliminary study for screening of 460 bacterial strains from Korean stored rice grains for their biological activity against Aspergillus and Penicillium spp. resulted in selection of several potential biocontrol candidates [77]. In another study, specific formulations of Pichia anomala exhibited biocontrol activity against P. verrucosum, which was capable of reducing the fungal growth as well as ochratoxin A production on moist grains [78]. In this section, the microbial effect on mycotoxigenic fungi was discussed; studies reporting the direct and microbial degradation of mycotoxins, including aflatoxin, will be discussed in the following section.
Microbial degradation of mycotoxins, including aflatoxins
Microbial degradation represents a promising method for management of mycotoxins contaminating grains and feed. Intensive researches have been conducted to find efficient and safe mycotoxin-detoxifying agents. Previously, rumen microbes could metabolize ochratoxin A, zearalenone, T-2 toxin, and deacetoxyscipenol [79]. Recently, several soil bacteria showed ochratoxin A degradation property; among them, Acinetobacter calcoaceticus strain 396.1 and Acinetobacter sp. strain neg1 showed consistently the toxin-degradation activity [80]. Similarly, several bacterial strains (belonging to the genera Anaerofilum, Collinsella, and Bacillus) capable of detoxification of deoxynivalenol were isolated and identified using polymerase chain reaction-denaturing gradient gel electrophoresis [81]. Besides, the extracellular extracts of Acinetobacter sp. exhibited biodegradation ability of zearalenone and this bacterium was able to grow rapidly on zearalenone as sole carbon source [82].
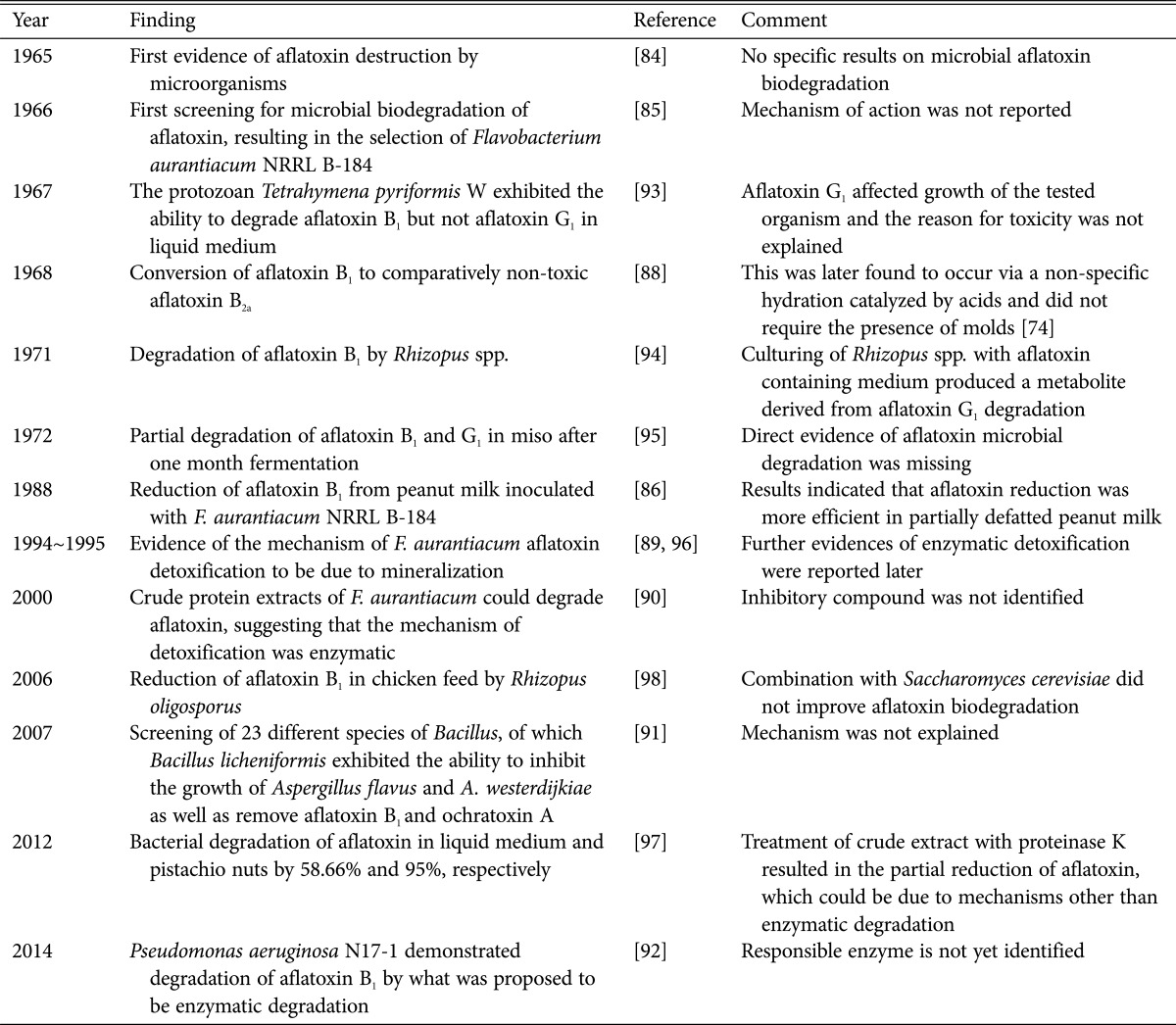
For aflatoxin detoxification, physical, chemical, and biological methods have been proposed, depending on the nature of the product being treated [83]. Several microbial agents have been shown to effectively degrade aflatoxins under laboratory conditions. The first report showing the possible microbial degradation of aflatoxins came in 1965, paving the way for more sophisticated screenings of microbes with similar properties [84]. Ciegler et al. [85] screened 1,000 different yeasts, molds, mold spores, actinomycetes, bacteria, and algae, of which only Flavobacterium aurantiacum NRRL B-184 could degrade aflatoxins in vitro. Other studies described a similar role for F. aurantiacum in irreversibly clearing aflatoxins from different foods (e.g., milk, oil, peanut butter, peanuts, and corn) [85,86,87], yet the underlying mechanism remained unexplained. Another significant observation by Ciegler and Peterson [88] revealed that aflatoxin B1 could be converted to the relatively non-toxic aflatoxin B2a by acid-producing molds. It was later found that the conversion was a nonspecific hydration catalyzed by acids and did not require the presence of molds [83]. Since at least part of the aflatoxin was metabolized to water-soluble degradation products, mineralization was proposed as the mechanism of aflatoxin detoxification by F. aurantiacum [89]. More recently, crude protein extracts from F. aurantiacum have been shown to successfully degrade the aflatoxin, suggesting that the mechanism of detoxification may be enzymatic [90]. In other studies [91], 23 different species of Bacillus were screened and B. licheniformis was identified as capable of inhibiting the growth of A. flavus and Aspergillus westerdijkiae, as well as degrading aflatoxin B1 and ochratoxin A. Recently, P. aeruginosa N17-1 was also shown to degrade aflatoxin B1, possibly through enzymatic degradation [92]. These findings demonstrated that antagonistic bacteria do not only inhibit the growth of deleterious storage fungal species, but can also degrade the mycotoxins present in food. Thus, the bacteria could be utilized as sources of efficient enzymes for mycotoxin degradation in food. The microbial degradation of aflatoxins is not the only way to directly reduce the toxins; other studies have indicated the ability of some bacteria to interfere with the aflatoxin-biosynthetic pathway in the toxin-producing fungi. A historical review [84,85,86,88,89,90,91,92,93,94,95,96,97,98] of microbial aflatoxin degradation is summarized in Table 3.
Table 3. Historical reviews on the biological degradation of aflatoxins.
Microbial interference in the aflatoxin-biosynthetic pathway
The discovery of the aflatoxin precursor, norsolorinic acid [99], characterization of the biosynthetic and regulatory genes, and the complete sequence of aflatoxin genes in A. parasiticus promoted interest in the aflatoxin-biosynthetic pathway [100]. Consequently, the biosynthetic pathway and the roles of different genes governing aflatoxin production are well understood and it is one of the best-characterized fungal metabolites [101]. Such understanding paved the way for further studies on the mechanisms of aflatoxin biosynthesis suppression in A. flavus and A. parasiticus, by co-cultivation with other microorganisms or physical factors such as water activity and temperature [75,102].
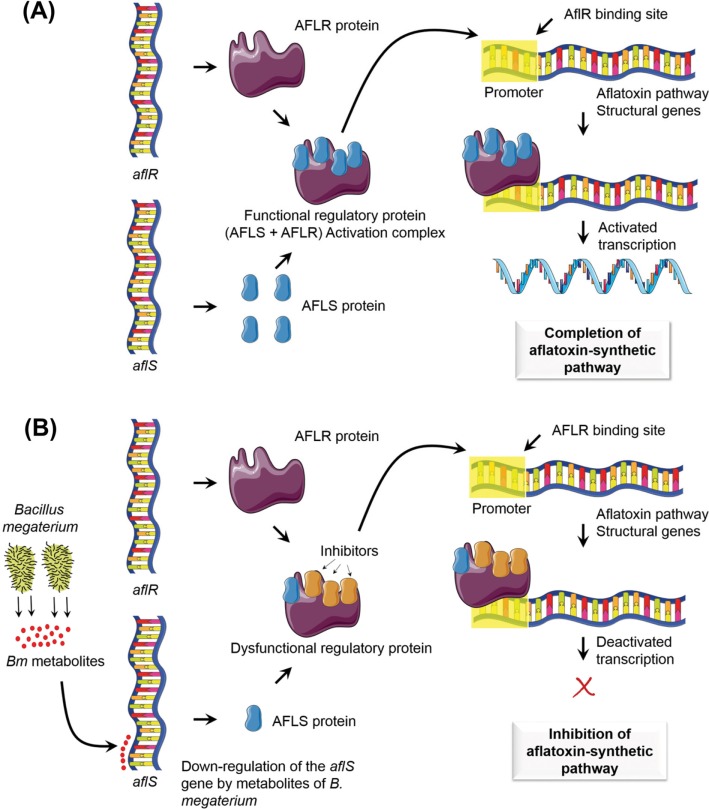
Since this review focuses the use of microbe-based approaches for aflatoxin suppression, specific details of aflatoxin biosynthesis will not be covered. However, to better understand how microbe-related inhibition of aflatoxin occurs, the aflatoxin-biosynthetic processes and the related genes will be discussed. To this end, there are interesting studies on the possible mechanisms responsible for the down-regulation of the transcription-activator genes aflS (formerly aflJ) and aflR [75,103] in Aspergillus spp. Changes in the aflR/aflS-expression ratio in response to temperature and water activity have also been reported [102]. The gene aflR encodes a sequence-specific DNA-binding transcription activator (AFLR) that regulates aflatoxin biosynthetic genes, while the accessory regulatory gene aflS encodes a transcription co-activator (AFLS) [104,105]. Both genes were reported as essential for aflatoxin production. Meyers et al. [106] reported that disrupting aflS prevented A. flavus from converting intermediates of the pathway to aflatoxins. According to Chang [104], the role of AFLS was to support the binding of AFLR to DNA, thereby activating the transcription of aflatoxin structural genes, and the lack of this interaction, prevented aflatoxin production. Similarly, over-expression of aflS resulted in increased aflatoxin production, suggesting a role in modulating early aflatoxin biosynthetic genes, while the over-expression of aflR resulted in higher aflS-transcription rates, indicating that aflS expression was regulated by aflR [107]. Therefore, the exact role of aflS remains elusive; however, it could be involved in not only the activation of early and mid aflatoxin pathway genes but also the regulation of structural genes through its association with aflR.
In light of this, co-cultivation of A. flavus with a strain of B. megaterium was proposed as a possible mechanism for the inhibition of aflatoxin biosynthesis through the down-regulation of the regulatory genes [75]. The co-cultivation resulted in the down-regulation of several genes in the aflatoxin-biosynthetic pathway, including aflS, as confirmed by qRT-PCR. Down-regulation of aflS would alter the aflR/aflS ratio, inhibiting the regulation of aflatoxin structural genes and blocking the aflatoxin-biosynthetic pathway. Interestingly, a similar effect of down-regulating aflS was previously obtained by Yu et al. [108] upon increasing the incubation temperature to 37℃. While it remains unknown exactly how the down-regulation of aflS leads to the inactivation of structural genes, it is speculated that it is mediated by potential inhibitors in fungal cells. The latter could be similar in size to AFLS and form a dysfunctional AFLS/AFLR activation complex that will bind to the aflatoxin pathway-structural genes, resulting in the inhibition or termination of their transcription (Fig. 1) [75]. In another recent study, Streptomyces spp. were used as mutual antagonists against A. flavus and A. parasiticus, and lowered the aflatoxin levels produced by the Aspergillus fungi. In the case of A. parasiticus, aflatoxin reduction was attributed reduction of fungal growth. No influence on fungal growth was reported in the case of A. flavus. Instead, Streptomyces spp. appeared to repress aflM and aflS by altering the aflR/aflS ratio in a species-specific manner [103]. Microbial interference with aflatoxin-biosynthetic pathway is an important feature of certain beneficial bacteria, which can be applied for prevention of aflatoxin occurrence on stored products, including rice.
Fig. 1. A hypothetical mechanism illustrating the role of the genes aflS and aflR in the aflatoxin-biosynthetic pathway in Aspergillus flavus: under the normal aflatoxin-producing culture condition (A) and under co-cultivation condition (B) with Bacillus megaterium, which inhibits aflatoxin biosynthesis by down-regulating the expression of the gene aflS. This figure was re-drawn, with appropriate modification, from Kong et al. [75], with permission of Springer, using Servier Medical Art (www.servier.com).
CONCLUSIONS
The increased understanding of the impact of rice-grain fungal species on human and animal health, whether in the form of disease-causing spores or dangerous mycotoxins (e.g., aflatoxins), has raised awareness about the control of fungi contaminated food and feed. An effective and safe method for controlling grain fungi during storage may be the microbe-mediated (biological) control measure that, as discussed in this review, can negatively affect grain fungi and detoxify (or biodegrade) mycotoxins or inhibit their biosynthesis. Therefore, further screening and selection of potential biocontrol agents should be encouraged and improved, as these will help finding novel active agents for antifungal compounds or detoxification enzymes and biosynthesis inhibitors of mycotoxins. Based on the reviewed literature, it appears that the selection of antagonistic bacteria that originated on rice grains has been largely neglected. Instead, such antagonistic bacteria from rice could serve as sources of broad-spectrum biocontrol agents against rice grain fungi, as they are more likely to survive and compete against pathogenic rice mycoflora. Taken together, further research should be directed towards the screening of rice-originated bacteria for effective biocontrol agents against mycotoxigenic rice-grain fungal species. In addition, it may be noted that most research on microbe-mediated mycotoxin (including aflatoxin) degradation or production inhibition was made under laboratory conditions. However, it is essential that a safe and effective method for aflatoxin detoxification should be devised on food commodities.
ACKNOWLEDGEMENTS
This work was supported by a Korea University Grant. We thank anonymous reviewers for valuable comments on this manuscript.
References
- 1.De Datta SK. Principles and practices of rice production. New York: John Wiley & Sons; 1981. [Google Scholar]
- 2.Oh JY, Jee SN, Nam Y, Lee H, Ryoo MI, Kim KD. Populations of fungi and bacteria associated with samples of stored rice in Korea. Mycobiology. 2007;35:36–38. doi: 10.4489/MYCO.2007.35.1.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams RJ, McDonald D. Grain molds in the tropics: problems and importance. Annu Rev Phytopathol. 1983;21:153–178. [Google Scholar]
- 4.Mew TW, Gonzales P. A handbook of rice seedborne fungi. Los Baños: International Rice Research Institute; 2002. [Google Scholar]
- 5.Chełkowski J. Cereal grain: mycotoxins, fungi and quality in drying and storage. Amsterdam: Elsevier Science Publishers; 1991. [Google Scholar]
- 6.Filtenborg O, Frisvad JC, Thrane U. Moulds in food spoilage. Int J Food Microbiol. 1996;33:85–102. doi: 10.1016/0168-1605(96)01153-1. [DOI] [PubMed] [Google Scholar]
- 7.Misra JK, Gergon EB, Mew TW. Storage fungi and seed health of rice: a study in the Philippines. Mycopathologia. 1995;131:13–24. doi: 10.1007/BF01103899. [DOI] [PubMed] [Google Scholar]
- 8.Oh JY, Sang MK, Oh JE, Lee H, Ryoo MI, Kim KD. Microbial population, aflatoxin contamination and predominant Aspergillus species in Korean stored rice. Plant Pathol J. 2010;26:121–129. [Google Scholar]
- 9.Park JW, Choi SY, Hwang HJ, Kim YB. Fungal mycoflora and mycotoxins in Korean polished rice destined for humans. Int J Food Microbiol. 2005;103:305–314. doi: 10.1016/j.ijfoodmicro.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Sawane A, Sawane M. Mycotoxigenicity of Aspergillus, Penicillium and Fusarium spp. isolated from stored rice. Int J Curr Microbiol Appl Sci. 2014;3:116–121. [Google Scholar]
- 11.Khosravi AR, Shokri H, Zaboli F. Grain-borne mycoflora and fumonisin B1 from fresh-harvested and stored rice in northern Iran. Jundishapur J Microbiol. 2013;6:e6414. [Google Scholar]
- 12.Oh JY, Sang MK, Lee H, Ryoo MI, Kim KD. First detection of Penicillium fellutanum from stored rice in Korea. Res Plant Dis. 2011;17:216–221. [Google Scholar]
- 13.Oh JY, Kim EN, Ryoo MI, Kim KD. Morphological and molecular identification of Penicillium islandicum isolate KU101 from stored rice. Plant Pathol J. 2008;24:469–473. [Google Scholar]
- 14.Taligoola HK, Ismail MA, Chebon SK. Mycobiota associated with rice grains marketed in Uganda. J Biol Sci. 2004;4:271–278. [Google Scholar]
- 15.Dvořáčková I. Aflatoxins and human health. Boca Raton(FL): CRC Press; 1989. [Google Scholar]
- 16.Agrios G. Plant pathology. 5th ed. Boston (MA): Elsevier Academic Press; 2005. [Google Scholar]
- 17.Krysińska-Traczyk E, Dutkiewicz J. Aspergillus candidus: a respiratory hazard associated with grain dust. Ann Agric Environ Med. 2000;7:101–109. [PubMed] [Google Scholar]
- 18.Klich MA. Identification of common Aspergillus species. Utrecht: Centraalbureau voor Schimmelcultures; 2002. [Google Scholar]
- 19.Abad A, Fernández-Molina JV, Bikandi J, Ramírez A, Margareto J, Sendino J, Hernando FL, Pontón J, Garaizar J, Rementeria A. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev Iberoam Micol. 2010;27:155–182. doi: 10.1016/j.riam.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Pitt JI, Hocking AD. Fungi and food spoilage. Boston (MA): Springer; 2009. [Google Scholar]
- 21.Pitt JI. A laboratory guide to common Penicillium species. North Ryde: CSIRO; 1988. [Google Scholar]
- 22.Bräse S, Encinas A, Keck J, Nising CF. Chemistry and biology of mycotoxins and related fungal metabolites. Chem Rev. 2009;109:3903–3990. doi: 10.1021/cr050001f. [DOI] [PubMed] [Google Scholar]
- 23.Zhelifonova VP, Antipova TV, Kozlovsky AG. Secondary metabolites in taxonomy of the Penicillium fungi. Microbiology. 2010;79:277–286. [Google Scholar]
- 24.Cabañes FJ, Bragulat MR, Castellá G. Ochratoxin A producing species in the genus Penicillium. Toxins (Basel) 2010;2:1111–1120. doi: 10.3390/toxins2051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tonon SA, Marucci RS, Jerke G, García A. Mycoflora of paddy and milled rice produced in the region of northeastern Argentina and southern Paraguay. Int J Food Microbiol. 1997;37:231–235. doi: 10.1016/s0168-1605(97)00066-4. [DOI] [PubMed] [Google Scholar]
- 26.Kozlovsky AG, Vinokurova NG, Adanin VM, Burkhardt G, Dahse HM, Gräfe U. New diketopiperazine alkaloids from Penicillium fellutanum. J Nat Prod. 2000;63:698–700. doi: 10.1021/np9903853. [DOI] [PubMed] [Google Scholar]
- 27.Vinokurova NG, Boichenko LV, Arinbasarov MU. Production of alkaloids by fungi of the genus Penicillium grown on wheat grain. Appl Biochem Microbiol. 2003;39:403–406. [Google Scholar]
- 28.Cole RJ, Cox RH. Handbook of toxic fungal metabolites. New York: Academic Press; 1981. [Google Scholar]
- 29.Flieger M, Wurst M, Shelby R. Ergot alkaloids - sources, structures and analytical methods. Folia Microbiol (Praha) 1997;42:3–29. doi: 10.1007/BF02898641. [DOI] [PubMed] [Google Scholar]
- 30.Gerhards N, Neubauer L, Tudzynski P, Li SM. Biosynthetic pathways of ergot alkaloids. Toxins (Basel) 2014;6:3281–3295. doi: 10.3390/toxins6123281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newberne PM. Mycotoxins: toxicity, carcinogenicity, and the influence of various nutritional conditions. Environ Health Perspect. 1974;9:1–32. doi: 10.1289/ehp.9-1475399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wawrzyniak J, Waśkiewicz A. Ochratoxin A and citrinin production by Penicillium verrucosum on cereal solid substrates. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2014;31:139–148. doi: 10.1080/19440049.2013.861933. [DOI] [PubMed] [Google Scholar]
- 33.Kostecki M, Wisniewska H, Perrone G, Ritieni A, Golinski P, Chelkowski J, Logrieco A. The effects of cereal substrate and temperature on production of beauvericin, moniliformin and fusaproliferin by Fusarium subglutinans ITEM-1434. Food Addit Contam. 1999;16:361–365. doi: 10.1080/026520399283849. [DOI] [PubMed] [Google Scholar]
- 34.Shotwell OL, Hesseltine CW, Stubblefield RD, Sorenson WG. Production of aflatoxin on rice. Appl Microbiol. 1966;14:425–428. doi: 10.1128/am.14.3.425-428.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy KR, Reddy CS, Abbas HK, Abel CA, Muralidharan K. Mycotoxigenic fungi, mycotoxins, and management of rice grains. Toxin Rev. 2008;27:287–317. [Google Scholar]
- 36.Liu Z, Gao J, Yu J. Aflatoxins in stored maize and rice grains in Liaoning Province, China. J Stored Prod Res. 2006;42:468–479. [Google Scholar]
- 37.Bansal J, Pantazopoulos P, Tam J, Cavlovic P, Kwong K, Turcotte AM, Lau BP, Scott PM. Surveys of rice sold in Canada for aflatoxins, ochratoxin A and fumonisins. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2011;28:767–774. doi: 10.1080/19440049.2011.559279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Begum F, Samajpati N. Mycotoxin production on rice, pulses and oilseeds. Naturwissenschaften. 2000;87:275–277. doi: 10.1007/s001140050720. [DOI] [PubMed] [Google Scholar]
- 39.Trung TS, Bailly JD, Querin A, Le Bars P, Guerre P. Fungal contamination of rice from south Vietnam, mycotoxinogenesis of selected strains and residues in rice. Rev Méd Vét. 2001;152:555–560. [Google Scholar]
- 40.Abd-Allah EF, Ezzat SM. Natural occurrence of citrinin in rice grains and its biocontrol by Trichoderma hamatum. Phytoparasitica. 2005;33:73–84. [Google Scholar]
- 41.Nguyen MT, Tozlovanu M, Tran TL, Pfohl-Leszkowicz A. Occurrence of aflatoxin B1, citrinin and ochratoxin A in rice in five provinces of the central region of Vietnam. Food Chem. 2007;105:42–47. [Google Scholar]
- 42.Sugimoto T, Minamisawa M, Takano K, Sasamura Y, Tsuruta O. Detection of ochratoxin A, citrinin and sterigmatocystin from stored rice by natural occurrence of Penicillium viridicatum and Aspergillus versicolor. J Food Hyg Soc Jpn. 1977;18:176–181. [Google Scholar]
- 43.Luk KC, Kobbe B, Townsend JM. Production of cyclopiazonic acid by Aspergillus flavus Link. Appl Environ Microbiol. 1977;33:211–212. doi: 10.1128/aem.33.1.211-212.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abbas HK, Cartwright RD, Shier WT, Abouzied MM, Bird CB, Rice LG, Ross PF, Sciumbato GL, Meredith FI. Natural occurrence of fumonisins in rice with Fusarium sheath rot disease. Plant Dis. 1998;82:22–25. doi: 10.1094/PDIS.1998.82.1.22. [DOI] [PubMed] [Google Scholar]
- 45.Chung SH, Kim YB. Natural occurrence of fumonisin B1 in Korean corn and rough rice. Food Sci Biotechnol. 1995;4:212–216. [Google Scholar]
- 46.Scott PM, Lawrence GA, Lombaert GA. Studies on extraction of fumonisins from rice, corn-based foods and beans. Mycotoxin Res. 1999;15:50–60. doi: 10.1007/BF02945215. [DOI] [PubMed] [Google Scholar]
- 47.Kushiro M, Nagata R, Nakagawa H, Nagashima H. Liquid chromatographic detection of fumonisins in rice seed. Rep Natl Food Res Inst. 2008;72:37–44. [Google Scholar]
- 48.Frisvad JC, Smedsgaard J, Samson RA, Larsen TO, Thrane U. Fumonisin B2 production by Aspergillus niger. J Agric Food Chem. 2007;55:9727–9732. doi: 10.1021/jf0718906. [DOI] [PubMed] [Google Scholar]
- 49.Richard JL, Lyon RL, Fichtner RE, Ross PF. Use of thin layer chromatography for detection and high performance liquid chromatography for quantitating gliotoxin from rice cultures of Aspergillus fumigatus Fresenius. Mycopathologia. 1989;107:145–151. doi: 10.1007/BF00707552. [DOI] [PubMed] [Google Scholar]
- 50.Abd Alla ES. Natural occurrence of ochratoxin A and citrinin in food stuffs in Egypt. Mycotoxin Res. 1996;12:41–44. doi: 10.1007/BF03192079. [DOI] [PubMed] [Google Scholar]
- 51.Pena A, Cerejo F, Lino C, Silveira I. Determination of ochratoxin A in Portuguese rice samples by high performance liquid chromatography with fluorescence detection. Anal Bioanal Chem. 2005;382:1288–1293. doi: 10.1007/s00216-005-3254-9. [DOI] [PubMed] [Google Scholar]
- 52.Uchiyama M, Isohata E, Takeda Y. A case report on the detection of ochratoxin-A from rice. Food Hyg Saf Sci. 1976;17:103–104. [Google Scholar]
- 53.Okeke B, Seigle-Murandi F, Steiman R, Benoit-Guyod JL, Kaouadji M. Identification of mycotoxin-producing fungal strains: a step in the isolation of compounds active against rice fungal diseases. J Agric Food Chem. 1993;41:1731–1735. [Google Scholar]
- 54.Tian H, Liu X. Survey and analysis on sterigmatocystin contaminated in grains in China. Wei Sheng Yan Jiu. 2004;33:606–608. [PubMed] [Google Scholar]
- 55.Llewellyn GC, Sherertz PC, Armstrong CW, Miller GB, Jr, Reynolds JD, Kimbrough TD, Bean GA, Hagler WM, Jr, Haney CA, Trempus CS, et al. Mycotoxigenic isolates and toxin production on buckwheat and rice hulls used as bedding materials. J Ind Microbiol. 1988;3:351–356. [Google Scholar]
- 56.Mateo JJ, Mateo R, Jiménez M. Accumulation of type A trichothecenes in maize, wheat and rice by Fusarium sporotrichioides isolates under diverse culture conditions. Int J Food Microbiol. 2002;72:115–123. doi: 10.1016/s0168-1605(01)00625-0. [DOI] [PubMed] [Google Scholar]
- 57.Lee T, Lee SH, Lee SH, Shin JY, Yun JC, Lee YW, Ryu JG. Occurrence of Fusarium mycotoxins in rice and its milling by-products in Korea. J Food Prot. 2011;74:1169–1174. doi: 10.4315/0362-028X.JFP-10-564. [DOI] [PubMed] [Google Scholar]
- 58.Sweeney MJ, Dobson AD. Mycotoxin production by Aspergillus, Fusarium and Penicillium species. Int J Food Microbiol. 1998;43:141–158. doi: 10.1016/s0168-1605(98)00112-3. [DOI] [PubMed] [Google Scholar]
- 59.Moore-Landecker E. Fundamentals of the fungi. Englewood Cliffs (NJ): Prentice-Hall; 1972. [Google Scholar]
- 60.Squire RA. Ranking animal carcinogens: a proposed regulatory approach. Science. 1981;214:877–880. doi: 10.1126/science.7302565. [DOI] [PubMed] [Google Scholar]
- 61.Trail F, Mahanti N, Linz J. Molecular biology of aflatoxin biosynthesis. Microbiology. 1995;141(Pt4):755–765. doi: 10.1099/13500872-141-4-755. [DOI] [PubMed] [Google Scholar]
- 62.Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev. 2002;66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Rensburg SJ, Cook-Mozaffari P, Van Schalkwyk DJ, Van der Watt JJ, Vincent TJ, Purchase IF. Hepatocellular carcinoma and dietary aflatoxin in Mozambique and Transkei. Br J Cancer. 1985;51:713–726. doi: 10.1038/bjc.1985.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Egmond HP, Jonker MA. Worldwide regulations for mycotoxins in food and feed in 2003. Rome: Food and Agriculture Organization; 2004. [Google Scholar]
- 65.Cottyn B, Regalado E, Lanoot B, De Cleene M, Mew TW, Swings J. Bacterial populations associated with rice seed in the tropical environment. Phytopathology. 2001;91:282–292. doi: 10.1094/PHYTO.2001.91.3.282. [DOI] [PubMed] [Google Scholar]
- 66.Hauptmann RM, Widholm JM, Paxton JD. Benomyl: a broad spectrum fungicide for use in plant cell and protoplast culture. Plant Cell Rep. 1985;4:129–132. doi: 10.1007/BF00571298. [DOI] [PubMed] [Google Scholar]
- 67.Naseer R, Sultana B, Khan MZ, Naseer D, Nigam P. Utilization of waste fruit-peels to inhibit aflatoxins synthesis by Aspergillus flavus: a biotreatment of rice for safer storage. Bioresour Technol. 2014;172:423–428. doi: 10.1016/j.biortech.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 68.Ruiqian L, Qian Y, Thanaboripat D, Thansukon P. Biocontrol of Aspergillus flavus and aflatoxin production. KMITL Sci J. 2004;4:1685–2044. [Google Scholar]
- 69.Blakeman JP, Fokkema NJ. Potential for biological control of plant diseases on the phylloplane. Annu Rev Phytopathol. 1982;20:167–190. [Google Scholar]
- 70.Ou SH. Rice diseases. 2nd ed. Wallingford: CABI Publishing; 1985. [Google Scholar]
- 71.Mew TW, Rosales AM. Bacterization of rice plants for control of sheath blight caused by Rhizoctonia solani. Phytopathology. 1986;76:1260–1264. [Google Scholar]
- 72.Santoyo G, Orozco-Mosqueda MD, Govindappa M. Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: a review. Biocontrol Sci Technol. 2012;22:855–872. [Google Scholar]
- 73.Kim YG, Kang HK, Kwon KD, Seo CH, Lee HB, Park Y. Antagonistic activities of novel peptides from Bacillus amyloliquefaciens PT14 against Fusarium solani and Fusarium oxysporum. J Agric Food Chem. 2015;63:10380–10387. doi: 10.1021/acs.jafc.5b04068. [DOI] [PubMed] [Google Scholar]
- 74.Kong Q, Shan S, Liu Q, Wang X, Yu F. Biocontrol of Aspergillus flavus on peanut kernels by use of a strain of marine Bacillus megaterium. Int J Food Microbiol. 2010;139:31–35. doi: 10.1016/j.ijfoodmicro.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 75.Kong Q, Chi C, Yu J, Shan S, Li Q, Li Q, Guan B, Nierman WC, Bennett JW. The inhibitory effect of Bacillus megaterium on aflatoxin and cyclopiazonic acid biosynthetic pathway gene expression in Aspergillus flavus. Appl Microbiol Biotechnol. 2014;98:5161–5172. doi: 10.1007/s00253-014-5632-8. [DOI] [PubMed] [Google Scholar]
- 76.Reddy KR, Reddy CS, Muralidharan K. Potential of botanicals and biocontrol agents on growth and aflatoxin production by Aspergillus flavus infecting rice grains. Food Control. 2009;20:173–178. [Google Scholar]
- 77.Lee SY, Oh JY, Ryoo ML, Kim KD. Biological control of the rice storage fungi Aspergillus and Penicillium species by antagonistic bacteria originated from rice. Plant Pathol J. 2007;23:328. [Google Scholar]
- 78.Mokiou S, Magan N. Physiological manipulation and formulation of the biocontrol yeast Pichia anomala for control of Penicillium verrucosum and ochratoxin A contamination of moist grain. Biocontrol Sci Technol. 2008;18:1063–1073. [Google Scholar]
- 79.Kiessling KH, Pettersson H, Sandholm K, Olsen M. Metabolism of aflatoxin, ochratoxin, zearalenone, and three trichothecenes by intact rumen fluid, rumen protozoa, and rumen bacteria. Appl Environ Microbiol. 1984;47:1070–1073. doi: 10.1128/aem.47.5.1070-1073.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Bellis P, Tristezza M, Haidukowski M, Fanelli F, Sisto A, Mule G, Grieco F. Biodegradation of ochratoxin A by bacterial strains isolated from vineyard soils. Toxins (Basel) 2015;7:5079–5093. doi: 10.3390/toxins7124864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu H, Zhou T, Gong J, Young C, Su X, Li XZ, Zhu H, Tsao R, Yang R. Isolation of deoxynivalenol-transforming bacteria from the chicken intestines using the approach of PCR-DGGE guided microbial selection. BMC Microbiol. 2010;10:182. doi: 10.1186/1471-2180-10-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu Y, Qiu L, Wu H, Tang Y, Yu Y, Li X, Liu D. Degradation of zearalenone by the extracellular extracts of Acinetobacter sp. SM04 liquid cultures. Biodegradation. 2011;22:613–622. doi: 10.1007/s10532-010-9435-z. [DOI] [PubMed] [Google Scholar]
- 83.Ciegler A. Detoxification of aflatoxin-contaminated agricultural commodities. In: Rosenberg P, editor. Toxins: animal, plant and microbial. New York: Pergamon Press; 1978. pp. 729–738. [Google Scholar]
- 84.Ashworth LJ, Jr, Schroeder HW, Langley BC. Aflatoxins: environmental factors governing occurrence in Spanish peanuts. Science. 1965;148:1228–1229. doi: 10.1126/science.148.3674.1228. [DOI] [PubMed] [Google Scholar]
- 85.Ciegler A, Lillehoj EB, Peterson RE, Hall HH. Microbial detoxification of aflatoxin. Appl Microbiol. 1966;14:934–939. doi: 10.1128/am.14.6.934-939.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hao YY, Brackett RE. Removal of aflatoxin B1 from peanut milk inoculated with Flavobacterium aurantiacum. J Food Sci. 1988;53:1384–1386. [Google Scholar]
- 87.Lillehoj EB, Ciegler A, Hall HH. Aflatoxin B1 uptake by Flavobacterium aurantiacum and resulting toxic effects. J Bacteriol. 1967;93:464–471. doi: 10.1128/jb.93.1.464-471.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ciegler A, Peterson RE. Aflatoxin detoxification: hydroxydihydro-aflatoxin B1. Appl Microbiol. 1968;16:665–666. doi: 10.1128/am.16.4.665-666.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Line JE, Brackett RE. Role of toxin concentration and second carbon source in microbial transformation of aflatoxin B1 by Flavobacterium aurantiacum. J Food Prot. 1995;58:1042–1044. doi: 10.4315/0362-028X-58.9.1042. [DOI] [PubMed] [Google Scholar]
- 90.Smiley RD, Draughon FA. Preliminary evidence that degradation of aflatoxin B1 by Flavobacterium aurantiacum is enzymatic. J Food Prot. 2000;63:415–418. doi: 10.4315/0362-028x-63.3.415. [DOI] [PubMed] [Google Scholar]
- 91.Petchkongkaew A, Taillandier P, Gasaluck P, Lebrihi A. Isolation of Bacillus spp. from Thai fermented soybean (Thua-nao): screening for aflatoxin B1 and ochratoxin A detoxification. J Appl Microbiol. 2008;104:1495–1502. doi: 10.1111/j.1365-2672.2007.03700.x. [DOI] [PubMed] [Google Scholar]
- 92.Sangare L, Zhao Y, Folly YM, Chang J, Li J, Selvaraj JN, Xing F, Zhou L, Wang Y, Liu Y. Aflatoxin B1 degradation by a Pseudomonas strain. Toxins (Basel) 2014;6:3028–3040. doi: 10.3390/toxins6103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Teunisson DJ, Robertson JA. Degradation of pure aflatoxins by Tetrahymena pyriformis. Appl Microbiol. 1967;15:1099–1103. doi: 10.1128/am.15.5.1099-1103.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cole RJ, Kirksey JW. Aflatoxin G1 metabolism by Rhizopus species. J Agr Food Chem. 1971;19:222–223. doi: 10.1021/jf60174a044. [DOI] [PubMed] [Google Scholar]
- 95.Manabe M, Matsuura S. Studies on the fluorescent compound in fermented foods. Part IV. Degradation of added aflatoxin during miso fermentation. Nihon Shokuhin Kōgyō Gakkai shi. 1972;19:275–279. [Google Scholar]
- 96.Line JE, Brackett RE, Wilkinson RE. Evidence for degradation of aflatoxin B1 by Flavobacterium aurantiacum. J Food Prot. 1994;57:788–791. doi: 10.4315/0362-028X-57.9.788. [DOI] [PubMed] [Google Scholar]
- 97.Farzaneh M, Shi ZQ, Ghassempour A, Sedaghat N, Ahmadzadeh M, Mirabolfathy M, Javan-Nikkhah M. Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control. 2012;23:100–106. [Google Scholar]
- 98.Kusumaningtyas E, Widiastuti R, Maryam R. Reduction of aflatoxin B1 in chicken feed by using Saccharomyces cerevisiae, Rhizopus oligosporus and their combination. Mycopathologia. 2006;162:307–311. doi: 10.1007/s11046-006-0047-4. [DOI] [PubMed] [Google Scholar]
- 99.Bennett JW. Loss of norsolorinic acid and aflatoxin production by a mutant of Aspergillus parasiticus. Microbiology. 1981;124:429–432. [Google Scholar]
- 100.Yu J, Bhatnagar D, Cleveland TE. Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus. FEBS Lett. 2004;564:126–130. doi: 10.1016/S0014-5793(04)00327-8. [DOI] [PubMed] [Google Scholar]
- 101.Georgianna DR, Payne GA. Genetic regulation of aflatoxin biosynthesis: from gene to genome. Fungal Genet Biol. 2009;46:113–125. doi: 10.1016/j.fgb.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 102.Schmidt-Heydt M, Abdel-Hadi A, Magan N, Geisen R. Complex regulation of the aflatoxin biosynthesis gene cluster of Aspergillus flavus in relation to various combinations of water activity and temperature. Int J Food Microbiol. 2009;135:231–237. doi: 10.1016/j.ijfoodmicro.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 103.Verheecke C, Liboz T, Anson P, Diaz R, Mathieu F. Reduction of aflatoxin production by Aspergillus flavus and Aspergillus parasiticus in interaction with Streptomyces. Microbiology. 2015;161(Pt5):967–972. doi: 10.1099/mic.0.000070. [DOI] [PubMed] [Google Scholar]
- 104.Chang PK. The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol Genet Genomics. 2003;268:711–719. doi: 10.1007/s00438-003-0809-3. [DOI] [PubMed] [Google Scholar]
- 105.Woloshuk CP, Foutz KR, Brewer JF, Bhatnagar D, Cleveland TE, Payne GA. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl Environ Microbiol. 1994;60:2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meyers DM, Obrian G, Du WL, Bhatnagar D, Payne GA. Characterization of aflJ, a gene required for conversion of pathway intermediates to aflatoxin. Appl Environ Microbiol. 1998;64:3713–3717. doi: 10.1128/aem.64.10.3713-3717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Du W, Obrian GR, Payne GA. Function and regulation of aflJ in the accumulation of aflatoxin early pathway intermediate in Aspergillus flavus. Food Addit Contam. 2007;24:1043–1050. doi: 10.1080/02652030701513826. [DOI] [PubMed] [Google Scholar]
- 108.Yu J, Fedorova ND, Montalbano BG, Bhatnagar D, Cleveland TE, Bennett JW, Nierman WC. Tight control of mycotoxin biosynthesis gene expression in Aspergillus flavus by temperature as revealed by RNA-Seq. FEMS Microbiol Lett. 2011;322:145–149. doi: 10.1111/j.1574-6968.2011.02345.x. [DOI] [PubMed] [Google Scholar]