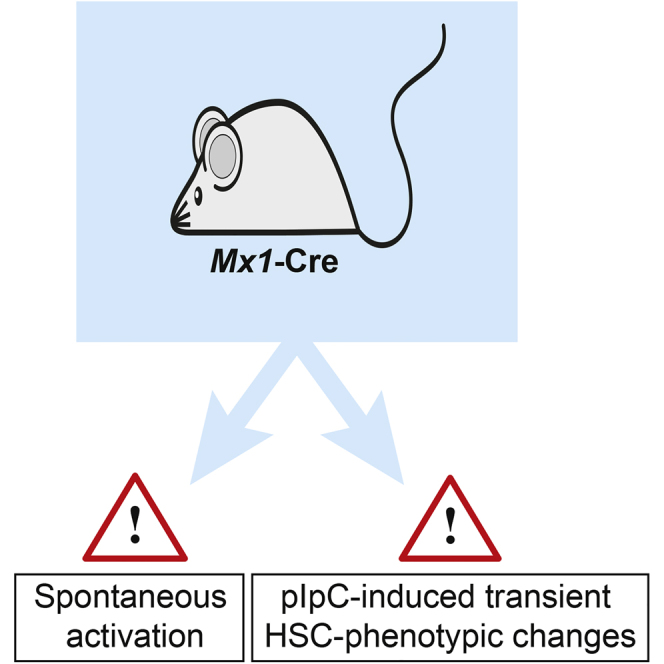
Summary
Conditional knockout mice are commonly used to study the function of specific genes in hematopoiesis. Different promoters that drive Cre expression have been utilized, with the interferon-inducible Mx1-Cre still being the most commonly used “deleter strain” in experimental hematology. However, different pitfalls associated with this system could lead to misinterpretation in functional studies. We present here two of these issues related to the use of Mx1-Cre: first, a high spontaneous recombination rate when applying commonly used techniques in experimental hematology, and second, undesired short-term consequences of the use of polyinosinic:polycytidylic acid, including changes in cellular phenotypes that, however, resolve within days. Our studies emphasize therefore that proper controls are crucial when modeling gene deletion using the Mx1-Cre transgene.
Graphical Abstract
Highlights
-
•
Common research methods induce uncontrolled activation of the Mx1-Cre system
-
•
The leakiness of the Mx1-Cre system can result in misinterpretation of results
-
•
pIpC alters the phenotype of HSPCs that resolves by day 8
-
•
pIpC has limited effects on the proliferation of HSPCs
In this article, Cammenga, Velasco-Hernandez, and colleagues describe technical issues associated with the Mx1-Cre model in hematopoiesis, including a high level of spontaneous activation induced by multiple experimental procedures. These pitfalls can lead to misinterpretation of results so proper controls are needed to avoid these problems.
Introduction
Conditional knockout (cKO) mice based on Cre-mediated excision (Jiang and Gridley, 1997) are frequently used when studying normal and malignant hematopoiesis. Most cKO models are generated by flanking a gene segment of interest with loxP sites, which allows for its removal in a cell- or tissue-specific manner by Cre recombinase. The Mx1-Cre model (Kuhn et al., 1995) represents the most commonly used “deleter strain” in experimental hematology. In this strain, the Mx dynamin-like GTPase 1 (Mx1) promoter is activated in an interferon-dependent manner following injection of polyinosinic:polycytidylic acid (pIpC), which results in downstream expression of Cre recombinase. Despite the wide use of the Mx1-Cre strain, caution has to be entertained due to caveats that have not been well described. These include spontaneous recombination and undesired side effects caused by the pIpC itself.
In experimental hematology, bone marrow (BM) transplantation represents a key approach in the investigation of the role of specific genes. Most often, this is performed with cells obtained from cKO mice. We show here that the transplantation assay and expression of an activated receptor tyrosine kinase (FLT-3ITD) can trigger spontaneous Mx1-Cre-driven deletion of floxed genes, which substantially exceeded the 2%–3% originally reported (Kemp et al., 2004, Kuhn et al., 1995, Mupo et al., 2013). In addition, we observed transient changes in phenotype and frequency of hematopoietic stem and progenitor cells (HSPCs) after pIpC injection, which could lead to incorrect identification of subsets of cells in short-term studies. Finally, we propose a potential alternative strategy for gene deletion using an ex vivo Tat-Cre-recombination approach. Awareness of these shortcomings has high relevance for hematopoietic research because it can aid in experimental design and avoid potential errors in the interpretation of results.
Results
Transplantation Induces Uncontrolled Recombination in Hematopoietic Cells from Mx1-Cre Mice
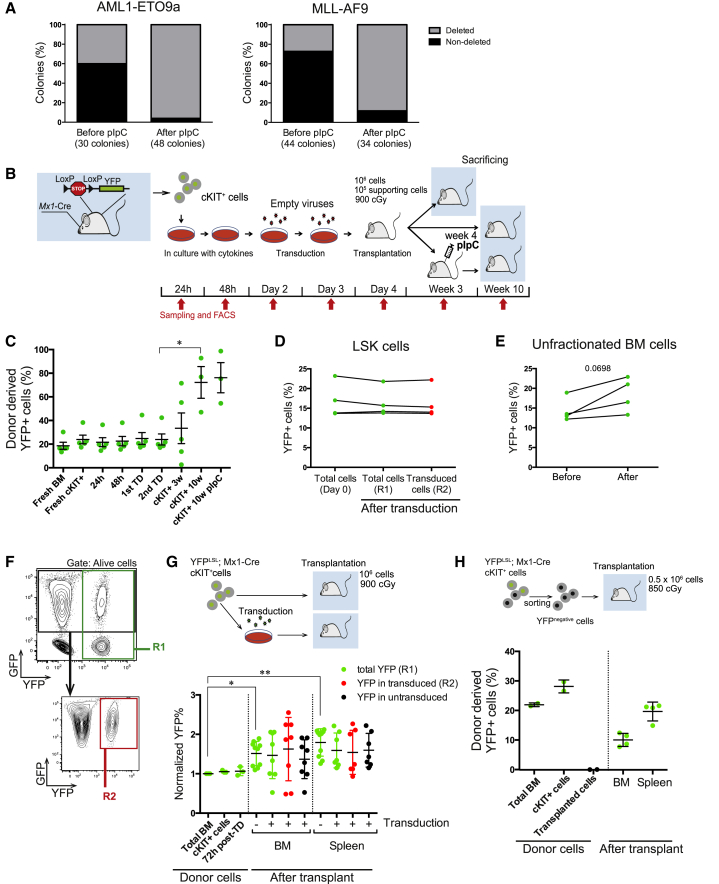
To study the influence of HIF-1α in the initiation of leukemia, we retrovirally introduced different oncogenes into Mx1-Cre; Hif-1αflox/flox cKIT+ cells and transplanted them into wild-type (WT) recipients (Velasco-Hernandez et al., 2014). Surprisingly, we observed an unexpectedly high deletion frequency of the floxed gene prior to pIpC injection, ranging between 30% and 50% (Figure 1A).
Figure 1.
Spontaneous Deletion of Floxed Genes in Hematopoietic Cells from Mx1-Cre Mice after Transplantation
(A) Frequency of HIF-1α-deleted colonies from peripheral blood cells derived from animals transplanted with transduced Mx1-Cre; Hif-1αflox/flox HSPCs before (week 3 post-transplantation) and after pIpC injection (pIpC injection at week 4 and analysis at week 7). Plots show two independent experiments, using either AML1-ETO9a or MLL-AF9 expressing donor cells.
(B) Experimental setup for analysis of spontaneous deletion in a commonly used transduction/transplantation protocol. Donor cells were collected from five separate animals.
(C) Variation in frequency of recombined cells (YFP+) along the different steps of the protocol described in (B). Percentage of recombined cells is calculated within the donor-derived population after the transplantation step. ∗p < 0.05.
(D) Recombined cell frequency in YFPLSL; Mx1-Cre LSK cells at harvesting/sorting time (day 0) or 48 hr after transduction with a GFP virus (MIGR1): YFP+ cells from total cells (R1 population) and YFP+ cells within transduced cells (R2 population) (n = 4). Transduction levels were 10%–20%.
(E) Recombined cell frequency in unfractionated YFPLSL; Mx1-Cre BM cells before and after 4 days culture (n = 4).
(F) Gating strategy used to analyze R1 and R2 population within transduced cells.
(G) Analysis of the effect of the transplantation alone on the spontaneous recombination. YFPLSL; Mx1-Cre cKIT+ cells were isolated and directly transplanted or transduced with GFP virus before transplantation. Recombination levels were analyzed 15 days after transplantation (donor mice, n = 3; transplanted mice, n = 10). Recombination levels are normalized to initial YFP+ cells values from each independent donor. Transduction levels were between 10% and 50%. Pooled data from two independent experiments. ∗p < 0.05, ∗∗p < 0.01.
(H) Recombined cell frequency of donor-derived BM and spleen cells 15 days after transplantation with sorted YFPLSL; Mx1-Cre cKIT+ YFP− (n = 4). Plots show mean ± SEM. TD, transduction. See also Figures S1 and S2.
To identify experimental procedures that contribute to the spontaneous Mx1-Cre activation, we crossed Rosa26-Lox-Stop-Lox-YFP (YFPLSL) reporter mice (Srinivas et al., 2001) with the Mx1-Cre mice. HSPCs from these mice were next subjected to a routine protocol for transduction/transplantation experiments, using retroviral vectors encoding fluorescence proteins (Figures 1B–1G). Isolated unmanipulated cells from donor mice displayed a spontaneous recombination close to 20% (Figures 1C, 1D, 1E, 1H, and S1). Analyses after transplantation indicated that the percentage of YFP+ (recombined) cells in the donor-derived compartment had increased significantly at week 10 (Figures 1C and S1). To address if this was due to the transduction or transplantation procedures, we performed the same protocol with or without transduction (Figures 1G and S1). Analysis of BM and spleen cells 15 days after transplantation indicated that the levels of recombination were similar in both groups. However, the frequency of recombined cells increased by 50% as a consequence of the transplantation, despite the lack of an evident positive selection pressure in favor of the recombined cells. Investigations into the effect of retroviral transduction in a more primitive and homogeneous population (Lin−SCA1+cKIT+, LSK cells) also failed to reveal increments in recombination 48 hr after transduction (Figures 1D and S2). Furthermore, no significant increase in recombination was observed when culturing unfractionated BM cells, including more mature populations that could include interferon-producing cells (Figure 1E). Finally, to conclusively exclude a potential positive selection of already recombined cells that could increase in vivo after transplantation, we sorted cKIT+ YFP− cells from YFPLSL; Mx1-Cre mice and transplanted them into WT recipients without pIpC injection (Figure 1H). This revealed 10%–20% of YFP+ donor cells 15 days after transplantation in donor BM and spleen cells of transplanted mice.
These results demonstrate that the major cause of the unexpectedly high levels of spontaneous gene deletion is caused by the transplantation procedure per se.
Constitutively Active Flt-3 Induces Spontaneous Recombination in Mx1-Cre Mice
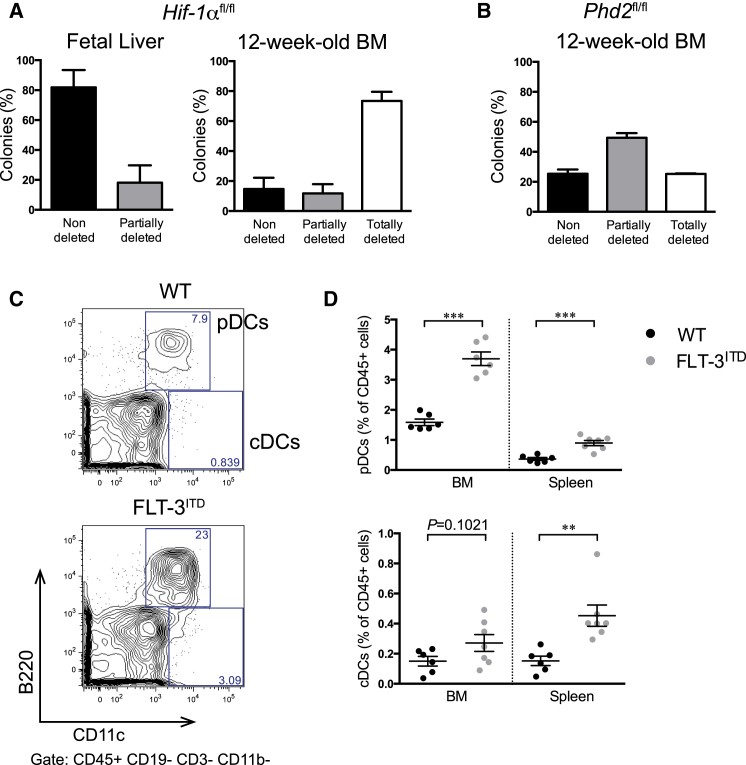
We recently crossed Hif-1α cKO mice with Flt-3ITD knockin mice. We observed that a large fraction of hematopoietic cells in these mice showed a spontaneous deletion prior to pIpC injection ((Velasco-Hernandez et al., 2015) and Figure 2A), consistent with previous publications (Mead et al., 2013, Mupo et al., 2013). To test if this was present also early in development, we analyzed deletion in fetal liver (FL) cells. FL hematopoietic cells showed either no deletion or only a minor portion of deleted cells in a few colonies (Figures 2A and S3), indicating that spontaneous Mx1-Cre-mediated excision is a cumulative effect during the lifespan of these animals. Since it has been reported that floxed alleles have different sensitivities to Cre-induced recombination (Liu et al., 2013), we investigated spontaneous recombination in a different (Phd2flox/flox; Mx1-Cre; Flt-3ITD/+) model. Using this, colonies derived from BM cells were analyzed and consistently presented a high rate of recombination in the absence of pIpC injection (Figures 2B and S3). Because FLT-3 signaling is critical for dendritic cell (DC) differentiation and proliferation (Gilliet et al., 2002, Karsunky et al., 2003, Maraskovsky et al., 1996, O'Keeffe et al., 2002), and plasmacytoid DCs (pDCs) are prominent producers of interferon (Colonna et al., 2004), we analyzed BM and spleen in Flt-3ITD mice for pDCs and classical DCs (cDCs) (Figures 2C, 2D, and S3). In both organs, we observed increments in pDCs and cDCs when comparing Flt-3ITD/+ with WT mice. These data imply that spontaneous recombination can be induced by FLT-3ITD expression, probably in an interferon-dependent manner, illustrating that this phenomenon is not unique to the transplantation system.
Figure 2.
Spontaneous Deletion of Different Floxed Genes in Hematopoietic Cells from Mx1-Cre Mice Expressing Flt-3ITD
(A) Evaluation of Hif-1α deletion in colonies derived from fetal liver (E14.5) (n = 4) or BM (12-week-old mice) (n = 3) cells from Flt-3ITD/+; Hif-1αflox/flox; Mx1-Cre embryos/mice.
(B) Evaluation of Phd2 deletion in colonies from BM (12-week-old mice) cells (n = 2) from Flt-3ITD/+; Phd2flox/flox; Mx1-Cre mice.
(C and D) Flow cytometry analysis of dendritic cells (pDCs [CD45+CD19−CD3−CD11b−B220+ CD11clow/int] and cDCs [CD45+CD19−CD3−CD11b−B220−CD11c+]) (C) in BM and spleen (D) from FLT-3ITD/+ and WT animals (n = 6). Plots show mean ± SEM. ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S3.
The Immunophenotype of HSPCs Is Transiently Perturbed Following pIpC Administration
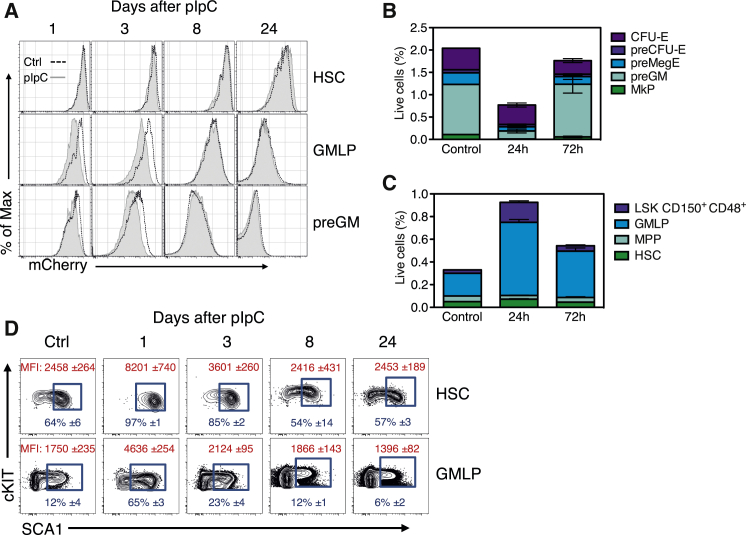
Interferon signaling has previously been shown to enhance proliferation of HSPCs in vivo (Baldridge et al., 2010, Essers et al., 2009). To evaluate this, we took advantage of an in vivo approach in which a histone 2B-mCherry fusion protein can be conditionally expressed in a doxycycline-inducible manner (Sawen et al., 2016).
Col1a1-tetO-H2B-mCherry mice were administered doxycycline to label the pool of HSPCs, followed by assessments of mCherry intensities in phenotypically defined subsets at different time points (days 1, 3, 8, and 24) after pIpC or PBS (control) injections. While proliferation of hematopoietic stem cells (HSCs, Lin−SCA1+cKIT+CD150+CD48−) was not appreciably altered at any time point after pIpC injections (Figure 3A), lower mCherry intensities suggested enhanced proliferation of granulocyte/macrophage lymphoid progenitors (GMLPs, Lin−SCA1+cKIT+CD150−CD48+) and more committed pre-granulocyte/macrophage progenitors (preGMs, Lin−cKIT+SCA1−CD150−CD105−) in the short term (days 1 and 3) after pIpC administration (Figure 3A). However, this effect was not observed at later time points. While this potentially could reflect an elimination of more rapidly proliferating cells at later times, the frequencies of different HSPC subsets (Figure S4) were dramatically altered early after pIpC injections, but largely normalized after 3 days (Figures 3B and 3C). We noted transient elevations in the levels of SCA1 1 day after pIpC injections, leading to higher expression and frequency of SCA1+ cells (Figure 3D). This declined to levels observed in untreated mice relatively rapidly. Therefore, pIpC doses typically used for Cre induction do not cause any pronounced effects on HSPC proliferation, but pIpC-mediated induction of SCA1 can complicate more refined phenotypic analyses of these cells by “pushing” SCA1−/low progenitor cells into the pool of more primitive LSK cells. These data highlight an additional and unwanted transient effect of pIpC administration on hematopoiesis that can lead to erroneous conclusions on cell identities in short-term studies (before day 8).
Figure 3.
Alterations in Phenotype of BM Stem and Progenitor Subsets after pIpC Injections
(A) Col1a1-tetO-H2B-mCherry mice fed with doxycycline-containing food were injected with pIpC and analyzed 1 (n = 4), 3 (n = 4), 8 (n = 3), and 24 days (n = 3) after the last injection. Representative histograms show H2B-mCherry label retention in HSPCs at the indicated time points after pIpC injection.
(B and C) Frequencies of (B) phenotypic myeloid progenitors and (C) LSK HSPC subsets 24 or 72 hr following one pIpC injection.
(D) SCA1 expression in phenotypically defined HSCs and GMLPs. In red, mean fluorescent intensity (MFI) values of SCA1 and, in blue, average percentage of SCA1+ cells (blue gating). Plots represent mean ± SD. Ctrl, control; HSC, hematopoietic stem cell; GMLP, granulocyte/macrophage lymphoid progenitors; preGM, pre-granulocyte/macrophage progenitors; MkP, megakaryocyte progenitor; preMegE, pre-megakaryocyte–erythrocyte progenitors; preCFU-E, pre-colony-forming units-erythrocytes; CFU-E, colony-forming units-erythrocytes; MPP, multipotent progenitor; LSK, Lin−SCA1+cKIT+. See also Figure S4.
An Alternative In Vitro Cre-Recombination Strategy Using Ex Vivo Tat-Cre Treatment
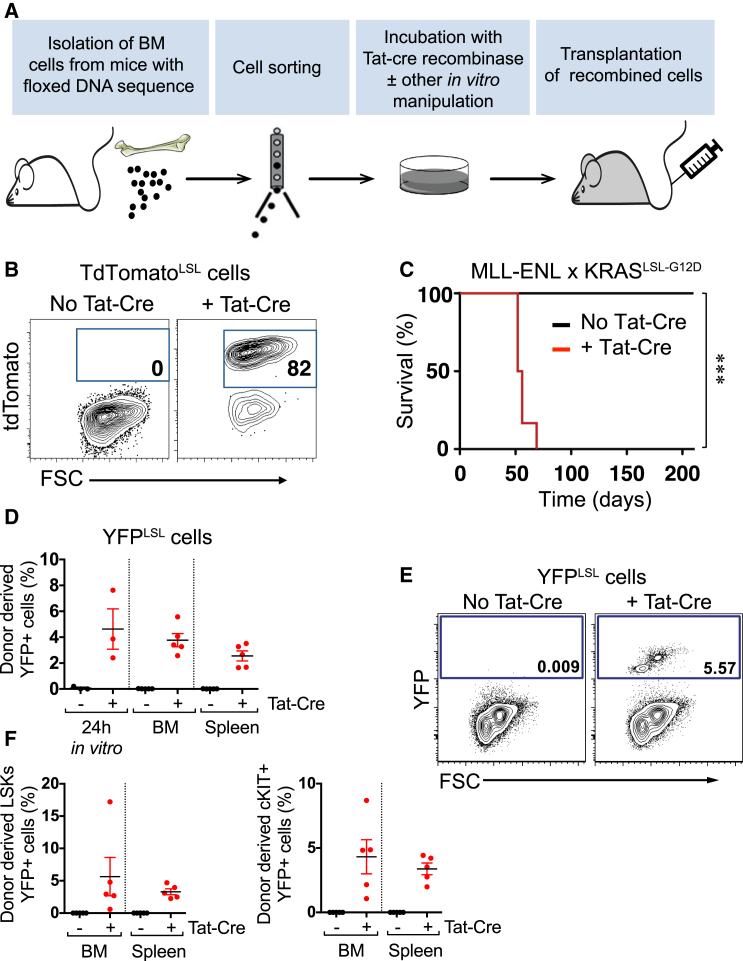
Finally, to explore an alternative approach to the Mx1-Cre-mediated recombination, we subjected HSPCs from Rosa26-LoxP-Stop-LoxP-RFP(TdTomato) (TdTomatoLSL) mice to a brief in vitro exposure to recombinant Tat-Cre. This revealed an efficient induction of RFP expression, with no spontaneous deletion in the absence of Tat-Cre (Figures 4A and 4B). We next applied this deletion strategy to investigate the impact of an activating KRASG12D mutation on MLL-ENL-driven leukemia in vivo. This KRASG12D model has previously been shown to associate with a relatively high level of spontaneous activation when crossed to Mx1-Cre mice (Chan et al., 2004, Sabnis et al., 2009). After crossing doxycycline-inducible MLL-ENL mice (Ugale et al., 2014) to the KRASLSL−G12D strain, we sorted HSCs and subjected these to Tat-Cre or sham recombination, followed by transplantation. As expected (Ugale et al., 2014), recipients of HSCs expressing only the MLL-ENL oncogene (no Tat-Cre) failed to develop leukemia, while all recipients with combined activation of KRASG12D and MLL-ENL (+ Tat-Cre) developed myeloid leukemia within 10 weeks (Figure 4C). To test this strategy in a non-leukemic model in which recombined cells have no selective advantage, we isolated LSK cells from the YFPLSL model (lacking Mx1-Cre) and applied the same protocol (Figures 4D and 4E). We observed recombination rates of 2%–8%, also present in BM and spleen cells 15 days after transplantation. We did not observe any undesired recombination in sham-recombined cells. These results demonstrate the utility of an alternative recombination strategy that can circumvent some of the technical caveats associated with the Mx1-Cre system, although we have not rigorously shown that recombination occurs in stem cells with this system.
Figure 4.
Alternative Strategy for Cre-Mediated Excision Using Ex Vivo Treatment with Tat-Cre Recombinase
(A) Schematic outline of the in vitro Cre-recombination alternative strategy using Tat-Cre recombinase.
(B) TdTomatoLSL HSCs were treated with Tat-Cre in vitro. Representative fluorescence-activated cell sorting (FACS) plots of TdTomato expression in cells after 3 days in culture are shown.
(C) Col1a1-tetO-MLL/ENL x KRASLSL−G12D HSCs were induced in vitro with Tat-Cre to promote the activation of KRASG12D. Kaplan-Meier curves depict the survival of mice transplanted with Tat-Cre-induced (n = 6) or uninduced (n = 7) HSCs, under continuous MLL-ENL activation with doxycycline-containing food. ∗∗∗p < 0.001.
(D) YFP expression in BM and spleen of transplanted mice with YFPLSL LSK cells, Tat-Cre or sham treated, 15 days after transplantation (donor mice, n = 3; transplanted mice, n = 5).
(E) Representative FACS plots of YFP expression in donor BM cells.
(F) YFP expression in LSK and cKIT+ cells in the transplanted mice shown in (D).
Plots show means ± SEM.
Discussion
In experimental hematopoiesis, the requirement of specific genes for normal HSC function, initiation, and maintenance of leukemia, using either transplantation alone or the retroviral transduction/transplantation model, has been tested extensively using specific cKO mice (Kocabas et al., 2012, Takeishi et al., 2013, Wang et al., 2010, Zhang et al., 2004). However, the requirement of a specific gene for leukemia initiation will be difficult to evaluate if the gene to be tested is deleted in an uncontrolled manner.
It is a general assumption that deletion of floxed genes using the Mx1-Cre system occurs by pIpC injections either in steady-state hematopoiesis or after engraftment into recipients. Yet, despite its widespread use, little information is available on baseline levels of recombination and the influence of transplantation itself on spontaneous gene deletion. When we investigated the role of HIF-1α in leukemia initiation by transducing HSPCs from Hif-1α cKO mice with leukemic oncogenes, we observed a 40% gene deletion frequency after transplantation, but prior to pIpC injections. This led us to further investigate this phenomenon because of its broader importance for hematopoiesis and leukemia research, in which these methods are commonly used.
In the literature, a spontaneous recombination rate of 2%–3% when using the Mx1-Cre deleter strain has been described (Kemp et al., 2004, Kuhn et al., 1995, Mupo et al., 2013). Even a low recombination rate can lead to a high gene deletion frequency if the cells with the deleted gene have a growth advantage; for example, when investigating leukemia initiation. Based on our findings, Mx1-Cre mice not injected with pIpC should be included in all experiments using this model and deletion assessed before initiation of experiments. This is not unique to the transplantation scenario, as we observed that activation of Mx1-Cre can also be triggered by an oncogene (constitutive active Flt-3). Activation of this pathway, which leads to excessive generation and activation of interferon-producing DCs, is likely induced also by other activated receptor tyrosine kinases, and might therefore be particularly problematic in combination with the Mx1-Cre system.
The Mx1-Cre system can be potentially associated with additional problems. For instance, pIpC administration has been suggested to trigger HSC cycling, similar to interferon itself (Essers et al., 2009, Gidali et al., 1981, Sugiyama et al., 2006). While we found less evidence for a dramatic effect of pIpC in inducing HSPC proliferation, we noted that pIpC induced rapid and transient changes (that resolved by day 8) in cellular phenotypes, which could affect the interpretation of experimental results.
Several alternatives to the Mx1-Cre model can be found in the literature (Joseph et al., 2013, Kemp et al., 2004, Lewandoski, 2001, Rossi et al., 2012). In addition, we provide evidence for the use of Tat-Cre recombinase (Peitz et al., 2002) as an alternative method (De Santa et al., 2007, Gordon et al., 2012, Li et al., 2014). Because Tat-Cre-mediated excision represents an exogenous expression system, cells are not susceptible to undesired recombination. However, this method also has limitations since it relies on an ex vivo step for the treatment with Tat-Cre, and is therefore not compatible with steady-state studies or for the study of genes necessary for homing. In addition, we have observed a significant variation in the recombination rate depending on the floxed gene, as described previously for Cre strains in general (Liu et al., 2013). Although the data we report herein displayed a rather low recombination rate, deletion efficiencies can be substantially increased in a cell-type-dependent and procedure-specific manner (data not shown). However, it is perhaps unlikely that the procedure will be able to reach 100% efficiencies. The titratable recombination rates can, however, also be beneficial, for instance in leukemogenesis studies, in which only a small percentage of recombined cells is desired.
In summary, we present evidence from multiple experimental conditions and procedures that can contribute to the induction of high spontaneous recombination in Mx1-Cre-based cKO mice. Being up to 20-fold higher than the ones reported in the literature for steady-state conditions, our studies highlight the importance of proper controls when applying this system to study gene function in the hematopoietic system.
Experimental Procedures
Mice
The following mouse strains were used: Flt-3ITD (Lee et al., 2007) (kindly provided by G. Gilliland), Mx1-Cre (Kuhn et al., 1995), Hif-1αflox/flox (Ryan et al., 2000), Phd2flox/flox (kindly provided by P. Carmeliet), YFPLSL (Srinivas et al., 2001) (JAX 006148), Col1a1-tetO-H2B-mCherry (Egli et al., 2007) (JAX 014602), TdTomatoLSL (Madisen et al., 2010) (JAX 007905), Col1a1-tetO-MLL/ENL (Ugale et al., 2014), and KRASLSL-G12D (Jackson et al., 2001) (JAX 008179). To activate Mx1-Cre in vivo, 400 μg of pIpC (Sigma-Aldrich) was either injected three times every second day or once (analyzed samples at days 1 and 3). Mice were maintained at the animal facilities of the Biomedical Center at Lund University. All experiments were performed with consent from a local ethics committee.
Statistical Analysis
Data were analyzed using Microsoft Excel (Microsoft) and Prism 6 (GraphPad Software). Data are expressed as means ± SEM or SD. “n” always stands for individual mice. Differences between groups were assessed by two-tailed Student's t test. Survival rates were plotted using the Kaplan-Meier method (significance values were calculated by Mantel-Cox log rank test). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Author Contributions
T.V. and P.S. performed the experiments, analyzed the data, and generated the figures. T.V., P.S., D.B., and J.C. designed the experiments and wrote the paper.
Acknowledgments
We would like to thank Gary Gilliland (Fred Hutchinson Cancer Center, Seattle, USA) and Peter Carmeliet (Flanders Institute for Biotechnology, VIB, KU Leuven, Belgium) for kindly providing Flt-3ITD and Phd2flox/flox mice, respectively. This study was financed by a post-doctoral fellowship grant and a project grant by Barncancerfonden to T.V. and J.C., respectively. Additional financing was provided by ERC consolidator grant 615068 to D.B. and the Hemato-Linne grant from Vetenskapsrådet to D.B. and J.C.
Published: June 30, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.06.002.
Contributor Information
Talia Velasco-Hernandez, Email: talia.velasco@med.lu.se.
Jörg Cammenga, Email: jorg.cammenga@med.lu.se.
Supplemental Information
References
- Baldridge M.T., King K.Y., Boles N.C., Weksberg D.C., Goodell M.A. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 2010;465:793–797. doi: 10.1038/nature09135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan I.T., Kutok J.L., Williams I.R., Cohen S., Kelly L., Shigematsu H., Johnson L., Akashi K., Tuveson D.A., Jacks T. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J. Clin. Invest. 2004;113:528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M., Trinchieri G., Liu Y.J. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- De Santa F., Totaro M.G., Prosperini E., Notarbartolo S., Testa G., Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Egli D., Rosains J., Birkhoff G., Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., Trumpp A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- Gidali J., Feher I., Talas M. Proliferation inhibition of murine pluripotent haemopoietic stem cells by interferon or poly I:C. Cell Tissue Kinet. 1981;14:1–7. doi: 10.1111/j.1365-2184.1981.tb00505.x. [DOI] [PubMed] [Google Scholar]
- Gilliet M., Boonstra A., Paturel C., Antonenko S., Xu X.L., Trinchieri G., O'Garra A., Liu Y.J. The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 2002;195:953–958. doi: 10.1084/jem.20020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.M., Chaix J., Rupp L.J., Wu J., Madera S., Sun J.C., Lindsten T., Reiner S.L. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E.L., Willis N., Mercer K., Bronson R.T., Crowley D., Montoya R., Jacks T., Tuveson D.A. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R., Gridley T. Gene targeting: things go better with Cre. Curr. Biol. 1997;7:R321–R323. doi: 10.1016/s0960-9822(06)00149-7. [DOI] [PubMed] [Google Scholar]
- Joseph C., Quach J.M., Walkley C.R., Lane S.W., Lo Celso C., Purton L.E. Deciphering hematopoietic stem cells in their niches: a critical appraisal of genetic models, lineage tracing, and imaging strategies. Cell Stem Cell. 2013;13:520–533. doi: 10.1016/j.stem.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Karsunky H., Merad M., Cozzio A., Weissman I.L., Manz M.G. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J. Exp. Med. 2003;198:305–313. doi: 10.1084/jem.20030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp R., Ireland H., Clayton E., Houghton C., Howard L., Winton D.J. Elimination of background recombination: somatic induction of Cre by combined transcriptional regulation and hormone binding affinity. Nucleic Acids Res. 2004;32:e92. doi: 10.1093/nar/gnh090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabas F., Zheng J., Thet S., Copeland N.G., Jenkins N.A., DeBerardinis R.J., Zhang C., Sadek H.A. Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood. 2012;120:4963–4972. doi: 10.1182/blood-2012-05-432260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R., Schwenk F., Aguet M., Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Tothova Z., Levine R.L., Anderson K., Buza-Vidas N., Cullen D.E., McDowell E.P., Adelsperger J., Frohling S., Huntly B.J. FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer Cell. 2007;12:367–380. doi: 10.1016/j.ccr.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M. Conditional control of gene expression in the mouse. Nat. Rev. Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- Li Y.F., Xu S., Ou X., Lam K.P. Shp1 signalling is required to establish the long-lived bone marrow plasma cell pool. Nat. Commun. 2014;5:4273. doi: 10.1038/ncomms5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Willet S.G., Bankaitis E.D., Xu Y., Wright C.V., Gu G. Non-parallel recombination limits Cre-LoxP-based reporters as precise indicators of conditional genetic manipulation. Genesis. 2013;51:436–442. doi: 10.1002/dvg.22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraskovsky E., Brasel K., Teepe M., Roux E.R., Lyman S.D., Shortman K., McKenna H.J. Dramatic increase in the numbers of functionally mature dendritic cells in Flt3 ligand-treated mice: multiple dendritic cell subpopulations identified. J. Exp. Med. 1996;184:1953–1962. doi: 10.1084/jem.184.5.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead A.J., Kharazi S., Atkinson D., Macaulay I., Pecquet C., Loughran S., Lutteropp M., Woll P., Chowdhury O., Luc S. FLT3-ITDs instruct a myeloid differentiation and transformation bias in lymphomyeloid multipotent progenitors. Cell Rep. 2013;3:1766–1776. doi: 10.1016/j.celrep.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mupo A., Celani L., Dovey O., Cooper J.L., Grove C., Rad R., Sportoletti P., Falini B., Bradley A., Vassiliou G.S. A powerful molecular synergy between mutant Nucleophosmin and Flt3-ITD drives acute myeloid leukemia in mice. Leukemia. 2013;27:1917–1920. doi: 10.1038/leu.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keeffe M., Hochrein H., Vremec D., Pooley J., Evans R., Woulfe S., Shortman K. Effects of administration of progenipoietin 1, Flt-3 ligand, granulocyte colony-stimulating factor, and pegylated granulocyte-macrophage colony-stimulating factor on dendritic cell subsets in mice. Blood. 2002;99:2122–2130. doi: 10.1182/blood.v99.6.2122. [DOI] [PubMed] [Google Scholar]
- Peitz M., Pfannkuche K., Rajewsky K., Edenhofer F. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl. Acad. Sci. USA. 2002;99:4489–4494. doi: 10.1073/pnas.032068699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L., Lin K.K., Boles N.C., Yang L., King K.Y., Jeong M., Mayle A., Goodell M.A. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 2012;11:302–317. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan H.E., Poloni M., McNulty W., Elson D., Gassmann M., Arbeit J.M., Johnson R.S. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- Sabnis A.J., Cheung L.S., Dail M., Kang H.C., Santaguida M., Hermiston M.L., Passegue E., Shannon K., Braun B.S. Oncogenic Kras initiates leukemia in hematopoietic stem cells. PLoS Biol. 2009;7:e59. doi: 10.1371/journal.pbio.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawen P., Lang S., Mandal P., Rossi D.J., Soneji S., Bryder D. Mitotic history reveals distinct stem cell populations and their contributions to hematopoiesis. Cell Rep. 2016;14:2809–2818. doi: 10.1016/j.celrep.2016.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Kohara H., Noda M., Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Takeishi S., Matsumoto A., Onoyama I., Naka K., Hirao A., Nakayama K.I. Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer Cell. 2013;23:347–361. doi: 10.1016/j.ccr.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Ugale A., Norddahl G.L., Wahlestedt M., Sawen P., Jaako P., Pronk C.J., Soneji S., Cammenga J., Bryder D. Hematopoietic stem cells are intrinsically protected against MLL-ENL-mediated transformation. Cell Rep. 2014;9:1246–1255. doi: 10.1016/j.celrep.2014.10.036. [DOI] [PubMed] [Google Scholar]
- Velasco-Hernandez T., Hyrenius-Wittsten A., Rehn M., Bryder D., Cammenga J. HIF-1alpha can act as a tumor suppressor gene in murine acute myeloid leukemia. Blood. 2014;124:3597–3607. doi: 10.1182/blood-2014-04-567065. [DOI] [PubMed] [Google Scholar]
- Velasco-Hernandez T., Tornero D., Cammenga J. Loss of HIF-1alpha accelerates murine FLT-3ITD-induced myeloproliferative neoplasia. Leukemia. 2015;29:2366–2374. doi: 10.1038/leu.2015.156. [DOI] [PubMed] [Google Scholar]
- Wang Y., Krivtsov A.V., Sinha A.U., North T.E., Goessling W., Feng Z., Zon L.I., Armstrong S.A. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Iwasaki-Arai J., Iwasaki H., Fenyus M.L., Dayaram T., Owens B.M., Shigematsu H., Levantini E., Huettner C.S., Lekstrom-Himes J.A. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.