Abstract
K-Ras4B is a membrane-bound small GTPase with a prominent role in cancer development. It contains a polybasic farnesylated C-terminus that is required for the correct localization and clustering of K-Ras4B in distinct membrane domains. PDEδ and the Ca2+-binding protein calmodulin (CaM) are known to function as potential binding partners for farnesylated Ras proteins. However, they differ in the number of interaction sites with K-Ras4B, leading to different modes of interaction, and thus affect the subcellular distribution of K-Ras4B in different ways. Although it is clear that Ca2+-bound CaM can play a role in the dynamic spatial cycle of K-Ras4B in the cell, the exact molecular mechanism is only partially understood. In this biophysical study, we investigated the effect of Ca2+/CaM on the interaction of GDP- and GTP-loaded K-Ras4B with heterogeneous model biomembranes by using a combination of different spectroscopic and imaging techniques. The results show that Ca2+/CaM is able to extract K-Ras4B from negatively charged membranes in a nucleotide-independent manner. Moreover, the data demonstrate that the complex of Ca2+/CaM and K-Ras4B is stable in the presence of anionic membranes and shows no membrane binding. Finally, the influence of Ca2+/CaM on the interaction of K-Ras4B with membranes is compared with that of PDEδ, which was investigated in a previous study. Although both CaM and PDEδ exhibit a hydrophobic binding pocket for farnesyl, they have different effects on membrane binding of K-Ras4B and hence should be capable of regulating K-Ras4B plasma membrane localization in the cell.
Introduction
Ras GTPases are key regulators of signal transduction pathways that control cell proliferation, differentiation, survival, and apoptosis. As binary molecular switches, they cycle between inactive and active nucleotide-bound states at the plasma membrane, linking extracellular signals through membrane receptors to intracellular signaling cascades (1, 2, 3, 4). Among the activating mutations in Ras genes, K-Ras mutations are predominant, with K-Ras4B mutations occurring in up to 90% of pancreatic tumors, 57% of colorectal tumors, and 50% of lung cancers (cf. Ref. (5) and references therein) (6, 7). The 4B splice variant of K-Ras (K-Ras4B) is localized to the cytoplasmic, negatively charged leaflet of the plasma membrane by a farnesylated and polybasic targeting sequence within its hypervariable region (2, 8, 9, 10, 11) (Fig. 1 A). Recent results revealed an essential role of the cytosolic, GDI-like solubilizing factor GMP phosphodiesterase 6 delta subunit (PDEδ) in the plasma membrane localization of K-Ras4B, whereby the Arl2-PDEδ perinuclear membrane delivery system seems to regulate a dynamic spatial cycle of K-Ras4B in the cell (12, 13, 14). The hydrophobic pocket of PDEδ binds and solubilizes farnesylated K-Ras4B proteins irrespective of the bound nucleotide state, thereby facilitating their cytoplasmic diffusion (12, 14). Moreover, it was suggested that K-Ras4B can be redirected from the plasma membrane to intracellular membranes through a Ca2+-calmodulin (CaM)-dependent pathway (15). Ca2+-loaded CaM (Ca2+/CaM) specifically binds the Ras isoform K-Ras4B and modulates its downstream signaling (15, 16, 17).
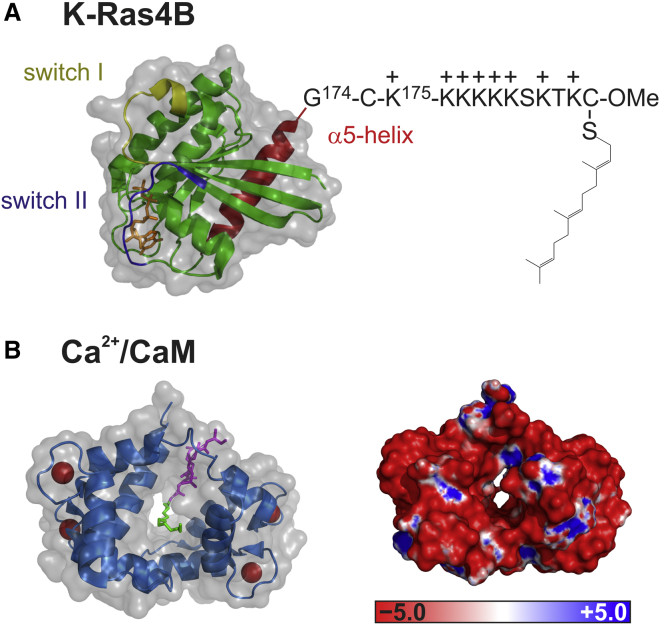
Figure 1.
Schematic representation of the semisynthetic K-Ras4B protein and Ca2+/CaM (PDB: 3GFT and 1L7Z, respectively). (A) The synthesized, lipidated K-Ras4B peptide was ligated to a truncated K-Ras4B protein expressed in Escherichia coli to yield the S-farnesylated K-Ras4B protein bearing an additional cysteine between Gly174 and Lys175 (30). (B) The left image shows the crystal structure of Ca2+/CaM in complex with a myristoylated peptide. The N- and C-terminal domains of CaM are shown in blue and the four bound Ca2+ ions are shown as red spheres. The myristoyl group (green) goes through the hydrophobic tunnel of Ca2+/CaM, and the peptide consisting of GGKLSK (purple) is located along the groove between the two lobes of Ca2+/CaM (24). On the right, an Adaptive Poisson-Boltzmann Solver-generated electrostatic surface of Ca2+/CaM is displayed with the use of PyMOL (58). The positive (blue) and negative (red) electrostatic potential isocontours were set to +5 and −5 kBT e−1, respectively.
The calcium-modulated protein CaM is a small, highly conserved, intracellular, acidic EF-hand protein that is present in many eukaryotic cells (18, 19, 20). Binding of Ca2+/CaM to its target proteins is enabled mainly by hydrophobic interactions of hydrophobic patches that become exposed in its Ca2+-loaded state (21). Electrostatic interactions contribute to the stability of the Ca2+/CaM target complex and have been suggested to play an important role in target recognition (18, 22, 23). However, the crystal structure of Ca2+/CaM bound to a myristoylated peptide revealed a different binding mode. Whereas the conformation of Ca2+/CaM is similar to those of other Ca2+/CaM-peptide complexes, the N-terminal myristoyl group was found to be firmly anchored to Ca2+/CaM by multiple hydrophobic interactions, i.e., it is accommodated in a large hydrophobic cavity created by the hydrophobic pockets of the N- and C-terminal domains of Ca2+/CaM (Fig. 1 B, left). In addition, intermolecular electrostatic interactions were shown to play a significant role. When the peptide is located along the groove between the two lobes of CaM, the C-terminal basic cluster of the peptide (myr-GGKLSKKKK) becomes surrounded by many acidic residues of CaM (24).
There is evidence that Ca2+/CaM binds to K-Ras4B and dissociates it from membranes independently of the nucleotide state of K-Ras4B (15, 25). In contrast, a recent study observed that inhibition of Ca2+/CaM reduced the rate of dissociation from the plasma membrane for active K-Ras4B only (26). In solution, an equimolar, GTP-dependent binding of K-Ras4B to Ca2+/CaM, with micromolar affinity, was detected (16, 17, 27). Previous studies revealed that at least three different regions of K-Ras4B are important for the interaction with Ca2+/CaM: 1) the hypervariable region as the primary binding site, with both the farnesyl group and polybasic sequence being essential for binding; 2) the amphipathic helix α5; and 3) the switch II region (which could explain the observed GTP dependence) (15, 17, 27, 28). Although the importance of farnesylation for binding of K-Ras4B to CaM is well established (15, 28), no crystal structure of such a complex is available so far. However, in a study by Matsubara et al. (24), the crystal structure of Ca2+/CaM in complex with a myristoylated peptide that also contains a polybasic stretch of lysines (myr-GGKLSKKKK) revealed an ellipsoidal, compact structure for CaM, whose hydrophobic cavity is mainly filled with the myristoyl moiety (cf. Fig. 1 B). These authors also demonstrated that CaM residues within 5 Å of the myristoyl group are mainly hydrophobic, and that multiple hydrophobic interactions are responsible for binding. The binding interface of the myristoyl group in the hydrophobic tunnel of the CaM complex is shown in Fig. S1 in the Supporting Material. The space model indicates that although the farnesyl residue is unsaturated and branched (in contrast to myristoyl), the myristoyl-binding pocket is large enough to also accommodate the bulkier farnesyl moiety.
Although it is clear that Ca2+/CaM can regulate K-Ras4B cycling, the exact molecular mechanism is only partially understood. In this study, we sought to determine whether Ca2+/CaM directly extracts K-Ras4B from membranes or binds K-Ras4B only after its dissociation from heterogeneous model membranes. In particular, we investigated the debated nucleotide dependence of the process. A combination of different biophysical techniques, namely, surface plasmon resonance (SPR), atomic force microscopy (AFM), infrared reflection absorption spectroscopy (IRRAS), and fluorescence anisotropy, was used. An analysis of the interaction of K-Ras4B with Ca2+/CaM in the absence and presence of heterogeneous model membranes revealed that Ca2+/CaM can extract membrane-bound K-Ras4B independently of its nucleotide-bound state, contrary to what was observed in a previous study of PDEδ (29).
Materials and Methods
Materials and sample preparation
The phospholipids 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) sodium salt (DOPG), 1,2-dipalmitoyl-sn-glycero-3-phospho-(1′-rac-glycerol) sodium salt (DPPG), and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) were purchased from Avanti Polar Lipids (Alabaster, AL) as lyophilized powders. Cholesterol (Chol) and 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (Hepes) were obtained from Sigma Aldrich (Deisenhofen, Germany). Magnesium chloride, tris(hydroxymethyl)-aminomethan (Tris), chloroform, and CaM (bovine brain) were obtained from Merck (Darmstadt, Germany); bovine serum albumin from Pierce (Bonn, Germany). The fluorescent marker 5-(dimethylamino)naphthalene-1-sulfonyl chloride (dansyl chloride) was purchased from Life Technologies (Darmstadt, Germany) and 1,4-bis(5-phenyloxazol-2-yl)benzene was obtained from Alfa Aesar (Karlsruhe, Germany). Details regarding the formation of large unilamellar vesicles (100 nm in size) can be found in Supporting Material.
Protein synthesis and purification
The synthesis of K-Ras4B proteins has been described in detail before (30). Briefly, the S-farnesylated K-Ras4B protein was synthesized by a combination of expressed protein ligation and lipopeptide synthesis. Nucleotide exchange (GppNHp-bound state as a nonhydrolyzable GTP analog) was carried out as described before (31). Dansyl labeling of CaM was performed by incubating 5 mg protein in 250 μL buffer (100 mM NaHCO3, 1 mM CaCl2, pH 8.5) with a fivefold molar excess of dansyl chloride for 2 h at 4°C. Unbound fluorophores were removed with the use of a 5 mL HiTrap desalting column (GE Healthcare, Freiburg, Germany). A degree of labeling of 46% was determined by absorbance measurements at 280 nm (ε280 = 3345 M−1 cm−1) and 340 nm (ε340 = 3400 M−1 cm−1).
SPR
SPR experiments were carried out with a Biacore 3000 system (Biacore, Uppsala, Sweden; now GE Healthcare). For the protein-membrane interaction studies, the L1 sensor chip (GE Healthcare) was used. Details on sample preparation, vesicle immobilization, performance of SPR measurements, regeneration of the chip surface, and analysis of the SPR sensorgrams have been described before (29, 32) and are given in Supporting Materials and Methods. For the curve-fitting procedure, BIAevaluation software 4.1 (Biacore, Uppsala, Sweden) and Origin 8.6 (OriginLab, Northampton, MA) were used.
IRRAS
IRRAS experiments were carried out on a setup consisting of two linked Teflon troughs and a Vertex 70 Fourier transform infrared (FT-IR) spectrometer connected to an A511 reflection attachment (both Bruker, Germany) with an MCT detector. The measurements were performed according to the setup, sample preparation, and spectra analysis described before (29, 31). The temperature of the subphase was maintained at 20°C ± 0.5°C and time-dependent measurements were performed in the small (reference) trough at a constant surface area. The resulting curve of surface pressure versus time is referred to as the π/t isotherm. Both troughs were filled with 20 mM Tris, 7 mM MgCl2, 1 mM CaCl2, pD 7.4. Monolayers of DOPC/DOPG/DPPC/DPPG/Chol 20:5:45:5:25 (mol%) were formed by directly spreading the lipid solution (1 mM) in a mixture of chloroform and methanol (3:1) onto the subphase. The lipid monolayer was equilibrated until a constant surface pressure of ∼20 mN m−1 was detected. Protein measurements were performed by careful injection of the concentrated protein solution (cK-Ras4B GDP ≈ 300 μM, cK-Ras4B GTP ≈ 150−200 μM in 20 mM Tris, 5 mM MgCl2, 1 mM dithioerythritol pH 7.5; cCaM ≈ 270 μM in 10 mM Hepes, 5 mM MgCl2, 150 mM NaCl, 1 mM CaCl2 pH 7.4) through the lipid monolayer into the D2O subphase to yield a final concentration of K-Ras4B and Ca2+/CaM of 200 and 300 nM, respectively. Further details are given in Supporting Materials and Methods.
AFM
The preparation of the supported lipid bilayers and the AFM setup is described in detail in Refs. (31, 33) and Supporting Materials and Methods. For protein-membrane interaction studies, 800 μL of K-Ras4B (0.2 μM), CaM (0.3 μM), or K-Ras4B/CaM (0.2 μM/0.3 μM) in Tris buffer (20 mM Tris, 7 mM MgCl2, 1 mM CaCl2, pH 7.4) was injected into the AFM fluid cell and allowed to incubate for 1 h at room temperature. Measurements were performed on a MultiMode scanning probe microscope with a NanoScope IIIa controller (Digital Instruments (now Bruker), Santa Barbara, CA) and a J-scanner (scan size 125 μm). Images were obtained by applying the tapping mode in liquid with sharp nitride lever probes mounted in a fluid cell (MTFML, both from Veeco (now Bruker), Mannheim, Germany).
Fluorescence anisotropy
Frequency-domain fluorescence anisotropy measurements were performed at 25°C with a K2 multifrequency phase and modulation fluorometer (ISS, Champaign, IL). Dansyl-labeled CaM was excited by a 370 nm laser diode (370 ± 10 nm excitation filter) directly connected to a function generator, yielding intensity-modulated excitation light over a frequency range of 2−173 MHz at a cross-correlation frequency of 400 Hz. The dansyl emission was collected through a 400 nm long-pass filter. Fluorescence lifetime measurements were carried out in 5 mm path-length quartz cuvettes under magic-angle conditions before anisotropy experiments were conducted. Experimental data were fitted with the use of VINCI analysis software (ISS). A more detailed description of the setup, sample preparation, and data analysis is given in Supporting Materials and Methods.
Results and Discussion
In the last few years, the importance of Ras localization in different compartments of the plasma membrane has become apparent and the impact of Ras clustering in distinct plasma membrane domains has been studied (34, 35). There is evidence that distinct regions in cell membranes, termed rafts, may play a role in a wide range of important biological processes (35, 36). Such liquid-ordered, sphingolipid- and cholesterol-enriched domains could also act as signaling platforms, coupling events outside of the cell to signal transduction pathways in its interior. The well-established anionic model raft membrane system used here consists of DOPC/DOPG/DPPC/DPPG/Chol at a molar ratio of 20:5:45:5:25 and segregates into liquid-ordered and liquid-disordered domains under ambient conditions, thus mimicking a heterogeneous plasma membrane with different degrees of membrane order (37, 38). Phosphatidylglycerol is widely used as a simplified model of negatively charged phospholipids to mimic the electrostatic effects of the monovalent acidic lipids present in mammalian membranes. Previous studies on N-Ras and K-Ras4B revealed a membrane interaction behavior that was independent of the heterogeneous membrane composition (31, 39, 40), justifying the use of the low-melting-temperature lipid DOPC in liquid phase coexistence model systems, although DOPC is rare in mammalian membranes. Semisynthetic, fully functional, lipidated GDP- and GTP-loaded (with the GppNHp-bound state as a nonhydrolyzable GTP analog) K-Ras4B (30) has been investigated in the absence and presence of Ca2+/CaM and/or anionic model raft membranes.
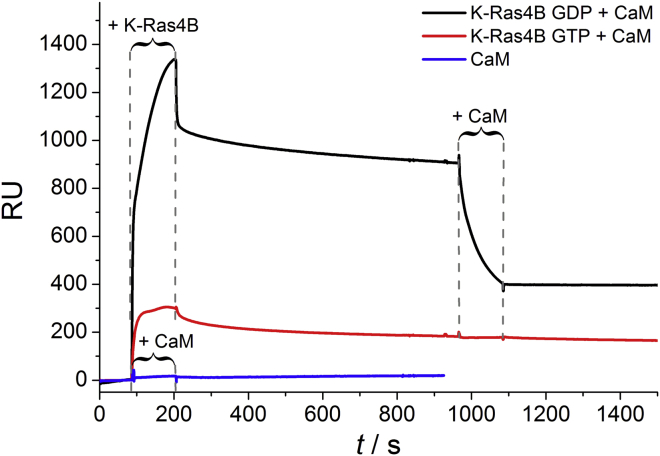
First, we carried out SPR experiments to test the ability of Ca2+/CaM to dissociate K-Ras4B from membranes. SPR allows one to detect the binding and dissociation kinetics of proteins to and from a membrane surface in real time without any labeling by measuring changes in the resonance angle (41). The sensorgram includes a plot of the SPR signal in resonance units (1 RU = 1 pg mm−2, i.e., surface coverage with protein in terms of mass protein/mm2 surface area) against time (42, 43). For Ca2+-loaded CaM, no significant increase in RUs could be observed upon protein injection (Fig. 2), demonstrating that Ca2+/CaM does not bind to the immobilized membrane. The primary cause of this is repulsive interactions between the strongly negatively charged Ca2+/CaM (net charge of −15 at pH 7 with four calcium ions bound; Fig. 1 B, right) and the anionic membrane. In contrast, both GDP- and GTP-bound K-Ras4B bound to anionic model raft membranes, as indicated by an increase in RUs upon protein injection (Fig. 2). Unfarnesylated K-Ras4B was used as a control and showed no significant membrane binding (Fig. S2). While changing the protein solution to buffer, we followed the dissociation of K-Ras4B from the membrane and observed amounts of quasi-irreversible bound protein of 72% and 65% for K-Ras4B GDP and K-Ras4B GTP, respectively (cf. Supporting Materials and Methods). These data are in good agreement with previous results (29). Subsequent addition of Ca2+/CaM resulted in a significant decrease in RUs, i.e., a release of membrane-anchored K-Ras4B GDP. In the Ca2+/CaM−K-Ras4B complex, the farnesyl anchor of K-Ras4B is buried in the hydrophobic tunnel of Ca2+/CaM. This prevents membrane binding of K-Ras4B, which requires insertion of its farnesyl anchor into the membrane. However, no such marked effect could be detected for active K-Ras4B (Fig. 2). By comparison, no membrane extraction was detected for either active or inactive K-Ras4B by PDEδ in a previous study (29), and in contrast to Ca2+/CaM, PDEδ itself exhibited a relatively strong affinity for heterogeneous membranes.
Figure 2.
SPR sensorgrams of the binding of GDP- and GTP-loaded K-Ras4B (c = 2 μM) to anionic lipid raft membranes and subsequent addition of Ca2+/CaM (c = 3 μM).
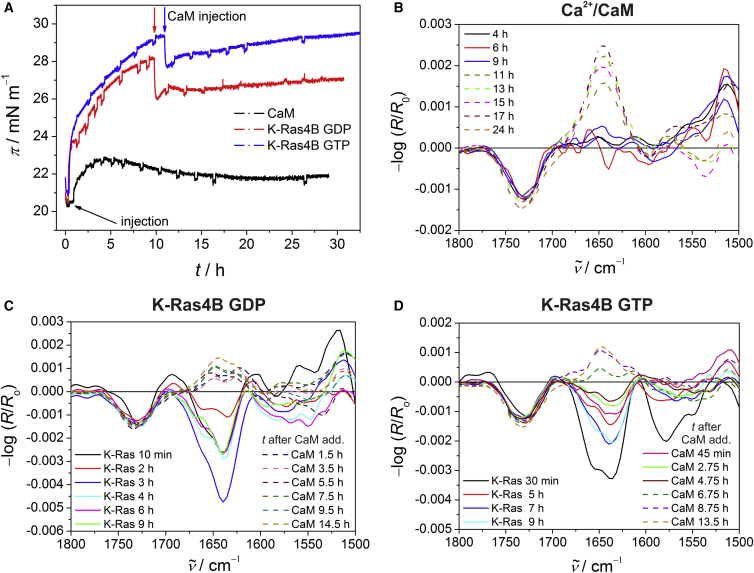
Second, by simultaneously recording IRRA spectra and surface pressure/time (π/t) isotherms, we studied the interaction of K-Ras4B and Ca2+/CaM with anionic lipid raft monolayers. In IRRAS, the slow kinetics of K-Ras distribution in the aqueous subphase is based on the absence of convection, so the process of K-Ras diffusion to the lipid interface is rather slow. Although these interaction processes occur on much faster timescales in vivo (which of course is due to different dynamics in the complex biological cell), the underlying biophysical mechanism will not be influenced by the different dynamics. Owing to the much slower process of diffusion to the lipid interface, even kinetic intermediates may be resolved by IRRAS. The long-term stability and integrity of the pure lipid monolayer on the D2O subphase was ensured in a control experiment (Fig. S3). Injection of the proteins underneath the lipid monolayer was accomplished at π ≈ 20 mN m−1. The minor changes in surface pressure together with the lack of an amide-I′ band in the corresponding IRRA spectra indicate that Ca2+/CaM does not insert into the lipid monolayer. Instead, an accumulation of Ca2+/CaM at the air/D2O interface in the reference trough was detected, with the IR band around 1646 cm−1 being characteristic of the α-helical structure of Ca2+/CaM (Fig. 3, A and B). Since this effect was not observed for membrane-binding proteins, one can assume that protein diffusion into the reference trough only occurs for non-membrane-binding proteins such as Ca2+/CaM, as no accumulation of the protein at the lipid membrane is possible. Again, these results can be explained by repulsive interactions between the strongly negatively charged Ca2+/CaM and the anionic membrane. By comparison, PDEδ exhibits a net charge of approximately −2 at pH 7.5. In a previous study (29), an interaction with the lipid headgroups together with a strong amide-I′ band intensity was detected for PDEδ in the IRRA spectra, indicating a parallel orientation of the β-sheets of PDEδ to the membrane interface.
Figure 3.
(A) Surface pressure profiles for K-Ras4B GDP, K-Ras4B GTP, and Ca2+/CaM upon interaction with the anionic lipid raft monolayer. After equilibration of the pure lipid monolayer, i.e., observation of a stable surface pressure of ∼20 mN m−1, injection of K-Ras4B and CaM alone was carried out at t = 0 h (indicated by black arrow). For GDP- and GTP-loaded K-Ras4B, a concentrated solution of Ca2+/CaM was injected into the D2O subphase after ∼10 h (indicated by red and blue arrows, respectively), resulting in a significant decrease in surface pressure within 15 min for both GDP- and GTP-loaded K-Ras4B. (B–D) Corresponding IRRA spectra for Ca2+/CaM (B), K-Ras4B GDP (C), and K-Ras4B GTP (D) inserted into or adsorbed at the anionic lipid raft monolayer (with an initial surface pressure of ∼20 mN m−1) were acquired with p-polarized light at an angle of incidence of 35°. On the left-hand side in (C) and (D), the legend for binding of K-Ras4B to the lipid monolayer is given in a time-dependent manner up to 9 h after addition of K-Ras4B to the lipid monolayer. After ∼10 h, CaM was injected into the D2O subphase of the Langmuir trough. The legend for the spectra collected after the addition of CaM to the membrane-bound K-Ras4B is given on the right. The dashed lines indicate an accumulation of proteins at the air/D2O interface in the reference trough.
Contrary to Ca2+/CaM, both active and inactive K-Ras4B showed an intensive amide-I′ band in the IRRA spectra, with band maxima around 1639 cm−1 (Fig. 3, D and C, respectively). The corresponding π/t profiles indicate an effective insertion of the K-Ras4B farnesyl anchor into the lipid monolayer, resulting in a marked increase in surface pressure (Fig. 3 A). An increase of ∼10 mN m−1 in membrane surface pressure upon injection of Ras protein is not unusual at such initial surface pressures, as we have shown previously for other Ras proteins (44). This increase results from insertion of the lipid anchors of the lipidated Ras proteins into the lipid monolayer, as nonlipidated Ras constructs are unable to insert into lipid monolayers and thus exhibit no increase in surface pressure (44). Moreover, the data presented here agree with previous studies on K-Ras4B in the presence of membranes (29, 31, 44). Corresponding transmission FT-IR spectroscopic measurements ensured the long-term stability of K-Ras4B proteins in the absence and presence of membranes by revealing no significant changes in secondary structure over the whole time range covered (Fig. S4) (31, 45). After Ca2+/CaM was injected into the D2O subphase of the Langmuir trough containing the membrane-bound K-Ras4B proteins, a significant decrease in surface pressure was observed within ∼15 min (Fig. 3 A), indicating a partial release of K-Ras4B from the lipid monolayer. This decrease was not due to a disturbance of the lipid monolayer during protein injection, since no such decrease was detected when solutions of proteins that have been shown to not interact with the lipid membrane were injected into the subphase underneath the lipid monolayer (44). In contrast to the surface pressure profiles, the intensity changes in the IRRA spectra are not caused only by varying concentrations of protein at the D2O/lipid interface. Since no changes in secondary structure take place for K-Ras4B upon membrane binding (31), contributions from orientational changes of the protein at the lipid interface are observed in the IRRA spectra. Moreover, the possibility that an accumulation of Ca2+/CaM in the reference trough modulates the IRRA spectra intensity of membrane-bound K-Ras4B cannot be ruled out. Hence, it is not possible to quantitatively interpret the intensity changes of the IRRAS amide-I′ band in terms of a decreased localization of K-Ras4B at the lipid monolayer. However, since the structures of the two proteins are quite different (predominantly α-helical for Ca2+/CaM and mixed α/β-type for K-Ras4B), the IR signatures for the two proteins are rather distinct. Thus, the broader amide-I′ band in the IRRA spectra of the reference trough after Ca2+/CaM addition to membrane-bound K-Ras4B (dashed lines in Fig. 3, C and D) as compared with Ca2+/CaM alone (Fig. 3 B), together with the disappearance of the amide-I′ band in the IRRA spectra of K-Ras4B at the lipid interface, points to a partial extraction of membrane-bound K-Ras4B from the lipid monolayer by Ca2+/CaM independently of nucleotide loading (Fig. 3, C and D), supporting the surface pressure results. In line with the electrostatic-repulsion model, binding of the strongly negatively charged Ca2+/CaM to the polybasic stretch of membrane-bound K-Ras4B would reverse its charge, leading to repulsion of the complex from anionic lipid monolayers and, in this case, diffusion of the K-Ras4B−Ca2+/CaM complex into the subphase and an accumulation at the air/D2O interface (dashed lines in Fig. 3, C and D). The observed delay in IR spectral changes as compared with the fast change in surface pressure upon Ca2+/CaM addition is due to the different timescales of data acquisition (i.e., ∼1 s for π and ∼40 min for IR spectra). Moreover, the slightly different timescales for K-Ras4B extraction might be caused by the different orientational flexibilities of GDP- and GTP-loaded K-Ras4B at the lipid membrane, which might modulate their initial interaction with Ca2+/CaM. We previously showed that membrane-bound K-Ras4B GDP adopts a rather fixed conformation at the membrane as compared with K-Ras4B GTP, which displays a much higher orientational flexibility (44). In contrast, the previous study with PDEδ (29) revealed a dissociation of the K-Ras4B−PDEδ complex in the presence of anionic monolayers, since the π profiles and IRRA spectra of the complex resembled those of the GDP- and GTP-loaded K-Ras4B membrane interaction.
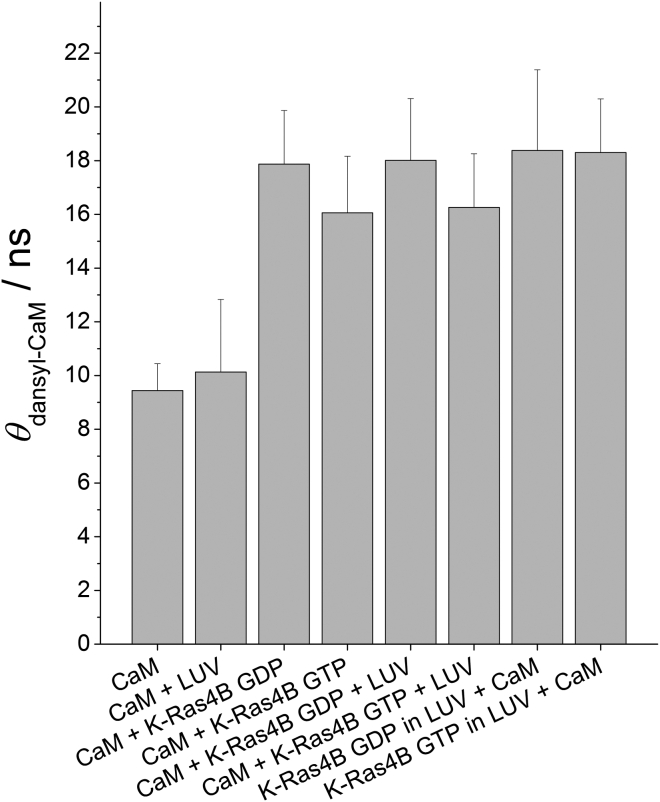
Third, we performed fluorescence anisotropy measurements to unambiguously determine whether or not Ca2+/CaM can release membrane-bound K-Ras4B in a GTP-dependent manner. Preceding lifetime experiments of dansyl-labeled Ca2+/CaM in buffer revealed a fluorescence lifetime of 15.9 ± 0.4 ns (cf. Supporting Materials and Methods; Fig. S5; Table S1) in accordance with published data (46). Subsequent anisotropy measurements identified an overall rotational correlation time, θ, of 9.4 ± 0.9 ns for Ca2+/CaM in buffer (Fig. S6; Table S2), in agreement with earlier studies (47, 48). Addition of lipid vesicles did not significantly affect θdansyl-CaM, which verifies the SPR and IRRAS results in showing no membrane binding of the acidic Ca2+/CaM (Fig. 4; Table S2). Since the global rotational motion of Ca2+/CaM is sensitive to the overall dimensions of the protein, complex formation with K-Ras4B is expected to lead to a considerable increase in θdansyl-CaM. This is confirmed by a detected overall rotational correlation time of 17.9 and 16.1 ns for Ca2+/CaM in complex with GDP- and GTP-loaded K-Ras4B, respectively, in solution. Further addition of lipid vesicles to the complex did not alter the rotational dynamics of Ca2+/CaM. Consequently, Ca2+/CaM binds to K-Ras4B in the absence and presence of membranes irrespective of nucleotide loading. Moreover, no membrane binding was detected for Ca2+/CaM-complexed K-Ras4B, since this would result in a marked increase in θdansyl-CaM, as shown in a previous study of K-Ras4B (29). Moreover, the Ca2+/CaM−K-Ras4B complex was stable in the presence of anionic membranes, because complex dissociation would result in the value detected for θdansyl-CaM in buffer. This result is in contrast to previously published data for PDEδ, which pointed to a dissociation of the K-Ras4B−PDEδ complex upon membrane interaction (29).
Figure 4.
Overall rotational correlation times of dansyl-labeled Ca2+/CaM at T = 25°C. Results are shown for Ca2+/CaM in the presence and absence of GDP- and GTP-loaded K-Ras4B and/or anionic model raft membranes (lipid/K-Ras4B ratio of 200:1). The underlying fitting parameters are given in Tables S1 and S2.
To verify a direct release of membrane-bound K-Ras4B by Ca2+/CaM, K-Ras4B was incubated with lipid vesicles followed by subsequent addition of Ca2+/CaM. Previous results confirmed stable membrane binding of K-Ras4B under these conditions (29, 49). Rotational correlation times of 15 ns and 25 ns were determined for BODIPY-labeled K-Ras4B GDP in the absence and presence of membranes, respectively. To verify that the change in θ was indeed due to the binding of K-Ras4B to large unilamellar vesicles, we performed the same experiment with unlipidated K-Ras4B, which showed a rotational correlation time of 13 ns, i.e., no membrane binding (29, 49). Once bound, K-Ras4B is known to be stably anchored in the membrane (31). The value of θdansyl-CaM = 18.3 ns determined here for both K-Ras4B GDP and GTP shows that Ca2+/CaM is able to bind to membrane-anchored K-Ras4B and extract it from anionic membranes irrespective of GDP/GTP (Fig. 4; Table S2).
Finally, to gain complementary spatial information on the single-molecule level, we carried out time-lapse AFM experiments. Protein-enriched domains were observed within the bulk liquid-disordered phase for both GDP- and GTP-bound K-Ras4B, in agreement with previous data that focused on the membrane localization of K-Ras4B (29, 31). Corresponding AFM images of the anionic raft membrane before and after addition of K-Ras4B are given in Figs. S7 and S8 (cf. Supporting Results). Due to electrostatic repulsion, Ca2+/CaM itself showed no significant membrane binding (Fig. S9). However, upon addition of Ca2+/CaM to membrane-bound K-Ras4B, no substantial changes were detected (Fig. S8). This may be due to the large K-Ras4B clusters observed in the AFM images, which may restrict efficient binding of Ca2+/CaM to membrane-anchored K-Ras4B and thus impede K-Ras4B membrane extraction. Moreover, steric constraints could be induced by the small separation of the lipid bilayer and solid support, which might impede the displacement of proteins. When the preformed K-Ras4B−Ca2+/CaM complex was added to anionic membranes, the AFM images showed a considerably lower amount of K-Ras4B clusters. This indicates that Ca2+/CaM is able to bind K-Ras4B in solution and obstructs binding of K-Ras4B to the membrane independently of its nucleotide state (Fig. S9). Again, in the previous study (29), addition of the K-Ras4B−PDEδ complex to anionic membranes led to AFM images that mirrored the membrane partitioning behavior of the single components, arguing for a dissociation of the complex.
Conclusions
Taken together, the IRRAS and fluorescence anisotropy data clearly demonstrate that Ca2+/CaM binds to membrane-anchored K-Ras4B and dissociates it from negatively charged membranes in a nucleotide-independent manner. In line with the electrostatic repulsion model, binding of the strongly negatively charged Ca2+/CaM to the polybasic stretch of K-Ras4B would reverse its charge, leading to repulsion of the complex from membranes containing anionic lipids. One farnesyl anchor alone is not sufficient to stably anchor Ras proteins in membranes. The combined data also reveal no binding of the K-Ras4B−Ca2+/CaM complex to anionic membranes, which can be easily explained by the farnesyl anchor buried in the hydrophobic pocket of Ca2+/CaM and repulsive electrostatic interactions, according to the model described above. This leads to the important conclusion that plasma membrane binding of K-Ras4B would not be possible in the CaM-complexed state. This would imply that binding of Ca2+/CaM to K-Ras4B inhibits its activity and thus signaling to Ras effectors (e.g., Raf) in vivo and consequently also ERK1/2 activation, since K-Ras4B membrane association is crucial for its function. Therefore, Ca2+/CaM would function as a negative regulator of K-Ras4B signaling in intact cells by releasing K-Ras4B from negatively charged membranes and preventing its plasma membrane localization. This is in agreement with previous results, which showed that Ca2+/CaM plays an essential role in the downregulation of the Ras/Raf/MEK/ERK pathway in cultured fibroblasts (50) and that inactivation of CaM enables activation of K-Ras4B (16). Thus, Ca2+/CaM seems to be an important element in the differential downregulation of Ras, since Ca2+/CaM binds specifically to K-Ras4B (but not N-Ras, K-Ras4A, or H-Ras) (16). Moreover, the polybasic-prenyl motif of K-Ras4B has been reported to act as a reversible Ca2+/CaM-regulated molecular switch that removes K-Ras4B from the plasma membrane and partially redistributes it to internal sites in neurons (15). Finally, a very recent study (51) revealed that binding of K-Ras to CaM leads to a sustained suppression of Wnt/Ca2+ signaling, which causes increased tumorigenicity. The authors of that study suggested that blocking of this Ras isoform-specific CaM interaction might represent a novel approach for selective K-Ras targeting.
A mechanism similar to that observed for the interaction of Ca2+/CaM with K-Ras4B has been detected for the binding of Ca2+/CaM to the myristoylated alanine-rich C-kinase substrate (MARCKS). Again, since one myristate anchor cannot hold the protein on the membrane, the binding of Ca2+/CaM to the central region of the basic cluster of MARCKS is expected to repel the complex from the membrane, thus leading to translocation of MARCKS from the plasma membrane to the cytoplasm (52, 53).
The reason for the discrepancies between the SPR data, that revealed membrane extraction by Ca2+/CaM only for K-Ras4B GDP, and the rest of the data, which showed no nucleotide dependency, most likely lies in the different extents of clustering for GDP- and GTP-loaded K-Ras4B in immobilized membranes. Protein clustering is more pronounced for the active form of Ras in supported bilayers, as seen in corresponding AFM experiments for both K-Ras and N-Ras (31, 33). Therefore, steric constraints introduced by strong clustering are a very likely reason for the observed nucleotide dependence of Ca2+/CaM binding to membrane-bound K-Ras4B in SPR. In IRRAS and fluorescence anisotropy experiments, no solid support is used for membrane immobilization (corresponding to a higher lateral mobility of the lipids at the air/D2O interface in IRRAS experiments and in freestanding lipid vesicles in anisotropy experiments). Therefore, the lack of significant extraction of membrane-bound K-Ras4B by Ca2+/CaM as detected by AFM is most probably due to the large K-Ras4B clusters observed in the AFM images, which restrict efficient binding of Ca2+/CaM to K-Ras4B bound to solid-supported membranes and thus largely impede K-Ras4B membrane extraction. Moreover, steric constraints could be induced by the small separation of the immobilized lipid bilayer and solid support (∼0.5−2.0 nm in buffer solution), which might impede easy displacement of proteins. Since Ras nanoclusters are thought to be much smaller in vivo (34) as compared with the strong clustering in microdomains observed in experiments using immobilized membranes, we conclude that membrane extraction by Ca2+/CaM occurs independently of GTP loading, as seen in the IRRAS and fluorescence anisotropy experiments presented here.
Contrary to the previously analyzed PDEδ, the Ca2+/CaM−K-Ras4B complex turned out to be stable in the presence of anionic membranes. In a recent study (29), we showed that PDEδ is unable to extract K-Ras4B from heterogeneous model membranes irrespective of GTP loading. The enhanced affinity of the K-Ras4B−PDEδ complex particularly for anionic membranes, together with the observation that K-Ras4B is released from PDEδ upon membrane interaction, led us to propose an effective delivery of PDEδ-solubilized K-Ras4B to the plasma membrane. This is in accordance with findings by Schmick et al. (13), which showed that binding of K-Ras4B to PDEδ involves a passive sequestration in the cytoplasm of dissociated K-Ras4B from any membrane.
By taking into account recent insights into the dynamic, spatially organizing cycle of Ras (54), one can envisage that PDEδ and Ca2+/CaM jointly control K-Ras4B plasma membrane localization, with Ca2+/CaM playing a different part than PDEδ in the regulation of K-Ras4B membrane binding (Fig. 5). Since the signaling activity of K-Ras4B is intrinsically tied to its level of enrichment at the plasma membrane, interference in K-Ras4B plasma membrane localization would constitute a possible means to impact cell growth and counter oncogenic K-Ras4B signaling.
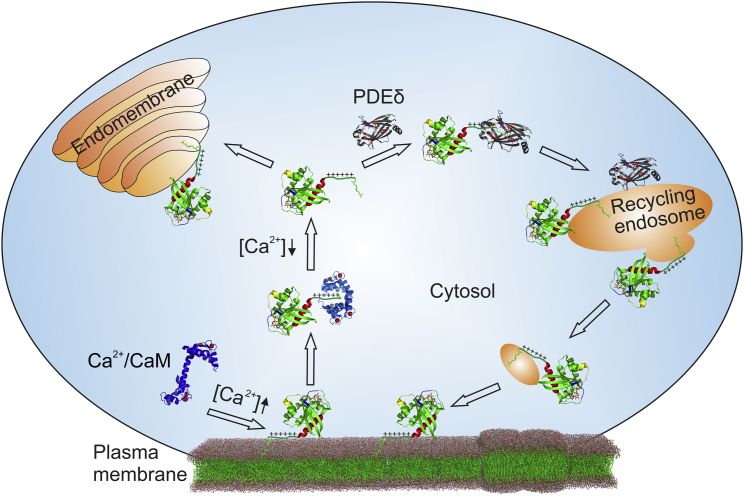
Figure 5.
Schematic illustration of the PDEδ and Ca2+/CaM-modulated K-Ras4B localization cycle. Ca2+/CaM extracts K-Ras4B from the plasma membrane at elevated levels of intracellular free Ca2+. This process might be facilitated by a decrease of the most-negative plasma membrane’s surface charge upon an increase in cytosolic Ca2+. After cell stimulation, the free Ca2+ concentration, [Ca2+], decreases, leading to a dissociation of the Ca2+/CaM−K-Ras4B complex. In the cytosol, K-Ras4B can bind to endomembranes or PDEδ, with the latter solubilizing K-Ras4B and facilitating cytoplasmic diffusion. When the PDEδ/K-Ras4B complex reaches the negatively charged recycling endosome, K-Ras4B can be released and its farnesyl anchor can be inserted into the membrane. Taking into account recent results from Schmick et al. (54), one can envisage a directed vesicular transport that brings K-Ras4B back to the plasma membrane, where it clusters in less-ordered domains.
Author Contributions
R.W. and K.W. designed research; B.S., S.K., and K.W. performed research and analyzed data; H.W. contributed reagents/analytic tools; and B.S., S.K., R.W., and K.W. wrote the article.
Acknowledgments
We thank Simone Möbitz for labeling of CaM and initial SPR experiments, Sasikala Thavam for help with the lipopeptide synthesis, Christine Nowak for technical assistance with Ras protein expression and ligation, and Andrea Gohlke for initial SPR experiments. We also thank Stefan M. Kast and Florian Mrugalla for providing Fig. S1 and helpful discussion.
This research was supported by the Deutsche Forschungsgemeinschaft (SFB 642).
Editor: Heiko Heerklotz
Footnotes
Supporting Materials and Methods, Supporting Results, nine figures, and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30389-7.
Contributor Information
Roland Winter, Email: roland.winter@tu-dortmund.de.
Katrin Weise, Email: katrin.weise@tu-dortmund.de.
Supporting Citations
References (55, 56, 57) appear in the Supporting Material.
Supporting Material
References
- 1.Barbacid M. ras genes. Annu. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 2.Hancock J.F. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 3.Wittinghofer A., Waldmann H. Ras—a molecular switch involved in tumor formation. Angew. Chem. Int. Ed. Engl. 2000;39:4192–4214. doi: 10.1002/1521-3773(20001201)39:23<4192::AID-ANIE4192>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Wittinghofer A., Pai E.F. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem. Sci. 1991;16:382–387. doi: 10.1016/0968-0004(91)90156-p. [DOI] [PubMed] [Google Scholar]
- 5.Friday B.B., Adjei A.A. K-ras as a target for cancer therapy. Biochim. Biophys. Acta. 2005;1756:127–144. doi: 10.1016/j.bbcan.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Bos J.L. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 7.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock J.F., Paterson H., Marshall C.J. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 9.Jackson J.H., Li J.W., Cochrane C.G. Polylysine domain of K-ras 4B protein is crucial for malignant transformation. Proc. Natl. Acad. Sci. USA. 1994;91:12730–12734. doi: 10.1073/pnas.91.26.12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall C.J. Ras effectors. Curr. Opin. Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 11.Willumsen B.M., Christensen A., Lowy D.R. The p21 ras C-terminus is required for transformation and membrane association. Nature. 1984;310:583–586. doi: 10.1038/310583a0. [DOI] [PubMed] [Google Scholar]
- 12.Chandra A., Grecco H.E., Bastiaens P.I. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 2011;14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 13.Schmick M., Vartak N., Bastiaens P.I. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell. 2014;157:459–471. doi: 10.1016/j.cell.2014.02.051. [DOI] [PubMed] [Google Scholar]
- 14.Ismail S.A., Chen Y.X., Wittinghofer A. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 2011;7:942–949. doi: 10.1038/nchembio.686. [DOI] [PubMed] [Google Scholar]
- 15.Fivaz M., Meyer T. Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J. Cell Biol. 2005;170:429–441. doi: 10.1083/jcb.200409157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villalonga P., López-Alcalá C., Agell N. Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol. Cell. Biol. 2001;21:7345–7354. doi: 10.1128/MCB.21.21.7345-7354.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abraham S.J., Nolet R.P., Gaponenko V. The hypervariable region of K-Ras4B is responsible for its specific interactions with calmodulin. Biochemistry. 2009;48:7575–7583. doi: 10.1021/bi900769j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crivici A., Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu. Rev. Biophys. Biomol. Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- 19.Klee C.B., Vanaman T.C. Calmodulin. Adv. Protein Chem. 1982;35:213–321. doi: 10.1016/s0065-3233(08)60470-2. [DOI] [PubMed] [Google Scholar]
- 20.Babu Y.S., Bugg C.E., Cook W.J. Structure of calmodulin refined at 2.2 A resolution. J. Mol. Biol. 1988;204:191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang M., Tanaka T., Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 22.Vetter S.W., Leclerc E. Novel aspects of calmodulin target recognition and activation. Eur. J. Biochem. 2003;270:404–414. doi: 10.1046/j.1432-1033.2003.03414.x. [DOI] [PubMed] [Google Scholar]
- 23.Hoeflich K.P., Ikura M. Calmodulin in action: diversity in target recognition and activation mechanisms. Cell. 2002;108:739–742. doi: 10.1016/s0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 24.Matsubara M., Nakatsu T., Taniguchi H. Crystal structure of a myristoylated CAP-23/NAP-22 N-terminal domain complexed with Ca2+/calmodulin. EMBO J. 2004;23:712–718. doi: 10.1038/sj.emboj.7600093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidhu R.S., Clough R.R., Bhullar R.P. Ca2+/calmodulin binds and dissociates K-RasB from membrane. Biochem. Biophys. Res. Commun. 2003;304:655–660. doi: 10.1016/s0006-291x(03)00635-1. [DOI] [PubMed] [Google Scholar]
- 26.Bhagatji P., Leventis R., Silvius J.R. Multiple cellular proteins modulate the dynamics of K-ras association with the plasma membrane. Biophys. J. 2010;99:3327–3335. doi: 10.1016/j.bpj.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L.J., Xu L.R., Liang Y. Both the C-terminal polylysine region and the farnesylation of K-RasB are important for its specific interaction with calmodulin. PLoS One. 2011;6:e21929. doi: 10.1371/journal.pone.0021929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Alcalá C., Alvarez-Moya B., Agell N. Identification of essential interacting elements in K-Ras/calmodulin binding and its role in K-Ras localization. J. Biol. Chem. 2008;283:10621–10631. doi: 10.1074/jbc.M706238200. [DOI] [PubMed] [Google Scholar]
- 29.Weise K., Kapoor S., Winter R. Dissociation of the K-Ras4B/PDEδ complex upon contact with lipid membranes: membrane delivery instead of extraction. J. Am. Chem. Soc. 2012;134:11503–11510. doi: 10.1021/ja305518h. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y.X., Koch S., Waldmann H. Synthesis of the Rheb and K-Ras4B GTPases. Angew. Chem. Int. Ed. Engl. 2010;49:6090–6095. doi: 10.1002/anie.201001884. [DOI] [PubMed] [Google Scholar]
- 31.Weise K., Kapoor S., Winter R. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J. Am. Chem. Soc. 2011;133:880–887. doi: 10.1021/ja107532q. [DOI] [PubMed] [Google Scholar]
- 32.Gohlke A., Triola G., Winter R. Influence of the lipid anchor motif of N-ras on the interaction with lipid membranes: a surface plasmon resonance study. Biophys. J. 2010;98:2226–2235. doi: 10.1016/j.bpj.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weise K., Triola G., Winter R. Influence of the lipidation motif on the partitioning and association of N-Ras in model membrane subdomains. J. Am. Chem. Soc. 2009;131:1557–1564. doi: 10.1021/ja808691r. [DOI] [PubMed] [Google Scholar]
- 34.Plowman S.J., Muncke C., Hancock J.F. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omerovic J., Prior I.A. Compartmentalized signalling: Ras proteins and signalling nanoclusters. FEBS J. 2009;276:1817–1825. doi: 10.1111/j.1742-4658.2009.06928.x. [DOI] [PubMed] [Google Scholar]
- 36.Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 37.Kapoor S., Werkmüller A., Winter R. Temperature-pressure phase diagram of a heterogeneous anionic model biomembrane system: results from a combined calorimetry, spectroscopy and microscopy study. Biochim. Biophys. Acta. 2011;1808:1187–1195. doi: 10.1016/j.bbamem.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Evers F., Jeworrek C., Winter R. Detection of lipid raft domains in neutral and anionic Langmuir monolayers and bilayers of complex lipid composition. Soft Matter. 2012;8:2170–2175. [Google Scholar]
- 39.Vogel A., Nikolaus J., Huster D. Interaction of the human N-Ras protein with lipid raft model membranes of varying degrees of complexity. Biol. Chem. 2014;395:779–789. doi: 10.1515/hsz-2013-0294. [DOI] [PubMed] [Google Scholar]
- 40.Vogel A., Reuther G., Huster D. The lipid modifications of Ras that sense membrane environments and induce local enrichment. Angew. Chem. Int. Ed. Engl. 2009;48:8784–8787. doi: 10.1002/anie.200903396. [DOI] [PubMed] [Google Scholar]
- 41.Green R.J., Frazier R.A., Tendler S.J. Surface plasmon resonance analysis of dynamic biological interactions with biomaterials. Biomaterials. 2000;21:1823–1835. doi: 10.1016/s0142-9612(00)00077-6. [DOI] [PubMed] [Google Scholar]
- 42.Mozsolits H., Thomas W.G., Aguilar M.I. Surface plasmon resonance spectroscopy in the study of membrane-mediated cell signalling. J. Pept. Sci. 2003;9:77–89. doi: 10.1002/psc.439. [DOI] [PubMed] [Google Scholar]
- 43.Besenicar M., Macek P., Anderluh G. Surface plasmon resonance in protein-membrane interactions. Chem. Phys. Lipids. 2006;141:169–178. doi: 10.1016/j.chemphyslip.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Kapoor S., Weise K., Winter R. The role of G-domain orientation and nucleotide state on the Ras isoform-specific membrane interaction. Eur. Biophys. J. 2012;41:801–813. doi: 10.1007/s00249-012-0841-5. [DOI] [PubMed] [Google Scholar]
- 45.Kapoor S., Triola G., Winter R. Revealing conformational substates of lipidated N-Ras protein by pressure modulation. Proc. Natl. Acad. Sci. USA. 2012;109:460–465. doi: 10.1073/pnas.1110553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kincaid R.L., Vaughan M., Tkachuk V.A. Ca2+-dependent interaction of 5-dimethylaminonaphthalene-1-sulfonyl-calmodulin with cyclic nucleotide phosphodiesterase, calcineurin, and troponin I. J. Biol. Chem. 1982;257:10638–10643. [PubMed] [Google Scholar]
- 47.Yao Y., Schöneich C., Squier T.C. Resolution of structural changes associated with calcium activation of calmodulin using frequency domain fluorescence spectroscopy. Biochemistry. 1994;33:7797–7810. doi: 10.1021/bi00191a007. [DOI] [PubMed] [Google Scholar]
- 48.Anderson S.R. Time-resolved fluorescence spectroscopy. Applications to calmodulin. J. Biol. Chem. 1991;266:11405–11408. [PubMed] [Google Scholar]
- 49.Werkmüller A., Triola G., Winter R. Rotational and translational dynamics of ras proteins upon binding to model membrane systems. ChemPhysChem. 2013;14:3698–3705. doi: 10.1002/cphc.201300617. [DOI] [PubMed] [Google Scholar]
- 50.Bosch M., Gil J., Agell N. Calmodulin inhibitor W13 induces sustained activation of ERK2 and expression of p21(cip1) J. Biol. Chem. 1998;273:22145–22150. doi: 10.1074/jbc.273.34.22145. [DOI] [PubMed] [Google Scholar]
- 51.Wang M.T., Holderfield M., McCormick F. K-ras promotes tumorigenicity through suppression of non-canonical Wnt signaling. Cell. 2015;163:1237–1251. doi: 10.1016/j.cell.2015.10.041. [DOI] [PubMed] [Google Scholar]
- 52.Kim J., Shishido T., McLaughlin S. Phosphorylation, high ionic strength, and calmodulin reverse the binding of MARCKS to phospholipid vesicles. J. Biol. Chem. 1994;269:28214–28219. [PubMed] [Google Scholar]
- 53.McLaughlin S., Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 54.Schmick M., Kraemer A., Bastiaens P.I.H. Ras moves to stay in place. Trends Cell Biol. 2015;25:190–197. doi: 10.1016/j.tcb.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Gratton E., Limkeman M. A continuously variable frequency cross-correlation phase fluorometer with picosecond resolution. Biophys. J. 1983;44:315–324. doi: 10.1016/S0006-3495(83)84305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gratton E., Jameson D.M., Hall R.D. Multifrequency phase and modulation fluorometry. Annu. Rev. Biophys. Bioeng. 1984;13:105–124. doi: 10.1146/annurev.bb.13.060184.000541. [DOI] [PubMed] [Google Scholar]
- 57.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 58.Baker N.A., Sept D., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.