Abstract
Composition of the gut microbiota has profound effects on intestinal carcinogenesis. Diet and host genetics play critical roles in shaping the composition of gut microbiota. Whether diet and host genes interact with each other to bring specific changes in gut microbiota that affect intestinal carcinogenesis is unknown. Ability of dietary fibre to specifically increase beneficial gut microbiota at the expense of pathogenic bacteria in vivo via unknown mechanism is an important process that suppresses intestinal inflammation and carcinogenesis. Free fatty acid receptor 2 (FFAR2 or GPR43) is a receptor for short-chain fatty acids (acetate, propionate and butyrate), metabolites of dietary fibre fermentation by gut microbiota. Here, we show FFAR2 is down modulated in human colon cancers than matched adjacent healthy tissue. Consistent with this, Ffar2−/− mice are hypersusceptible to development of intestinal carcinogenesis. Dietary fibre suppressed colon carcinogenesis in an Ffar2-dependent manner. Ffar2 played an essential role in dietary fibre-mediated promotion of beneficial gut microbiota, Bifidobacterium species (spp) and suppression of Helicobacter hepaticus and Prevotellaceae. Moreover, numbers of Bifidobacterium is reduced, whereas those of Prevotellaceae are increased in human colon cancers than matched adjacent normal tissue. Administration of Bifidobacterium mitigated intestinal inflammation and carcinogenesis in Ffar2−/− mice. Taken together, these findings suggest that interplay between dietary fibre and Ffar2 play a key role in promoting healthy composition of gut microbiota that stimulates intestinal health.
Introduction
Decreased intake of dietary fibre in modern era is associated with increased risk of colon cancers. Dietary fibre is fermented in the colon by gut microbiota into short-chain fatty acids (SCFAs; acetate, propionate and butyrate). SCFAs, specifically butyrate is well known for its anti-inflammatory and anti-carcinogenic effects in the gut.1 Butyrate enemas are known to suppress inflammation during ulcerative colitis, a risk factor for development of colorectal cancers.1 Moreover, recent studies demonstrate a reduction in butyrate-producing bacteria in feces of individuals with ulcerative colitis and colon cancers than healthy individuals.2, 3 This comparison may lead to inadequate information because there are several variables that are known to influence composition of gut microbiota that differ between healthy and colon cancer subjects such as genetic make up, lifestyle, age of individuals, dietary habits and therapeutic treatment. Therefore, an alternative method such as comparison between cancerous tissue and adjacent normal tissue may yield better information regarding role of different gut bacteria in promotion or suppression of colon cancers.
SCFAs interact with G-protein-coupled receptors 41 (GPR41 or FFAR1), FFAR2 and HCAR2 (also known as NIACR1 or GPR109A). FFAR1 and FFAR2 interact with all three SCFAs, whereas HACR2 interacts with butyrate only.4, 5 SCFAs are involved in homeostasis of regulatory T cells (Treg cells) in colon and modulation of intestinal carcinogenesis.6, 7, 8, 9, 10, 11 Ffar2−/− mice reveal altered susceptibility to allergic airway inflammation in lung and colonic inflammation induced by dextran sulfate sodium (DSS), ethanol or trinitrobenzoic sulfonic acid.12, 13, 14, 15 FFAR2 expression in colon cancer cell lines promotes their apoptosis.16 However, the role of Ffar2 in regulation of intestinal carcinogenesis and underlying mechanism in vivo has not been investigated.
Fermentable dietary fibre (prebiotics) and Bifidobacterium spp (probiotics) improve gut epithelial barrier function, prevent apoptosis of intestinal epithelial cells, and suppress intestinal inflammation and carcinogenesis.17, 18, 19, 20, 21, 22, 23 In vitro studies using mixed cultures have shown that dietary fibre support growth of divergent groups of colonic bacteria such as Clostridium, Bifidobacteria, Bacteroides, Enterococcus and Escherichia.24, 25 In contrast, numerous human and animal studies show that dietary fibre consumption specifically and consistently promote only one group of gut bacteria, which is Bifidobacterium spp, and repress others such as Enterococcus, Clostridium and Eubacterium24, 25 suggesting that other factors regulate bifidogenic activity of dietary fibre in vivo.
It has been hypothesized that butyrate-producing gut bacteria play a critical role in dietary fibre-mediated suppression of colon carcinogenesis. However, studies show conflicting data about abundance of butyrate-producing bacteria in feces from colon cancers subjects than healthy individuals.2, 26 Similarly, both colon cancer-promoting and -suppressive effects of butyrate have been observed in vivo and this phenomenon has been termed as ‘butyrate paradox'.10, 11 Fusobacterium nucleatum, which is normally present in oral cavity of nearly all humans, has been shown to promote colon carcinogenesis.27, 28, 29 F. nucleatum is a butyrate producer.30 Butyrate-producing bacteria poorly utilize dietary fibre for their growth.24, 31 This may be the reason that a recent human study found that dietary fibre failed to significantly increase any single butyrate-producing bacterial species despite reducing the markers associated with colon carcinogenesis.32 On the other hand, Bifidobacterium spp are the best fermenters of dietary fibre.24, 31 Furthermore, Bifidobacterium ferment dietary fibre into acetate and lactate, which are utilized by butyrate-producing bacteria for growth and butyrate production and this process is termed as cross-feeding.24, 31 Therefore, if fermentation of dietary fibre is essential in suppression of colon carcinogenesis, being the primary fermenter of dietary fibre, Bifidobacterium must play an important role in it. In this report, we investigated whether Ffar2 regulates dietary fibre-mediated changes in gut microbiota and what is the impact of these changes on intestinal carcinogenesis. Our findings demonstrate the critical role of Ffar2 in maintaining a healthy composition of gut microbiota leading to suppression of intestinal carcinogenesis and suggest that Ffar2 signaling could be utilized as a potential target for therapeutic correction of gut microbiota to suppress intestinal carcinogenesis.
Results
Ffar2 is downregulated in human colon cancers
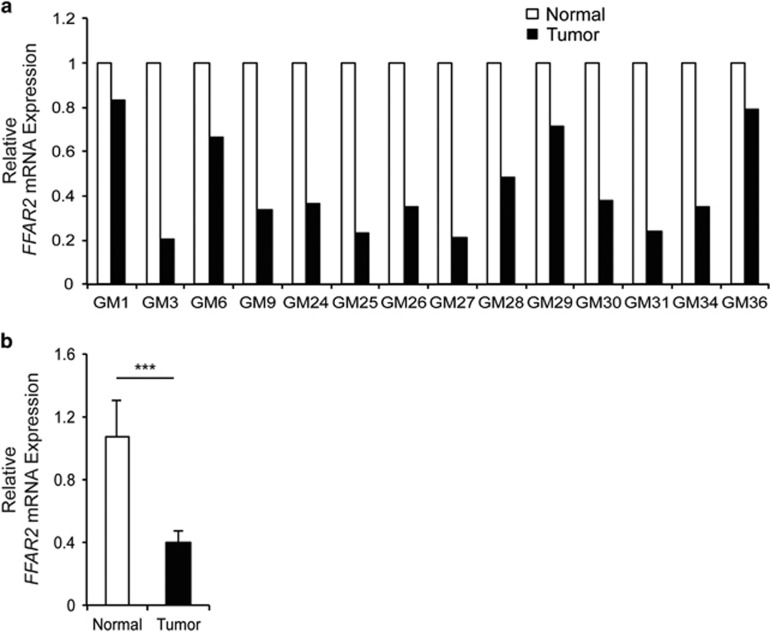
To test the relevance of FFAR2 in colon cancers, FFAR2 mRNA expression was analyzed in paired colon cancer and matched adjacent healthy tissue by quantitative polymerase chain reaction (qPCR). Figure 1a shows that expression of FFAR2 mRNA was reduced by more than twofold in 71% (10 out of 14) of the colon cancers than adjacent normal tissue. When data was pooled for these samples, FFAR2 expression was still significantly downregulated (by ~60%) in colon cancers than adjacent normal tissue (Figure 1b) (P<0.0001). Similarly, expression of FFAR3 mRNA was also downregulated in colon cancer samples (Supplementary Figure 1). Same set of colon cancer samples as used in current study also exhibits decreased expression of HCAR2 (GPR109A).33
Figure 1.
Expression of FFAR2 is diminished in human colon cancers. (a) Expression of FFAR2 was assessed by quantitative polymerase chain reaction in 14 colon cancer and matched adjacent normal tissue. (b) Data from a was pooled and re-plotted. ***P<0.0001.
Ffar2 suppresses inflammation-associated colon carcinogenesis
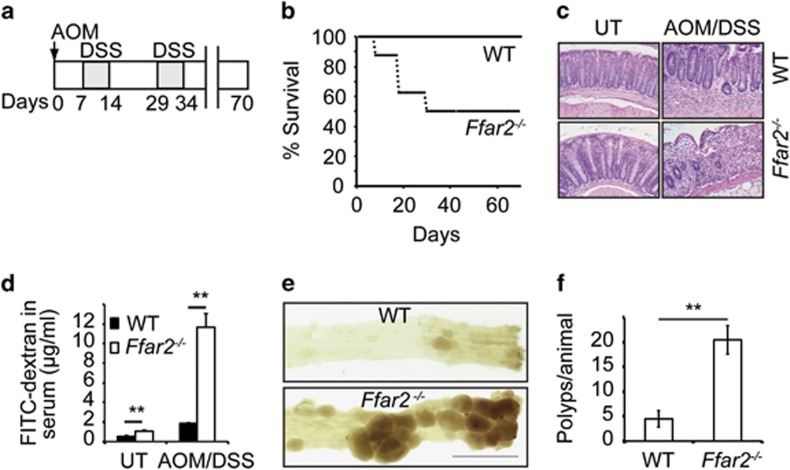
Since Ffar2 was downregulated in colon cancers, an inflammation-associated colon carcinogenesis model was used to test the role of Ffar2 in colonic inflammation and carcinogenesis. Ffar2−/− mice and their wild-type (WT) littermates were injected with the colon-specific carcinogen azoxymethane (AOM) intraperitoneally (i.p.) followed by two cycles of DSS in drinking water as shown in Figure 2a. All the WT littermates treated with AOM and DSS (AOM/DSS) survived until the experimental endpoint. However, 50% of the Ffar2−/− mice died within 30 days of treatment (Figure 2b). In contrast to WT littermates, Ffar2−/− mice exhibited drastic reduction in body weight, and severe diarrhea during the first cycle of DSS (Supplementary Figures 2a and b). Similarly, higher weight loss was also observed in Ffar2−/− mice than WT counterparts during second cycle of DSS treatment (Supplementary Figure 2a). Following AOM/DSS treatment, colons of Ffar2−/− mice exhibited drastically increased loss of epithelium, crypt structure and heightened infiltration of colonic mucosa by immune cells relative to WT littermates resulting in higher histopathological scores in the former group (Figure 2c and Supplementary Figure 2c). Following oral gavage of fluorescein isothiocyanate (FITC)-dextran to AOM/DSS treated animals, sixfold more FITC-dextran was translocated to systemic circulation of Ffar2−/− mice than WT littermates, revealing increased gut epithelial barrier dysfunction in former group (Figure 2d). Consistent with these findings, staining with claudin-3, a tight junction protein revealed denudation of epithelium following AOM/DSS treatment in Ffar2−/− mice (Supplementary Figure 2d). In addition, colons of AOM/DSS treated Ffar2−/− mice exhibited significantly higher weight per centimeter length than WT littermates, presumably due to thickening of colon in Ffar2−/− mice (Supplementary Figure 2e). Expression of several pro-inflammatory molecules such as IL-1β, IL-17a, Ccl-2, Cox-2, Cxcl-1, Cxcl-2 and IL-12 that promote colonic inflammation was significantly increased in colons of Ffar2−/− mice than WT mice after AOM/DSS treatment (Supplementary Figure 3). At the experimental endpoint, colons of mice were harvested, opened longitudinally and analyzed for number of polyps. Colons of AOM/DSS treated Ffar2−/− mice had ~4.5 time more number of polyps than WT littermates mice (Figures 2e–f). In addition, polyps in colons of Ffar2−/− mice were larger in size than WT littermates (Supplementary Figure 2f). Collectively, these data demonstrate that Ffar2 is a key receptor that suppresses inflammation-associated colon carcinogenesis.
Figure 2.
Ffar2 suppresses colon carcinogenesis. WT and Ffar2−/− mice (littermates) were fed with conventional mouse chow. (a) Experimental model to induce inflammation-associated colon carcinogenesis. (b) Survival of WT and Ffar2−/− mice subjected to AOM/DSS treatment as described in a (n=8 mice). (c) A representative photograph of H&E stained cross-section of colons of untreated (UT) or AOM/DSS treated WT and Ffar2−/− mice. Original magnification 200 ×. (d) After the completion of first cycle of DSS, mice were gavaged with FITC-dextran and 6 h later FITC was quantified in serum (n=5 mice per genotype). (e) A representative photographs of luminal side of colons from WT and Ffar2−/− animals on day 70 after AOM/DSS treatment. (f) Polyp burden in WT and Ffar2−/− mice after AOM/DSS treatment (n=6 mice). A representative or pooled data from 2 experiments are shown. Error bars represent standard deviation. **P<0.002.
Ffar2 controls intestinal carcinogenesis in Apc Min/+ mice
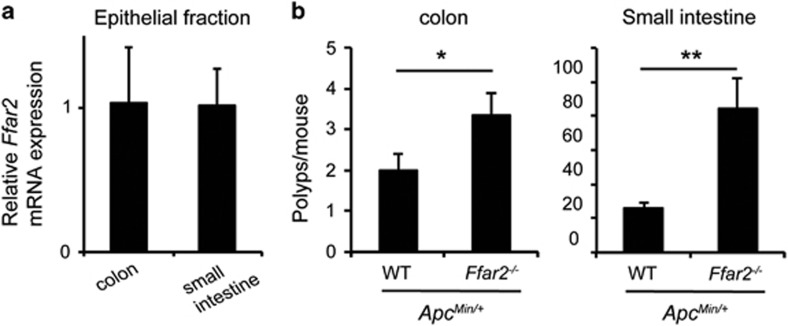
Next, we analyzed the role of Ffar2 in a genetic model of intestinal carcinogenesis. Approximately 85% of the sporadic human colon cancers harbor mutations in the adenomatous polyposis coli (APC) gene.34 Heterozygous mutation in Apc that is present in ApcMin/+ mice results in expression of a truncated protein. These mice develop of numerous polyps in small intestine and colon. We found that Ffar2 is expressed in epithelial cells throughout the small intestine (Figure 3a). Ffar2−/−ApcMin/+ mice developed significantly more polyps in colon than ApcMin/+ mice (Figure 3b). SCFAs are generated in the cecum and colon, and therefore it has been hypothesized that they will inhibit carcinogenesis only in the cecum and colon. However, the concentrations of SCFAs in ileum is ~10 mM.35 Ec50 of SCFA to induce Ca++ flux or GDP to GTP exchange via Ffar2 is <1 mM,4 therefore it is highly possible that at steady state there is spontaneous signaling via Ffar2 in small intestine. At 5 month of age, Ffar2−/−ApcMin/+ mice developed significantly more polyps in small intestine than age matched ApcMin/+ mice (Figure 3b). These data demonstrate that Ffar2 signaling regulates development of intestinal carcinogenesis induced by germ line mutation of Apc.
Figure 3.
Ffar2 regulates tumorigenesis in ApcMin/+ mice. (a) Ffar2 mRNA expression by colonic and small intestinal epithelium (n=3 mice). A representative of two experiments is shown. (b) Polyp burden in colon and small intestine of ApcMin/+ (n=7) and Ffar2−/− ApcMin/+ mice (n=6). *P<0.01, **P<0.002.
Decreased numbers of Bifidobacterium spp and increased numbers of Prevotellaceae and H. hepaticus in gut microbiota of Ffar2 −/− mice
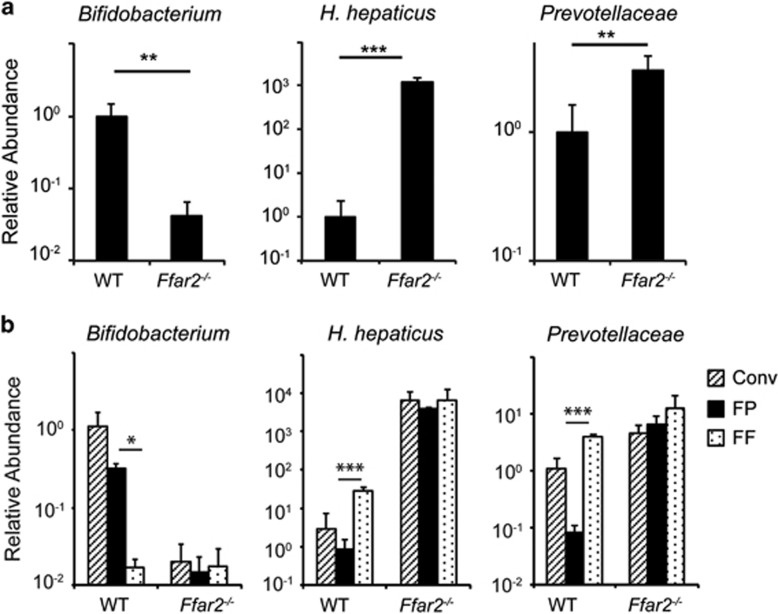
Numerous studies have documented that gut microbiota dysbiosis enhances risk for development of intestinal inflammation and cancers.2, 3, 19, 36 We hypothesized that gut microbiota dysbiosis enhances risk of colon carcinogenesis in Ffar2−/− mice. To test our hypothesis, we analyzed the presence of different bacterial groups among gut microbiota in Ffar2−/− mice and their WT littermates. Feces of WT littermates contained ~24-fold more Bifidobacterium spp among gut microbiota than Ffar2−/− mice (Figure 4a). Compared with feces of WT littermates, those of Ffar2−/− mice exhibited ~1200-fold more H hepaticus (Figure 4a). Similarly, members belonging to the Prevotellaceae family (Phylum, Bacteroidetes) were present in significantly higher numbers in feces of Ffar2−/− mice than WT littermates (Figure 4a). Bacterial members from phyla Firmicutes and Bacteroidetes dominate gut microbiota.37 Firmicutes in gut microbiota are mainly represented by members belonging to Lactobacillus spp, Clostridium leptum, segmented filamentous bacteria and Eubacterium rectales groups.38 Feces from WT littermates and Ffar2−/− mice contained similar frequencies of bacteria from these groups (Supplementary Figure 4a). Similarly, Bacteroides and mouse intestinal Bacteroides groups, both from phylum Bacteroidetes38 were present at comparable levels in feces of WT and Ffar2−/− littermates (Supplementary Figure 4a). In accordance with fecal microbiota, number of Bifidobacterium was decreased, whereas those of H hepaticus and Prevotellaceae were increased among microbiota attached to colons of Ffar2−/− mice than WT littermates (Supplementary Figure 4b). Comparable numbers of bacteria belonging to Bacteroides, mouse intestinal Bacteroides, C leptum, E rectales and Lactobacillus groups were attached to the colons of WT and Ffar2−/− littermates (Supplementary Figure 4b). Taken together, these data demonstrate that the composition of gut microbiota from Ffar2−/− mice is enriched in favor of bacteria that promote intestinal inflammation and cancers, whereas the numbers of bacteria that suppress intestinal inflammation and cancers are reduced.
Figure 4.
An essential role of Ffar2 in dietary fibre-mediated promotion of Bifidobacterium and suppression of Prevotellaceae and H. hepaticus. (a) Fecal DNA from Ffar2−/− mice and their WT littermates were evaluated for abundance of indicated gut microbiota. (b) Ffar2−/− mice and their WT littermates maintained on conventional diet (Conv) were fed with FF or FP diets. The abundance of indicated bacterial groups in feces was quantified 1 day before (Conv) switching the diet or after 1 week on experimental diets (FF or FP). Shown is the relative abundance of indicated bacterial groups to that of total bacteria. *P<0.05, **P<0.01, ***P<0.0001. (n=4 mice). A representative of two experiments is shown.
Essential role of Ffar2 in dietary fibre-mediated promotion of Bifidobacterium spp and decrease of Prevotellaceae and H hepaticus among gut microbiota
Human as well as animal studies have shown that dietary fibre consumption specifically and consistently increases number of only Bifidobacterium spp in gut.24 We hypothesized that Ffar2 plays an obligatory role in dietary fibre-mediated promotion of Bifidobacterium spp. To test this hypothesis, we fed Ffar2−/− mice and their WT littermates with a fibre free (FF) or fibre plus (FP) diets. FP diet contained 2% inulin, 2% pectin and 1% cellulose as source of dietary fibre, whereas FF diet completely lacks dietary fibre, otherwise all other components between these two diets are exactly same. The composition of gut microbiota in feces of Ffar2−/− mice and their WT littermates were evaluated 1 week later. Fecal microbiota changed between conventional diet and FF or FP diet (Figure 4b). Conventional diet contains many components such as soyabean meal, ground corn, flaked corn, molasses, wheat middling, ground wheat etc. Compositions of these components are not exactly defined. On the other hand, FF and FP diets are made from purified and defined components. Thus, conventional diet differs from FF or FP with respect to several food components. Therefore, it is difficult to assign any changes in gut microbiota from conventional diet to FF or FP diet to a particular food component. Feces of FF diet-fed WT animals contained ~17-fold less number of Bifidobacterium spp than FP diet-fed WT littermates (Figure 4b). In sharp contrast, feces of Ffar2−/− mice fed with either FF or FP diet contained similar numbers of Bifidobacterium spp, which were significantly lower than numbers present in feces of FP-fed WT littermates (Figure 4b). Prevotellaceae and H hepaticus were present in similar number in feces of FF-fed WT, FF- or FP-fed Ffar2−/− mice, which was significantly higher than that present in feces of FP-fed WT littermates (Figure 4b). In contrast, dietary fibre increased Bacteroides in gut microbiota in an Ffar2-independent manner (Supplementary Figure 5). Numbers of mouse intestinal Bacteroides and Lactobacillus spp among gut microbiota were unaffected by dietary fibre content (Supplementary Figure 5). Consistent with previous studies,39, 40 abundance of E. rectales and C. leptum group was modestly decreased by dietary fibre in WT mice (Supplementary Figure 5). Collectively, these data demonstrate that Ffar2 is indispensable for dietary fibre-mediated promotion of Bifidobacterium spp and suppression of H hepaticus and Prevotellaceae among gut microbiota.
An essential role of Ffar2 in dietary fibre-mediated suppression of colonic inflammation and carcinogenesis
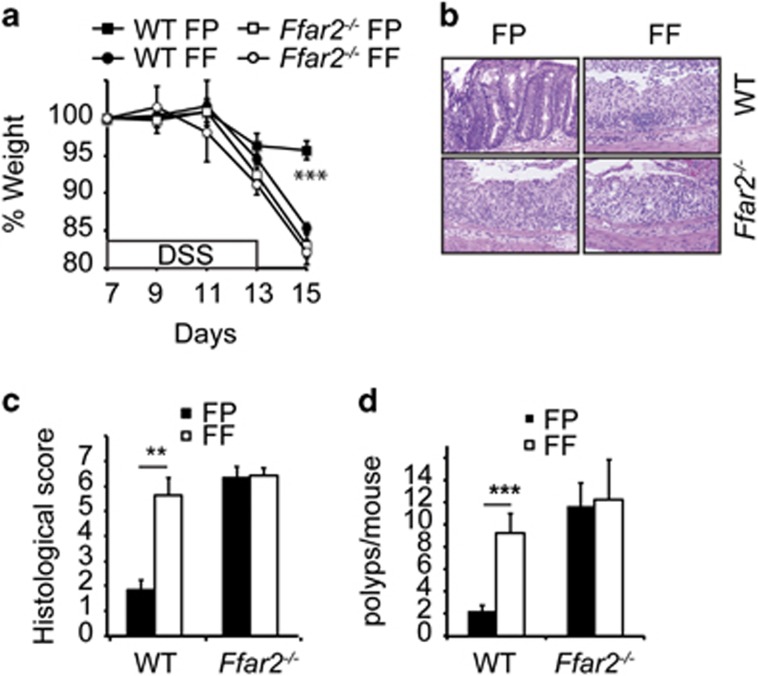
Next, role of Ffar2 in dietary-mediated suppression of colon carcinogenesis in AOM/DSS model was tested. FF diet-fed WT littermates exhibited dramatically more weight loss and diarrhea than FP diet-fed counterparts, demonstrating an essential role for dietary fibre in AOM/DSS-mediated disease (Figure 5a and Supplementary Figure 6a). Colons of WT mice fed with FF diet showed severe pathology with erosion of epithelial layer, ulceration, loss of crypt structures and higher histopathological scores than FP diet-fed counterparts (Figures 5b and c). Consistent with this, depletion of dietary fibre led to the development of significantly higher numbers of colonic polyps in AOM/DSS-treated WT littermates (Figure 5d). The ability of dietary fibre to suppress AOM/DSS-induced weight loss, diarrhea and development of colonic polyps was dependent on Ffar2, because Ffar2−/− mice fed with either FF or FP diets exhibited weight loss, diarrhea and colonic polyps, which were comparable with outcomes observed in WT littermates fed with FF diet (Figures 5a–d). Taken together these data demonstrates a critical role of Ffar2 in dietary fibre-mediated suppression of colonic inflammation and carcinogenesis.
Figure 5.
An essential role of Ffar2 in dietary fibre-mediated suppression of colonic inflammation and carcinogenesis. WT and Ffar2−/− mice (littermates) were fed with FF and FP diets and 1 month later were injected with AOM (i.p.). After 1 week, all the mice were given 1.5% DSS in drinking water for next 6 days. (a) Weight loss during and after DSS treatment. (b) A representative photograph of H&E stained cross-section of colons of mice after first cycle of DSS as treated in a. (c) Histopathological score (inflammation+epithelial damage) of colons from mice 4 days after completion of DSS treatment. (d) Polyp burden in colons of indicated mice at the end of experiment. A representative of two experiments is shown (n=4 mice). **P<0.01, ***P<0.0001.
Bifidobacterium suppresses colonic inflammation and carcinogenesis in Ffar2 −/− mice
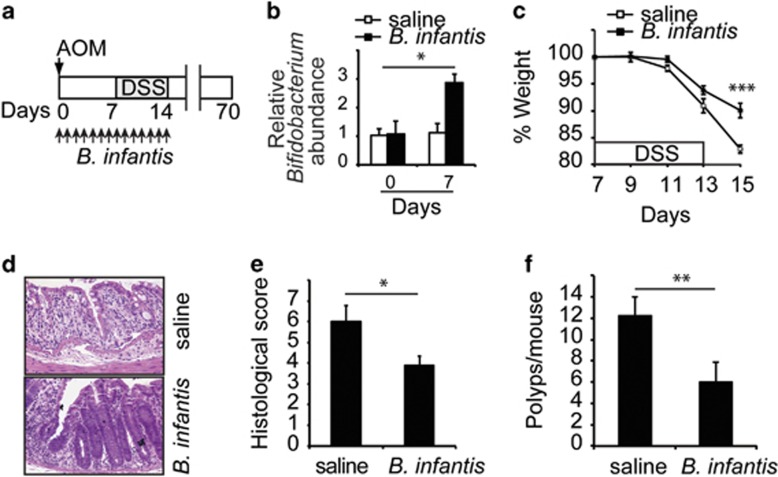
A recent study demonstrates that co-housing of Ffar2−/− mice with WT mice attenuate DSS-induced colonic inflammation in Ffar2−/− mice.41 This finding suggested that (1) an unknown beneficial gut bacteria that got transferred from WT to Ffar2−/− mice during co-housing protected Ffar2−/− mice from colonic inflammation and (2) Ffar2−/− mice are deficient in this gut bacteria. However, identity of this beneficial gut bacterium is unknown. Data presented above argues in favor of Bifidobacterium spp being these beneficial gut bacteria. Therefore, the ability of Bifidobacteria to suppress colonic inflammation and carcinogenesis in Ffar2−/− mice was tested. FP-fed Ffar2−/− mice were gavaged orally with Bifidobacterium longum subsp. infantis (ATCC 15697, B infantis) and challenged with AOM/DSS as described in Figure 6a. Number of Bifidobacterium in feces significantly increased in mice gavaged with B. infantis than saline (Figure 6b). B infantis significantly suppressed weight loss, diarrhea, inflammation and development of polyps in colon of Ffar2−/− mice (Figures 6c–f and Supplementary Figure 6b). Collectively, these data demonstrate that Bifidobacterium deficiency enhances risk for development of colonic inflammation and carcinogenesis in Ffar2−/− mice.
Figure 6.
Bifidobactreia suppress colonic inflammation and carcinogenesis in Ffar2−/− mice. (a) Ffar2−/− mice fed with FP diet and 1 month later were challenged with AOM/DSS and B. infantis as shown. B. infantis was gavaged everyday (2 × 108 cfu/mouse) as indicated from day of AOM injection till the completion of DSS treatment. (b) Relative abundance of Bifidobacterium in mice gavaged with B. infantis or saline as in a. Shown is the weight loss (c) during and after DSS challenge. (d) A representative photograph of H&E stained cross-section of colons of mice 4 days after cessation of DSS treatment. (e) Histopathological score (inflammation+epithelial damage) of colons. (f) Polyp burden in colons of mice treated as described in a at experimental endpoint. A representative of two experiments is shown (n=4 mice). *P<0.05, **P<0.01, ***P<0.0001.
Decreased numbers of Bifidobacterium and increased numbers of Prevotellaceae in human colon cancers
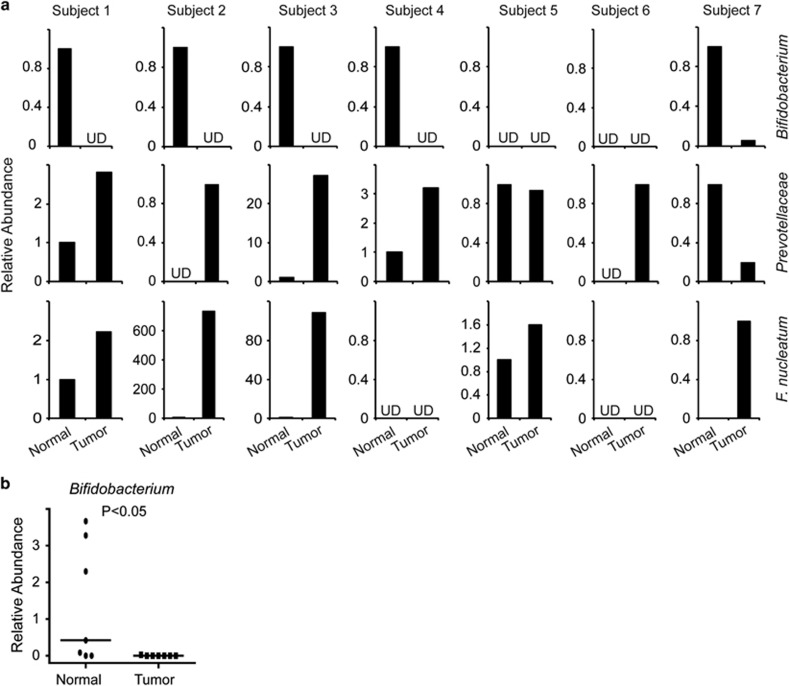
Based on our findings above, we evaluated the presence of Bifidobacterium and Prevotellaceae in human colon cancers and adjacent normal tissue. Bifidobacterium spp were readily detectable in most (5 out of 7) of the normal tissue, whereas it was either undetectable or present at drastically reduced numbers in cancerous tissue than corresponding adjacent normal tissue (Figure 7a). When all the seven paired samples were analyzed together, significantly higher number of Bifidobacterium was present in matched adjacent normal tissue than tumor (Figure 7b). In contrast, Prevotellaceae was present in higher numbers in most of the cancerous tissue than matched adjacent normal tissue (Figure 7). Consistent with previous studies,27, 28, 29 F nucleatum was present in higher numbers in colon cancer tissues than corresponding adjacent normal tissue (Figure 7a). Analysis of feces from healthy and colon cancer subjects have shown a decrease of butyrate-producing bacteria in later. However, F nucleatum is a butyrate producer and its numbers are increased in colon cancers than normal tissue.27, 28, 29, 30 Therefore, we analyzed the presence of other butyrate-producing groups, such as C leptum and C coccoides/E rectale in colon cancers and matched adjacent normal tissue. Surprisingly, we found that numbers of C coccoides/E rectale group was drastically increased in four out of seven colon cancers than adjacent normal tissue (Supplementary Figure 7). However, we could not find a clear correlation for abundance of C leptum or F prausnitzii between cancerous and matched normal adjacent tissue (Supplementary Figure 7). Collectively these data demonstrate that the number of Bifidobacterium is decreased, whereas that of Prevotellaceae is increased in colon cancers than matched normal tissue.
Figure 7.
Decreased abundance of Bifidobacterium spp in human colon cancers. (a) Relative abundance of indicated bacterial groups in DNA extracted from colon cancers and matched adjacent normal tissue was measured by quantitative polymerase chain reaction. (b) Relative levels of Bifidobacterium in colon cancer and matched adjacent normal tissue. The horizontal line represents the median value. *P<0.05. UD, undetectable. Statistical significance was calculated using Mann–Whitney test with two-tailed analysis.
Discussion
In the current study, we identified decreased numbers of Bifidobacterium spp and increased numbers of Prevotellaceae in colon cancers than matched adjacent normal tissue. One of the most important findings of this study is that Ffar2 is indispensable for dietary fibre-mediated promotion of Bifidobacterium spp and inhibition of H hepaticus and Prevotellaceae among gut microbiota. Increase in Bifidobacterium spp in gut is not specific to the mixture of inulin, pectin and cellulose as source of dietary fibre used in our study, because different types of dietary fibre such as lactulose, trans-galactooligosaccharides and fructooligosaccharides increase Bifidobacterium spp in human gut.24, 25 The effect of dietary fibre in enhancing numbers of Bifidobacterium spp varies among individuals.25 Based on our findings, it will be important to investigate whether altered expression or polymorphism of Ffar2 relates to dietary fibre-mediated promotion of Bifidobacterium spp in humans. Although host genetics and diet can independently affect gut microbiota,42, 43 our data demonstrate that interaction between both diet and host genes also play a critical role in influencing the composition of gut microbiota.
In contrast to comparisons between feces of healthy and colon cancer subjects, our data comparing colon cancer and adjacent normal tissue found an increase in butyrate-producing bacteria belonging to E rectale group in most of the colon cancers, which is in line with a previous study.44 F nucleatum, which is associated with colon cancer tissue and promotes colon carcinogenesis, is a butyrate producer. Evidences have been presented for both tumor-promoting and -suppressing effect of butyrate in colon.10, 11 Tumor-suppressing activity of butyrate has been largely attributed to its ability to inhibit HDAC activity. At single cell level, butyrate can induce apoptosis in colon cancer cells; however, at tissue or organ level, continuous exposure to butyrate will lead to toxicity towards other cells such as underlying connective tissue in colon, which may induce inflammation and/or promote metastasis. In fact, it has been proposed that F nucleatum produced butyrate plays a key role in induction of plaques and pathologies in dental tissues by inhibiting growth of fibroblast.30 Therefore, a lot of care should be taken in to considerations, such as effects of butyrate at tissue or organ level, effects of other cellular, molecular, metabolic or virulence factors of butyrate-producing bacteria in evaluating their role in colon carcinogenesis.
In summary, data presented in this study demonstrate that Ffar2 signaling promotes intestinal health by stimulating growth of beneficial bacteria such as Bifidobacterium and by decreasing the number of harmful gut microbiota such as H hepaticus and Prevotellaceae. It is evident that SCFA receptors Ffar2 and Hcar2 induce non-redundant mechanisms such as inflammasome activation, homeostasis of colonic Treg cells and promoting a healthy composition of gut microbiota,8, 9, 10, 41 which could explain how a defect in even one of the these receptors may result in reduced efficacy of dietary fibre to suppress intestinal carcinogenesis. A better understanding of all the signaling pathways induced by SCFA and dietary fibre will be useful in designing precise and better therapeutic strategies to prevent and/or treat intestinal inflammation and cancers.
Materials and methods
Mice and diets
C57BL/6J mice were obtained from Jackson laboratory. Ffar2+/− mice on C57BL/6 background were obtained from Deltagen. Ffar2+/− mice were bred to obtain Ffar2−/− mice. Both males and females of ages between 3 and 6 months were used in study. Animals were maintained on conventional rodent chow, TD-8604 (Harlan Laboratories, Madison, WI, USA) unless stated. FF (TD-00278) and FP (TD-130715) mouse chows were purchased from Harlan Laboratories. Formulation for FF diet is as follows: Lactalbumin 20.5%, DL-Methionine 0.22%, Dextrose monohydrate 52.839%, Maltodextrin 15%, Soybean Oil 5%, Mineral Mix AIN-93G 3.5%, Vitamin Mix AIN-93 1.5%, Potassium phosphate monobasic 0.84%, Calcium carbonate 0.3%, Choline Bitartrate 0.3% and TBHQ antioxidant 0.001%. FP diet is similar to FF diet, with 5% dietary fibre (1% cellulose, 2% pectin and 2% inulin) added. Bifidobacterium longum subsp. Infantis ATCC 15697 (B infantis) was purchased from American Type Culture Collection (ATCC). Where indicated, mice were gavaged orally with 2 × 108 CFU of B infantis in 100 μl of sterile saline buffered with 5% sodium bicarbonate. The Institutional Animal Care and Use Committee, Georgia Regents University approved all animal procedures.
Colitis-associated colon cancer model
Mice were injected intraperitoneally (i.p.) with AOM, (Sigma, St Louis, MO, USA) at dose of 10 mg/kg body weight in phosphate buffered saline. After 7 days, mice were fed 2% DSS, (MP Biochemicals, Santa Ana, CA, USA) molecular mass 36 000–50 000 Da in the drinking water for 7 days, followed by 15 days of regular water. This cycle was repeated twice. In experiments involving FP or FF diet, mice received a single cycle of 1.5% DSS in drinking water. Mice were monitored for weight changes, diarrhea and rectal bleeding. Diarrhea was scored as (0) normal stool, (1) soft but formed pellet, (2) very soft pellet, (3) diarrhea (no pellet) or (4) dysenteric diarrhea. Rectal bleeding was recorded as (0) no bleeding, (2) presence of occult blood in stool or (4) gross macroscopic bleeding. Seventy days following AOM injection, colonic lumens of mice were evaluated for polyp number and size.
Measurement of gut epithelial barrier function
Gut epithelial barrier function was evaluated using FITC-dextran, 4kD (Sigma). Mice were administered an oral gavage of FITC-dextran as a permeability tracer at a dose of 50 mg/100 g of body weight. Six hours later, mice were bled, serum was collected and FITC-dextran in serum was measured by fluorescence spectrophotometry (SpectraMax, Molecular Devices, Sunnyvale, CA, USA). FITC-dextran concentration was determined from standard curves generated by serial dilution of known concentrations of FITC-dextran in control serum ran in parallel.
Immunohistochemistry
Colons were excised and cleaned with phosphate buffered saline followed by fixation in neutral buffered formalin (Thermo Fisher, Waltham, MA, USA). The fixed colon tissues were embedded in paraffin and 5 μm thick sections were sliced and placed on glass microscope slides. Immunohistochemical staining was performed as described previously.8 Briefly, sections were deparaffinized, followed by antigen retrieval using the antigen retrieval solution (Agilent Technologies, Santa Clara, CA, USA). Endogenous peroxidase activity was quenched with 3% H2O2 in phosphate buffered saline for 10 min. Sections were stained using specific primary antibodies and Vectastain ABC kit and diaminobenzidine (Vector Laboratories, Burlingame, CA, USA). Counterstaining was performed with hematoxylin and stained sections were visualized by Leica DM550B microscope. Alternatively, in some experiments, fluorescent dye-labeled secondary antibody was used and sections were visualized by LSM 510 (Zeiss) confocal microscopy.
Isolation of RNA, DNA and quantitative PCR
Total RNA was extracted from colon tissue using TRIzol reagent (Thermo Fisher). The high capacity cDNA synthesis kit (Thermo Fisher) was used for synthesis of cDNA. Relative amounts of mRNAs were measured using polymerase chain reaction (PCR) and StepOne Real-time PCR system (Thermo Fisher) on cDNA samples. Gene expression was quantified using ΔΔCT method.
Colon cancer and matched adjacent normal tissue
Anonymous human colon cancer tissue and matched adjacent normal tissue from all demographics were obtained after informed consent and approval from the Institutional Review Board. The samples were collected over a period of time at Georgia Regents Medical Center Augusta, GA, USA. PCR primer for detection of FFAR2 has been described.45
Quantification of gut microbiota
Feces (~100 mg) were suspended with 710 μl of disruption buffer (200 mM NaCl, 200 mM Tris pH 8.0, 20 mM EDTA pH 8.0 and 6% SDS) and 500 μl of phenol/chloroform/isoamyl alcohol, pH 8.0 inside tubes containing Zirconium beads (0.1 mm diameter, Benchmark Scientific, Edison, NJ, USA). The mixture was homogenized using a Beadbeater (BioSpec, Bartlesville, OK, USA) for 2 cycles of 2 min each. Sample was centrifuged at 7000 g for 3 min and aqueous phase was collected and a second round of phenol/chloroform/isoamyl extraction was performed. DNA from the clear aqueous phase was precipitated using sodium acetate and isopropanol. Dried DNA pellet was dissolved in TE buffer. Quantitative polymerase chain reaction using group-specific PCR primers were used to quantify relative abundance of different groups to the total gut microbiota as reported previously38, 46, 47 and the sequences of PCR primers are provided in Supplementary Table 1.
Statistical analysis
Statistical significance was calculated using T-test with two-tailed analysis unless stated otherwise. No method was used to predetermine the sample size. Experiments were performed and analyzed in non-randomized and non-blinded fashion.
Acknowledgments
We thank Dr Leszek Ignatowicz for helpful discussions and reviewing the manuscript. This research was supported by National Institutes of Health grants R21AI085440 and R01DK103576 to NS.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogenesis website (http://www.nature.com/oncsis)
Supplementary Material
References
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 2008; 27: 104–119. [DOI] [PubMed] [Google Scholar]
- Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 2012; 6: 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Amand AL St, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 2007; 104: 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem 2003; 278: 11312–11319. [DOI] [PubMed] [Google Scholar]
- Taggart AK, Kero J, Gan X, Cai TQ, Cheng K, Ippolito M et al. (D)-beta-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J Biol Chem 2005; 280: 26649–26652. [DOI] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013; 504: 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504: 446–450. [DOI] [PubMed] [Google Scholar]
- Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014; 40: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, Hillhouse A et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov 2014; 4: 1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 2014; 158: 288–299. [DOI] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009; 461: 1282–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014; 20: 159–166. [DOI] [PubMed] [Google Scholar]
- Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013; 145: 396–406 e1-10. [DOI] [PubMed] [Google Scholar]
- Sina C, Gavrilova O, Forster M, Till A, Derer S, Hildebrand F et al. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J Immunol 2009; 183: 7514–7522. [DOI] [PubMed] [Google Scholar]
- Tang Y, Chen Y, Jiang H, Robbins GT, Nie D. G-protein-coupled receptor for short-chain fatty acids suppresses colon cancer. Int J Cancer 2011; 128: 847–856. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan F, Quigley EM. Manipulation of the microbiota for treatment of IBS and IBD-challenges and controversies. Gastroenterology 2014; 146: 1554–1563. [DOI] [PubMed] [Google Scholar]
- Slavin J. Fiber and prebiotics: mechanisms and health benefits. Nutrients 2013; 5: 1417–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011; 469: 543–547. [DOI] [PubMed] [Google Scholar]
- Albenberg LG, Wu GD. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology 2014; 146: 1564–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013; 13: 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Kim JY, Lee KW, Lee HJ. Cancer chemopreventive effects of lactic acid bacteria. J Microbiol Biotechnol 2007; 17: 1227–1235. [PubMed] [Google Scholar]
- Gibson GR, Scott KP, Rastall RA, Tuohy KM, Hotchkiss A, Dubert-Ferrandon A et al. Dietary prebiotics: current status and new definition. Food Sci Tech Bull Funct Foods 2010; 7: 1–19. [Google Scholar]
- Davis LM, Martinez I, Walter J, Goin C, Hutkins RW. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS ONE 2011; 6: e25200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE 2011; 6: e16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013; 14: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015; 42: 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut (e-pub ahead of print 26 August 2015; doi:10.1136/gutjnl-2015-310101). [DOI] [PMC free article] [PubMed]
- Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A et al. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol 2002; 184: 2005–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G, Vlachou A, Verbrugghe K, De Vuyst L. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl Environ Microbiol 2006; 72: 7835–7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun 2015; 6: 6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res 2009; 69: 2826–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell 1996; 87: 159–170. [DOI] [PubMed] [Google Scholar]
- Le Bourgot C, Ferret-Bernard S, Le Normand L, Savary G, Menendez-Aparicio E, Blat S et al. Maternal short-chain fructooligosaccharide supplementation influences intestinal immune system maturation in piglets. PLoS ONE 2014; 9: e107508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ. The microbiome revolution. J Clin Invest 2014; 124: 4162–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature 2012; 489: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 2010; 11: 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslinger AJ, Eller LK, Reimer RA. Yellow pea fiber improves glycemia and reduces Clostridium leptum in diet-induced obese rats. Nutr Res 2014; 34: 714–722. [DOI] [PubMed] [Google Scholar]
- Koleva PT, Valcheva RS, Sun X, Ganzle MG, Dieleman LA. Inulin and fructo-oligosaccharides have divergent effects on colitis and commensal microbiota in HLA-B27 transgenic rats. Br J Nutr 2012; 108: 1633–1643. [DOI] [PubMed] [Google Scholar]
- Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun 2015; 6: 6734. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R et al. Human genetics shape the gut microbiome. Cell 2014; 159: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol 2015; 6: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang Z, Er JZ, Ding JL. The short-chain fatty acid receptor GPR43 is transcriptionally regulated by XBP1 in human monocytes. Sci Rep 2015; 5: 8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011; 145: 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011; 331: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.