Abstract
In this study, we assessed the contributions of plasma membrane (PM) microdomain targeting to the functions of H-Ras and R-Ras. These paralogs have identical effector-binding regions, but variant C-terminal targeting domains (tDs) which are responsible for lateral microdomain distribution: activated H-Ras targets to lipid ordered/disordered (Lo/Ld) domain borders, and R-Ras to Lo domains (rafts). We hypothesized that PM distribution regulates Ras-effector interactions and downstream signaling. We used tD swap mutants, and assessed effects on signal transduction, cell proliferation, transformation and tumorigenesis. R-Ras harboring the H-Ras tD (R-Ras-tH) interacted with Raf, and induced Raf and ERK phosphorylation similar to H-Ras. R-Ras-tH stimulated proliferation and transformation in vitro, and these effects were blocked by both MEK and PI3K inhibition. Conversely, the R-Ras tD suppressed H-Ras-mediated Raf activation and ERK phosphorylation, proliferation and transformation. Thus, Ras access to Raf at the PM is sufficient for MAPK activation and is a principal component of Ras mitogenesis and transformation. Fusion of the R-Ras extended N-terminal domain to H-Ras had no effect on proliferation, but inhibited transformation and tumor progression, indicating that the R-Ras N-terminus also contributes negative regulation to these Ras functions. PI3K activation was tD independent; however, H-Ras was a stronger activator of PI3K than R-Ras, with either tD. PI3K inhibition nearly ablated transformation by R-Ras-tH, H-Ras and H-Ras-tR, whereas MEK inhibition had a modest effect on Ras-tH-driven transformation but no effect on H-Ras-tR transformation. R-Ras-tH supported tumor initiation, but not tumor progression. While H-Ras-tR-induced transformation was reduced relative to H-Ras, tumor progression was robust and similar to H-Ras. H-Ras tumor growth was moderately suppressed by MEK inhibition, which had no effect on H-Ras-tR tumor growth. In contrast, PI3K inhibition markedly suppressed tumor growth by H-Ras and H-Ras-tR, indicating that sustained PI3K signaling is a critical pathway for H-Ras-driven tumor progression, independent of microdomains.
Introduction
The Ras superfamily comprises over 150 small GTPases, which can bind and hydrolyze guanine triphosphate (GTP). The proto-oncogenic homologs most prominently associated with cancers, H-, N- and K-Ras, are ubiquitously expressed and have overlapping yet non-redundant functions.1, 2, 3, 4 Ras propagates growth factor signaling pathways regulating cell proliferation, differentiation, angiogenesis and survival.1, 2, 3 Ras proteins cycle between an active GTP-bound and an inactive GDP-bound state. Constitutively active (CA) (that is, GTP-locked) Ras mutations are highly transforming and induce tumor formation,5 and the combined set of activating Ras mutations altogether are associated with as high as ~30% of human malignancies.6 However, each isotype displays tissue type specificities in cancers, indicating context-dependent modes of action.6 Thus, the molecular mechanisms of isotypic Ras oncogenesis are still not completely understood.1, 2, 3
Related-Ras 1 (R-Ras) is a highly conserved Ras paralog with limited mitogenic signaling properties. R-Ras functions include cell motility, survival and vascular quiescence, distinct from other Ras paralogs.7, 8, 9, 10, 11, 12 CA R-Ras weakly transforms select cell lines, and activating mutations are not prominent in human malignancies.13, 14 However, despite identical effector-binding domains, H- and R-Ras support distinct effector signal outputs, suggesting contributions from other domains in the regulation of Ras-effector interactions and signaling. H- and R-Ras paralogs are most divergent in the termini: a 26 amino-acid N-terminal extension in R-Ras which is absent from H-Ras, and in the C-terminal hypervariable domains (HVR), which harbor membrane targeting domains (tDs) at the C-termini. The R-Ras N-terminal domain regulates R-Ras-dependent cell migration, but has not been associated with mitogenic signaling.15 Activated H-Ras interacts in cells with Raf kinase and facilitates its activation, which propagates phosphorylation of cytosolic kinases MEK and ERK, driving cell proliferation.16, 17, 18 R-Ras is capable of binding Raf in vitro; however this interaction either does not occur or is of low affinity in cells, believed to account for the weak transforming function of R-Ras.19, 20 The mechanism for these distinct behaviors is not understood but may reflect distinct properties based on the HVRs.
Ras proteins are initially synthesized as globular, cytoplasmic polypeptides, which undergo a series of irreversible lipid modifications within the C-terminal tDs that increase the hydrophobicity of the C-termini and support anchorage of the Ras proteins to ER membranes and subsequent transport to the Golgi, before delivery to the plasma membrane (PM).21 The K-Ras tD contains Lys-rich sequences, which enhance charge-based PM interactions.22 In contrast, the tDs in H-, N- and R-Ras also contain adjacent target sites for reversible palmitoylation by a thioester linkage (C181 and C184 in H-Ras, C213 in R-Ras).23, 24 Palmitoylation occurs at the Golgi,25, 26 and is required for sorting into post-Golgi vesicles and vesicular transport to the PM; depalmitoylation at the PM by cognate palmitoyl (acyl) thioesterases completes the cycle by driving retrograde recycling to the Golgi.12, 27, 28, 29, 30, 31 Ras isotypes also have distinct distributions in microdomains at the PM, driven by guanine nucleotide loading as well as palmitoylation.30, 31, 32, 33 H-Ras is anchored to the lipid ordered (Lo, that is, lipid raft) membrane while GDP loaded, and is shuttled to the lipid ordered/lipid disordered (Lo/Ld) border upon GTP loading.34, 35, 36 R-Ras preferentially anchors within the Lo domain, regardless of activation state.34, 37 These membrane microdomain preferences are supported by electron microscopy and in silico simulations.34, 36, 37 Efficient activation of Raf by H-Ras requires signaling from the Ld membrane.36, 38, 39 We hypothesized that the lateral membrane distributions of H-Ras and R-Ras are key determinants of their distinct mitogenic and oncogenic properties. In this study, we investigated the spatial regulation of Ras/Raf interaction and signal propagation by PM microdomain Ras targeting, and the contributions of microdomain-dependent signaling to Ras-induced cell proliferation and tumorigenesis.
Results
The H-Ras tD is both necessary and sufficient for palmitoylated Ras/Raf interaction, Raf-1 activation and MAPK signal propagation in cells
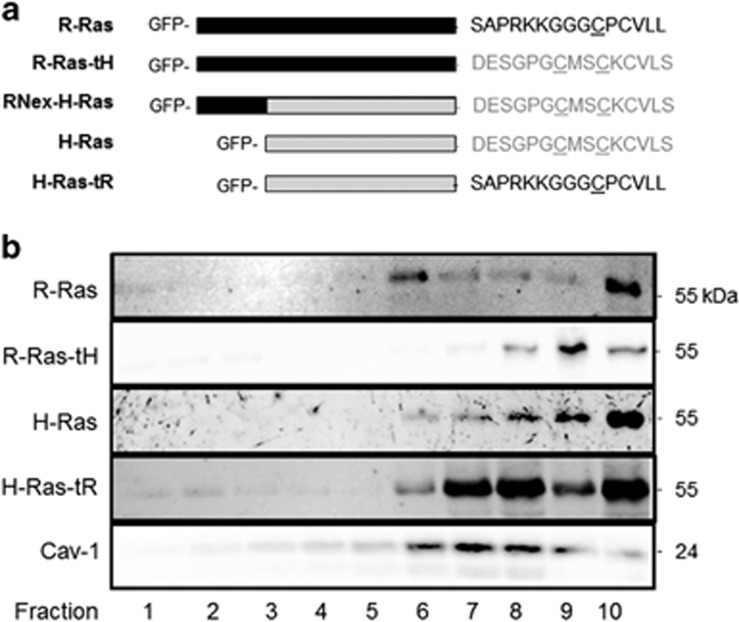
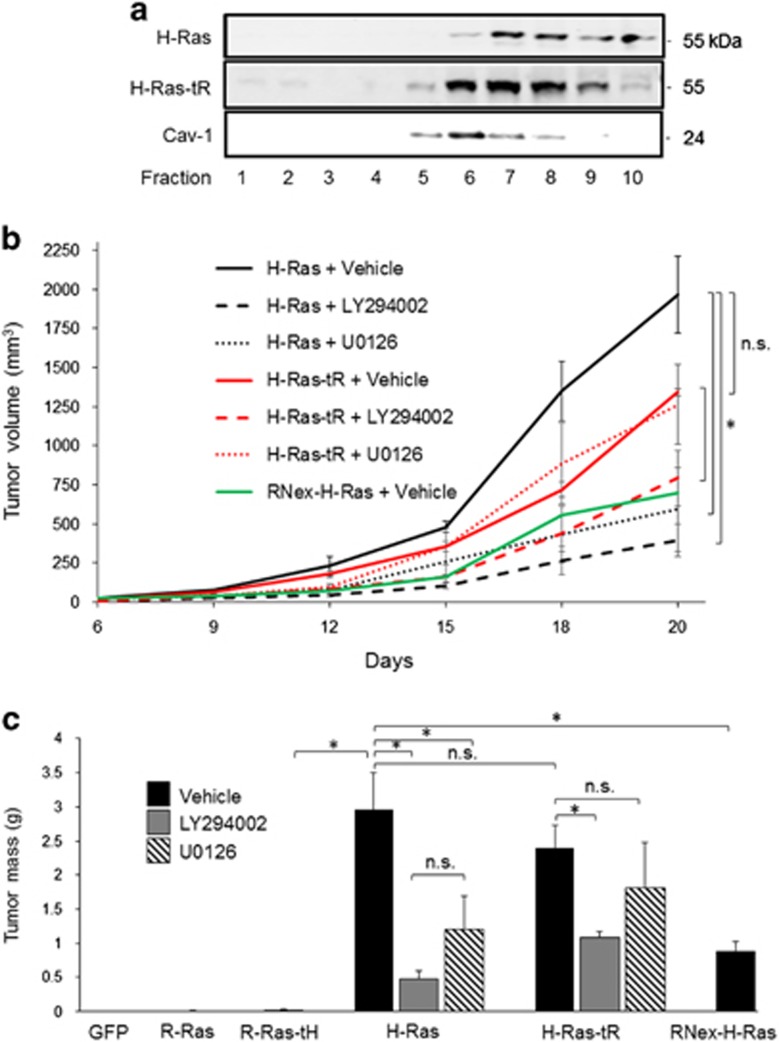
We created tD switch mutants between H- and R-Ras on the CA background (H-Ras(G12V), R-Ras(G38V)), in which the final 15 amino acids were swapped (Figure 1a), as GFP fusions at the Ras N-terminus. We confirmed the tD-dependent membrane microdomain distributions of these variants by sucrose fractionation.40 As predicted, active H-Ras was only partially enriched in Cav-1-positive (Lo domain) fractions, while highly enriched in dense Cav-1-negative fractions. R-Ras was enriched in Cav-1-positive fractions, confirming that activated H-Ras targets to the Lo/Ld border whereas activated R-Ras is primarily sequestered in the Lo domain. H-Ras harboring the R-Ras tD (hereafter referred to as H-Ras-tR) was highly enriched in Cav-1-positive fractions as observed previously,37 whereas R-Ras harboring the H-Ras tD (R-Ras-tH) was minimally in Cav-1 fractions, and was highly enriched in the dense Ld fractions (Figure 1b). Thus, the H-Ras targeting domain (tH) and the R-Ras targeting domain (tR) enforced isotypic lateral targeting of Ras proteins to the Lo/Ld border or Lo domain, respectively.
Figure 1.
Ras targeting domain swap mutants determine PM microdomain localization. (a) Schematic representation of GFP-Ras expression constructs used in these studies. R-Ras-tH, R-Ras(1–203)H-Ras(175–189); H-Ras-tR, H-Ras(1–174)-R-Ras(204–218); RNex-H-Ras, R-Ras(1–26)-H-Ras(1–189). Black bar, R-Ras G domain; gray bar, H-Ras G domain; underline, palmitoylation sites; lower case, geranylgeranylation site; *, farnesylation site. (b) NIH3T3 cells stably expressing the indicated GFP-Ras variants were fractionated by ultracentrifugation using a step-gradient from 10 to 45% sucrose followed by immunoblotting with α-GFP antibodies. α-Cav-1 was used as a lipid raft marker. All blots representative of three independent experiments.
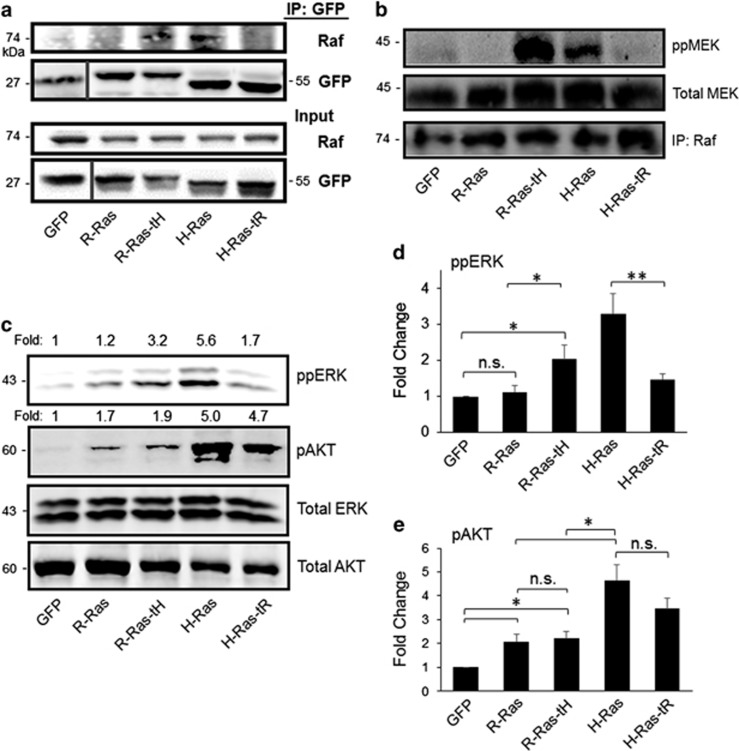
R-Ras can bind Raf-1 in vitro; however, R-Ras/Raf-1 interaction and downstream signaling has not been observed in cells.19, 20 To test whether membrane microdomain localization was responsible for this property of R-Ras, we investigated the effect of mistargeted H- and R-Ras on Raf interaction in cells. NIH3T3 cells stably-expressing GFP-Ras variants at similar protein levels (Figure 2a) were lysed and subjected to immunoprecipitation (IP) with GFP antibodies. Endogenous Raf-1 co-precipitated with H-Ras but was absent from R-Ras IP fractions, confirming that H-Ras but not R-Ras interacts with Raf-1 in these cells.19, 41 In contrast to R-Ras, Raf-1 was enriched in GFP-R-Ras-tH precipitate fractions. Conversely, the interaction of H-Ras with Raf-1 was abrogated with H-Ras harboring the tR (Figure 2a). Thus, PM microdomain localization is a critical determinant in Ras/Raf-1 interaction in cells, and the tH supports Ras/Raf-1 interaction. To address whether interaction of Ras with Raf correlated with Raf-1 activation, we assessed Raf-1 kinase activity in serum-deprived cells expressing Ras variants. Endogenous Raf-1 isolated (by IP) from cells expressing H-Ras and R-Ras-tH, but not R-Ras or H-Ras-tR, was capable of phosphorylating recombinant MEK in vitro (Figure 2b). Thus, the tH is both necessary and sufficient for Ras-mediated Raf-1 recruitment to the membrane and interaction of a Ras protein with Raf-1 in cells, and this interaction facilitates Raf-1 activation.
Figure 2.
Ras targeting domains dictate access to Raf and MAPK signaling. (a) NIH3T3 murine fibroblasts were stably transfected with GFP-tagged Ras variants as indicated, and GFP fusion proteins were immunoprecipitated (IP) from cell lysates (Input) with α-GFP antibodies, followed by immunoblotting with α-Raf or α-GFP antibodies. (b) Raf kinase assay. Cells were serum-starved, and Raf activity was assessed as described in Materials and methods. Immunoblotting of the IP fraction with α-Raf antibodies is shown in the lower panel. (c) ERK and AKT activation in Ras-expressing cells after 72 h serum starvation, as assessed by immunoblotting of cell lysates with the indicated antibodies. Phospho:total ratios are shown above the respective blots as a ratio to GFP. (d, e) Fold change in phospho-ERK:total ERK or phospho-AKT:total AKT ratios compared with GFP control+s.e.m. *P<0.05; **P<0.003, n.s., not significant. All blots representative of five independent experiments.
To investigate downstream signaling of the tD mutants, we assessed phosphorylation of ERK and AKT (ppERK (T202/Y204) and pAKT (S473), indicating MEK and PI3K activation, respectively) in serum-starved cells expressing Ras-tD variants. R-Ras did not stimulate ERK phosphorylation, consistent with previous reports.19 Conversely, H-Ras as well as R-Ras-tH stimulated phosphorylation of ERK in serum-starved cells. H-Ras-tR yielded a marked reduction in ppERK compared with H-Ras, although these phosphorylation events were not completely blocked (Figures 2c and d). Thus, H-Ras stimulation of ERK phosphorylation is tD dependent. We observed similar results in HEK293 cells transiently transfected with GFP or the GFP-Ras variants, indicating that Ras-tD-dependent MAPK signaling is not cell-type specific (Supplementary Figures 1a and b). Moreover, R-Ras, R-Ras-tH, H-Ras and H-Ras-tR each promoted robust AKT phosphorylation, regardless of the tD, but H-Ras was a more potent activator of PI3K than R-Ras (Figures 2c and e; Supplementary Figures 1a and c). Thus, the Ras-tH domain regulates Ras access to Raf-1 in cells, and facilitates activation of the MAPK pathway. In contrast, both H-Ras and R-Ras can activate PI3K independent of microdomain localization.
The H-Ras tD is necessary for Ras-induced cell proliferation
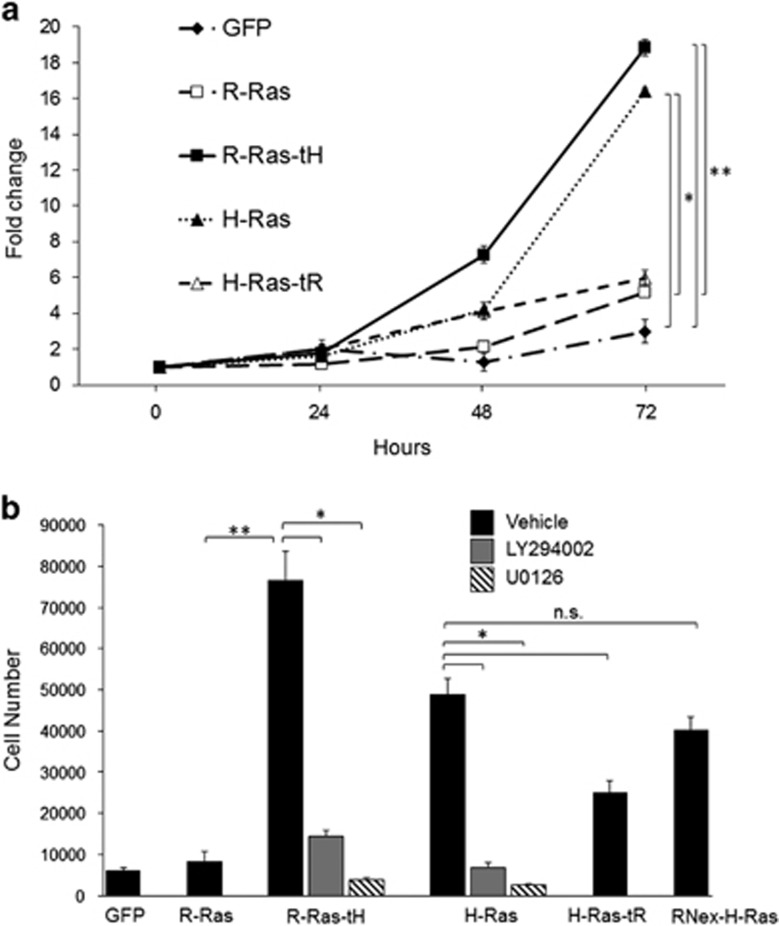
We investigated the effect of Ras targeting on cell proliferation. Cells stably expressing the Ras chimeras were maintained in low serum conditions, and cell growth was assessed over 72 h. These conditions resulted in inhibited cell growth and maintenance in G0/G1 in GFP- and R-Ras-expressing cells, as well as H-Ras-tR cells, as evidenced by DNA labeling and FACS, whereas H-Ras and R-Ras-tH cells showed population shifts toward S/G2, suggesting induction of mitogenesis in these cells. Each population had few dead cells, indicating that the Ras variants combined with low serum growth conditions did not result in substantial cell death (Supplementary Figure 2). To assess the ability of the Ras variants to promote cell proliferation, Ras-expressing cells were maintained in low serum conditions and cell population counts were monitored over time. Cell growth was significantly retarded by low serum conditions in control cells expressing vector (GFP) alone, whereas both H-Ras- and R-Ras-tH-expressing cells proliferated rapidly under these conditions. R-Ras and H-Ras-tR expression had little effect on stimulating cell growth over control (Figure 3a). To investigate whether these proliferation effects were cell-type specific, we monitored proliferation of HEK293 cells in low serum, after transient transfection with Ras variants. These cells showed similar trends in proliferation as the stably-expressing Ras NIH3T3 cells: R-Ras-tH and H-Ras potently stimulated proliferation, whereas R-Ras and H-Ras-tR did not (Supplementary Figure 3). Thus, H-Ras-typic targeting supports Ras-induced cell proliferation, while R-Ras-typic targeting does not support proliferation.
Figure 3.
The tH is necessary for a Ras protein to promote cell proliferation. 1 × 104 NIH3T3 cells stably expressing the indicated Ras mutants were seeded in triplicate wells in media with low serum. (a) Cells were harvested and counted at the indicated intervals. Cells were harvested and counted 5 h after seeding for 0-h time points. (b) Cells were plated in the presence of either 20 μm LY294002 or 30 μm U0126 as indicated and counted at 72 h. *P<0.01; **P<0.002, n.s., not significant. Representative of five independent experiments.
Next, we addressed effector pathways important for Ras-tH-induced cell growth. Cultured cells expressing the Ras chimeras were placed in low serum conditions in the presence of either 30 μm U0126 or 20 μm LY294002 (MEK and PI3K inhibitors, respectively). We confirmed that these inhibitors blocked their respective targets in these cells: LY294002 inhibited pAKT but not ERK, and conversely, U0126 inhibited ppERK but not pAKT, in serum-starved Ras cells (Supplementary Figure 4). Although R-Ras-tH strongly promoted proliferation, we observed a marked reduction in R-Ras-tH-driven cell growth with either LY294002 or U0126 treatment (Figure 3b). Similarly, H-Ras-induced proliferation was drastically reduced by blockade of either PI3K or MEK, indicating that both pathways are critical for Ras-induced cell proliferation.
In addition to the tDs, R-Ras and H-Ras are primarily distinguished by a non-conserved 26 amino-acid N-terminal extended domain in R-Ras (‘RNex'). To address a potential contribution of this R-Ras domain to Ras-induced cell growth, we stably expressed a GFP-Ras fusion harboring the RNex domain fused at the N-terminus to H-Ras(G12V) (hereafter referred to as RNex-H-Ras) (Figure 1a). RNex-H-Ras stable transfectants proliferated at a rate similar to H-Ras cells under low serum conditions, indicating that the RNex domain is not a major contributor to Ras-induced cell growth (Figure 3b). Thus, the Ras-tD is the critical domain dictating Ras-induced cell growth, which requires both PI3K and MAPK pathway activation by Ras.
The H-Ras tD modulates Ras-induced cell transformation
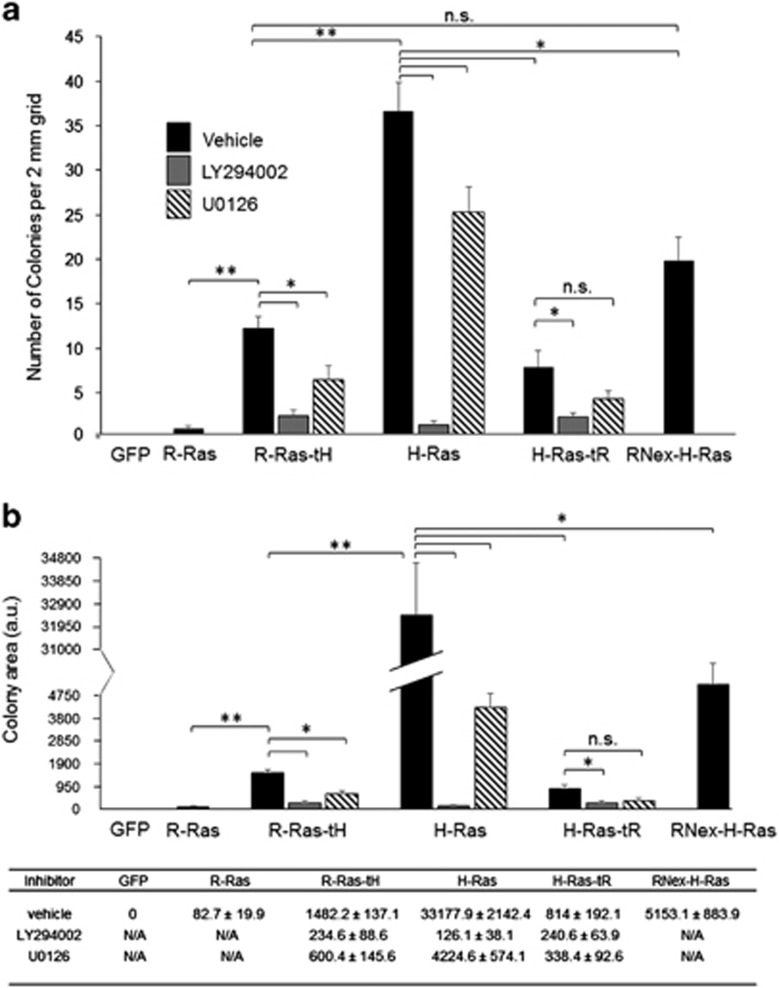
To investigate the contributions of Ras targeting to cellular transformation, we evaluated anchorage-independent growth by assessing colony formation of Ras stably transfected cells in soft agar. R-Ras stable transfectants produced minimal colony formation, similar to the baseline control. H-Ras supported formation of many large colonies, whereas H-Ras-tR expression yielded significantly attenuated colony formation (Figures 4a and b). Thus, sequestration of H-Ras in the Lo domain attenuated but did not ablate its ability to promote cellular transformation. R-Ras-tH stimulated colony formation, indicating a gain of function for R-Ras by fusion of the tH domain (Figure 4a). However, although R-Ras-tH expression yielded an increase in colony formation compared with parental R-Ras, with both the number and size of the colonies greater than those produced by R-Ras cells, the R-Ras-tH colonies were fewer in number and smaller than those formed by H-Ras (Figures 4a and b). Colony formation by RNex-H-Ras-expressing cells was similar to that in R-Ras-tH cells, indicating that the RNex domain negatively regulates tH-targeted Ras-induced colony formation (Figures 4a and b).
Figure 4.
The H-Ras tD modulates Ras-induced cell transformation. 5 × 104 cells were seeded into 0.3% agar for 14 days, and cells were fixed and imaged. (a) Number of colonies per 2 mm grid are shown. (b) Area of colonies per 2 mm grid were measured using ImageJ software. Average total areas per grid are shown+s.e.m. *P<0.001; **P<2 × 10−6. n.s., not significant. n=6. The values for the histogram in (b) are shown below in tabular form, ±s.e.m.
MEK inhibition partially inhibited colony formation in H-Ras and Ras-tH cells, but had no effect on the already diminished colony growth by H-Ras-tR cells. In contrast, PI3K inhibition ablated Ras-induced colony formation by H-Ras, R-Ras-tH and H-Ras-tR cells (Figure 4). Together, these data demonstrate that Lo/Ld targeting by the tH domain facilitates Ras-induced cellular transformation through both PI3K and MEK signaling, and sustained PI3K signaling is a major determinant for Ras-driven transformation.
Roles of the tDs in Ras-mediated tumorigenesis and tumor progression
The distinct effects of Ras lateral membrane distribution on MAPK signaling, cell proliferation and transformation in vitro suggested a critical role of Ras microdomain localization at the PM in Ras-induced tumorigenesis. To address this possibility, we employed a tumor allograft model in which athymic mice were injected subcutaneously in each flank with Ras stable cells, and resultant tumors were resected after 20 days.42 We assessed membrane microdomain distribution of H-Ras variants in tumor lysates ex vivo, and found that the tR shifted H-Ras to the Lo domain, indicating that the predicted membrane microdomain distributions of these Ras variants were upheld in the tumor cells in vivo (Figure 5a). Cells expressing GFP alone produced no detectable tumors in any mice (Figure 5b, Table 1). R-Ras-expressing cells produced few tumors (that is, tumors only formed at 2 out of 13 allograft sites), and tumors that did form were <2 mm in diameter and appeared avascular, consistent with previous observations with R-Ras (Figure 5b, Table 1).13, 14, 43, 44 In contrast, R-Ras-tH cells produced tumors at 50% of injection sites, and average tumor mass in these cases was ~2.3-fold higher than those generated by cells expressing R-Ras, indicating that fusion of the tH domain enhances tumorigenesis by R-Ras. However, tumors produced by R-Ras-tH cells were small—one-tenth the size of tumors produced by H-Ras cells (Table 1). H-Ras cells formed large tumors at every injection site. RNex-H-Ras cell tumors grew at a slower rate than H-Ras cell tumors (Figures 5a and b), reflecting diminished colony formation in soft agar by the RNex fusion (Figure 4), and indicating that the RNex domain negatively regulates both H-Ras-driven transformation and tumor growth. Interestingly, H-Ras-tR cell lines formed tumors at every injection site, all of which were significantly larger than those generated by either of the R-Ras counterparts, and which more closely resembled H-Ras tumors (Figure 5b, Table 1). This was a surprising result, as we anticipated that tumor promotion by H-Ras-tR would be attenuated compared with H-Ras. Thus, Ras targeting is a critical factor for Raf-1 interaction, activation, MAPK signaling and cell transformation in vitro as well as tumor initiation in vivo; however, another determinant outside the Ras targeting domain is necessary for Ras-induced in vivo tumor progression.
Figure 5.
H-Ras is tumorigenic even with the R-Ras tD. (a) Cells stably expressing the indicated Ras variants were injected as a bolus allograft into the flanks of athymic mice. After 20 days, tumors were excised, and tumor lysates were fractionated by ultracentrifugation using a step-gradient from 10 to 45% sucrose. α-Cav-1 was used as a lipid raft marker. (b) Upon detection, tumor dimensions were measured every 72 h for volume calculation. (c) Average tumor masses of resected tumors at 20 days are shown+s.e.m. *P<0.05, n.s., not significant. n=10.
Table 1. Summary of tumor allografts derived from Ras-tD chimera cells.
| Stable cell line | Number of tumors/allografts | Average tumor mass (g) |
|---|---|---|
| GFP | 0/14 | 0 |
| R-Ras | 2/13 | 0.008±0.007 |
| R-Ras-tH | 6/12 | 0.019±0.012 |
| H-Ras | 18/18 | 2.09±0.51 |
| H-Ras-tR | 12/12 | 1.75±0.27 |
Mice were allografted with a bolus injection of cells stably expressing each Ras chimera into each flank, and the number of solid tumors formed per allograft is shown. Masses of resected tumors are shown±s.e.m.
To investigate signaling pathways important for Ras-driven tumorigenesis, either U0126 or LY294002 was administered intraperitoneally every 72 h starting at the time of seeding Ras cells, and tumor growth was monitored using calipers. Inhibition of MEK by U0126 administration resulted in ~70% attenuation of tumor growth by H-Ras cells, but had no effect on tumor growth by H-Ras-tR cells. However, blockade of PI3K by administration of LY294002 substantially inhibited growth of both H-Ras and H-Ras-tR tumors (Figures 5b and c). Thus, MAPK and PI3K signaling contributes to H-Ras-driven transformation and tumorigenesis, whereas PI3K activation is the major pathway driving tumor progression by lipid raft-sequestered H-Ras, which is deficient in MAPK signaling.
The effects of the tD swaps on Ras-induced signaling, proliferation, transformation and tumor progression are summarized in Table 2. Taken together, these data indicate that Ras membrane microdomain distribution regulates Raf pathway signaling and Ras-induced cell proliferation, and both MAPK and PI3K pathways are important for Ras-induced proliferation and transformation. However, Ras-induced PI3K signaling is tD independent, and this pathway is required to mediate H-Ras-induced tumor progression.
Table 2. Summary of signaling, cellular and tumorigenic effects of Ras-tD chimeras.
| Ras variant | PI3K | MAPK | Proliferation | Transformation | Tumorigenesis |
|---|---|---|---|---|---|
| GFP | − | − | − | − | − |
| R-Ras | + | − | + | − | − |
| R-Ras-tH | + | + | +++ | + + | + |
| H-Ras | +++ | +++ | +++ | +++ | +++ |
| H-Ras-tR | +++ | + | + | + + | +++ |
| RNex-H-Ras | ND | ND | +++ | + + | + + |
Abbreviations: −, undetectable activity; +, weak, but detectable activity; +++, very strong activity; ND, not determined.
Discussion
The results of this study demonstrate key roles of membrane microdomain targeting of Ras in signal transduction and stimulation of cell proliferation, transformation and tumor progression. H-Ras-typic targeting to the Lo/Ld border at the PM was sufficient to support R-Ras/Raf interaction, and to facilitate activation of Raf-1 and ERK, providing a molecular mechanism for distinct MAPK signaling by H- and R-Ras. Moreover, Ras-induced proliferation was tD dependent, indicating that Ras signaling from the Lo/Ld border is a critical determinant of Ras mitogenesis, and this function required both MAPK and PI3K pathway activation. However, PI3K activation by H-Ras was tD independent, and H-Ras with repressed MAPK activation by Lo sequestration was competent to promote tumor growth comparable to H-Ras, in a PI3K-dependent manner. Together, these studies demonstrate that the tD of H-Ras supports the ability of a Ras protein to drive MAPK signaling and cell proliferation, while PI3K activation is Ras-tD independent and is a major determinant of Ras-induced tumor progression.
Microdomain targeting of H-Ras and R-Ras was the critical determinant for Ras/Raf interaction. Early reports established that R-Ras was capable of binding Raf-1 in vitro,20 though later studies indicated that this interaction does not occur in vivo,19 alluding to spatial segregation of R-Ras and Raf-1 at the PM. Indeed, mistargeting R-Ras with swapped regions from the H-Ras HVR abrogates many of R-Ras' unique biological responses, including modulation of integrin activation, focal adhesion targeting and reactive oxygen species production, indicating a dominant role of the HVRs for functional distinctions between these proteins.37, 38, 39, 45, 46 In the current study, H-Ras sequestered within rafts via the tR domain did not robustly induce Raf-1 kinase activity or ERK phosphorylation, consistent with previous observations.36, 39 The marked reduction but incomplete ablation of H-Ras-induced MAPK signaling by the tR may reflect a scenario in which Ras-tR is enriched in Lo domains, but is not completely sequestered from the Lo/Ld border where Raf can be accessed, which may account for conflicting reports on whether R-Ras supports minimal ERK activation, or none.19, 47, 48 This notion is supported by sucrose gradient fractionation data, which show that a small portion of tR-targeted Ras (R-Ras or H-Ras-tR) localized to the Lo/Ld border or Ld domain. In an earlier study, H-Ras harboring an N-terminal raft-targeting domain from Lck was shown to activate ERK comparable to H-Ras(V12) in NIH3T3 cells.49 This discrepancy may reflect distinct orientation of the Ras-effector binding loop by membrane association with an N-terminal, as opposed to a C-terminal tag, which could alter effector interactions.50, 51 Thus, microdomain distribution appears to be the major determinant of Raf interaction and activation by H-Ras, and is sufficient for a Ras protein to propagate MAPK signaling.
Distinct from MAPK signaling, PI3K activation by H-Ras was tD independent, and these distinctions correlated with different pathway dependencies for H-Ras-induced proliferation, transformation and tumor progression. Ras microdomain-dependent Raf interaction and MAPK signaling corresponded to Ras mitogenesis, consistent with a requirement for Ras in Raf membrane recruitment and MEK/ERK and its roles in proliferation.52, 53, 54, 55, 56 However, PI3K pathway activation was also required for Ras-induced mitogenesis, indicating that both effector pathways are important for Ras stimulation of proliferation. Unlike cell proliferation, however, transformation by Ras reflected a much stronger dependence on PI3K signaling rather than MAPK signaling. The RNex domain, which is not conserved and is unique to R-Ras, negatively regulated H-Ras-driven transformation and tumor progression, demonstrating that microdomain localization is not the only factor separating H- and R-Ras functions. In a recent study, mice harboring Ras-binding mutations in the p110α subunit of PI3K showed diminished H-Ras-induced AKT phosphorylation and transformation of cells ex vivo despite no reduction in ERK phosphorylation, supporting the notion that PI3K signaling is a major pathway for Ras-induced transformation, consistent with our results.57, 58 These mice also showed defective K-Ras-driven tumorigenesis, which may reflect K-Ras-specific mechanisms of tumor promotion.58 Similarly, depletion of RalA, a major effector of RalGDS, diminished H-Ras-induced transformation and inhibited tumorigenesis, although RalA depletion had no effect on cell growth in vitro,59 underscoring that Ras-induced transformation and tumor promotion rely on distinct pathways from Ras mitogenesis. Moreover, we found that weak MAPK signaling was sufficient to support tumor initiation (R-Ras-tH), but not tumor progression. Ras-induced tumor progression was supported by MAPK-deficient, PI3K-competent H-Ras-tR, but not by R-Ras-tH, which showed weak activation of PI3K, and Ras tumor progression was blocked in all cases by inhibition of PI3K. Together, our data indicate that pathway activation driving Ras-induced cell proliferation does not correspond directly to Ras-mediated tumor progression, which specifically required strong PI3K pathway activation by Ras. Thus, while PM microdomain localization of Ras is critical for Ras-driven MAPK signaling and proliferation, and contributes to transformation, PI3K activation is Ras microdomain independent and is a key pathway in Ras-induced tumor progression.
Misregulated PI3K signaling occurs frequently in human cancers. Overstimulation of PI3K signaling may promote resistance to clinical therapies, which has sparked interest to improve current therapies or find new drug targets.60, 61, 62, 63 Notably, EGFR mutations, which are also common in human cancers, can stimulate PI3K signaling directly by Ras-independent pathways,64, 65, 66 which adds to the importance of misregulated PI3K signaling in oncogenesis. Our data support the notion that PI3K activation is a dominant pathway of Ras tumor progression, and is therefore an attractive target for increased efficacy in therapeutic intervention of H-Ras- and EGFR-mutant cancers.
Materials and methods
Antibodies, reagents and cDNAs
GFP (sc-9996), Raf (sc-133, sc-227) antibodies and purified recombinant MEK-1 (sc-4025) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). AKT (AH01112) and Phospho-AKT (44623G) were from Invitrogen (Waltham, MA, USA). p42/44 MAPK (ERK1/2), pp44/42 MAPK (ERK T202/Y204), ppMEK (S217/221) and pAKT (S473) antibodies were from Cell Signaling Technology (Danvers, MA, USA). Fluorophore-conjugated secondary antibodies were from LI-COR Biosciences (Lincoln, NE, USA). LY294002 was purchased from LC laboratories (Woburn, MA, USA). U0126 was purchased from Alfa Aesar (Ward Hill, MA, USA). Restriction endonucleases were obtained from New England Biolabs (Ipswich, MA, USA). pEGFP-C1 was from Clontech Laboratories (Mountain View, CA, USA). GFP-R-Ras constructs were made as described in Wurtzel et al.12 GFP-H-Ras G12V was a gift from K Svoboda (Addgene plasmid 18666). GFP-H-Ras (1–174)G12V- R-Ras (204–218) was generated from GFP-H-Ras G12V by PCR. GFP-RNex-H-Ras was generated by insertion of the first 26 amino acids of R-Ras from an R-Ras N-terminal domain construct originally described in Silver et al.,67 into the GFP-H-Ras background.
Cell culture and transfection
NIH3T3 and HEK293 cells were obtained from American Type Culture Collection (Manassas, VA, USA) and tested for mycoplasma in our laboratory before use. NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Cellgro, Manassas, VA, USA) supplemented with 10% Bovine Calf Serum (BCS), 4 mm l-glutamine, 4500 mg/ml glucose, 50 U/ml penicillin, 50 μg/ml streptomycin sulfate and 1% non-essential amino acids (Sigma-Aldrich, St Louis, MO, USA) at 37 °C in 5% CO2. HEK293 cells were maintained in DMEM supplemented with 10% Fetal Bovine Serum (FBS). Cells were analyzed 24–48 h after transfection. For stable transfectants, transfected NIH3T3 cells were selected in media containing 2 μg/ml G418 Sulfate (Geneticin). Colonies were subcloned after 2 weeks in the continued presence of Geneticin with cloning cylinders, and transgene expression was confirmed by western blot analysis. Stable transfectants were maintained in 200 ng/ml Geneticin.
Protein precipitation with perchloric acid
Cells lysates were collected and Bradford protein assays (Thermo Scientific, Waltham, MA, USA) were used to determine protein concentration. Total protein was precipitated with 6.6 m perchloric acid added to each sample for a final concentration of 0.66 m. Samples were incubated for 20 min at −20 °C and centrifuged at max r.p.m. for 15 min. Pellets were rinsed with water and centrifuged again, and resuspended in SDS buffer for SDS–PAGE.
Immunoprecipitations and western blotting
Cells were cultured in complete DMEM and, in some cases, serum-starved by culturing in DMEM/0.2% serum. Cells were rinsed 2 × in PBS, and cell lysates were harvested by scraping in lysis buffer (10 mm Tris-Cl, pH 7.5, 100 mm NaCl, 2 mm MgOAc, 0.5% Nonidet P-40, 10 μm GTP, 1 mm Na3VO4, 20 μm β-glycerophosphate, 1 mm NaF, plus a cocktail of protease inhibitors; Roche, Madison, WI, USA). Insoluble material was removed by centrifugation. Fractions of the lysates were separated by SDS–PAGE, followed by western blotting (immunoblotting) with specific primary antibodies, followed by detection with infrared fluoriphore-conjugated secondary antibodies using fluorescence laser scanning (LI-COR Biosciences, Lincoln, NE, USA). For IPs, supernatants were pre-cleared by incubation with Protein G-coupled sepharose beads (Roche) for 1 h at 4 °C. Cleared lysates were incubated with 2 μg of antibody suspensions for 16 h at 4 °C, followed by antibody capture on protein G-sepharose beads for 1 h. Antibody-bound complexes were precipitated by centrifugation, washed and separated by SDS–PAGE, followed by western blotting with relevant antibodies.
Raf kinase assay
NIH3T3 transfectants were maintained in serum-starved conditions and lysed as described above, followed by Raf-1 IP with specific antibodies. G-sepharose beads were washed 2 × with kinase buffer (20 mm MOPS, pH 7.2, 25 mm β-glycerophosphate, 5 mm EGTA, 1 mm Na3VO4, 1 mm dithiothreitol) before addition of kinase buffer containing 75 mm MgCl2, 0.5 mm ATP, and 0.4 μg recombinant MEK. Samples were incubated at 30°C on a rocker. Kinase reaction was stopped at 30 min by being boiled in SDS sample buffer.
Cell proliferation
Cells were seeded at 1 × 104 cells per well in 1% serum. Cells were collected by trypsinization and counted at 5 h for the initial time point, and represented as a ratio of total/initial cells for all other indicated time points. In some cases, 20 μm LY294002 or 30 μm U0126 was added to the culture medium. Cells were harvested and counted at the indicated times.
Sucrose gradient cell fractionation
NIH3T3 cells were scraped and washed with PBS (with protease inhibitors), resuspended and lysed in 0.5 m Na2CO3 (pH 11.0) as described previously.36 Briefly, cells were passed through a 23-gauge needle 15 times, sonicated and centrifuged at 39 000 r.p.m. in a SW41TI rotor in a 10–45% (w/v) step sucrose gradient. Ten 950-μl fractions were collected from the top sucrose layer, and equivalent total protein fractions were separated by SDS–PAGE and examined by western blotting. Tumor lysates were Dounce homogenized in buffer containing protease inhibitors, passed 10 times through an 18-G needle, and processed as described above.
Cell transformation
In all, 5 × 104 cells were mixed with Noble agar such that the final agar concentration was 0.3%, supplemented with 10% FBS. This mixture was loaded into a 60-mm dish containing 0.6% Noble agar. Cells were fed every 4 days with 100 μl of medium containing 10% calf serum. In some cases, either 20 μm LY294002 or 30 μm U0126 was added to both the top and bottom layer of agar, and media was replaced every 4 days with media containing 10% FBS with the same concentration of inhibitor. After 14 days, cells were fixed with 10% MeOH/10% acetic acid for 10 min. Plates were stained with 0.01% crystal violet for 1 h, and colonies were visualized after light washes.
Tumor growth in nude mice
Female NU(NCR)-Foxn1nu mice (Charles River, Wilmington, MA, USA) at 6–7 weeks of age were divided into five groups of two animals each. Cells for allografts were harvested and suspended in Hanks balanced salt solution (Thermo Scientific) at a density of 5 × 106/ml, and 200 μl of the cell suspension was injected subcutaneously into each flank. In some cases, 75 mg/kg LY294002, 30 mg/kg U0126, or vehicle only was administered every 72 h. Vehicle mice were given 200 μl of 5% DMSO. Tumor dimensions were measured using calipers, and tumor volume was calculated using the formula Volume=long axis × short axis2 × 0.52.68 Mice were killed at 20 days post injection, and resected tumors were fixed in 4% paraformaldehyde, and stored in PBS with 0.02% sodium azide. All animal experiments followed protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Temple University, which requires compliance with NIH ethical regulations.
Statistical analysis
One-way ANOVA followed by Fisher protected least significant difference analysis was used, using StatView (SAS Institute, Cary, NC, USA). A 5% probability was considered as significant. For in vivo studies, to detect a 30% difference between four groups using ANOVA with Bonferroni post hoc test (four tests), a minimum of five animals in each group would be needed, based on anticipated standard deviation=0.35, power=0.8 for P<0.05. The following web page was used for power calculation: http://www.stat.uiowa.edu/~rlenth/Power/index.html. Exclusion criteria included animal morbidity or premature death (none were observed). Tumor measurements were carried out by observers blinded to the allograft genotypes. Results are representative of three independent experiments with at least three replicates where possible (for example, cell assays) unless indicated otherwise.
Acknowledgments
This work was supported by AHA Grant 16GRNT27260319 to LEG.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogenesis website (http://www.nature.com/oncsis)
Supplementary Material
References
- Adjei A. Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst 2001; 93: 1062–1074. [DOI] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 2003; 3: 11–22. [DOI] [PubMed] [Google Scholar]
- Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes and Cancer 2011; 2: 344–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev 1997; 11: 2468–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CM, Li H, Gasman S, Huang J, Schiff R, Chang EC. Compartmentalized Ras proteins transform NIH 3T3 cells with different efficiencies. Mol Cell Biol 2010; 31: 983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011; 11: 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrier AL, Mastrangelo AM, Downward J, Ginsberg M, LaFlamme SE. Activated R-ras, Rac1, PI 3-kinase and PKCepsilon can each restore cell spreading inhibited by isolated integrin beta1 cytoplasmic domains. J Cell Biol 2000; 151: 1549–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin MW, Ada-Nguema A, Parsons M, Riching KM, Keely PJ. R-Ras regulates beta1-integrin trafficking via effects on membrane ruffling and endocytosis. BMC Cell Biol 2010; 11: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely PJ, Rusyn EV, Cox AD, Parise LV. R-Ras signals through specific integrin alpha cytoplasmic domains to promote migration and invasion of breast epithelial cells. J Cell Biol 1999; 145: 1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong L, Wozniak MA, Collins AS, Wilson SD, Keely PJ. R-Ras promotes focal adhesion formation through focal adhesion kinase and p130(Cas) by a novel mechanism that differs from integrins. Mol Cell Biol 2003; 23: 933–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi T, Ginsberg MH, Downward J, Hughes PE. The small GTP-binding protein R-Ras can influence integrin activation by antagonizing a Ras/Raf initiated integrin suppression pathway. Mol Biol Cell 1999; 10: 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel JG, Kumar P, Goldfinger LE. Palmitoylation regulates vesicular trafficking of R-Ras to membrane ruffles and effects on ruffling and cell spreading. Small GTPases 2012; 3: 139–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Brtva TR, Lowe DG, Der CJ. R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells. Oncogene 1994; 9: 3281–3288. [PubMed] [Google Scholar]
- Saez R, Chan AM, Miki T, Aaronson SA. Oncogenic activation of human R-ras by point mutations analogous to those of prototype H-ras oncogenes. Oncogene 1994; 9: 2977–2982. [PubMed] [Google Scholar]
- Holly SP, Larson MK, Parise LV. The unique N-terminus of R-ras is required for Rac activation and precise regulation of cell migration. Mol Biol Cell 2005; 16: 2458–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodie S, Willumsen BM, Weber MJ, Wolfman A. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science 1993; 260: 1658–1661. [DOI] [PubMed] [Google Scholar]
- Morrison D, Cutler RE. The complexity of Raf-1 regulation. Curr Opin Cell Biol 1997; 9: 174–179. [DOI] [PubMed] [Google Scholar]
- Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Complex formation between RAS and RAF and other protein kinases. Proc Natl Acad Sci USA 1993; 90: 6213–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marte BM, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr Biol 1997; 7: 63–70. [DOI] [PubMed] [Google Scholar]
- Spaargaren M, Martin GA, McCormick F, Fernandez-Sarabia MJ, Bischoff JR. The Ras-related protein R-ras interacts directly with Raf-1 in a GTP-dependent manner. Biochem J 1994; 300: 303–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol 2012; 13: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 1990; 63: 133–139. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 1989; 57: 1167–1177. [DOI] [PubMed] [Google Scholar]
- Schmittberger T, Waldmann H. Synthesis of the palmitoylated and prenylated C-terminal lipopeptides of the human R- and N-Ras proteins. Bioorg Med Chem 1999; 7: 749–762. [DOI] [PubMed] [Google Scholar]
- Baumgart F, Corral-Escariz M, Perez-Gil J, Rodriguez-Crespo I. Palmitoylation of R-Ras by human DHHC19, a palmitoyl transferase with a CaaX box. Biochim Biophys Acta 2010; 1798: 592–604. [DOI] [PubMed] [Google Scholar]
- Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ et al. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem 2005; 280: 31141–31148. [DOI] [PubMed] [Google Scholar]
- Ahearn IM, Tsai FD, Court H, Zhou M, Jennings BC, Ahmed M et al. FKBP12 binds to acylated H-ras and promotes depalmitoylation. Mol Cell 2011; 41: 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol 2000; 20: 2475–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR et al. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol 2005; 170: 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaki R, Morimatsu M, Uemura T, Waguri S, Miyoshi E, Taniguchi N et al. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol 2010; 191: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 2005; 307: 1746–1752. [DOI] [PubMed] [Google Scholar]
- Agudo-Ibanez L, Herrero A, Barbacid M, Crespo P. H-ras distribution and signaling in plasma membrane microdomains are regulated by acylation and deacylation events. Mol Cell Biol 2015; 35: 1898–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S, Laude AJ, Beckett AJ, Mageean CJ, Aran V, Hernandez-Valladares M et al. The role of palmitoylation in regulating Ras localization and function. Biochem Soc Trans 2013; 41: 79–83. [DOI] [PubMed] [Google Scholar]
- Janosi L, Li Z, Hancock JF, Gorfe AA. Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proc Natl Acad Sci USA 2012; 109: 8097–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallanas D, Arozarena I, Berciano MT, Aaronson David S, Pellicer A, Lafarga M et al. Differences on the inhibitory specificities of H-Ras, K-Ras, and N-Ras (N17) dominant negative mutants are related to their membrane microlocalization. J Biol Chem 2002; 278: 4572–4581. [DOI] [PubMed] [Google Scholar]
- Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat Cell Biol 2001; 3: 368–375. [DOI] [PubMed] [Google Scholar]
- Hansen M, Prior IA, Hughes PE, Oertli B, Chou FL, Willumsen BM et al. C-terminal sequences in R-Ras are involved in integrin regulation and in plasma membrane microdomain distribution. Biochem Biophys Res Commun 2003; 311: 829–838. [DOI] [PubMed] [Google Scholar]
- Hansen M, Rusyn EV, Hughes PE, Ginsberg MH, Cox AD, Willumsen BM. R-Ras C-terminal sequences are sufficient to confer R-Ras specificity to H-Ras. Oncogene 2002; 21: 4448–4461. [DOI] [PubMed] [Google Scholar]
- Jaumot M, Yan J, Clyde-Smith J, Sluimer J, Hancock JF. The linker domain of the Ha-Ras hypervariable region regulates interactions with exchange factors, Raf-1 and phosphoinositide 3-kinase. J Biol Chem 2002; 277: 272–278. [DOI] [PubMed] [Google Scholar]
- Parton RG, Hancock JF. Caveolin and Ras function. Methods Enzymol 2001; 333: 172–183. [DOI] [PubMed] [Google Scholar]
- Self AJ, Paterson HF, Hall A. Different structural organization of Ras and Rho effector domains. Oncogene 1993; 8: 655–661. [PubMed] [Google Scholar]
- Lee S, Wurtzel JG, Singhal SS, Awasthi S, Goldfinger LE. RALBP1/RLIP76 depletion in mice suppresses tumor growth by inhibiting tumor neovascularization. Cancer Res 2012; 72: 5165–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff SY, Quilliam LA, Cox AD, Der CJ. R-Ras is regulated by activators and effectors distinct from those that control Ras function. Oncogene 1997; 14: 133–143. [DOI] [PubMed] [Google Scholar]
- Rey I, Taylor-Harris P, van Erp H, Hall A. R-ras interacts with rasGAP, neurofibromin, and c-raf but does not regulate cell growth or differentiation. Oncogene 1994; 9: 685–692. [PubMed] [Google Scholar]
- Cole AL, Subbanagounder G, Mukhopadhyay S, Berliner JA, Vora DK. Oxidized phospholipid-induced endothelial cell/monocyte interaction is mediated by a cAMP-dependent R-Ras/PI3-kinase pathway. Arterioscler Thromb Vasc Biol 2003; 23: 1384–1390. [DOI] [PubMed] [Google Scholar]
- Furuhjelm J, Peranen J. The C-terminal end of R-Ras contains a focal adhesion targeting signal. J Cell Sci 2003; 116: 3729–3738. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol 2004; 24: 4943–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self AJ, Caron E, Paterson HF, Hall A. Analysis of R-Ras signalling pathways. J Cell Sci 2001; 114: 1357–1366. [DOI] [PubMed] [Google Scholar]
- Matallanas D, Sanz-Moreno V, Arozarena I, Calvo F, Agudo-Ibanez L, Santos E et al. Distinct utilization of effectors and biological outcomes resulting from site-specific Ras activation: Ras functions in lipid rafts and Golgi complex are dispensable for proliferation and transformation. Mol Cell Biol 2006; 26: 100–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S, Weise K, Erlkamp M, Triola G, Waldmann H, Winter R. The role of G-domain orientation and nucleotide state on the Ras isoform-specific membrane interaction. Euro Biophys J 2012; 41: 801–813. [DOI] [PubMed] [Google Scholar]
- Mazhab-Jafari MT, Marshall CB, Smith MJ, Gasmi-Seabrook GM, Stathopulos PB, Inagaki F et al. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc Natl Acad Sci USA 2015; 112: 6625–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent P, Haser W, Haystead TA, Vincent LA, Roberts TM, Sturgill TW. Activation of mitogen-activated protein kinase kinase by v-Raf in NIH 3T3 cells and in vitro. Science 1992; 257: 1404–1407. [DOI] [PubMed] [Google Scholar]
- Kyriakis J, App H, Zhang XF, Banerjee P, Brautigan DL, Rapp UR et al. Raf-1 activates MAP kinase-kinase. Nature 1992; 358: 417–421. [DOI] [PubMed] [Google Scholar]
- Leevers S, Paterson HF, Marshall CJ. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature 1994; 369: 411–414. [DOI] [PubMed] [Google Scholar]
- Oikonomou E, Koustas E, Goulielmaki M, Pintzas A. BRAF vs RAS oncogenes: are mutations of the same pathway equal? Differential signalling and therapeutic implications. Oncotarget 2014; 5: 11752–11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokoe D, Macdonald SG, Cadwallader K, Symons M, Hancock JF. Activation of Raf as a result of recruitment to the plasma membrane. Science 1994; 264: 1469–1467. [DOI] [PubMed] [Google Scholar]
- Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: a new perspective. Cancer 2014; 120: 3446–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell 2007; 129: 957–968. [DOI] [PubMed] [Google Scholar]
- Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 2005; 7: 533–545. [DOI] [PubMed] [Google Scholar]
- Fruman D, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov 2014; 13: 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Echeverria C, Sellers WR. Drug discovery approaches targeting the PI3K/AKT pathway in cancer. Oncogene 2008; 27: 5511–5526. [DOI] [PubMed] [Google Scholar]
- Lim HJ, Crowe P, Yang JL. Current clinical regulation of PI3K/PTEN/AKT/mTOR signalling in treatment of human cancer. J Cancer Res Clin Oncol 2015; 141: 671–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Rexer BN, Garrett JT, Arteaga CL. Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res 2011; 13: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011; 6: 49–69. [DOI] [PubMed] [Google Scholar]
- Lynch T, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- Paez J, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- Silver L, Michael JV, Goldfinger LE, Gallo G. Activation of PI3K and R-ras signaling promotes the extension of sensory axons on inhibitory chondroitin sulfate proteoglycans. Dev Neurobiol 2014; 74: 918–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Johnson BW, Williams SM, Chan AW, Reczek EE, Lynch RC et al. TORC1 is essential for NF1-associated malignancies. Curr Biol 2008; 18: 56–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.