Abstract
The purpose of this article is to inform readers about technical challenges that we encountered when assembling exome sequencing data from the ‘Simplifying Complex Exomes' (SIMPLEXO) consortium—whose mandate is the discovery of novel genes predisposing to breast and ovarian cancers. Our motivation is to share these obstacles—and our solutions to them—as a means of communicating important technical details that should be discussed early in projects involving massively parallel sequencing.
Keywords: genomics, exome
Introduction
The study of common genetic variants using genome-wide association studies (GWAS) have revealed multiple loci predisposing to several complex diseases including common human cancers. These genetic markers predominantly map outside of known functional genes [1]. They have added modestly to the overall narrow-sense heritability [2]. Advances in the sequencing technologies and rapid reduction in costs have helped in the identification of rare causal variants using next-generation sequencing techniques in Mendelian phenotypes. It is widely believed that rare variants with moderate to large effect sizes will contribute to the missing heritability of common diseases [3–6]. However, because such variants are presumed rare, successful statistical approaches for agnostic discovery may require many thousands of samples for the primary analysis of a disease type [7]. Additional samples are required when variance is partitioned among subtypes, treatment endpoints, ethnic/racial subgroups, geographic strata and environmental exposures. Because exome sequencing is still relatively expensive compared with array-based genotyping, it is less likely that a single center will procure sufficient funding to perform sequencing to achieve robust power required for novel discoveries. Combining and sharing exome sequencing data from multiple groups—or consortia—provides the resources necessary to identify novel genes for genetically complex phenotypes. So far, many of the common disease variants were discovered through GWAS, coordinated by large multinational consortia [8–10]. While some standards of compatibility and interoperability have been established for data such as FASTQ, BAM and VCF files produced from next-generation sequencing, many challenges still remain.
Even though several guidelines have been recently established regarding the design and interpretation of human exome sequencing data [11], other significant limitations remain and have not been effectively disseminated to the community. In this article, we discuss the technical challenges that we encountered when assembling exome sequencing data from the ‘Simplifying Complex Exomes' (SIMPLEXO) consortium whose mandate is the discovery of novel genes predisposing to breast and ovarian cancers and resolving the missing heritability of breast cancer. In contrast to efforts in other consortiums to identify candidate breast cancer susceptibility genes from aggregate findings of next-generation sequencing [12], SIMPLEXO aims to combine and harmonize primary sequencing data from multiple centers, and then take these findings on to validation in other data sets. We detail relevant postsequencing steps at multiple centers with the aim of germ line gene discovery in a common human cancer.
Step 1: Alignment
In many cases, the BAM file is considered the raw data from exome sequencing. It contains all necessary information about each read including mapping location, quality scores and raw sequence. Multiple methods exist to generate BAM files, and sequencing biases may be introduced depending on the method. Therefore, an early discussion of the methods to create the BAM files is essential.
The first element to decide on is which reference genome to use, as several ‘flavors’ of the reference genome are available for the same organism. A reference genome may include alternative haplotypes, unplaced contigs and decoy sequences, whereas others may mask the pseudo-autosomal region of chromosome Y (to reduce false-positive variant calls) or be generated from a ‘patched’ (i.e. updated from the initial release) version of the genome. Differences between patched versions can cause nucleotides in the patched regions to become different. For instance, in GRCh37.p10 there is an updated sequence (ch17_ctg5_hap1), which changed 532 bases in a 330 kb region. If data were aligned to GRCh37.p10 and combined with hg19 (which is effectively patch 0), then a conflict would exist as to what the reference nucleotide should be. Such conflicts often cause conflicting results with downstream annotation tools until such errors are resolved. Even simple inconsistencies such as a reference genome prefixed with/without ‘chr' can cause problems. The latest version GRCh38, for example, offers completed annotation for ABO gene. A detailed overview is available at the Genome Reference Consortium (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/), and additional resources are available at Genome in a Bottle Consortium (https://sites.stanford.edu/abms/giab).
The second element in creating the BAM file is to decide up-front which aligner to use and the specific parameters to be used when performing the alignment. The Burrows-Wheeler Aligner (BWA) [13] and Novoalign [14] were the two aligners that were used in SIMPLEXO. Extensive comparisons between these aligners have been done previously [15, 16], and their comparison is outside the scope of this article. Differential sensitivities for detecting indels may lead to substantial downstream effects in data interpretation, perhaps owing to the different algorithms used in these programs. While BWA and Novoalign are some of the most accurate aligners, there are slight differences in their accuracy for insertions and deletions of different sizes [17]. Variants observed in data from one center may not be present in other centers because the aligner used had an advantage in that particular sequence context. For example, say a frameshift deletion was observed in a case population aligned with Novoalign, and it was not observed in a large control population that was aligned with BWA. Without considering the differences in aligner, the investigator may trigger a large-scale validation study only to find out that the frequency of the deletion is in equal proportion in cases and controls. Such validation experiments involving large numbers of samples increase the overall cost in time, effort and resources available to the project. If an indel is observed by multiple aligners, or if it validates using the variant calling approaches we detail below, then it is likely real.
Step 2: Variant calling
Despite using similar tools, variants may be identified separately in single samples (i.e. single-sample calling) or in all samples simultaneously (i.e. multi-sample calling), which leverages information across samples. A number of tools are available for these purposes, and each can result in somewhat different results [18, 19]. However, the current consensus is that multi-sample calling is almost always preferred [20] and is often referred to as best practices [21].
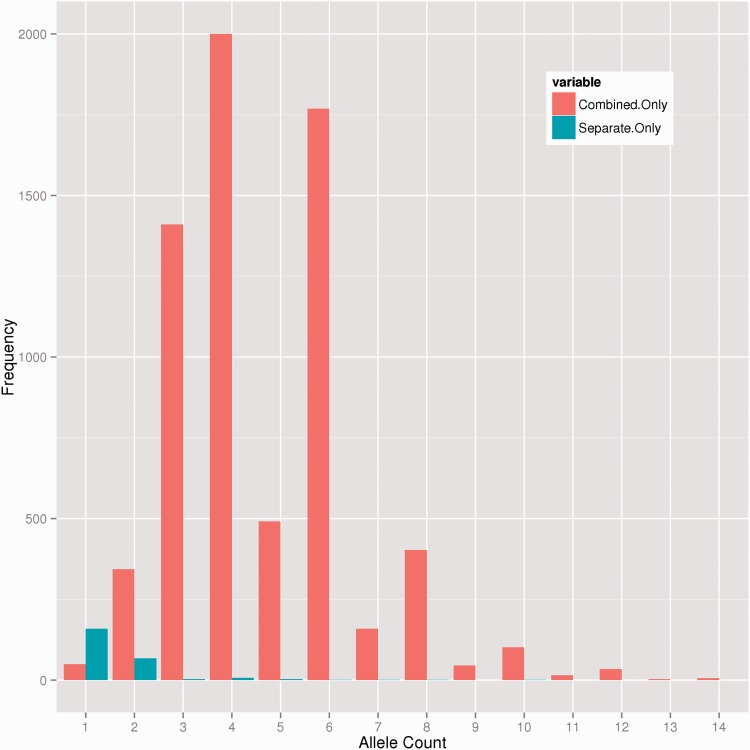
To gauge the impact of single- versus multi-sample calling in the SIMPLEXO data, we ran the Genome Analysis Toolkit (GATK) UnifiedGenotyper (2.4-3-g2a7af43) [22] in a single-sample and in multi-sample modes on a kindred of seven related individuals. Reassuringly, 335 049 variants were called between both groups; however, multi-sample calling provided many more variants. After removing variants that were intronic, intergenic and multi-allelic, single-sample analysis contained 244 variants not observed in the multi-sample analysis. The combined analysis called an additional 12 755 variants not found in the single analysis (Figure 1). Multi-sample calling integrates per sample likelihoods to jointly estimate allele frequency of variation, which helps to call rarer variants in a population, in addition to better error modeling using the joint estimation. Importantly, the combined analysis was not enriched for false positives, as all but ∼200 variants were observed in more than one of these related individuals (Figure 1). Only 399 from the combined analysis and 62 from the individual analyses were predicted to affect the protein (e.g. stop codon, frameshift, non-synonymous or canonical splice site). Therefore, we concluded that multi-sample variant calling in exome sequencing data calls more off-target (e.g. outside the coding exon) variants with less supporting reads. This can, however, be remedied by using a genomic interval file for variant calling, which is more efficient computationally. From the single-sample analyses, 93% of the variants that were unique to single-sample calling were found in either one or two individuals. Because our samples are related and nearly all the heterozygous variants were shared with at least one other individual, it is reasonable to assume that the majority of the variant calls were real. It is unknown whether (1) the single-sample analysis overcalls mutations; (2) the multi-sample analysis penalizes private or rare alleles; or (3) some combination of both.
Figure 1.
Distribution of variant alleles unique to single-sample or multi-sample genotyping. From seven members of a kindred, we genotyped them individually or as a whole as described in the text. For any variant, there are 14 alleles that could harbor the variant. More variants found heterozygous in a single individual (allele count = 1) were found only when single-sample genotyping was performed. When joint calling was performed, alleles shared among the kindred were detected at a higher rate. Note: The y-axis is truncated at 2000 variants to show the disparity between single- and multi-sample calling, because in the combined analysis, 7926 variants had an allele count of 4.
Step 3: Combining data from different institutions
For a number of reasons, it is not always possible to run multi-sample variant calling for every sample in a consortium. Sequence data processing and bioinformatic analyses require sufficient computational power and adequate allocation to store and retrieve large volumes of data. Assuming that data use agreements are in place, an average of 15 GB per exome, one would need 1.5 TB just for data from 100 exomes. The BAM files must be stored and fast-writing hard drives directly tied to a large compute cluster, which are expensive and sometimes a precious limited resource. Even if there is space and enough computational power available for such an analysis, full access and participation of other centers is limited by the host institution’s firewall blocking access of the data to collaborators. Common data set and resources-based analysis on ‘big-data' is becoming common among large academic centers and is encouraged by funding agencies such as NIH; however, cloud-based computing is yet to be mainstream in human disease research.
There are alternative ways to bring together disparate BAM files, which may achieve similar outcomes as single-institution multi-sample variant calling. Rather than merging BAM files together and running multi-sample variant calling, we exploited another feature of GATK, namely, Genotype Given Alleles (GGA). In this strategy, all participating centers run multi-sample variant calling on the BAM files stored at each institution. Then, the resulting VCF files are centrally merged and sent back to collaborators to run the GGA feature on their BAM files. This allows counts of the number of reference and alternate alleles, regardless of whether one of those alternate alleles were output in that center’s original discovery run. Once all centers complete the GGA step, the genotyped VCF are re-centralized and merged to create a single VCF that contains information about the number of reference and alternate reads from each sample. This is helpful in dealing the issues of costs related to data transfer, data security and space limitations. It should, however, be emphasized that several of the quality control parameters delivered by the initial variant calling run should be identical across centers to have parity in treatment of the data during postprocessing quality control. A drawback of this method is its inability to cope with newer data produced at centers because it will require all of the processing to be repeated.
A newer approach is now possible through an upgraded release of GATK, which contains several new tools including HaplotypeCaller. The HaplotypeCaller solves the N + 1 problem for re-genotyping variants across an entire data set without needing access to all of the BAM files simultaneously. Instead, the HaplotypeCaller emits a genomic VCF (gVCF) that contains data on variant and non-variant positions. gVCF files are comparably much smaller in size than native BAM files, hence easier to share. Once created, gVCF files from multiple samples can undergo the GenotypeGVCF step (also part of the GATK), which genotypes variants and produces the final VCF output. For consortia, each member institution could send gVCFs instead of BAM files to a central location to be jointly genotyped. At any time, more samples can be added to the consortium pool, with subsequent repeating of the re-genotyping step. At this time, this approach also suffers from incomplete annotations without the primary BAMs. When GVCFs are made from BAMs by GATK, it extracts some annotations from the BAMs such as GT, DP, GQ, PL. However, re-annotation using the VariantAnnotator, a GATK walker may be required to populate and harmonize all the fields required to test QC/QA processes downstream. It is important to mention that these methods are limited to use of GATK, as this was the platform of choice for our consortium. There are a plethora of other genotyping methods from exome data—each having different metrics of sensitivity and specificity [23]. As of now, HaplotypeCaller is the only method we are aware of that can use gVCF files as input, while the rest require direct access to BAM files. Regardless of the method or methods used to call variants, it is necessary that the same approaches to variant calling are applied universally to all samples so as not to bias the results coming from one center’s different genotyping algorithm.
Step 4: Technical filtering
Variant quality score recalibration (VQSR) is a popular way to annotate variants as to their likelihood of being real or artifactual. It categorizes variants as either ‘PASS' or lower quality bins (truth-tranches) using certain attributes present in the VCF file’s INFO field. VQSR is a representation of the data set from which the variant was called, meaning that the same variant can be in a lower quality tranche in one investigator’s data set but PASS in another. When using the GGA method described above, or when combining VCF files generated through other means, a simple solution is to specify the ‘-mergeInfoWithMaxAC' option in the GATK’s CombineVariants walker. This option gives priority to the VCF file that has the largest number of alternate alleles, and consequently the best joint probability. Without specifying this option, tranches can become mixed within the Filter field of the VCF, causing unexpected errors when using downstream tools. Importantly, those variants not marked as PASS are not necessarily false positives, rather the GATK algorithm cannot assign a 100% probability that they are not. By centralizing the VCF and running VQSR on only one VCF, these types of conflicts are mitigated.
Step 5: Variant annotation
Once variants are deemed to be of high quality (e.g. high VQSR score, alternate supporting reads), the next step for downstream filtering is annotation. There are no ‘one-stop shops' for annotation, rather it involves cobbling together disparate annotation tools and sources. There are two levels of annotations that are of great interest, variant-specific annotations and interval-based annotations on features such as genes and transcripts. One of the most informative types of variant annotation is population allele frequencies. Reference frequencies like those provided by the 1000 Genomes project [24], Exome Sequencing Project (ESP) (http://evs.gs.washington.edu/EVS/) and the Exome Aggregation Consortium (http://exac.broadinstitute.org/) are powerful tools to remove common variants from studies seeking to find novel or rare alleles. However, we caution that the absence of evidence is not evidence of absence. It is always a good practice to also consider the depth of coverage of the region harboring the variant in public databases. We and collaborators have observed that variants at times present in ESP may be much common in specific cohorts. These discrepancies may be owing to population origin (ethnicity) or owing to insufficiencies in the data caused by the sequencing platform, insufficient coverage or differences in bioinformatics analysis.
SnpEFF [25], VEP [26], ANNOVAR [27] and CAVA (formerly SAVANT [http://www.well.ox.ac.uk/cava]) are a popular tools to annotate the effect variants on their corresponding transcripts (e.g. frameshift, missense, stop gain). Recently, McCarthy et al. [28] demonstrated that variant annotations from these annotation tools will disagree on what effect to assign to a particular transcript 14% of the time, even when accounting for the differences in gene annotation sources like ENSEMBL, RefSeq and GENCODE, which can also introduce differences [29]. Regardless of annotation tool used, it is important to note that different transcripts can also lead to different amino acid changes. Normalization of transcripts to be reported is the focus of the Locus Reference Genomic initiative [30], though few genes have accessions at this time.
An alternative and transparent method is to present the data at the genomic DNA coordinate level with corresponding reference and alternate alleles. Surprisingly, there is discrepancy between reference genomes (e.g. hg19) and transcript annotation sources (e.g. RefSeq). For instance, the reference genome at chr13:32, 929, 387 is a ‘T’ in the reference genome suggesting a valine at cDNA position 2466, but a ‘C’ in RefSeq, causing that amino acid to appear as alanine instead. This causes confusion because the T allele of this single nucleotide polymorphism (SNP) has an associated dbSNP identifier (rs169547) but would not be identified as a variant because it matches the reference genome. This type of error is present in 5210 transcripts from 2993 genes. These types of errors are still present in the newest version of the human genome (GRCh38) in 4948 transcripts from 2924 genes. None of the existing annotation tools we are aware of can correct this problem systematically.
Conclusions
Aggregating genomic sequencing data from multiple centers is a complex, multifaceted challenge. Hopefully, this viewpoint will provide other consortia, a frame of reference and dialog to assist groups in avoiding potential obstacles inherent in multi-institutional projects involving massively parallel sequencing. We have not addressed other equally important topics such as variant prioritization or classification methods, as many of the underlying hypotheses or methodological approaches may be substantially different than our interest in identifying new breast cancer predisposition genes. Rather, we have focused instead on what is sometimes referred to as the ‘data janitor’ work [31], an often oversimplified yet essential component to collaborative study design. We have highlighted several technical challenges faced in the simPLEXO consortium and offer our own solutions in hopes to encourage discussion and development of a best practices guideline for future multi-institutional collaborations. Looking forward, we expect more diversity among tools, but also more unification and integration of pipelines in the cloud from both academic and industries that cater to groups that may not have specialization in simplifying and harmonizing next-generation sequencing data.
Key Points
Integrating sequencing data from large-scale collaborations is fraught with challenges.
Discrepancies arise throughout the entire process from alignment to variant annotation.
We provide the solutions we took to overcome several of the technical challenges.
It is imperative to discuss fine-grained detail of bioinformatics analysis early in the process of organizing multi-institutional collaborations.
Funding
This work was supported by the National Institutes of Health R01CA176785 and the Breast Cancer Research Foundation [F.J.C., K.N. and K.O], R21-CA139396 [J.V]. K.N.M. was supported by the Department of Defense (W81XWH-13-1-0338); views and opinions of, and endorsements by, the authors do not reflect those of the US Army or the Department of Defense. Other funding was provided by The Niehaus Clinical Genetics Initiative and The Miele family initiative [K.O.]. City of Hope Clinical Cancer Genetics Community Research Network and the Hereditary Cancer Research Registry, supported in part by Award Number RC4CA153828 (PI: J. Weitzel) from the National Cancer Institute and the Office of the Director, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The work was also supported by the MSKCC Core grant NIH P30CA008748.
Biographies
Steven N. Hart, PhD, is an assistant professor of Biomedical Informatics at Mayo Clinic. His research interests are in analyzing large-scale genomics data to understand complex diseases.
Kara N. Maxwell, MD, PhD, a fellow in the division of Hematology-Oncology in the Perelman School of Medicine at the University of Pennsylvania.
Tinu Thomas is a bioinformatics specialists in the Clinical Genetics Research Lab at Memorial Sloan Kettering Cancer Center. She is involved in the development and running of NGS analytic pipelines.
Vignesh Ravichandran is a bioinformatics specialists in the Clinical Genetics Research Lab at Memorial Sloan Kettering Cancer Center. He is involved in the development and running of NGS analytic pipelines.
Bradley Wubberhorst is a bioinformatician working with the Nathanson lab on hereditary breast cancer predisposition.
Robert J. Klein is interested in understanding the role inherited genetic variation plays in the development, progression, and outcome of cancer at the Icahn School of Medicine at Mount Sinai.
Kasmintan Schrader recently joined the University of British Columbia and the BC Cancer Agency as a clinical assistant professor in the Department of Medical Genetics and a staff physician at the Hereditary Cancer Program. Dr. Schrader’s research will focus on understanding the genetic basis of pancreatic cancer susceptibility in individuals and families affected by the disease.
Csilla Szabo is an investigator at the National Human Genome Research Institute, National Institutes of Health. Her work is focused on identifying new risk factors and genetic modifiers of breast cancer risk.
Jeffrey N. Weitzel, MD, is Chief of the Division of Clinical Cancer Genetics and the Cancer Screening & Prevention Program at the City of Hope Comprehensive Cancer Center in Duarte, California. Dr. Weitzel is Board Certified in clinical genetics and medical oncology, and he is a Professor of Oncology and Population Sciences at the City of Hope.
Susan L. Neuhusen is a molecular and genetic epidemiologist whose research focus is to identify genes and environmental stressors that increase risk of developing breast and ovarian cancers and to uncover what factors are important for disease-free survival in women who develop these cancers.
Katherine Nathanson, MD, an associate professor in the division of Translational Medicine and Chief Oncogenomics Physician for the Abramson Cancer Center.
Kenneth Offit, MD, MPH, is Chief of the Clinical Genetics Service at Memorial Hospital and a Member of the Cancer Biology and Genetics Program of the Sloan Kettering Institute at Memorial Sloan Kettering Cancer Center. Dr. Offit's laboratory focuses on utilizing genomic approaches to discover novel mechanisms associated with increased risk for common malignancies, or which modify the risks of known hereditary predispositions.
Fergus J. Couch is the Zbigniew and Anna M. Scheller Professor of Medical Research and Chair of the Division of Experimental Pathology in the Department of Laboratory Medicine and Pathology at Mayo Clinic.
Joseph Vijai, PhD, is an assistant attending geneticist in the Department of Medicine and the Lab Director of the Clinical Genetics Research Lab at the Memorial Sloan Kettering Cancer Center. His primary focus is to identify genes that predispose and affect outcomes in cancer by using approaches such as genome-wide associations and next-generation sequencing.
References
- 1.Nicolae DL, Wen X, Voight BF, et al. Coverage and characteristics of the Affymetrix GeneChip Human Mapping 100K SNP set. PLoS Genet 2006;2:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009;461:747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Do R, Stitziel NO, Won HH, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature 2015;518:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schick UM, Auer PL, Bis JC, et al. Association of exome sequences with plasma C-reactive protein levels in >9000 participants. Hum Mol Genet 2015;24:559–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purcell SM, Moran JL, Fromer M, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014;506:185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang H, Jin X, Li Y, et al. A large-scale screen for coding variants predisposing to psoriasis. Nat Genet 2014;46:45–50. [DOI] [PubMed] [Google Scholar]
- 7.Flannick J, Thorleifsson G, Beer NL, et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet 2014;46:357–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007;447:1087–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaudet MM, Kuchenbaecker KB, Vijai J, et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet 2013;9:e1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michailidou K, Beesley J, Lindstrom S, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet 2015;47:373–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014;508:469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Southey MC, Park DJ, Nguyen-Dumont T, et al. COMPLEXO: identifying the missing heritability of breast cancer via next generation collaboration. Breast Cancer Res 2013;15:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paila U, Chapman BA, Kirchner R, et al. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput Biol 2013;9:e1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatem A, Bozdag D, Toland AE, et al. Benchmarking short sequence mapping tools. BMC Bioinformatics 2013;14:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu X, Guda K, Willis J, et al. How do alignment programs perform on sequencing data with varying qualities and from repetitive regions?. BioData Min 2012;5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruffalo M, LaFramboise T, Koyuturk M. Comparative analysis of algorithms for next-generation sequencing read alignment. Bioinformatics 2011;27:2790–6. [DOI] [PubMed] [Google Scholar]
- 18.Pabinger S, Dander A, Fischer M, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform 2014;15:256–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar P, Al-Shafai M, Al Muftah WA, et al. Evaluation of SNP calling using single and multiple-sample calling algorithms by validation against array base genotyping and Mendelian inheritance. BMC Res Notes 2014;7:747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Should I analyze my samples alone or together? https://www.broadinstitute.org/gatk/guide/article?id=4150 (14 July 2015, date last accessed).
- 21.Calling variants on cohorts of samples using the HaplotypeCaller in GVCF mode. https://www.broadinstitute.org/gatk/guide/article?id=3893 (July 14 2015, date last accessed).
- 22.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Rawe J, Jiang T, Sun G, et al. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med 2013;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLaren W, Pritchard B, Rios D, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 2010;26:2069–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy DJ, Humburg P, Kanapin A, et al. Choice of transcripts and software has a large effect on variant annotation. Genome Med 2014;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frankish A, Uszczynska B, Ritchie GR, et al. Comparison of GENCODE and RefSeq gene annotation and the impact of reference geneset on variant effect prediction. BMC Genomics 2015;16(Suppl 8):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacArthur JA, Morales J, Tully RE, et al. Locus Reference Genomic: reference sequences for the reporting of clinically relevant sequence variants. Nucleic Acids Res 2014;42:D873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohr S. For Big-Data Scientists, ‘Janitor Work’ Is Key Hurdle to Insights. The New York Times. 2014.