Abstract
TRIM5α from the rhesus macaque (TRIM5αRh) is a restriction factor that shows strong activity against HIV-1. TRIM5αRh binds specifically to HIV-1 capsid (CA) through its B30.2/PRYSPRY domain shortly after entry of the virus into the cytoplasm. Recently, three putative SUMO interacting motifs (SIMs) have been identified in the PRYSPRY domain of human and macaque TRIM5α. However, structural modeling of this domain suggested that two of them were buried in the hydrophobic core of the protein, implying that interaction with SUMO was implausible, while the third one was not relevant to restriction. In light of these results, we re-analyzed the TRIM5αRh PRYSPRY sequence and identified an additional putative SIM (435VIIC438) which we named SIM4. This motif is exposed at the surface of the PRYSPRY domain, allowing potential interactions with SUMO or SUMOylated proteins. Introducing a double mutation in SIM4 (V435K, I436K) did not alter stability, unlike mutations in SIM1. SIM4-mutated TRIM5αRh failed to bind HIV-1CA and lost the ability to restrict this virus. Accordingly, SIM4 undergoes significant variation among primates and substituting this motif with naturally occurring SIM4 variants affected HIV-1 restriction by TRIM5αRh, suggesting a direct role in capsid recognition. Interestingly, SIM4-mutated TRIM5αRh also failed to activate NF-κB and AP-1-mediated transcription. Although there is no direct evidence that SIM4 is involved in direct interaction with SUMO or a SUMOylated protein, mutating this motif strongly reduced co-localization of TRIM5αRh with SUMO-1 and with PML, a SUMOylated nuclear protein. In conclusion, this new putative SIM is crucial for both direct interaction with incoming capsids and for NF-κB/AP-1 signaling. We speculate that the latter function is mediated by interactions of SIM4 with a SUMOylated protein involved in the NF-κB/AP-1 signaling pathways.
Keywords: Cellular aspects of innate immunity, Proteins, Virology, Viruses, Innate immune system
1. Introduction
Proteins from the TRIM5 family are known for their restriction activity against retroviruses [1, 2, 3, 4]. Restriction mediated by TRIM5α relies on its ability to recognize the viral CA core shortly after entry of the virus in the cell, in a virus-specific and species-specific manner [5]. TRIM5α contains RING, B-Box, coiled-coil (RBCC) domains [6, 7] and the C-terminal SPRY domain (also called B30.2 or PRYSPRY [8]). TRIM5α orthologs differ mostly in the 3 variable regions (V1,V2 and V3) of PRYSPRY [9] which confer specificity for the retroviral CA [10]; for instance, HIV-1 is inhibited by rhesus macaque TRIM5α (TRIM5αRh), but generally not by the human version of the protein [11]. The B-Box and coiled-coil domains are involved in TRIM5αRh dimerization and higher-order multimerization, properties that are likely important for the formation of cytoplasmic bodies (CBs) observable by light microscopy [12, 13, 14]. The RING domain has E3-ubiquitin ligase activity essential for auto-ubiquitylation of the protein and efficient restriction [15].
Interactions between TRIM5α and the retroviral CA N-terminal domain destabilize the CA lattice [16, 17, 18, 19, 20]. As a result, the CA core undergoes disassembly and some of its components are degraded, as well as TRIM5α itself [20, 21, 22]. TRIM5α also impairs viral transport to the nucleus [23, 24]. Another function has recently been attributed to TRIM5α: the activation of innate immunity pathways dependent on AP-1 and NF-κB [25, 26]. The RING domain recruits the E2-ubiquitin conjugating enzyme heterodimer Ubc13 (Ube2N)/Ube2V2 (Uev2) to generate lysine 63 (K63)-linked polyubiquitin chains that can be anchored onto TRIM5α through the action of another E2 enzyme, Ebe2W [27]. The presence of K63-linked ubiquitin chains has been proposed to be important for AP-1 and NF-κB transactivation [26, 28]. Activation of NF-κB and AP-1 by TRIM5α is triggered by contact with a restriction sensitive virus [26] or by transient transfection of TRIM5α [25, 26, 29, 30], but not by stable lentiviral transduction [31].
The human genome contains three functional SUMO genes encoding small-ubiquitin-like modifier (SUMO) -1/2/3 [32]. SUMOylation is a post-translational modification involved in many cellular pathways [32, 33]. SUMO modifies targeted proteins through its covalent attachment to lysines present in a specific consensus site: Ψ-K-X-D/E [32]. SUMO interacting motifs (SIMs) are necessary for the non-covalent interaction of proteins with free SUMO [34] or with SUMOylated proteins [35] and consist of a short domain rich in hydrophobic residues [34]. TRIM5α proteins have a consensus SUMOylation site at a position immediately upstream of the RING domain and three putative motifs in the PRYSPRY domain were proposed to function as SIMs (SIM1, SIM2 and SIM3) [36]. Although TRIM5α was recently found to be SUMOylated [37], the contribution of SUMO to TRIM5α functions has been controversial.
Nonetheless, recent publications have proposed a role for SUMO-1 in retroviral TRIM5α-mediated restriction [36, 37, 38]. The predicted SUMOylated lysine present in TRIM5αRh promotes AP-1 and NF-κB signaling pathways through modulation of the RING domain activity [30]. Moreover, it has been recently found that mutating the putative SIM1 and SIM2 impairs TRIM5α-mediated NF-κB activation and retroviral restriction, while mutating SIMs had no significant effect [36, 38, 39]. However, structural data revealed that the phenotypes of SIM1 and SIM2 mutants were probably a consequence of PRYSPRY misfolding. Indeed, these motifs appear to be buried deep inside the hydrophobic core of the PRYSPRY domain, making interactions with other proteins unlikely [39], although it cannot be excluded that contact with CA might result in structural changes that expose SIM1 and SIM2. In light of these results, we re-analyzed the PRYSPRY sequence for the presence of other possible SIMs and uncovered a novel putative SIM which we named SIM4. Unlike putative SIMs 1 and 2, SIM4 residues are present at the surface of the protein, making this motif more suitable for interaction with other proteins. Here we demonstrate the importance of this motif in HIV-1 restriction and in the activation of NF-κB and AP-1. We propose that this signaling function is achieved by mediating interactions with an unidentified SUMOylated protein.
2. Results
2.1. Identification of a new putative SUMO interacting motif in TRIM5αRh
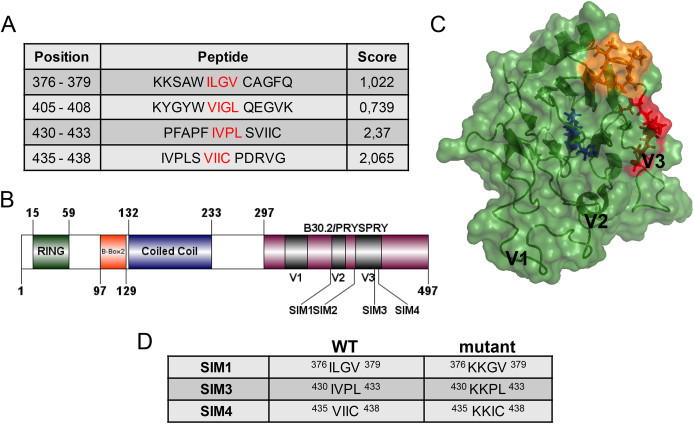
New insights into TRIM5αRh PRYSPRY structure suggested that the previously proposed SIMs 1 and 2 were located inside the hydrophobic core of this domain, implying that direct interactions with SUMO (or any other protein) were unlikely [39]. Analysis of the TRIM5αRh PRYSPRY domain using a SUMO-binding motif prediction software (GPS-SUMO [40]) yielded several putative SUMO interacting motifs with different scores (Fig. 1A). SIM1 (376ILGV379), SIM2 (405VIGL408) and SIM3 (430IVPL433) were confirmed as possible SIMs for TRIM5αRh, but the analysis revealed the presence of another putative SIM (435VIIC438) which we named SIM4. Unlike SIM1 and SIM2, SIM4 residues are mostly exposed at the surface of the protein, next to SIM3 and within the variable region 3 (V3) (Fig. 1B, C). The side chain of Val435 is not oriented toward the solvent, but the variable regions are present as flexible loops, and thus this residue is potentially available for protein-protein interactions.
Fig. 1.
Putative SUMO interacting motifs in TRIM5αRh. (A) Table showing scores of predicted SIMs (shown in red) and their position within TRIM5αRh. (B) Schematic of the full-length TRIM5αRh protein depicting its main domains and the position of predicted SIMs. V1-V3, variable regions 1-3. (C) Structural model of the TRIM5αRh PRYSPRY domain (PDB 2ML3) showing the positions of SIM1 (blue), SIM3 (red) and SIM4 (orange). The localization of V1, V2 and V3 regions is also shown (D) Table showing mutations introduced in TRIM5αRh SIMs.
To investigate the potential role of SIM4 in TRIM5αRh-mediated HIV-1 restriction, the two first aminoacids were mutated to lysines, thus mimicking the mutations that had been introduced in the other putative SIMs (Fig. 1D). The mutants of putative SIMs 1 and 3 described by others [36, 38, 39] were also included in this study, while SIM2 was not included since its phenotype was identical to that of SIM1 [36, 38, 39].
2.2. V435K-I436K TRIM5αRh does not bind HIV-1CA and does not restrict HIV-1
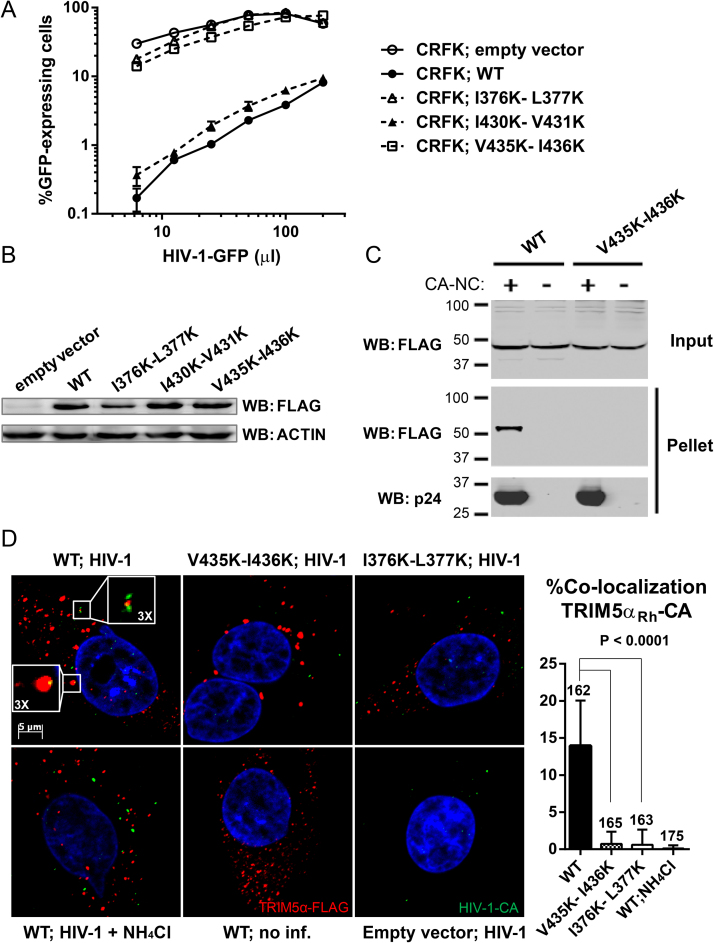
To analyze the ability of SIM4-mutated TRIM5αRh to restrict HIV-1, CRFK cells stably expressing WT, SIM1 mutant I376K-L377K, SIM2 mutant I430K-V431K or SIM4 mutant V435K-I436K TRIM5αRh were challenged with increasing amounts of VSV G-pseudotyped HIV-1-GFP. Cells transduced with the empty vector were used as a non-restrictive control. As expected [38, 39], both WT and I430K-V431K TRIM5αRh strongly restricted HIV-1, by up to ∼100-fold, while cells expressing I376K-L377K TRIM5αRh were about as much permissive to HIV-1 infection as cells expressing the empty vector (Fig. 2A). Expression of V435K-I436K TRIM5αRh resulted in a very small decrease in HIV-1 permissiveness (less than 2-fold), showing that the ability of this TRIM5αRh mutant to restrict HIV-1 was almost completely lost (Fig. 2A). Levels of protein expression were similar for all mutants, except for I376K-L377K whose expression level was decreased (Fig. 2B), as previously seen by others [38, 39]. Therefore, the V435K-I436K mutation affects restriction of incoming HIV-1 seemingly without altering protein levels, unlike I376K-L377K which reduces both.
Fig. 2.
The V435K-I436K mutation in SIM4 suppresses binding of TRIM5αRh to HIV-1CA and abolishes HIV-1 restriction. (A) Functional restriction assay. CRFK cells stably expressing WT TRIM5αRh or the various indicated mutants, or transduced with the “empty” vector were challenged with increasing amounts of VSV G-pseudotyped HIV-1-GFP. Infections were performed in triplicates and the percentage of cells expressing GFP was analyzed by FACS two days later. Standard deviation bars are shown. (B) TRIM5αRh expression levels. WT and mutant TRIM5αRh proteins expression levels in stably transduced CRFK cells were determined by Western blotting with an anti-FLAG antibody. Actin expression was analyzed as a loading control. (C) CA-binding assay. HIV-1CA-NC complexes were assembled in vitro and a binding assay was then performed using FLAG-tagged WT or V435K-I436K TRIM5αRh expressed in HEK293T cells. Lysates from the transfected HEK293T cells were incubated with HIV-1CA-NC complexes, then the mixtures were applied to a 70% sucrose cushion and centrifuged. “Input” shows a Western blot analysis of pre-centrifugation lysates using a FLAG antibody, while “Pellet” shows post-centrifugation pellets analyzed using antibodies against FLAG or CA. A single experiment representative of three independent experiments is presented. (D) TRIM5α association with incoming HIV-1CA in cells. CRFK cells stably expressing WT, I376K-L377K (SIM1 mutant) or V435K-I436K (SIM4 mutant) TRIM5αRh-FLAG were infected for 2 h with HIV-1-GFP. A rabbit polyclonal anti-FLAG antibody was used for the detection of TRIM5αRh (red) while HIV-1CA was stained using a mouse monoclonal antibody (green). DNA was stained using Hoeschst33342 (blue). Examples of co-localization between WT TRIM5αRh cytoplasmic bodies and HIV-1CA foci are magnified in white boxes. A quantitative analysis of co-localization frequency was performed and is shown in the graph on the right. Each bar represents the mean % of co-localization observed in 15 randomly chosen fields, each field considered as a replicate for statistical analysis. P values were calculated using the one-way ANOVA test. The total number of CA foci in the 15 fields is shown on top of each bar.
We investigated the ability of WT and V435K-I436K TRIM5αRh to physically interact with HIV-1CA-NC complexes in a CA-binding assay in vitro [41, 42]. Briefly, purified and assembled HIV-1CA-NC complexes were mixed with TRIM5αRh proteins, which were over-expressed in and isolated from HEK293T cells. The mixture was then applied to a 70% sucrose cushion and following centrifugation, pellets and supernatants were analyzed by Western blotting. In this assay, binding of TRIM5αRh to HIV-1CA is evidenced when both are present in the pellet fraction. This analysis had previously been conducted for I376K-L377K and I430K-V431K TRIM5αRh, and results obtained were consistent with their restriction phenotype since I430K-V431K was found to bind HIV-1CA-NC complexes while I376K-L377K did not [39]. Similarly to I376K-L377K, V435K-I436K TRIM5αRh failed to bind HIV-1CA-NC complexes (Fig. 2C), hence explaining its inability to restrict this virus.
HIV-1 restriction by TRIM5α correlates with co-localization of both proteins following infection, suggesting the existence of a “sequestration” mechanism of restriction [23]. To investigate TRIM5α-CA interactions in a cellular context, we assessed the co-localization of FLAG-tagged WT, I376K-L377K or V435K-I436K TRIM5αRh with incoming HIV-1CA. Cells were infected with VSV G-pseudotyped HIV-1-GFP for 2 h and then stained with antibodies against FLAG and CA. MG132 was used as a proteasome inhibitor to prevent the degradation of TRIM5αRh [22]. Fig. 2D shows representative photographs; in this assay, CA foci observed likely are single viruses [43], but the assay does not distinguish between fused and unfused particles. WT, I376K-L377K and V435K-I436K TRIM5αRh were all able to form cytoplasmic bodies (CBs). Ammonium chloride (NH4Cl) treatment was used as a no-entry control as it inhibits the acidification of endosomes [44], thus preventing the release of VSV G-pseudotyped viruses in the cytoplasm and making TRIM5α-CA co-localization improbable. CA foci were bigger when cells were treated with NH4Cl (Fig. 2D), probably a consequence of CA accumulation in endosomes. As expected [23], a significant fraction (14%) of HIV-1CA foci co-localized with WT TRIM5αRh CBs (Fig. 2D). In contrast, I376K-L377K and V435K-I436K TRIM5αRh showed very little co-localization with HIV-1CA (0.6% and 0.7%, respectively). CA-TRIM5αRh co-localization dropped to 0.1% when cells were treated with NH4Cl, suggesting that the co-localization events observed in absence of this drug were specific and involved post-entry virus cores. Taken together, these results show that mutating SIM4 can disrupt physical interactions of TRIM5αRh with its viral CA target, hence impairing restriction.
2.3. V435K-I436K TRIM5αRh is stable and able to shuttle to the nucleus
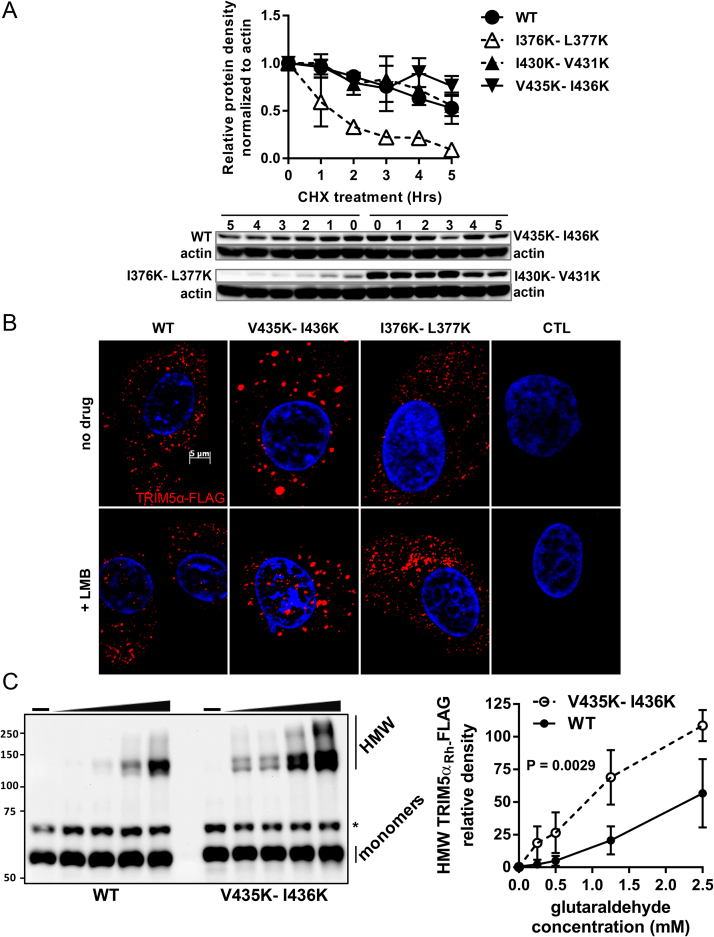
As shown in Fig. 2B, I376K-L377K but not V435K-I436K altered steady-state levels of TRIM5αRh, suggesting that protein folding was affected by the former but not the latter mutation. To further investigate protein stability, CRFK cells stably expressing WT, I376K-L377K, I430K-V431K or V435K-I436K TRIM5αRh were treated with the translation inhibitor cycloheximide (CHX) and reduction in protein levels was then monitored over time by Western blotting coupled with densitometry analysis. As shown in Fig. 3A, the I376K-L377K TRIM5αRh half-life was <1.5 h, compared to >5 h for WT, I430K-V431K and V435K-I436K TRIM5αRh. Of note, other studies reported a shorter half-life for WT TRIM5αRh than in our assay [15, 22], but they used different cell lines. The rate of decay for a given protein can vary depending on the cell type used and growth conditions [45, 46]. Using a two-way ANOVA followed by Dunnett's multiple comparison test, we found no statistically significant differences for I430K-V431K and V435K-I436K when compared to WT TRIM5αRh in this assay (P values = 0.8832 and 0.3038, respectively). On the other hand, I376K-L377K stability was significantly decreased (P < 0.0001), supporting the misfolding hypothesis.
Fig. 3.
Characterization of V435K-I436K TRIM5αRh stability, nuclear shuttling and multimerization. (A) CRFK cells stably expressing WT, I376K-L377K (SIM1 mutant), I430K-V431K (SIM3 mutant) or V435K-I436K (SIM4 mutant) TRIM5αRh were treated with cycloheximide and protein lysates were then prepared at the indicated time points. The graph shows mean values of relative protein densities from two independent experiments normalized to actin at different time points following cycloheximide treatment, and then normalized to the value obtained at T0. Error bars are shown. Western blots from one experiment are presented below the graph. (B) IF microscopy analysis of subcellular distribution. CRFK cells stably expressing FLAG-tagged WT, I376K-L377K, V435K-I436K TRIM5αRh and empty vector-transduced cells were treated with leptomycin B (LMB) for 12 h or left untreated prior to fixation and staining with an anti-FLAG antibody (red). DNA was stained using Hoeschst33342 (blue). Representative images from multiple fields analyzed are shown. (C) Multimerization assay. Lysates were prepared from CRFK cells stably expressing FLAG-tagged WT or V435K-I436K TRIM5αRh. The soluble fraction of each lysate was divided in aliquots that were treated with different glutaraldehyde concentrations (0, 0.25, 0.5, 1.25 and 2.5 mM) before performing Western blots using an anti-FLAG antibody. A single experiment representative of three independent experiments is presented. High molecular weight (HMW) bands correspond to apparent TRIM5αRh multimers according to the size of the bands. The graph on the right shows the average (from three experiments) of relative HMW protein density (ranging from 120 to >250 kDa) calculated at each glutaraldehyde concentration. Standard deviation bars are shown on the graph and the P value was calculated using a two-way ANOVA statistical test. * points to a non-relevant band detected by the antibody used.
TRIM5α has the capacity to shuttle between the nucleus and the cytoplasm, and the fraction of the protein present in the nucleus can be enriched by treatment with the CRM-1/exportin 1 inhibitor leptomycin B (LMB) [39, 47, 48]. In the presence of this drug, TRIM5αRh forms nuclear bodies (NBs) that co-localize with promyelocytic leukemia (PML) bodies [47]. CRFK cells transduced with I376K-L377K, V435K-I436K or WT TRIM5αRh were treated with LMB for 12 h and the presence of TRIM5αRh NBs was analyzed (Fig. 3B). In absence of the drug, WT, V435K-I436K and I376K-L377K TRIM5αRh formed CBs but not NBs, as expected [1]. I376K-L377K TRIM5αRh CBs were similar to their WT counterpart, unlike V435K-I436K TRIM5αRh CBs which were noticeably bigger (a 78.8% increase in diameter as averaged from 150 randomly chosen CBs). In presence of LMB, WT TRIM5αRh could be found in both cytoplasmic and nuclear compartments as expected (Fig. 3B). I376K-L377K TRIM5αRh was not observed in the nucleus, as previously reported [39]. Interestingly, the distribution of V435K-I436K was similar to that of the WT protein, showing that mutating this motif did not affect nuclear translocation (Fig. 3B).
The relatively large size of V435K-I436K TRIM5αRh CBs (Fig. 3B) prompted us to analyze the potential of this mutant to multimerize. CRFK cells stably expressing WT or V435K-I436K TRIM5αRh were lysed and the lysates were treated with increasing concentrations of glutaraldehyde, a crosslinking agent previously used in similar experiments [49, 50]. As shown in Fig. 3C, in the absence of glutaraldehyde we observed only monomers of TRIM5αRh-FLAG (approximately 54 kDa). Multimers of V435K-I436K TRIM5αRh were present in greater relative amounts, regardless of the glutaraldehyde concentration used, compared to the WT (Fig. 3C). This suggests that this mutation increases TRIM5αRh multimerization potential, explaining the enlarged CBs. Altogether, the results shown in Fig. 2 and Fig. 3 suggest that V435K-I436K disrupts restriction by affecting surface determinants of interaction with CA, although we cannot exclude that its effects on multimerization patterns also play a role. In contrast, I376K-L377K probably caused major misfolding of the PRYSPRY domain as also proposed by others [39].
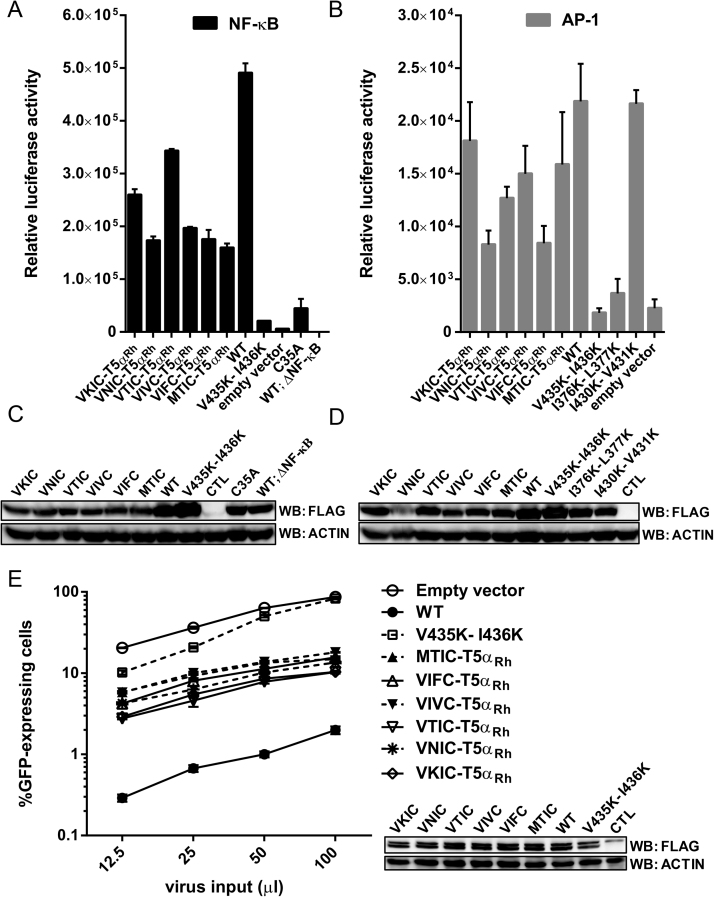
2.4. V435K-I436K abolishes the ability of TRIM5αRh to activate AP-1 and NF-κB
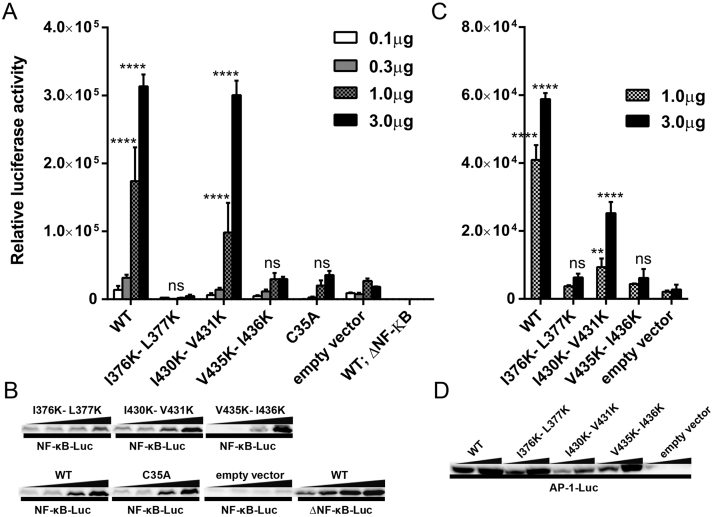
TRIM5α is a pattern recognition receptor that promotes the activation of innate immune signaling pathways dependent on transcription factors AP-1 and NF-κB [25, 26, 29, 30]. These innate immune pathways result in the production of various inflammatory cytokines and chemokines [51]. A previous study proposed that specific interactions between TRIM5 proteins and HIV-1CA lead to the activation of AP-1 and NF-κB [26]. However, TRIM5αRh can also activate both NF-κB and AP-1 when it is transiently transfected and in the absence of infection [25, 29, 30, 38]. To determine whether the mutations in the various putative SIMs could interfere with this TRIM5αRh function, HEK293T cells were co-transfected with increasing amounts of WT, I376K-L377K, I430K-V431K or V435K-I436K TRIM5αRh, and a constant concentration of a plasmid expressing luciferase under control of the NF-κB response element [25]. The TRIM5αRh RING domain mutant C35A, which has no ubiquitin ligase activity and is unable to activate NF-κB, was used as a negative control [25]. Another control consisted of cells co-transfected with WT TRIM5αRh and a version of the luciferase expression plasmid bearing a deletion in the NF-κB response element. As shown in Fig. 4A, transfection of 3 μg of WT and I430K-V431K TRIM5αRh resulted in strong NF-κB activation (∼17-fold and ∼16-fold compared to the empty vector control, respectively; P < 0.0001). Transfection of 1 μg of these plasmids had similar effects, but using smaller amounts (0.1–0.3 μg) activated NF-κB only weakly (Fig. 4A), as seen previously [30]. C35A and I376K-L377K TRIM5αRh were unable to activate NF-κB, as recently described by others [38]. Interestingly, V435K-I436K TRIM5αRh did not induce significant NF-κB activation since levels of luciferase expression were similar to those of the C35A control and the empty vector control (Fig. 4A). Levels of luciferase expression in cells transfected with 3 μg of I376K-L377K and V435K-I436K TRIM5αRh were significantly decreased compared to cells transfected with the WT plasmid (∼65-fold for I376K-L377K and ∼11-fold for V435K-I436K TRIM5αRh; P < 0.0001). Levels of TRIM5α expression were similar for the WT and the various mutants, with the exception of I376K-L377K which showed reduced expression levels (Fig. 4B). Therefore, the inability of V435K-I436K TRIM5αRh to activate NF-κB was not caused by decreased expression levels. On the other hand, I376K-L377K TRIM5αRh low expression levels partly explain the very low levels of NF-κB induction for this mutant (Fig. 4A). However, higher protein amounts were observed when 3 μg of I376K-L377K TRIM5αRh were transfected than when 0.3 μg of either WT or I430K-V431K TRIM5αRh were used (Fig. 4B), while NF-κB activation was readily detected for WT and I430K-V431K but not for I376K-L377K TRIM5αRh in these same conditions (Fig. 4A). Thus, we conclude that both I376K-L377K and V435K-I436K mutants are deficient for NF-κB activation.
Fig. 4.
The V435K-I436K mutation in SIM4 affects TRIM5αRh-mediated activation of NF-κB and AP-1. (A) HEK293T cells were transfected with increasing amounts of either WT or the indicated mutants of pMIP-TRIM5αRh and co-transfected with a constant amount (0.6 μg) of a reporter plasmid expressing luciferase from a NF-κB-dependent promoter. As controls, cells were co-transfected with the empty pMIP plasmid and the NF-κB reporter construct, or were co-transfected with WT TRIM5αRh and the activation-deficient mutant of the reporter construct (ΔNF-κB-Luc). Two days later, cells were lysed and luciferase activity was measured. Bars show averages from triplicate transfections with standard deviations. P values obtained when comparing each of the TRIM5α to the empty vector (1 or 3 μg transfected) were calculated using a two-way ANOVA test followed by a Tukey's multiple comparisons test: ****<0.0001; ns, non-significant. (B) The cellular lysates prepared to quantify luciferase activity in (A) were also used to analyze TRIM5α expression. For this, the three lysates from each triplicate transfection were pooled and analyzed by Western blotting using an anti-FLAG antibody. Samples were loaded on more than one gel due to space limitations but all gels and blotting membranes were handled at the same time and in the exact same way. (C) Same as (A), using a reporter plasmid expressing luciferase from an AP-1-dependent promoter. Two different DNA amounts (1 and 3 μg) of the different pMIP plasmids expressing WT, I376K-L377K, I430K-V431K or V435K-I436K TRIM5αRh were co-transfected with a constant amount of AP-1-Luc (0.6 μg). The empty vector was used as a negative control. The graph shows relative luciferase activity 48 h post transfection. The experiment was performed in triplicate and standard deviations are shown. P values obtained when comparing each of the TRIM5α to the empty vector were calculated using a two-way ANOVA test followed by the Tukey's multiple comparisons test: **<0.01; ***<0.001; ****<0.0001. (D) TRIM5α protein expression was analyzed as described in (B).
To evaluate the effects of mutating the TRIM5αRh SIMs on AP-1-dependent transactivation, HEK293T cells were co-transfected with 1 or 3 μg of WT, I376K-L377K, I430K-V431K or V435K-I436K TRIM5αRh and with 0.6 μg of a reporter plasmid expressing luciferase under the control of the AP-1 response element [26]. WT and I430K-V431K TRIM5αRh significantly activated AP-1 at both DNA concentrations compared to the empty vector control (Fig. 4C). On the other hand, I376K-L377K and V435K-I436K TRIM5αRh were very weak activators of AP-1, as evidenced by low luciferase signal in both cases. Specifically, when 3 μg of the TRIM5αRh-expressing plasmid were used, WT and I430K-V431K TRIM5αRh increased luciferase activity by ∼21-fold and ∼9-fold relative to the empty vector control, respectively (P < 0.0001) while I376K-L377K and V435K-I436K TRIM5αRh increased it by only ∼2.3-fold (P = 0.1957) and ∼2.2-fold (P = 0.2304), respectively. Thus, these two mutants did not activate AP-1 to significant levels, while they resulted in a very significant (P < 0.0001) decrease in activation levels when compared to the WT (Fig. 4C). Levels of protein expression were determined in the transfected cells by Western blot analysis (Fig. 4D) and we found that V435K-I436K TRIM5αRh was expressed at WT-like levels while I430K-V431K TRIM5αRh was expressed at smaller levels (a 2- to 3-fold decrease). This explains that in the experiment shown in Fig. 4C, AP-1 activation by I430K-V431K TRIM5αRh is similarly less efficient than for the WT control. On the other hand, the inability of I376K-L377K and V435K-I436K to activate AP-1 does not result from poor expression levels in HEK293T cells. Altogether, we conclude that V435K-I436K TRIM5αRh is unable to activate both AP-1 and NF-κB signaling pathways, suggesting that this motif is a determinant for innate immune signaling.
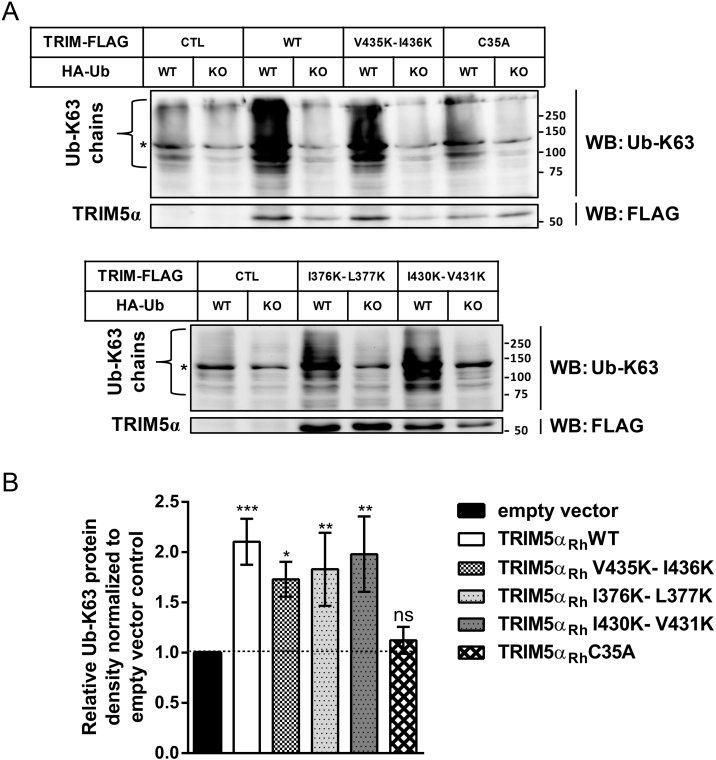
2.5. Mutating putative SIMs does not hinder the capacity of TRIM5αRh to trigger K63-linked polyubiquitin chains formation
E3 ubiquitin ligase activity associated with the RING domain is essential for the ability of TRIM5α to promote formation of K63-linked ubiquitin chains [26, 52] that can then mediate the activation of AP-1 and NF-κB signaling through phosphorylation of TAK1 [51, 53, 54]. To study the ability of I376K-L377K, I430K-V431K and V435K-I436K mutants to generate Ub-K63 chains, FLAG-tagged TRIM5αRh, WT or mutated, was transfected in HEK293T cells along with a construct expressing ubiquitin. To detect K63-linked polyubiquitin, Western blot analysis was performed using an antibody specific for this type of ubiquitin chains [55] (Fig. 5A). As controls, cells were co-transfected with KO-Ubiquitin, a polyubiquitylation-deficient version of ubiquitin in which all potentially ubiquitylated lysines have been substituted to arginines [56]. K63-linked ubiquitin chains were detected between ∼70 and 300 kDa, consistent with previous reports [26, 30], and such polyubiquitylation products were almost undetectable in cells transfected with the KO ubiquitin mutant (Fig. 5A). The C35A TRIM5αRh RING domain mutant was used as a negative control. As expected, K63-linked ubiquitin chains were almost undetectable in cells transfected with this mutant (Fig. 5A). Co-transfection of WT, I376K-L377K, I430K-V431K and V435K-I436K TRIM5αRh with WT ubiquitin resulted in strong stimulation of K63-linked polyubiquitylation, compared to cells co-transfected with TRIM5αRh and KO ubiquitin or compared to cells co-transfected with the empty vector and WT ubiquitin (Fig. 5A, B). A densitometry-based quantitative analysis was performed using Western blots from 3 independent experiments (Fig. 5B). Values were obtained by normalizing relative K63-linked ubiquitin chains densities from the indicated TRIM5αRh to the one obtained with the empty vector control (CTL) when co-transfected with WT ubiquitin. The mean levels of TRIM5α protein expression in the 3 experiments were similar for all FLAG-tagged proteins utilized (data not shown). Results show that only C35A TRIM5αRh was unable to significantly induce K63-linked ubiquitin chains (P = 0.916 when compared to the empty vector). In contrast, WT, I376K-L377K, I430K-V431K and V435K-I436K TRIM5αRh all promoted the formation of K63-linked ubiquitin chains at WT levels (Fig. 5B). Specifically, all three SIM mutants showed statistically significant induction of K63-linked polyubiquitin when compared to the empty control while none of them showed statistically significant differences with the WT (Fig. 5B). Therefore, the putative SIMs 1 and 4 are dispensable for the formation of K63-linked ubiquitin chains even though they are required for NF-κB and AP-1 activation.
Fig. 5.
Mutating putative PRYSPRY SIMs does not affect TRIM5αRh ability to trigger K63-linked ubiquitin chains formation. (A) HEK293T cells were transfected with either the empty pMIP (CTL) or with pMIP expressing FLAG-tagged WT, I376K-L377K (SIM1 mutant), I430K-V431K (SIM3 mutant) or V435K-I436K (SIM4 mutant) TRIM5αRh, and co-transfected with WT or KO ubiquitin. KO ubiquitin has all its lysines mutated to arginine to prevent polyubiquitylation. C35A TRIM5αRh was used as a negative control for its incapacity to significantly promote K63-linked chains formation. Two days later, whole cell lysates were analyzed by Western blotting using rabbit antibodies directed at K63-linked ubiquitin chains or FLAG. A single experiment representative of three independent experiments is presented. * points to a non-relevant band detected by the antibody used. Samples were loaded on two gels due to space limitations but gels and blotting membranes were handled at the same time and in the exact same way. (B) Mean densities of bands ranging from 75 to >250 kDa detected using the K63 chains-specific antibody, normalized to the control consisting of cells co-transfected with WT-Ub and empty pMIP. Data were obtained in three independent experiments and standard deviations are shown. P values were calculated using a one-way ANOVA test for each TRIM5α variant compared to the empty vector control: *<0.05, **<0.01, ***<0.001.
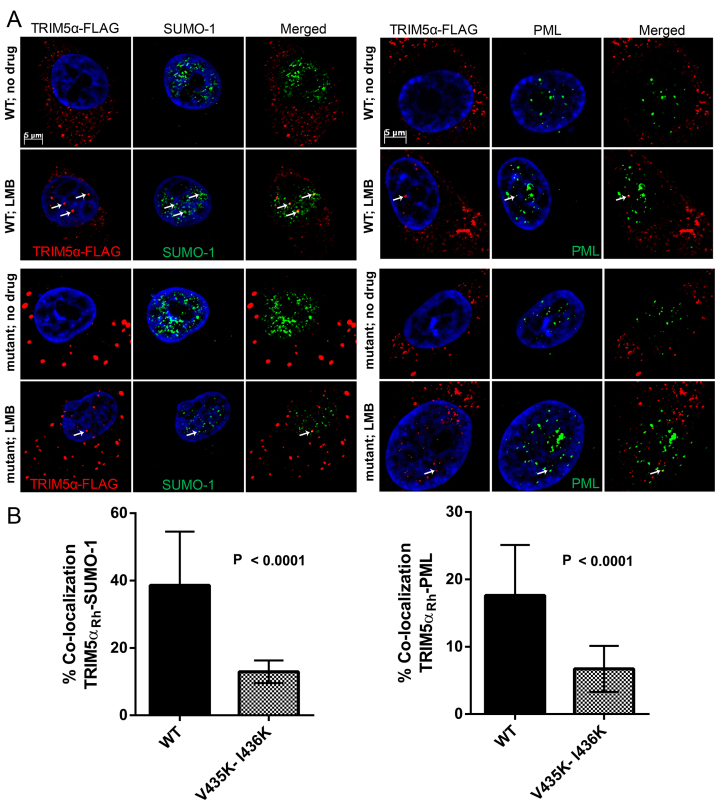
2.6. V435K-I436K TRIM5αRh shows reduced co-localization with PML and SUMO-1 in the nucleus
PML/TRIM19, a member of the TRIM family of proteins, is the main determinant for the formation of PML bodies, nuclear structures that can harbor multiple proteins and have known roles in antiviral defense [57]. PML is heavily SUMOylated and SUMO is a major contributing factor to the formation of PML NBs [58]. TRIM5αRh is able to shuttle to the nucleus and co-localize with PML NBs [47]. We hypothesized that direct interactions between SIM4 and either SUMO or a SUMOylated protein may determine the localization of TRIM5α at PML NBs. Thus, we reasoned that mutating SIM4 would affect the co-localization of TRIM5α with PML or SUMO or both. To test this possibility, IF microscopy was performed on CRFK cells stably expressing WT or V435K-I436K TRIM5αRh and in the presence or absence of LMB. As shown in Fig. 6A, LMB treatment resulted in a fraction of WT and V435K-I436K TRIM5αRh being present in the nucleus, while no nuclear TRIM5α staining was observed in the absence of the drug, as expected [47]. A significant fraction of TRIM5αRh NBs co-localized with SUMO-1 (left panel) and PML (right panel) in LMB-treated cells, and this was observed for both WT and V435K-I436K TRIM5αRh. However, co-localization between V435K-I436K TRIM5αRh NBs and both SUMO-1 and PML was less frequent than for WT TRIM5αRh NBs. Specifically, an analysis of multiple randomly selected fields (Fig. 6B) showed that in presence of LMB, 38.6% of WT TRIM5αRh NBs but only 12.9% of V435K-I436K TRIM5αRh NBs co-localized with SUMO-1 (P < 0.0001). Similarly, 17.6% of WT TRIM5αRh NBs but only 6.7% of V435K-I436K TRIM5αRh NBs co-localized with PML (P < 0.0001). Therefore, mutating putative SIM4 affects TRIM5αRh association with SUMO-1 and PML NBs, suggesting that this motif is a determinant for the interaction with SUMO or a SUMOylated protein.
Fig. 6.
Decreased nuclear co-localization of V435K-I436K TRIM5αRh with endogenous SUMO-1 and PML. (A) CRFK cells stably expressing FLAG-tagged WT or V435K-I436K (SIM4 mutant) TRIM5αRh were treated or not with LMB for 12 h and then processed for IF microscopy. A rabbit anti-FLAG antibody was used for the detection of TRIM5αRh (red) and mouse antibodies were used for the detection of endogenous SUMO-1 and PML (green). DNA was stained with Hoechst33342 (blue). White arrows point to examples of co-localizations of nuclear TRIM5αRh with SUMO-1 (left panels) or PML (right panels). (B) Quantitative analysis of the frequency of WT or V435K-I436K TRIM5αRh NBs co-localization with either SUMO-1 (left panel) or PML (right panel) in LMB-treated cells. The percentage of WT and V435K-I436K TRIM5αRh NBs co-localizing with SUMO-1 or PML is shown. Each bar represents the average % of co-localization observed in 15 randomly chosen fields considered as replicates. P values were calculated using the Student T-test and standard deviations are shown. More than 150 TRIM5αRh NBs were analyzed for each condition.
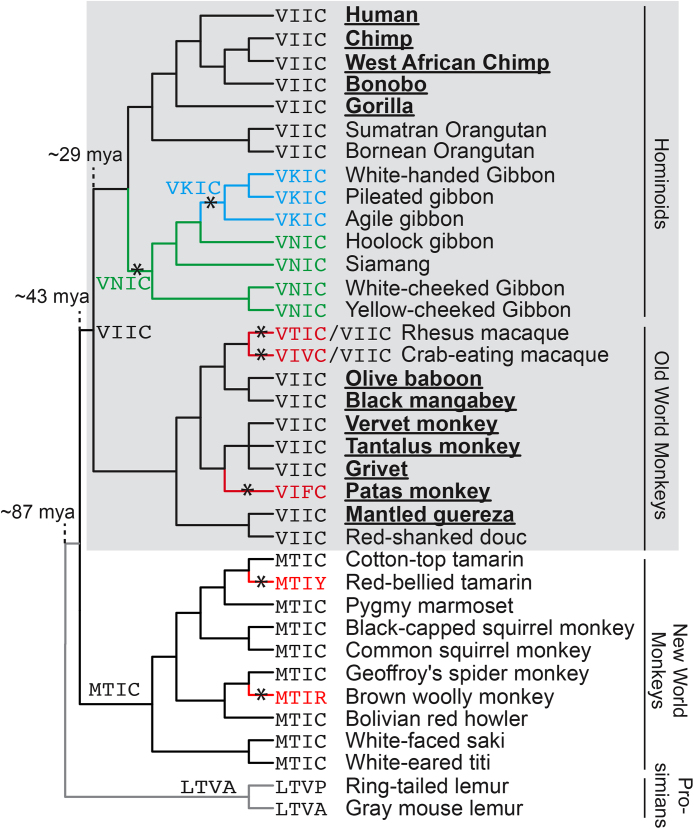
2.7. Natural simian variants of SIM4 activate both NF-KB and AP-1 signaling pathways and exhibit partial HIV-1 restriction
Our results suggest that SIM4 (435VIIC438) is an important motif for the ability of TRIM5αRh to inhibit HIV-1 and activate NF-κB/AP-1. We wondered whether this motif was well conserved among primates given that TRIM5α is known to have evolved under positive selection in order to restrict different retroviruses [59]. For this purpose, we aligned the TRIM5α coding sequence from 36 primate species. The relationship between these species is shown in Fig. 7. None of the New World monkeys (NWM) or the prosimians encode an intact VIIC motif. On the other hand, all representatives of the great apes (human, chimpanzee, West African chimpanzee, bonobo, gorilla, Sumatran orangutan and Bornean orangutan) and a majority of the Old World monkeys (OWM) have conserved SIM4 VIIC motifs. Based on these observations, we propose that SIM4 VIIC did not appear independently in these two groups but, instead, arose before the split of Old World monkeys and hominoids which occurred approximately 29 million years ago. Within the OWM and hominoids lineages, SIM4 has been mutated at 5 independent times throughout evolution (stars on branches in Fig. 7). Analysis of TRIM5α exon 8 sequences from 34 individual crab-eating macaques was done using BLAST and revealed 2 individuals from the Mauritian population which encode VIVC. For rhesus macaque, sequences from 15 individuals were analyzed and one individual from Guangsi was found to encode VTIC. These data demonstrate that SIM4 is quite variable both between and within primate species.
Fig. 7.
Divergence of the TRIM5α putative SIM4 motif during primate speciation. The SIM4 motif was analyzed in 36 primate species (bases 1291-1302 of the TRIM5 gene, encoding residues 431-434 in the human TRIM5α protein). These primate species represent ∼87 million years of primate evolution and the four major primate groups are shown. Amino acid residues encoded in this motif by each primate genome are indicated. Ancestral forms of the SIM4 motif were predicted with the Program PAML [79]. Asterisks indicate positions where mutations in this motif are predicted to have occurred. The gray box indicates the clade of primates whose ancestral SIM4 motif is VIIC. SNPs within the SIM4 domain have been reported in both rhesus and crab-eating macaque populations (M. mulatta from Guangxi province and individuals from the Mauritian M. fascicularis population, respectively) [70, 71]. These polymorphisms are indicated on the tree with hyphens, illustrating the alternate alleles that are co-circulating. Underlined names indicate primates that live in Continental Africa.
All natural SIM4 variants were tested for their ability to activate NF-κB/AP-1 and restrict HIV-1. Using mutagenesis, we substituted TRIM5αRh-SIM4 (VIIC) with all the natural variants (VKIC, VNIC, VTIC, VIVC, VIFC and MTIC), resulting in the production of 6 new proteins. First, their capacity to activate the NF-κB and AP-1 signaling pathways was tested, as described above, using co-transfections in HEK293T cells. All the SIM4 variants were able to activate NF-κB (Fig. 8A) and AP-1 (Fig. 8B) since significant luciferase activity was observed in both experiments. The SIM4 variants seemed to show some level of reduction (∼15% to 60%) in their capacity to activate both pathways, compared to the WT, but this could largely be attributed to decreased expression levels (Fig. 8C, D). In contrast, V435K-I436K TRIM5αRh was unable to activate both signaling pathways, as expected. Taken together, these results suggest that, despite the emergence of variations within the SIM4 sequence among primates, its capacity to activate NF-κB and AP-1 is conserved.
Fig. 8.
Natural simian variants of SIM4 activate both NF-KB and AP-1. (A) HEK293T cells were co-transfected with either WT or the indicated mutants of pMIP-TRIM5αRh (2 μg) and a reporter plasmid expressing luciferase from a NF-κB-dependent promoter (0.6 μg). As controls, cells were co-transfected with the empty pMIP plasmid and the NF-κB reporter construct, or were co-transfected with WT TRIM5αRh and the activation-deficient mutant of the reporter construct (ΔNF-κB-Luc). Cells were lysed 48 h post transfection and luciferase activity was measured. Bars show averages from triplicate transfections with standard deviations. (B) Similar experiment using a reporter plasmid expressing luciferase from an AP-1-dependent promoter. The empty vector was used as a negative control. The graph shows relative luciferase activity 48 h post transfection. The experiment was performed in triplicate and standard deviations are shown. (C) The cellular lysates prepared to quantify luciferase activity in (A) were also used to analyze TRIM5αRh expression. For this, the three lysates from each triplicate transfection were pooled and analyzed by Western blotting using an anti-FLAG antibody. (D) TRIM5αRh expression levels in cells from the experiment shown in (B). The analysis was done as in (C). (E) The HIV-1 restriction activity of all TRIM5αRh SIM4 variant constructs was assessed by infecting CRFK cells stably expressing the indicated TRIM5αRh with increasing amount of HIV-1-GFP. Percentages of GFP-positive cells were measured by FACS two days later. The experiment was performed in triplicate and standard deviation bars are shown. WT and mutant TRIM5αRh protein expression levels in stably transduced CRFK cells were determined by Western blotting with an anti-FLAG antibody. Actin expression was analyzed as a loading control.
The second function to be analyzed was TRIM5α capacity to inhibit HIV-1 infection. CRFK cells stably expressing all the SIM4 variants, V435K-I436K or WT TRIM5αRh were challenged with increasing amounts of HIV-1-GFP and further analyzed by flow cytometry. As expected, WT TRIM5αRh strongly inhibited HIV-1 (Fig. 8E) and V435K-I436K TRIM5αRh failed to restrict HIV-1 and was only slightly less permissive than cells expressing the empty vector control. All the SIM4 variants restricted HIV-1 at an intermediate level: ∼8-fold less than WT and ∼6-fold more than the empty vector control, on average. Levels of protein expression were determined by Western blotting and were found to be comparable in all transduced cells (Fig. 8E). An upper unspecific band was detected by the anti-FLAG antibody, as previously observed in CRFK cells (Fig. 3C). Thus, mutating VIIC into naturally occurring sequences found in other species affects TRIM5αRh ability to optimally restrict HIV-1, consistent with this motif being involved in interactions with CA in addition to being important for NF-κB/AP-1 signaling. The fact that, for some of the mutants tested, the effect on NF-κB activation is much milder than the effect on restriction supports a model in which the two functions can have distinct determinants.
3. Discussion
In this study, we identified a new putative SIM (435VIIC438) in TRIM5αRh PRYSPRY that is, on one hand, involved in specific interactions with the retroviral capsid, and on the other hand, is also essential for the activation of NF-κB/AP-1 at least in the infection-free model employed here. Mutating this motif impaired restriction (Fig. 2) and NF-κB/AP-1 signaling (Fig. 4), while a similar mutation in the adjacent SIM3 (430IVPL433) affected neither function. As also proposed by Brandariz-Nuñez et al. [39], our results support the hypothesis that mutating the previously proposed SIM1 causes gross misfolding of the PRYSPRY domain, preventing the nuclear translocation of TRIM5αRh (Fig. 3) and resulting in its proteasomal degradation (Nepveu-Traversy and Berthoux, unpublished data). Misfolded proteins are generally degraded by the ubiquitin-proteasome system to ensure cellular homeostasis [46]. Although the SIM1 mutant (I376K-L377K) did not show an unstable phenotype upon transfection in HEK293T cells (see Fig. 4, Fig. 5), this is probably due to over-expression conditions in this particular cell line. In contrast to I376K-L377K, the SIM4 mutant (V435K-I436K) was stable and able to shuttle to the nucleus (Fig. 3) indicating that this mutant was probably not misfolded, consistent with SIM4 being mostly a surface motif. However, V435K-I436K TRIM5αRh shows increased potential for multimerization and CB formation (Fig. 3). The TRIM5α coiled-coil and Linker2 domains upstream of SPRY form an anti-parallel homodimer which is expected to bring the SPRY domains in relatively close proximity [60, 61], and it is conceivable that mutations in the proposed SIM4 could modulate interactions between the two PRYSPRY domains with negative consequences on restriction. In addition, it is possible that the larger size of the SIM4-mutated TRIM5α CBs means that less TRIM5α is available to intercept incoming HIV-1, contributing to the loss of restriction.
The CA protein of the N-tropic murine leukemia virus (N-MLV), which is restricted by human TRIM5α (TRIM5αHu) [11] is likely modified by SUMO-1 [62]. It has been proposed that TRIM5αHu restricts N-MLV efficiently through the binding of TRIM5αHu SIM1 and/or SIM2 to the incoming SUMOylated N-MLV CA [36]. Recent structural data suggesting that SIM1 and SIM2 are internal motifs [39] seem to invalidate this theory, although other structural data indicate that the PRYSPRY domain can adopt several conformations [63] and that the variable loops at its surface probably undergo conformational changes upon binding to CA [64], opening the possibility that SIM1 or SIM2 could be brought at the surface of the protein upon CA interactions. This, however, does not appear to be a likely possibility. The putative SIM identified here, SIM4, is clearly a surface motif and thus could potentially bind a SUMOylated CA. However, HIV-1CA has not been found to be SUMOylated, and in addition, the evidence linking SUMO to direct restriction of incoming retroviruses is not overwhelming. Arriagada and colleagues showed that over-expression of SUMO-1 in 293T cells restricts N-MLV and only in conditions in which this virus is also restricted by TRIM5αHu [36], suggesting that SUMO is a co-factor for TRIM5α. However, over-expression of SUMO-1 had much smaller effects on the permissiveness of other human cell lines to N-MLV infection, and a follow-up study focusing this time on TRIM5αRh did not show that over-expression of SUMO-1 inhibited HIV-1 in restrictive conditions [38]. Instead, the authors reported that SUMO-1 depletion canceled HIV-1 restriction by TRIM5αRh, but this conclusion was weakened by inefficient SUMO-1 knockdown and abnormally low levels of HIV-1 restriction by TRIM5αRh in the control cells [38]. Similarly, another recent study reported a reduction in restriction by transfection of siRNAs targeting SUMO-1, Ubc9 or PIAS1 [37]. However, restriction of HIV-1 by TRIM5αRh in this transient transfection-based setting was similarly much weaker than in stably transduced cells (less than 4-fold compared to the usual ∼100-fold). A more recent study reported that perturbing the SUMOylation pathway by over-expression of Gam1 [65, 66] had no impact on the restriction of N-MLV in human cells [39], and we found that Gam1 does not influence the restriction of HIV-1 by TRIM5αRh expressed in human cells (Nepveu-Traversy and Berthoux, unpublished data). Thus, there is altogether neither strong nor even consensual evidence that the SUMOylation pathway is important for the direct restriction of incoming retroviruses, especially for HIV-1. On the other hand, the presence of a conserved putative SUMOylation site and of SIMs indicates a role for SUMO in TRIM5α functions, and we propose that SUMO and the SUMOylation pathway are involved in the activation of NF-κB/AP-1 by TRIM5α. Recently, we have shown that mutating the putatively SUMOylated lysine (lysine 10) reduces the capacity of TRIM5αRh to generate K63-linked polyubiquitin and to activate NF-κB and AP-1 [30]. Here we show that the SIM4 double substitution mutant V435K-I436K completely disrupts the NF-κB/AP-1 activation potential of TRIM5αRh, while mutating SIM3 has no effect (Fig. 4).
It has been previously suggested that activation of NF-κB and AP-1 by TRIM5α is the consequence of a signaling cascade that includes the formation of K63-linked ubiquitin chains. Similarly to TRAF6, a RING domain protein, TRIM5α recruits a E2 heterodimer, Ubc13/Ube2V2 [27], to catalyze the formation of K63-linked ubiquitin chains [26] which are important for the activation of TAK1 [52], a key factor for the downstream activation of both NF-κB and AP-1 [53]. Interestingly, all the PRYSPRY mutants tested in this study were able to trigger significant formation of K63-linked ubiquitin chains (Fig. 5). This result suggests that the various putative SIMs, and probably the PRYSPRY domain itself, are irrelevant to this function, at least in this over-expression experimental model. This conclusion is also supported by the fact that the SIM1 mutant can promote K63-linked ubiquitin despite its PRYSPRY domain being possibly misfolded. In contrast, mutating the RING zinc finger abolished this function (Fig. 5), confirming that this domain is an essential determinant for the generation of K63-linked ubiquitin by TRIM5α [26]. Moreover, the fact that mutations in the putative SIM1 and SIM4 motifs abolish the stimulation of NF-κB and AP-1 indicates that presence of K63-linked polyubiquitin chains is not the sole requirement for activation of these innate immunity pathways. Thus, the SIM4 (435VIIC438) motif seems to be a determinant for an as yet unidentified activation step that is required for activation of NF-κB and AP-1 pathways. We cannot exclude, however, that the K63-linked ubiquitin chains generated by V435K-I436K TRIM5αRh are defective in a way that could not be revealed by our Western blot analysis, or that they are not properly located in the cell to activate the NF-κB and AP-1 pathways. A direct role for SIM4 in activating NF-κB and AP-1 appears more likely, however, and we propose that SIM4 interacts with SUMO or an unidentified SUMOylated protein and that this interaction contributes to the activation of NF-κB/AP-1. The existence of such interactions was suggested by IF microscopy results (Fig. 6).
The exact nature of the involvement of SUMO and the SUMOylation pathway in the activation of NF-κB/AP-1 by TRIM5α remains an open question. TAK-1 binding protein 2 (TAB2) was recently proposed to be SUMOylated, and mutating its SUMOylated lysine enhanced the activity of TAB2 and resulted in increased AP-1 activation [67]. Also, NF-κB essential modulator (NEMO/IKKγ) is SUMOylated in response to various stress signals, allowing NF-κB activation by translocation of SUMOylated NEMO to the nucleus [68]. On the other hand, the inhibitor of NF-κB (IκBα), which hides the nuclear localization signal, is degraded by the proteasome to allow translocation of NF-κB to the nucleus, but SUMOylation stabilizes it and consequently inhibits NF-κB activation [69]. Taken together, these findings show that activation of NF-κB is extensively controlled by SUMOylation, providing several speculative models for TRIM5α interacting with a SUMOylated factor in order to promote innate immune activation. For instance, TRIM5α could interact, through the SIM4 motif, with SUMOylated IκBα to disrupt its interaction with NF-κB and thus promote NF-κB translocation to the nucleus. It is possible that the shuttling of TRIM5α in the nucleus and its transient association with PML bodies are important in its capacity to activate NF-κB and AP-1.
A phylogenetic analysis of the proposed SIM4 revealed some significant changes in some of the 36 primates analyzed (Fig. 7). Six naturally occurring variants were tested for their ability to restrict HIV-1 when placed in a TRIM5αRh context and all were less efficient than their WT counterpart in this assay (Fig. 7E), confirming that this motif is important for CA interactions in addition to being important for the activation of NF-κB/AP-1. Interestingly, intra-species polymorphism was found at this position in rhesus and crab-eating macaques. The analysis of NCBI TRIM5α sequences from 34 crab-eating macaques revealed 2 isolates bearing the motif VIVC, and 1 of 15 rhesus macaques had the motif VTIC. These species are already known for their polymorphism due to the occasional presence of a supplementary allele coding for TRIMCyp [70, 71]. Interestingly, individuals expressing the modified SIM4 (VIVC and VTIC) did not have the additional TRIMCyp allele. It is possible that TRIMCyp and SIM4 variants emerged in response to different viruses. The high level of polymorphism in SIM4 also supports a role for this motif in interacting with incoming retroviral CA cores. Of note, the VIIC motif is found in great apes and in OWM, the two primate groups living in Africa where one finds the most cases of SIV infections. It is possible that primates in other parts of the world do not undergo a strong selective pressure in order to make TRIM5α ultra-responsive towards lentiviruses. Interestingly, the ability to activate NF-κB and AP-1 was maintained for the six naturally occurring SIM4 variants tested here (Fig. 7), suggesting that this function is essential and is evolutionarily conserved despite variations in the sequence that affect the capacity of these variants to restrict HIV-1 (Fig. 8). This observation supports the idea that the two functions are not necessarily coupled, even though interaction of TRIM5α with restriction sensitive viruses was shown to activate NF-κB/AP-1 [26].
In conclusion, we have characterized a new putative SIM (435VIIC438) in TRIM5αRh that is important for TRIM5α-dependent innate immune signaling, possibly through its capacity to interact with SUMO or a SUMOylated protein, and which also has a role in the retroviral tropism of TRIM5α.
4. Materials and methods
4.1. Plasmid DNAs and mutagenesis
pMIP-TRIM5αRh expresses a C-terminal FLAG-tagged version of TRIM5αRh and has been described before [72, 73]. We designed the following primers to introduce mutations in pMIP-TRIM5αRh: I376K- L377K, 5′-AAGTGCTTGGAAGAAGGGGGTATGTGCTGG-3′ (forward) and 5′-CATACCCCCTTCTTCCAAGCACTTTTCTT-3′ (reverse); I430K-V431K, 5′-TGCTCCTTTCAAGAAGCCCCTCTCTGTGAT-3′ (forward) and 5′-CAGAGAGGGGCTTCTTGAAAGGAGCAAAAG-3′(reverse); V435K-I436K, 5′CTTTCATTGTGCCCCTCTCTAAGAAGATTTGTCCTGA
TCGTGTTG-3′ (forward) and 5′-CAACACGATCAGGACAAATCTTCTTAGAGAG
GGGCACAATGAAAG-3′ (reverse). SIM4 variants were constructed using pMIP-TRIM5αRh as template and mutations were introduced using the following primers: VKIC, 5′-GTGCCCCTCTCTGTGAAAATTTGTCCTGATCGTGTTG-3′ (forward) and 5′-GATCAGGACAAATTTTCACAGAGAGGGGCACAATGAAAG-3′ (reverse); VNIC, 5′-GTGCCCCTCTCTGTGAATATTTGTCCTGATCGTGTTG-3′ (forward) and 5′-GATCAGGACAAATATTCACAGAGAGGGGCACAATGAAAG-3′ (reverse); VTIC, 5′-GTGCCCCTCTCTGTGACTATTTGTCCTGATCGTGTTG-3′ (forward) and 5′-GATCAGGACAAATAGTCACAGAGAGGGGCACAATGAAAG-3′ (reverse); VIVC, 5′-GTGCCCCTCTCTGTGATTGTTTGTCCTGATCGTGTTG-3′ (forward) and 5′-GATCAGGACAAACAATCACAGAGAGGGGCACAATGAAAG-3′ (reverse); VIFC, 5′-GTGCCCCTCTCTGTGAAATTTTGTCCTGATCGTGTTG-3′ (forward) and 5′-GATCAGGACAAAATTTCACAGAGAGGGGCACAATGAAAG-3′ (reverse); MTIC, 5′-GTGCCCCTCTCTATGACTATTTGTCCTGATCGTGTTG-3′ (forward) and 5′-GATCAGGACAAATAGTCATAGAGAGGGGCACAATGAAAG-3′ (reverse). All mutations were confirmed by Sanger sequencing. The vector production plasmids pMD-G, pΔR8.9, pCL-Eco and pTRIP-CMV-GFP have all been extensively described elsewhere [44, 74, 75, 76). pRK5-HA-Ubiquitin WT and KO [56] were obtained from Ted Dawson (Johns Hopkins University, Baltimore, MD) through Addgene. The KO ubiquitin version of this plasmid bears the following mutations eliminating all possibilities of ubiquitin chain formation: K6R, K11R, K27R, K29R, K33R, K48R and K63R. pCEP4-NF-κB-Luc expresses luciferase under the control of an NF-κB-dependent promoter, while pCEP4-ΔNF-κB-Luc is transcriptionally deficient due to deletion of the NF-κB binding site [25]. Both constructs were kind gifts from M. Emerman (University of Washington, Seattle, WA). pHTS-AP1-Luc expresses luciferase under the control of an AP-1-dependent promoter and was a kind gift from J. Luban (University of Massachusetts Medical School, Worcester, MA).
4.2. Cell lines
Human embryonic kidney (HEK) 293T cells and Crandell-Rees feline kidney (CRFK) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics at 37 °C, 5% CO2. All cell culture reagents were from HyClone (Thermo Scientific, Logan, UT, USA).
4.3. Transfections and virus production
MLV and HIV-1-based vectors were produced through transient transfection of HEK293T cells using polyethylenimine (MW 25,000; Polysciences, Warrington, PA) and collected as previously described [72, 77]. To produce the MLV-based MIP vectors, cells were transfected with the relevant pMIP plasmid and co-transfected with pCL-Eco and pMD-G. All stably transduced cell lines were produced as previously described [72, 77]. Successfully transduced CRFK cells were selected with 4 μg/ml of puromycin. To produce the GFP-expressing HIV-1 vector HIV-1TRIP-CMV-GFP (nicknamed HIV-1-GFP), cells were co-transfected with pΔR8.9, pMD-G and pTRIP-CMV-GFP.
4.4. Viral challenges
CRFK cells were plated in 24-well plates at 30,000 cells per well and infected the next day with different amounts of HIV-1-GFP or using a defined multiplicity of infection (MOI). Two days post-infection, cells were trypsinized and fixed in 2% formaldehyde in a PBS solution. The % of GFP-positive cells was then determined by analyzing 10,000 cells using a FC500 MPL cytometer with the CXP software (Beckman Coulter).
4.5. Western blotting
Cells were lysed in RIPA buffer (150 mM NaCl, 1% Triton, 0.1% SDS, 50 mM Tris pH 8.0, 0.5% sodium deoxycholate and Complete protease inhibitor cocktail (Roche, Bale, Switzerland). Whole cell lysates were then boiled in protein sample buffer (60 mM Tris-HCl pH 6.8, 10% glycerol, 0.002% bromophenol blue, 2% SDS, 2% beta-mercaptoethanol) and resolved by SDS-PAGE. After transfer to nitrocellulose membranes, blots were probed with rabbit anti-FLAG (1:2000, Cell Signaling, Danvers, Massachusetts) followed by a secondary antibody coupled to horseradish peroxydase (Santa Cruz, Dallas, TX), or with a HRP-conjugated mouse anti-actin (1:20,000, EMD Millipore, Billerica, MA), and revealed using a chemiluminescence detection system (SuperSignal West Femto, Thermo Scientific, Waltham, MA). Images were recorded on a UVP (Upland, California) EC3 imaging system, and densitometry analyses were performed using the area density tool of the VisionWorks LS software (UVP).
4.6. Stability assay
CRFK cells expressing TRIM5αRh were plated in 6-well plates at a cell density leading to 80% confluent cells on the next day. Cells were then treated with 100 μM cycloheximide (Sigma, Saint Louis, MO) and harvested at different time points as previously described [15]. Cells were lysed in stability lysis buffer [NaCl 100 mM, NP-40 0.5%, SDS 0.1%, Tris 100 mM (pH 8.0) and Complete protease inhibitor cocktail (Roche, Bale, Switzerland)] and further processed for Western blotting. Protein density was measured for each time point and normalized to actin, and then normalized to time zero density.
4.7. Multimerization assay
CRFK cells stably expressing either FLAG-tagged WT or V435K-I436K TRIM5αRh were plated in three 10-cm plates at a cell density leading to 80% confluent cells on the next day. Cells from the 3 dishes were then pooled together and lysed with 0.05% NP-40 on ice for 15 min. After centrifugation, supernatants were divided in 5 aliquots and treated for 5 min with different glutaraldehyde (J.T. Baker, Phillipsburg, NJ) concentrations (0, 0.25, 0.5, 1.25, 2.5 mM). Tris-HCl 0.1 M pH 7.5 was added to stop reaction and all samples were denaturated and further processed for Western blotting as described above.
4.8. NF-κB and AP-1 reporter assays
HEK293T cells were plated in 12-well plates at a cell density leading to 80% confluent cells at the time of transfection and were then transfected with increasing amounts (0.1, 0.3, 1 and 3 μg) of FLAG-TRIM5α-expressing pMIP-TRIM5α supplemented with empty pMIP to reach a total of 3 μg, or transfected with fixed amount (2 μg) of FLAG-tagged WT or mutant TRIM5α. Cells were simultaneously transfected with 0.6 μg of pCEP4-NF-κB-Luc, pCEP4-ΔNF-κB-Luc or pHTS-AP1. Cells were lysed with RIPA buffer 48 h post-transfection and assessed for luciferase activity using the BrightGlow Luciferase kit (Promega). Luminescence was measured with a Synergy HT multi-detection microplate reader (BioTek, Winooski, VT) and analyzed using the Gen5 software (BioTek).
4.9. K63 ubiquitin chains formation assay
HEK293T cells were seeded in 12-well plates at a cell density leading to 80% confluency. On the next day, cells were co-transfected with either WT or mutant FLAG-tagged TRIM5α (2 μg) and either WT or KO pRK5-HA-ubiquitin (0.5 μg). The “empty” plasmid pMIP was used as a negative control. Cells were lysed in RIPA buffer 48 h post-transfection and processed for Western blotting. A monoclonal human/rabbit chimeric K63-linked ubiquitin chains-specific antibody (clone Apu, Millipore) was used (1:1000 dilution) to detect K63 ubiquitin chains.
4.10. Immunofluorescence (IF) microscopy
60,000 CRFK cells were plated on microscope glass coverslips (Fisherbrand) in 6-well plates. Treatments with the nuclear export inhibitor leptomycin B (LMB, Enzo LifeScience) were done at 20 ng/ml for 12 h prior to fixation. For the experiment involving HIV-1 infection, cells were treated with MG132 (1 μg/ml) for 6 h and infected with HIV-1-GFP (MOI of 4) for 2 h prior to fixation. Control infections were done in presence of ammonium chloride (NH4Cl, 20 mM, Sigma). Cells were washed with PBS, fixed for 10 min in 4% formaldehyde-PBS at 37 °C, washed three times in PBS and permeabilized with 0.1% Triton X-100 for 2 min on ice. Cells were then washed twice with PBS and treated with 10% normal goat serum (Vector laboratories) in PBS for 30 min at room temperature. This saturation step was followed by a 4 h incubation with a rabbit polyclonal FLAG antibody (Cell Signaling) and an antibody against endogenous SUMO-1 (mouse monoclonal antibody, Invitrogen), both at a 1:200 dilution in PBS with 10% normal goat serum. To stain endogenous promyelocytic leukemia protein (PML), primary mouse monoclonal antibody (Enzo Life Sciences, Farmingdale, NY) was used at a 1:50 dilution. HIV-1CA was stained using a mouse monoclonal anti CAp24 antibody purified in our laboratory from the 183 hybridoma clone, AIDS Research and reference Reagent Program, NIAID, NIH. Fluorescent staining was done by incubating with an Alexa594-conjugated goat anti-rabbit antibody and an Alexa488-conjugated goat anti-mouse antibody (each used at 1:200 dilution). Cells were washed 4 times in PBS before mounting in Vectashield (Vector Laboratories, Burlington, Ontario). Hoechst33342 (0.8 μg/ml; Molecular Probes) was added along with the penultimate PBS wash to reveal DNA. Pictures were generated using a Zeiss AxioObserver microscope equipped with an Apotome module and the Axiovision software. Imaging parameters were set to identical values across samples.
4.11. Binding of TRIM5αRh to HIV-1 capsid-nucleocapsid (CA-NC) complexes
HIV-1CA-NC was expressed and purified as previously described [39, 41] and further used for the binding assay. HEK293T cells were transfected with plasmids expressing either WT or V435K-I436K FLAG-tagged TRIM5αRh. Two days post transfection, cells were lysed as previously described [39]. CA-NC complexes assembled in vitro were incubated with cell lysates for 1 h at room temperature. The mixture was then applied to a 70% sucrose cushion and centrifuged at 100,000 g for 1 h at 4 °C. Following centrifugation, the supernatant was removed and pellet was resuspended in protein sample buffer. Western blotting was performed to detect TRIM5αRh (anti-FLAG antibody, Cell Signaling) and HIV-1CA-NC (anti-CAp24 antibody, Immunodiagnostics).
4.12. Phylogenetic analysis
Primate gene sequences for TRIM5 were obtained from GenBank. TRIM5 exon 8 was sequenced from 5 additional primate species (bonobo, pileated gibbon, agile gibbon, yellow-cheeked gibbon and black mangabey). Primary and immortalized primate cell lines were grown and genomic DNA was harvested (detailed description of cell lines can be found in Demogines et al. [78]). TRIM5 sequence was amplified by PCR using the following primer pairs: 5′-CTCCTTCCAAGACACACATAACTTACCC-3′ with 5′-AAGAGGTGCTGTACAGAAGGGGC-3′. The PCR fragments were then sequenced using the PCR primers listed above or the following primers: 5′-GCTACTGGGTTATA
GGGTTAGAGGAAGG-3′ or 5′-CTCCTTATAACTTCTAAACAAGGTTCCTCCC-3′. Primate TRIM5 exon 8 gene sequences have been deposited in GenBank (accession numbers: KJ701422-KJ701426).
Declarations
Author contribution statement
Marie-Édith Nepveu-Traversy: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Ann Demogines: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
Thomas Fricke, Mélodie B. Plourde, Kathleen Riopel, Maxime Veillette: Performed the experiments.
Felipe Diaz-Griffero, Sara L. Sawyer, Lionel Berthoux: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by the Canadian Institutes of Health Research (MOP-102712, Lionel Berthoux), and National Institute of Health (R01-GM-093086, Sara L. Sawyer; R01-AI-087390, Thomas Fricke and Felipe Diaz-Griffero). Ann Demogines was supported by an American Cancer Society Postdoctoral Fellowship. Maxime Veillette was supported by FRQS Master's Training Award #23560.
Conflict of interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We are grateful to Natacha Mérindol and Paulina Pawlica for a critical reading of this manuscript, and Owen Pornillos for a fruitful discussion. We thank Michael Emerman and Ted Dawson for sharing reagents.
References
- 1.Stremlau M., Owens C.M., Perron M.J., Kiessling M., Autissier P., Sodroski J. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 2.Towers G.J. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 2007;4:40. doi: 10.1186/1742-4690-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sayah D.M., Sokolskaja E., Berthoux L., Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 4.Merindol N., Berthoux L. Restriction Factors in HIV-1 Disease Progression. Curr. HIV Res. 2015;13:448–461. doi: 10.2174/1570162x13666150608104412. [DOI] [PubMed] [Google Scholar]
- 5.Sebastian S., Luban J. TRIM5α selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Micale L., Chaignat E., Fusco C., Reymond A., Merla G. The tripartite motif: structure and function. Adv. Exp. Med. Biol. 2012;770:11–25. [PubMed] [Google Scholar]
- 7.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., Guffanti A., Minucci S., Pelicci P.G., Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Cruz A.A., Babon J.J., Norton R.S., Nicola N.A., Nicholson S.E. Structure and function of the SPRY/B30.2 domain proteins involved in innate immunity. Protein Sci.: a publication of the Protein Society. 2013;22:1–10. doi: 10.1002/pro.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song B., Gold B., O'Huigin C., Javanbakht H., Li X., Stremlau M., Winkler C., Dean M., Sodroski J. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohkura S., Yap M.W., Sheldon T., Stoye J.P. All three variable regions of the TRIM5α B30.2 domain can contribute to the specificity of retrovirus restriction. J. Virol. 2006;80:8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yap M.W., Nisole S., Lynch C., Stoye J.P. Trim5α protein restricts both HIV-1 and murine leukemia virus. P. Natl. Acad. Sci. USA. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz-Griffero F., Qin X.R., Hayashi F., Kigawa T., Finzi A., Sarnak Z., Lienlaf M., Yokoyama S., Sodroski J. A B-box 2 surface patch important for TRIM5α self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 2009;83:10737–10751. doi: 10.1128/JVI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Javanbakht H., Yuan W., Yeung D.F., Song B., Diaz-Griffero F., Li Y., Li X., Stremlau M., Sodroski J. Characterization of TRIM5α trimerization and its contribution to human immunodeficiency virus capsid binding. Virology. 2006;353:234–246. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Campbell E.M., Dodding M.P., Yap M.W., Wu X., Gallois-Montbrun S., Malim M.H., Stoye J.P., Hope T.J. TRIM5α cytoplasmic bodies are highly dynamic structures. Mol. Biol. cell. 2007;18:2102–2111. doi: 10.1091/mbc.E06-12-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz-Griffero F., Li X., Javanbakht H., Song B., Welikala S., Stremlau M., Sodroski J. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349:300–315. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 16.Stremlau M., Perron M., Lee M., Li Y., Song B., Javanbakht H., Diaz-Griffero F., Anderson D.J., Sundquist W.I., Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. P. Natl. Acad. Sci. USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Black L.R., Aiken C. TRIM5α disrupts the structure of assembled HIV-1 capsid complexes in vitro. J. Virol. 2010;84:6564–6569. doi: 10.1128/JVI.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perron M.J., Stremlau M., Lee M., Javanbakht H., Song B., Sodroski J. The human TRIM5α restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J. Virol. 2007;81:2138–2148. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganser-Pornillos B.K., Chandrasekaran V., Pornillos O., Sodroski J.G., Sundquist W.I., Yeager M. Hexagonal assembly of a restricting TRIM5α protein. P. Natl. Acad. Sci. USA. 2011;108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutluay S.B., Perez-Caballero D., Bieniasz P.D. Fates of Retroviral Core Components during Unrestricted and TRIM5-Restricted Infection. PLoS Pathog. 2013;9:e1003214. doi: 10.1371/journal.ppat.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukic Z., Hausmann S., Sebastian S., Rucci J., Sastri J., Robia S.L., Luban J., Campbell E.M. TRIM5α associates with proteasomal subunits in cells while in complex with HIV-1 virions. Retrovirology. 2011;8:93. doi: 10.1186/1742-4690-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rold C.J., Aiken C. Proteasomal degradation of TRIM5α during retrovirus restriction. PLoS Pathog. 2008;4:e1000074. doi: 10.1371/journal.ppat.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell E.M., Perez O., Anderson J.L., Hope T.J. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5α. J. Cell Biol. 2008;180:549–561. doi: 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson J.L., Campbell E.M., Wu X., Vandegraaff N., Engelman A., Hope T.J. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J. Virol. 2006;80:9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tareen S.U., Emerman M. Human Trim5α has additional activities that are uncoupled from retroviral capsid recognition. Virology. 2011;409:113–120. doi: 10.1016/j.virol.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pertel T., Hausmann S., Morger D., Zuger S., Guerra J., Lascano J., Reinhard C., Santoni F.A., Uchil P.D., Chatel L., Bisiaux A., Albert M.L., Strambio-De-Castillia C., Mothes W., Pizzato M., Grutter M.G., Luban J. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fletcher A.J., Christensen D.E., Nelson C., Tan C.P., Schaller T., Lehner P.J., Sundquist W.I., Towers G.J. TRIM5α requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription. EMBO J. 2015;34:2078–2095. doi: 10.15252/embj.201490361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Wijk S.J., de Vries S.J., Kemmeren P., Huang A., Boelens R., Bonvin A.M., Timmers H.T. A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol. Syst. Biol. 2009;5:295. doi: 10.1038/msb.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uchil P.D., Hinz A., Siegel S., Coenen-Stass A., Pertel T., Luban J., Mothes W. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J. Virol. 2013;87:257–272. doi: 10.1128/JVI.01804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nepveu-Traversy M.E., Berthoux L. The conserved sumoylation consensus site in TRIM5α modulates its immune activation functions. Virus Res. 2014;184C:30–38. doi: 10.1016/j.virusres.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Jung U., Urak K., Veillette M., Nepveu-Traversy M.E., Pham Q.T., Hamel S., Rossi J.J., Berthoux L. Preclinical Assessment of Mutant Human TRIM5α as an Anti-HIV-1 Transgene. Hum. Gene Ther. 2015;26:664–679. doi: 10.1089/hum.2015.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su H.L., Li S.S. Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene. 2002;296:65–73. doi: 10.1016/s0378-1119(02)00843-0. [DOI] [PubMed] [Google Scholar]
- 33.Hay R.T. Decoding the SUMO signal. Biochem. Soc. T. 2013;41:463–473. doi: 10.1042/BST20130015. [DOI] [PubMed] [Google Scholar]
- 34.Namanja A.T., Li Y.J., Su Y., Wong S., Lu J., Colson L.T., Wu C., Li S.S., Chen Y. Insights into high affinity small ubiquitin-like modifier (SUMO) recognition by SUMO-interacting motifs (SIMs) revealed by a combination of NMR and peptide array analysis. J. Biol. Chem. 2012;287:3231–3240. doi: 10.1074/jbc.M111.293118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sriramachandran A.M., Dohmen R.J. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta. 2014;1843:75–85. doi: 10.1016/j.bbamcr.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 36.Arriagada G., Muntean L.N., Goff S.P. SUMO-interacting motifs of human TRIM5α are important for antiviral activity. PLoS Pathog. 2011;7:e1002019. doi: 10.1371/journal.ppat.1002019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutrieux J., Portilho D.M., Arhel N.J., Hazan U., Nisole S. TRIM5α is a SUMO substrate. Retrovirology. 2015;12:28. doi: 10.1186/s12977-015-0155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukic Z., Goff S.P., Campbell E.M., Arriagada G. Role of SUMO-1 and SUMO interacting motifs in rhesus TRIM5α-mediated restriction. Retrovirology. 2013;10:10. doi: 10.1186/1742-4690-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brandariz-Nunez A., Roa A., Valle-Casuso J.C., Biris N., Ivanov D., Diaz-Griffero F. Contribution of SUMO-interacting motifs and SUMOylation to the antiretroviral properties of TRIM5α. Virology. 2013;435:463–471. doi: 10.1016/j.virol.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., Liu Z., Zhao Y., Xue Y., Ren J. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;42:W325–330. doi: 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lienlaf M., Hayashi F., Di Nunzio F., Tochio N., Kigawa T., Yokoyama S., Diaz-Griffero F. Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5α(rh): structure of the RING domain of TRIM5α. J. Virol. 2011;85:8725–8737. doi: 10.1128/JVI.00497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veillette M., Bichel K., Pawlica P., Freund S.M., Plourde M.B., Pham Q.T., Reyes-Moreno C., James L.C., Berthoux L. The V86M mutation in HIV-1 capsid confers resistance to TRIM5α by abrogation of cyclophilin A-dependent restriction and enhancement of viral nuclear import. Retrovirology. 2013;10:25. doi: 10.1186/1742-4690-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonald D., Vodicka M.A., Lucero G., Svitkina T.M., Borisy G.G., Emerman M., Hope T.J. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuertes G., Villarroya A., Knecht E. Role of proteasomes in the degradation of short-lived proteins in human fibroblasts under various growth conditions. Int. J. Biochem. Cell B. 2003;35:651–664. doi: 10.1016/s1357-2725(02)00382-5. [DOI] [PubMed] [Google Scholar]
- 46.Ding W.X., Yin X.M. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 47.Diaz-Griffero F., Gallo D.E., Hope T.J., Sodroski J. Trafficking of some old world primate TRIM5α proteins through the nucleus. Retrovirology. 2011;8:38. doi: 10.1186/1742-4690-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E.P., Wolff B., Yoshida M., Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. P. Natl. Acad. Sci. USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nepveu-Traversy M.E., Berube J., Berthoux L. TRIM5α and TRIMCyp form apparent hexamers and their multimeric state is not affected by exposure to restriction-sensitive viruses or by treatment with pharmacological inhibitors. Retrovirology. 2009;6:100. doi: 10.1186/1742-4690-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mische C.C., Javanbakht H., Song B., Diaz-Griffero F., Stremlau M., Strack B., Si Z., Sodroski J. Retroviral restriction factor TRIM5α is a trimer. J. Virol. 2005;79:14446–14450. doi: 10.1128/JVI.79.22.14446-14450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 52.Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z.J. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 53.Adhikari A., Xu M., Chen Z.J. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene. 2007;26:3214–3226. doi: 10.1038/sj.onc.1210413. [DOI] [PubMed] [Google Scholar]
- 54.Kawai T., Akira S. Signaling to NF-κB by Toll-like receptors. Trends Mol. Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 55.Wang H., Matsuzawa A., Brown S.A., Zhou J., Guy C.S., Tseng P.H., Forbes K., Nicholson T.P., Sheppard P.W., Hacker H., Karin M., Vignali D.A. Analysis of nondegradative protein ubiquitylation with a monoclonal antibody specific for lysine-63-linked polyubiquitin. P. Natl. Acad. Sci. USA. 2008;105:20197–20202. doi: 10.1073/pnas.0810461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim K.L., Chew K.C., Tan J.M., Wang C., Chung K.K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C.A., Dawson V.L., Dawson T.M. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci.: the official journal of the Society for Neuroscience. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geoffroy M.C., Chelbi-Alix M.K. Role of promyelocytic leukemia protein in host antiviral defense. J. Interf. Cytok. Res.: the official journal of the International Society for Interferon and Cytokine Research. 2011;31:145–158. doi: 10.1089/jir.2010.0111. [DOI] [PubMed] [Google Scholar]
- 58.Muller S., Matunis M.J., Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldschmidt V., Ciuffi A., Ortiz M., Brawand D., Munoz M., Kaessmann H., Telenti A. Antiretroviral activity of ancestral TRIM5α. J. Virol. 2008;82:2089–2096. doi: 10.1128/JVI.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldstone D.C., Walker P.A., Calder L.J., Coombs P.J., Kirkpatrick J., Ball N.J., Hilditch L., Yap M.W., Rosenthal P.B., Stoye J.P., Taylor I.A. Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. P. Natl. Acad. Sci. USA. 2014;111:9609–9614. doi: 10.1073/pnas.1402448111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanchez J.G., Okreglicka K., Chandrasekaran V., Welker J.M., Sundquist W.I., Pornillos O. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. P. Natl. Acad. Sci. USA. 2014;111:2494–2499. doi: 10.1073/pnas.1318962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yueh A., Leung J., Bhattacharyya S., Perrone L.A., de los Santos K., Pu S.Y., Goff S.P. Interaction of moloney murine leukemia virus capsid with Ubc9 and PIASy mediates SUMO-1 addition required early in infection. J. Virol. 2006;80:342–352. doi: 10.1128/JVI.80.1.342-352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kovalskyy D.B., Ivanov D.N. Recognition of the HIV capsid by the TRIM5α restriction factor is mediated by a subset of pre-existing conformations of the TRIM5α SPRY domain. Biochemistry. 2014;53:1466–1476. doi: 10.1021/bi4014962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang H., Ji X., Zhao G., Ning J., Zhao Q., Aiken C., Gronenborn A.M., Zhang P., Xiong Y. Structural insight into HIV-1 capsid recognition by rhesus TRIM5α. P. Natl. Acad. Sci. USA. 2012;109:18372–18377. doi: 10.1073/pnas.1210903109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colombo R., Boggio R., Seiser C., Draetta G.F., Chiocca S. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 2002;3:1062–1068. doi: 10.1093/embo-reports/kvf213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boggio R., Chiocca S. Gam1 and the SUMO pathway. Cell Cycle. 2005;4:533–535. doi: 10.4161/cc.4.4.1605. [DOI] [PubMed] [Google Scholar]
- 67.Wang X., Jiang J., Lu Y., Shi G., Liu R., Cao Y. TAB2, an important upstream adaptor of interleukin-1 signaling pathway, is subject to SUMOylation. Mol. Cell Biochem. 2014;385:69–77. doi: 10.1007/s11010-013-1815-3. [DOI] [PubMed] [Google Scholar]
- 68.Sebban H., Yamaoka S., Courtois G. Posttranslational modifications of NEMO and its partners in NF-κB signaling. Trends Cell Biol. 2006;16:569–577. doi: 10.1016/j.tcb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 69.Desterro J.M., Rodriguez M.S., Hay R.T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 70.Dietrich E.A., Brennan G., Ferguson B., Wiseman R.W., O'Connor D., Hu S.L. Variable prevalence and functional diversity of the antiretroviral restriction factor TRIMCyp in Macaca fascicularis. J. Virol. 2011;85:9956–9963. doi: 10.1128/JVI.00097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu C.Q., Na L., Lv X.L., Liu J.D., Liu X.M., Ji F., Zheng Y.H., Du H.L., Kong X.G., Zhou J.H. The TRIMCyp genotype in four species of macaques in China. Immunogenetics. 2013;65:185–193. doi: 10.1007/s00251-012-0670-9. [DOI] [PubMed] [Google Scholar]
- 72.Bérubé J., Bouchard A., Berthoux L. Both TRIM5α and TRIMCyp have only weak antiviral activity in canine D17 cells. Retrovirology. 2007;4:68. doi: 10.1186/1742-4690-4-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sebastian S., Sokolskaja E., Luban J. Arsenic counteracts human immunodeficiency virus type 1 restriction by various TRIM5 orthologues in a cell type-dependent manner. J. Virol. 2006;80:2051–2054. doi: 10.1128/JVI.80.4.2051-2054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Naviaux R.K., Costanzi E., Haas M., Verma I.M. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zufferey R., Nagy D., Mandel R.J., Naldini L., Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 76.Berthoux L., Sebastian S., Sokolskaja E., Luban J. Lv1 inhibition of human immunodeficiency virus type 1 is counteracted by factors that stimulate synthesis or nuclear translocation of viral cDNA. J. Virol. 2004;78:11739–11750. doi: 10.1128/JVI.78.21.11739-11750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pham Q.T., Bouchard A., Grutter M.G., Berthoux L. Generation of human TRIM5α mutants with high HIV-1 restriction activity. Gene Ther. 2010;17:859–871. doi: 10.1038/gt.2010.40. [DOI] [PubMed] [Google Scholar]
- 78.Demogines A., Truong K.A., Sawyer S.L. Species-specific features of DARC, the primate receptor for Plasmodium vivax and Plasmodium knowlesi. Mol. Biol. Evol. 2012;29:445–449. doi: 10.1093/molbev/msr204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]