Abstract
Patterns of DNA methylation (DNAm) that track with aging have been identified. However, the relevance of these patterns for aging outcomes remains unclear. Longitudinal epigenome-wide DNAm information was obtained from the InCHIANTI study, a large representative European population. DNAm was evaluated using the Illumina HumanMethylation450 array on blood samples collected at baseline and 9-year follow-up: observations from 499 participants with paired longitudinal blood sample and information on differential blood count were included in analyses. A total of 56,579 markers were significantly associated with age in cross-sectional analysis of DNAm at year 9, 31,252 markers were changed significantly over the 9-year follow-up, and 16,987 markers were both cross-sectionally associated with age and significantly changed over time. Rates of change at 76 markers and year 9 level of DNAm at 88 markers were identified as strongly associated with mortality in Cox proportional hazard models adjusted for age and relevant covariates (mean follow-up time 4.4 years). Less than 0.05% of markers associated with age or that changed over time were also associated with mortality after adjusting for chronological age. Although the influence of DNAm on health and longevity remains unclear, these findings confirm that aging is associated cross-sectionally and longitudinally with robust and consistent patterns of methylation change.
Keywords: Epigenome-wide DNA methylation, Chronological age, Survival
Age is the strongest risk factor for many chronic diseases, as well as physical and cognitive impairment in adults. Understanding the mechanisms that underlie the aging process could improve health in later life beyond the capacity of disease-specific treatments. A recent landmark paper outlined 9 biological mechanisms that, hypothetically, control aging and its consequences, including age-associated epigenetic modifications (1). Yet, whether these mechanisms play a role in the aging of complex organisms or contribute to the heterogeneity in the phenotypic manifestation of chronological aging is still not known.
The success of genetic studies that have addressed aging from the perspective of longevity has been limited: with APOE and FOXO3 as the only genes consistently associated with longer life (2). Studies of the epigenome may be more fruitful. In theory, epigenetic modifications as transcriptional regulators and points of integration for multiple biological signals (3) represent a tangible juncture where genetics and life course exposures contribute to the variability in the physical manifestation of aging.
DNA methylation (DNAm), the addition of a methyl group to cytosine at CpG dinucleotides, is the most commonly studied epigenetic marker. Family and twin studies have indicated that changes in DNAm with age may be influenced by both genetic and environmental factors (4–6). Characteristic methylation patterns have been observed in cancer, autoimmune diseases, as well as in other common chronic diseases (7–9). These observations suggest that differences in methylation could explain part of the wide variability of health and longevity experience by aging individuals.
Cross-sectional analyses of genome-wide DNAm have shown age-associated methylation differences in multiple cohorts and tissues (10–12), and these associations are robust enough for the development of methylation scores that closely predict chronological aging (11,13). However, the cross-sectional nature of these studies makes it impossible to differentiate changes in methylation with aging from those induced by secular trends in exposures across age cohorts.
Here, we use data from a large representative population dispersed over a wide age range to describe rate of change in genome-wide methylation over a 9-year follow-up. We contrast markers that change longitudinally in the same individuals with markers that are associated with age in cross-sectional analyses. Our primary aim is to identify sites where variability in DNAm may be relevant to aging processes. Subsequently, we verify whether independent of age, the level of DNAm, or change in DNAm predicts longevity over a 4.5-year follow-up period.
Methods
Study Population
DNAm was evaluated in samples from participants of the InCHIANTI study at two time points 9 years apart. InCHIANTI is a population-based prospective cohort study of residents from two areas in the Chianti region of Tuscany, Italy. Study participants were enrolled between 1998 and 2000 and were followed at 3-year intervals for 9 years. Selection of study participants and data collection procedures have been previously described (14); a brief summary and criteria specific to the present analyses are described later. Overall, 1,326 participants donated a blood sample at baseline (1998–2000) and, of these, 784 also donated a blood sample at the 9-year follow-up (2007–2009). Genome-wide DNAm was assayed on DNA samples corresponding to participants with sufficient DNA at both visits. The study population for the present analysis includes individuals with DNAm data meeting quality control criteria and complete covariate information at both baseline and 9-year follow-up (n = 499). InCHIANTI protocols were approved by the Instituto Nazionale Riposo e Cura Anziani institutional review board in Italy and study participants provided informed consent.
DNA Collection and Genome-Wide Methylation Scan
Genomic DNA was extracted from buffy coat samples using an AutoGen Flex and quantified on a Nanodrop1000 spectrophotometer prior to bisulfite conversion. Genomic DNA was bisulfite converted using Zymo EZ-96 DNA Methylation Kit (Zymo Research Corp., Irvine, CA) as per the manufacturer’s protocol. CpG methylation status of 485,577 CpG sites was determined using the Illumina Infinium HumanMethylation450 BeadChip (Illumina Inc., San Diego, CA) as per the manufacturer’s protocol and as previously described (15). Initial data analysis was performed using GenomeStudio 2011.1 (Model M Version 1.9.0, Illumina Inc.). Threshold call rate for inclusion of samples was 95%. Quality control of sample handling included comparison of clinically reported sex versus sex of the same samples determined by analysis of methylation levels of CpG sites on the X chromosome (16).
Quality filtering and normalization was performed on the pooled set of 1,022 samples (baseline and 9-year follow-up) using the DASEN method in the R package “wateRmelon” (17). Markers were removed if the bead count was less than 3 in ≥5% of samples (n CpG = 251). Samples and markers were also excluded if ≥5% of detection p values were greater than .01 (n sample = 3, n CpG = 1,893). A background adjustment and quantile normalization were applied to the filtered data set; the selected method normalizes both methylated and unmethylated probes as well as type I and II assays (the 450k array includes both paired probe and single probe assay designs) separately. Locations were annotated using the FDb.InfiniumMethylation.hg19 database. Methylation markers on the X and Y chromosome, as well as markers with potentially cross-reactive probes and probes that may be polymorphic in European populations (allele frequency ≥ .01) (18) were excluded from analyses. After filtering, 429,527 markers in 1,019 participant samples remained. Nine hundred and ninety-eight paired samples, representing 499 participants, with complete white blood cell differential count were included in analyses.
Other Independent Variables
Demographic and health behavior information were obtained through a structured interview and anthropometric measurements and fasting blood samples were collected using standardized protocols during InCHIANTI study visits. White blood cell differential count was assessed on ethylenediaminetetraacetic acid anticoagulated whole blood using a Coulter Counter (LH 750 Hematology Autoanalyzer, Beckman Coulter Inc., Brea, CA) and expressed as percentages of neutrophils, lymphocytes, monocytes, eosinophils, and basophils. DNA samples were processed in 13 experimental batches, and variability by batch was represented by covariates as described later. Vital status and dates of death were confirmed through a systematic search of municipality records through June 2013. IL6, gp130, sIL6R, IL18, TNFαR1, and TNFαR2 were measured using ELISA with commercial assays (R&D Systems, Inc., Minneapolis, MN); TNFα was measured using a multiplex panel (Millipore, Billerica, MA). C-reactive protein (CRP) was measured using enzyme-linked immunosorbent assay and a colorimetric competitive immunoassay that uses purified protein and polyclonal antibodies.
Statistical Analysis
Analyses were conducted on residuals after adjustment for experimental batch and white blood cell differential count in linear regression models including all samples (n = 998).
The associations between age and methylation marker residuals at 9-year follow-up were estimated in single linear regression models. The direction and magnitude of age coefficients from these models and the cross-sectional estimates of difference in methylation with a 1-year difference in age were qualitatively compared with mean rates of change in DNAm estimated from DNAm measured at baseline and year 9. Rates of change were calculated as: (residual9-year follow-up − residualbaseline)/years. Markers with statistically significant longitudinal change in methylation were identified based upon one-sample t tests indicating nonzero mean rates of change. We identified the set of markers that were both significantly correlated with cross-sectional age and significantly changed between baseline and the 9-year follow-up (Bonferroni corrected p value ~ 1.16×10−7).
In addition to evaluation of the rate of change in methylation, the associations of year 9 DNAm residual and of rate of change of DNAm over 9 years with subsequent mortality were estimated in Cox proportional hazard models for all markers passing filtering criteria. Subsequent to the 9-year follow-up visit, study participants were followed for an average of 4.4 years, during this period 79 deaths were observed. In these analyses, DNAm residuals and rates of change were z-score transformed and models were adjusted for age, sex, and study site. Because some methylation markers changed across both time and age in the primary analyses and age is the strongest known predictor of mortality age was treated as a potential confounder of the relationship between DNAm and mortality. Both the magnitude of the hazard ratio (HR) and the statistical significance of the p value were considered in evaluating survival analyses.
Because genes in close proximity to the selected sites appeared to relate to inflammatory pathways, correlations between rates of change in methylation at these sites and markers of inflammation at 9-year follow-up were also evaluated. Associations with markers of inflammation were estimated in multivariate linear regression models adjusted for age, sex, and study site. Rates of change were z-score transformed and inflammatory markers were log transformed as necessary. All analyses were performed using R 3.0 (R Foundation for Statistical Computing, Vienna, Austria).
Results
The study population included 499 InCHIANTI participants, 45% men, mean age at 9-year follow-up was 71.9 (range: 30–100; Table 1). The mean time between baseline sample collection and 9-year follow-up was 9.1 years. The mean body mass index was 26.9kg/m2 (SD: 4.2) and 56.7% had no history of smoking.
Table 1.
Characteristics of InCHIANTI Study Participants Included in the Study Sample (n = 499) at Year 9
| Characteristic | Mean (SD) or n (%) |
|---|---|
| Male | 225 (45.1) |
| Age, y* | 71.9 (30,100) |
| BMI, kg/m2† | 26.9 (4.2) |
| Smoking status | |
| Former | 166 (33.3) |
| Current | 50 (10.0) |
| Cancer‡ | 50 (10.0) |
| Neutrophil, % | 57.1 (8.8) |
| Lymphocyte, % | 31.3 (8.5) |
| Monocyte, % | 8.0 (2.1) |
| Eosinophil, % | 3.1 (1.9) |
| Basophil, % | 0.5 (0.2) |
| Time between visits, y | 9.1 (0.2) |
| Follow-up time, y | 4.4 (1.0) |
Notes: *Mean (range).
† n = 491.
‡Self-report of cancer at any study visit.
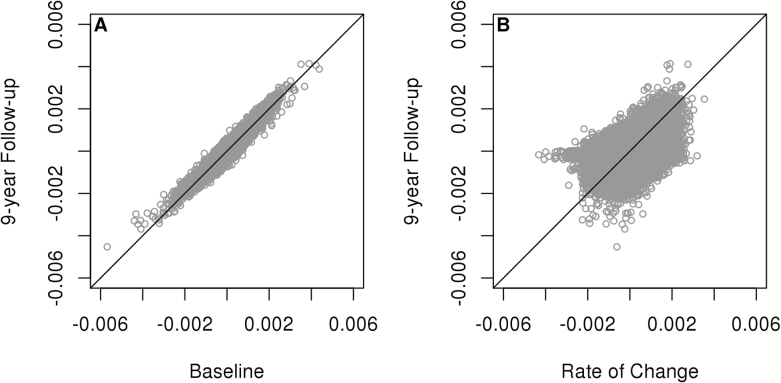
Age coefficients in single linear regression models at baseline and the 9-year follow-up visit were highly correlated (r = .957; Figure 1A). After adjustment for confounding factors and multiple testing, as per methods, methylation in 56,579 markers was significantly associated with age at the year 9 visit. The mean rate of change was negative at 50.6% of evaluated DNAm markers, the average across all mean rates of change was −9.12×10−6 proportion methylated per year. Overall, statistically significant changes in methylation were observed at 31,252 CpG. Rates of change of greatest positive and negative magnitude were observed for markers cg27526665 [THRB] (mean rate: 0.0035, SD: 0.0081) and cg18036763 [PHF21B] (mean rate: −0.0043, SD: 0.0132). The correlation between estimated rate of change and cross-sectional age coefficient was moderate (r = .576; Figure 1B). Thirty percent (16,987) of CpG sites where DNAm was significantly associated with age in cross-sectional analyses also demonstrated significant mean rates of change over the follow-up. However, the direction of change over time was consistent with the direction of association with cross-sectional age for 69.2% of markers.
Figure 1.
(A) Age coefficients from linear regression models estimating the association of methylation at each marker with age at baseline vs coefficients estimated in samples corresponding to 9-year follow-up. (B) Estimated mean rate of change in methylation between baseline and 9-year follow-up for each marker vs age coefficient from single linear regression models estimating the association of methylation at each marker with age at 9-year follow-up.
Adjusting for age at year 9, sex, and study site, rate of change of five markers, cg03735531 [PSMB9] (HR = 0.74; p = 8.33×10−8), cg16197272 [FAM120B] (HR = 0.75; p = 3.25×10−8), cg19409060 [SRCIN1] (HR = 0.70; p = 7.78×10−9), cg24437429 [ABR] (HR = 0.76; p = 6.92×10−8), and cg26833395 [HM13] (HR = 0.74; p = 4.02×10−8), was associated with survival in Cox proportional hazard models at a Bonferroni corrected p value threshold (p ~ 1.16×10−7). The most extreme HR was observed for marker cg01646019 (HR = 2.29; p = .002). There were 76 markers with p value less than .001 and moderately high HRs in survival analyses (; Table 2). In similar analyses, the year 9 level of DNAm, at 88 markers predicted mortality with p values less than .001 and moderate HRs (Supplementary Table 1). Four DNAm markers, namely cg06281205 [FANCL], cg12379145 [PRDM2], cg18741439 [CSGALNACT1], and cg23015983 [OSTM1] predicted mortality when measured at year 9 and also their 9-year change predicted mortality.
Table 2.
Cox Proportional Hazard Models Estimating the Association Between the Estimated Rate of Change in DNAm Markers of Interest and Mortality After 9-Year Follow-Up (n = 499, events = 79) Ordered by the Magnitude of the Hazard Ratio (HR)
| Marker | Chr | Position | Rate of Change (SD) | HR (95% CI) | Gene* |
|---|---|---|---|---|---|
| cg14575484 | 2 | 217557616 | 0.0002 (0.0017) | 0.60 (0.45, 0.78) | IGFBP5 |
| cg18120323 | 1 | 1978732 | 0.0008 (0.0053) | 1.67(1.34, 2.10) | PRKCZ |
| cg03402235 | 15 | 42749336 | −0.0004 (0.0030) | 1.66 (1.29, 2.15) | ZNF106 |
| cg16197857 | 18 | 75402282 | −0.00003 (0.0017) | 0.60 (0.47, 0.77) | |
| cg27635330 | 20 | 57020935 | 0.0001 (0.0019) | 0.61 (0.49, 0.77) | VAPB |
| cg16781880 | 16 | 58329007 | −0.0003 (0.0019) | 0.61 (0.48, 0.79) | PRSS54 |
| cg00292432 | 12 | 3393872 | −0.0001 (0.0028) | 1.62 (1.27, 2.06) | TSPAN9 |
| cg03623394 | 4 | 1005876 | 0.0002 (0.0023) | 0.62 (0.50, 0.77) | IDUA, FGFRL1 |
| cg03576411 | 14 | 99172002 | −0.0001 (0.0032) | 1.61 (1.25, 2.05) | C14orf177 |
| cg21158502 | 5 | 74348187 | 0.0004 (0.0033) | 1.60 (1.28, 2.01) | |
| cg24319508 | 6 | 394966 | −0.0006 (0.0030) | 1.60 (1.28, 2.00) | IRF4 |
| cg01723148 | 12 | 58021295 | 0.0002 (0.0018) | 1.60 (1.27, 2.00) | BC073932, SLC26A10, B4GALNT1 |
| cg17835356 | 8 | 41387960 | −0.0006 (0.0033) | 1.59 (1.26, 2.01) | GINS4 |
| cg07922411 | 16 | 34429609 | 0.00003 (0.0030) | 1.59 (1.25, 2.01) | |
| cg14403741 | 12 | 126465336 | −0.0001 (0.0020) | 0.63 (0.51, 0.79) | LINC00939 |
| cg19733534 | 5 | 1841548 | 0.0003 (0.0035) | 1.58 (1.26, 1.98) | |
| cg18533397 | 1 | 110186005 | −0.0001 (0.0013) | 1.58 (1.24, 2.01) | |
| cg20220255 | 12 | 95228096 | −0.0004 (0.0025) | 0.63 (0.51, 0.78) | KRT19P2 |
| cg26106417 | 4 | 3425381 | 0.00003 (0.0027) | 0.63 (0.49, 0.81) | RGS12 |
| cg11808581 | 1 | 1950725 | 0.0003 (0.0023) | 0.63 (0.50, 0.80) | AK054708, GABRD |
| cg01093854 | 10 | 70362422 | 0.0001 (0.0015) | 0.63 (0.50, 0.81) | TET1 |
| cg27601809 | 11 | 72091020 | 0.0001 (0.0023) | 1.57 (1.24, 1.99) | CLPB |
| cg22927043 | 1 | 11919699 | −0.0008 (0.0038) | 1.57 (1.27, 1.94) | NPPB |
| cg26160180 | 1 | 1822883 | −0.00004 (0.0015) | 0.64 (0.51, 0.81) | GNB1 |
| cg01514859 | 1 | 6557734 | −0.00003 (0.0011) | 1.57 (1.23, 2.00) | PLEKHG5 |
| cg12844117 | 7 | 155275418 | −0.0002 (0.0032) | 1.57 (1.25, 1.96) | |
| cg16054184 | 10 | 78944582 | 0.0002 (0.0024) | 1.56 (1.24, 1.98) | KCNMA1 |
| cg15505642 | 1 | 77836400 | 0.0001 (0.0019) | 0.64 (0.51, 0.80) | AK5 |
| cg26989202 | 8 | 99076837 | 0.0001 (0.0015) | 1.56 (1.31, 1.86) | C8orf47 |
| cg01065605 | 7 | 27702796 | 0.00004 (0.0016) | 0.64 (0.52, 0.79) | HIBADH |
| cg23608075 | 10 | 112272213 | 0.0001 (0.0015) | 0.64 (0.52, 0.80) | DUSP5 |
| cg24197330 | 19 | 47551642 | −0.0001 (0.0014) | 0.64 (0.51, 0.81) | NPAS1,TMEM160 |
| cg18741439 | 8 | 19310134 | 0.0003 (0.0026) | 1.55 (1.25, 1.92) | CSGALNACT1 |
| cg26318321 | 19 | 15446617 | 0.0000002 (0.0033) | 1.55 (1.24, 1.93) | BRD4 |
| cg23015983 | 6 | 108396139 | 0.00003 (0.0014) | 0.65 (0.51, 0.82) | OSTM1 |
| cg03967329 | 6 | 26030329 | 0.0001 (0.0030) | 1.55 (1.23, 1.95) | HIST1H3A, HIST1H4A, HIST1H4B, HIST1H3B, HIST1H2AB |
| cg24259629 | 3 | 45883796 | 0.0002 (0.0016) | 1.55 (1.23, 1.95) | LZTFL1 |
| cg11950982 | 21 | 46269126 | 0.0003 (0.0024) | 1.54 (1.20, 1.99) | PTTG1IP |
| cg19662895 | 14 | 69074455 | 0.0013 (0.0049) | 1.54 (1.25, 1.91) | RAD51B |
| cg25468681 | 1 | 9428106 | −0.0002 (0.0021) | 0.65 (0.51, 0.82) | SPSB1 |
| cg01310875 | 8 | 27938995 | −0.0002 (0.0030) | 1.54 (1.20, 1.98) | NUGGC, ELP3 |
| cg07161062 | 8 | 142204334 | −0.0004 (0.0028) | 1.54 (1.21, 1.96) | DENND3 |
| cg12379145 | 1 | 14030611 | 0.0001 (0.0017) | 1.54 (1.21, 1.95) | PRDM2 |
| cg06765172 | 22 | 36714496 | −0.0003 (0.0023) | 1.54 (1.24, 1.90) | MYH9 |
| cg02330352 | 16 | 20975912 | −0.0003 (0.0023) | 0.65 (0.52, 0.81) | DNAH3 |
| cg06281205 | 2 | 58462331 | 0.0003 (0.0019) | 1.53 (1.24, 1.90) | FANCL |
| cg11308319 | 2 | 240291426 | −0.0006 (0.0036) | 1.53 (1.23, 1.91) | HDAC4 |
| cg13665021 | 7 | 1717324 | −0.0003 (0.0031) | 0.65 (0.53, 0.81) | |
| cg08529529 | 13 | 31309799 | 0.0002 (0.0033) | 1.53 (1.23, 1.90) | ALOX5AP |
| cg23515619 | 17 | 61623700 | 0.00001 (0.0017) | 0.66 (0.52, 0.83) | KCNH6, DCAF7 |
| cg13009365 | 22 | 24093455 | −0.0001 (0.0014) | 0.66 (0.52, 0.82) | ZNF70, VPREB3 |
| cg27224642 | 2 | 39665105 | −0.0001 (0.0021) | 1.52 (1.24, 1.88) | MAP4K3, LOC728730 |
| cg02898994 | 20 | 2361440 | −0.0001 (0.0015) | 1.52 (1.19, 1.94) | TGM6 |
| cg25649038 | 6 | 6546777 | 0.0001 (0.0014) | 1.52 (1.25, 1.85) | LY86-AS1 |
| cg27257822 | 3 | 184292880 | −0.0012 (0.0033) | 1.52 (1.21, 1.91) | EPHB3 |
| cg09693464 | 7 | 41744949 | −0.0002 (0.0021) | 1.52 (1.20, 1.93) | INHBA, INHBA-AS1 |
| cg23524537 | 14 | 95036219 | −0.0001 (0.0027) | 1.52 (1.20, 1.92) | SERPINA4 |
| cg23324787 | 11 | 36619694 | −0.0004 (0.0037) | 1.51 (1.22, 1.89) | RAG1, RAG2, C11orf74 |
| cg03490711 | 3 | 187459194 | −0.0001 (0.0014) | 1.51 (1.22, 1.88) | LOC100131635, BCL6 |
| cg12484411 | 8 | 110099683 | 0.0001 (0.0025) | 1.51 (1.26, 1.81) | TRHR |
| cg10916429 | 15 | 64799784 | 0.0008 (0.0033) | 1.51 (1.22, 1.87) | ZNF609 |
| cg04373937 | 14 | 30552825 | 0.0003 (0.0022) | 0.66 (0.53, 0.82) | |
| cg16096311 | 1 | 151693261 | 0.0002 (0.0022) | 1.51 (1.20, 1.90) | CELF3, RIIAD1 |
| cg24737067 | 17 | 34432601 | −0.0002 (0.0021) | 0.66 (0.54, 0.81) | CCL4 |
| cg19174658 | 22 | 45018513 | −0.0006 (0.0021) | 0.66 (0.53, 0.83) | LINC00229 |
| cg03976645 | 7 | 16724981 | −0.0003 (0.0036) | 1.51 (1.20, 1.88) | BZW2 |
| cg02772928 | 3 | 18277949 | 0.0002 (0.0024) | 1.50 (1.22, 1.86) | LOC339862 |
| cg07457727 | 8 | 131451983 | 0.0005 (0.0026) | 1.50 (1.19, 1.90) | ASAP1 |
| cg09544380 | 10 | 27703486 | 0.0002 (0.0024) | 0.67 (0.55, 0.80) | PTCHD3 |
| cg05376185 | 1 | 47082869 | −0.0001 (0.0035) | 1.50 (1.20, 1.88) | MKNK1, MOB3C |
| cg13735018 | 6 | 158404121 | −0.0008 (0.0027) | 1.50 (1.18, 1.91) | SYNJ2 |
| cg20585841 | 8 | 102729926 | 0.0006 (0.0034) | 1.50 (1.20, 1.88) | NCALD |
| cg26078407 | 2 | 240225062 | 0.0006 (0.0034) | 1.50 (1.19, 1.89) | HDAC4, MIR4269 |
| cg07735969 | 16 | 68418473 | −0.0001 (0.0030) | 1.50 (1.21, 1.86) | SMPD3 |
| cg16989443 | 1 | 1849195 | 0.0003 (0.0019) | 0.67 (0.53, 0.84) | CALML6, TMEM52, C1orf222 |
| cg16248783 | 2 | 38304146 | 0.0002 (0.0019) | 1.50 (1.23, 1.83) | RMDN2, CYP1B1 |
Notes: CI = confidence interval. Markers were identified based upon p value < .001 and a HR > 3/2 or < 2/3.
*Gene symbols corresponding to transcripts within 10K, bold text indicates the closest gene where more than one transcript.
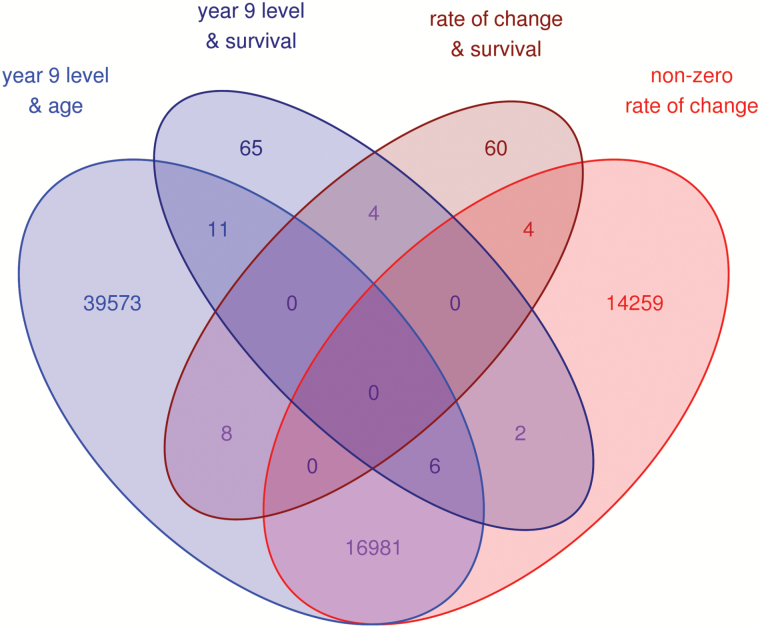
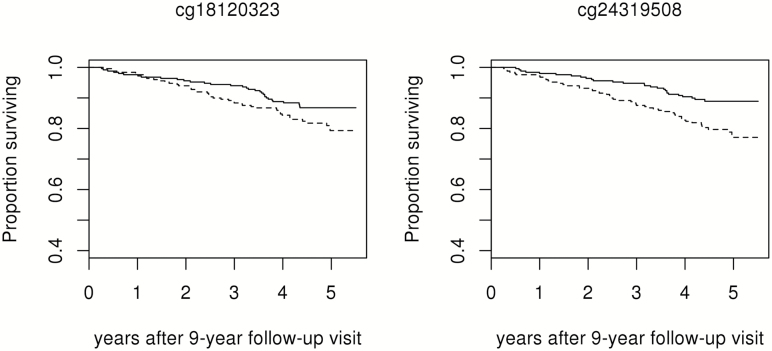
Among the set of 76 markers whose rate of change predicted mortality, 4 also had rates of change consistent with statistically significant longitudinal change and 8 were significantly associated with age in cross-sectional analyses (Figure 2). Ninety-one gene identifiers correspond to transcripts within 10kb of this group of methylation markers (Table 2). Two markers with relatively strong effect estimates and more extreme p values are also of interest based upon transcripts of genes related to inflammatory pathways in close proximity: cg18120323 is approximately 3kb upstream of a transcript variant of PRKCZ on chromosome 1 and cg24319508 is within an exon of IRF4 on chromosome 6 (Figure 3). Because proximity to these genes does not imply a role in inflammation, the association of rate of change at these markers with indicators of inflammation at 9-year follow-up was evaluated. In multivariate linear regression models, cg18120323 was associated with serum levels of gp130, TNFαR1, and TNFαR2 (p < .05; Table 3).
Figure 2.
Overlap between selected markers from survival analyses and markers with statistically significant rates of change or cross-sectional associations with age.
Figure 3.
Estimated survival after 9-year follow-up by age adjusted rate of change in methylation below (solid lines) or above (dashed lines) the median at markers of interest.
Table 3.
Coefficients Estimating the Association Between a Standard Deviation Difference in Rate of Change in Methylation With Markers of Inflammation at 9-Year Follow-Up in Multivariate Linear Regression Models Adjusted for Age, Sex, and Study Site
| Inflammatory Marker | n | Cg18120323 | Cg24319508 | ||
|---|---|---|---|---|---|
| β (SE) | p | β (SE) | p | ||
| log(CRP) | 499 | −0.05 (0.05) | .348 | 0.01 (0.05) | .808 |
| log(IL6) | 476 | 0.04 (0.02) | .122 | 0.03 (0.02) | .237 |
| gp130 | 499 | 3.57 (1.76) | .043 | 0.17 (1.76) | .923 |
| sIL6R | 499 | 0.34 (0.61) | .571 | −0.52 (0.60) | .389 |
| log(IL18) | 499 | 0.003 (0.02) | .865 | −0.01 (0.02) | .585 |
| log(TNFα) | 499 | 0.02 (0.03) | .368 | 0.002 (0.03) | .947 |
| log(TNFαR1) | 499 | 0.03 (0.01) | .005 | 0.01 (0.01) | .577 |
| log(TNFαR2) | 497 | 0.02 (0.01) | .024 | −0.004 (0.01) | .695 |
Note: Bold values indicate p < .05.
Discussion
We assessed genome-wide DNAm at two time points in 499 participants of the InCHIANTI study. We found that the magnitude and direction of coefficients that described cross-sectional relationships between methylation and age were moderately correlated with observed rates of change of methylation in the same marker over a 9-year follow-up. Thirty percent of markers where level of methylation was significantly associated with age also showed significant change over 9-year follow-up. These findings confirm that aging is associated with changes in DNAm and that these changes are robust and predictable. We also found a meaningful number of markers that were cross-sectionally associated with age but did not change over time, suggesting that findings from cross-sectional analysis may reflect a mixture of the effects of time and cohort-specific exposures, transient exposures, or random error.
Previous studies have suggested that composite DNAm measures may be used as indicators of accelerated aging (11). Accordingly, we hypothesized that individuals with larger or smaller than average changes in markers that systematically and significantly change over time would have shorter life, using survival as a proxy measure of the rate of aging. Contrary to this hypothesis, we observed almost no overlap between markers where methylation changes with aging and markers where level or rate of change in methylation appeared consistently associated with mortality within our study sample even though lenient thresholds were used to select markers of interest in analyses of mortality. We acknowledge that the survival analyses lacked substantial power because of the short follow-up and the limited number of death available for this analysis. Thus, is quite possible that important methylation markers associated with mortality remained undetected. Although this possibility should be tested in future large studies with longer follow-up, we propose two additional interpretations for this lack of concordance. DNAm change over time may indicate a loss of control where random drift in DNAm appears directional because the initial state was completely methylated or unmethylated at these sites. Alternatively, given the robustness of variability in methylation across age groups revealed by many studies, it is possible that consistent shifts in patterns of DNAm with time reflect expected change: direct preprogrammed modification of the genome or modification that is a consequence of other perpetual processes that have little or no influence on the development and clinical progression of diseases that are major causes of death in older persons. The true evolutionary scope of this “biological clock” is unclear but would an important objective of future research.
Of note, among the markers identified in survival analyses, we observe markers of DNAm in close proximity to factors that modulate inflammatory pathways. For example, IRF4 (cg24319508) and PRKCZ (cg18120323) were in close proximity to markers where a HR of moderate magnitude and statistical significance was linked to a shift in the rate of change in methylation. IRF4 is a member of the IRF family of transcription factors that play diverse roles in innate and adaptive immune responses (19). IRF4 is expressed only in cells of the immune system and has been implicated in the differentiation and development of several cell types including dendritic, B, plasma, and Th2 cells; a role in Th17 cell differentiation has also been indicated (19,20). IRF4 is also implicated in moderation of pro-inflammatory pathways as a negative regulator of Toll-like receptor signaling, and through a suggested role in suppression of Th2 responses by regulatory T cells (19,21). PRKCZ may also play a role in moderating the immune response, as one of several enzymes that phosphorylate RelA it plays a role in the regulation of NF-κB (22). PRKCZ is a member of the atypical subgroup of the protein kinase C family of serine-threonine kinases (23). It has been proposed that the role of PRKCZ in the immune system extends beyond NF-κB to IL4-STAT6 and Akt signaling pathways (23,24).
Markers of interest were selected from the results of the survival analyses based upon relaxed significance criteria; however, further validation of potential pathways was attempted through evaluation with available biomarkers. Because of many connections between aging and inflammation, we assessed whether these two candidate methylation markers were associated with serum levels of inflammatory markers. We observed a significant association between the rate of change in methylation at cg18120323 and gp130, TNFαR1, and TNFαR2 level at 9-year follow-up, further supporting a role in dysregulation of inflammation a key contributor to aging syndromes and adverse outcomes in older adults (25).
Our study is a first step toward evaluating the role of methylation in biological aging. Limitations of the current study should be addressed in future studies. The study sample was limited and follow-up time for mortality after year 9 was brief: power to detect association with mortality is low in the current study sample. The association of markers with small to moderate effect sizes may not have been recognized and markers identified in this study should be confirmed in larger populations with extended follow-up and/or through meta-analyses. Evaluation of DNA methylation with alternative indicators of biological aging may also help to determine the robustness of our observations. Analyses were limited to a single tissue: the observed changes in methylation may be unique to peripheral blood cells and methylation changes in other tissues may have stronger relationships with mortality. White blood cells also represent a heterogeneous mixture of cells and it was not possible to account for all common subtypes, some of which are known to change with age (eg, naïve T-lymphocytes): there may be residual confounding by cell type. Fuller and more refined measurement of methylation within white blood cells and across different tissues may help to further clarify the relationships of interest. Also, change in DNAm markers was estimated by only two points in time and may be affected by “regression to the mean.” Finally, we recognize that while the commercial array used to evaluate DNAm interrogates methylation across the genome it is far from comprehensive and likely does not capture a substantial portion of change in DNAm that occurs with age.
In spite of the limitations, our study also has unique characteristics that contribute to the current literature. These data confirm longitudinally the existence of a robust pattern of age-associated changes in DNAm. Our findings also contribute to the discussion on whether the pattern of methylation change can be considered a proxy measure of biological aging or used to measure “accelerated aging.” Future studies should continue to investigate whether “age-methylation signatures” are a biomarker of biological aging not only by analyzing their prognostic value for mortality in larger populations with longer follow-up but also exploring their correlation with other proxy measures, such as physical performance, disability, comorbidity, sarcopenia, and cognitive impairment.
Supplementary Material
Please visit the article online at http://gerontologist.oxfordjournals.org/ to view supplementary material.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging. The InCHIANTI study baseline (1998–2000) was supported as a “targeted project” (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336); the InCHIANTI Follow-up 1 (2001–2003) was funded by the U.S. National Institute on Aging (Contracts: N.1-AG-1-1 and N.1-AG-1-2111); the InCHIANTI Follow-ups 2 and 3 studies (2004–2010) were financed by the U.S. National Institute on Aging (Contract: N01-AG-5-0002); supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Baltimore, Maryland.
Supplementary Material
References
- 1. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi:10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brooks-Wilson AR. Genetics of healthy aging and longevity. Hum Genet. 2013;132(12):1323–1338. doi:10.1007/s00439-013-1342-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20(8):350–358. doi:10.1016/j.tig.2004.06.009 [DOI] [PubMed] [Google Scholar]
- 4. Talens RP, Christensen K, Putter H, et al. Epigenetic variation during the adult lifespan: cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell. 2012;11(4):694–703. doi:10.1111/j.1474-9726.2012.00835.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bjornsson HT, Sigurdsson MI, Fallin MD, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299(24):2877–2883. doi:10.1001/jama.299.24.2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102(30):10604–10609. doi:10.1073/pnas.0500398102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu Q. The critical importance of epigenetics in autoimmunity. J Autoimmun. 2013;41:1–5. doi:10.1016/j.jaut.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 8. Baccarelli A, Ghosh S. Environmental exposures, epigenetics and cardiovascular disease. Curr Opin Clin Nutr Metab Care. 2012;15(4):323–329. doi:10.1097/MCO.0b013e328354bf5c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–440. doi:10.1038/nature05919 [DOI] [PubMed] [Google Scholar]
- 10. Heyn H, Li N, Ferreira HJ, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109(26):10522–10527. doi:10.1073/pnas.1120658109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi:10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hernandez DG, Nalls MA, Gibbs JR, et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20(6):1164–1172. doi:10.1093/hmg/ddq561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi:10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferrucci L, Bandinelli S, Benvenuti E, et al. Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc. 2000;48(12):1618–1625. doi:10.1111/j.1532-5415.2000.tb03873.x [DOI] [PubMed] [Google Scholar]
- 15. Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6(5):e1000952. doi:10.1371/journal.pgen.1000952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holly AC, Pilling LC, Hernandez D, et al. Splicing factor 3B1 hypomethylation is associated with altered SF3B1 transcript expression in older humans. Mech Ageing Dev. 2014;135:50–56. doi:10.1016/j.mad.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi:10.1186/1471-2164-14-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. doi:10.4161/epi.23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi:10.1146/annurev.immunol.26.021607.090400 [DOI] [PubMed] [Google Scholar]
- 20. Brüstle A, Heink S, Huber M, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8(9):958–966. doi:10.1038/ni1500 [DOI] [PubMed] [Google Scholar]
- 21. Zheng Y, Chaudhry A, Kas A, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458(7236):351–356. doi:10.1038/nature07674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neumann M, Naumann M. Beyond IkappaBs: alternative regulation of NF-kappaB activity. FASEB J. 2007;21(11):2642–2654. doi:10.1096/fj.06-7615rev [DOI] [PubMed] [Google Scholar]
- 23. Diaz-Meco MT, Moscat J. The atypical PKCs in inflammation: NF-κB and beyond. Immunol Rev. 2012;246(1):154–167. doi:10.1111/j.1600-065X.2012.01093.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moscat J, Rennert P, Diaz-Meco MT. PKCzeta at the crossroad of NF-kappaB and Jak1/Stat6 signaling pathways. Cell Death Differ. 2006;13(5):702–711. doi:10.1038/sj.cdd.4401823 [DOI] [PubMed] [Google Scholar]
- 25. Singh T, Newman AB. Inflammatory markers in population studies of aging. Ageing Res Rev. 2011;10(3):319–329. doi:10.1016/j.arr.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.