Abstract
Although various parts of J. curcas (Jatropha curcas L., Euphorbiaceae) have long been used as traditional folk medicines for their antiviral, analgesic, and/or antidotal efficacies, we are the first to investigate the role of anti-carcinogenicity of isoamericanol A (IAA) from the seed extract. Our results showed that IAA is capable of inhibiting cell proliferation in a dose-dependent manner on the human cancer cell lines of MCF-7, MDA-MB231, HuH-7, and HeLa. Flow cytometry analysis showed IAA significantly induces cell cycle arrest at G2/M on MCF-7 cells. At both protein and mRNA levels examined by western blot and real-time PCR, the results revealed increased expression of BTG2 (B-cell translocation gene 2), p21 (p21WAF1/CIPI), and GADD45A (growth arrest and DNA-damage-inducible, alpha) after IAA treatment, but inversed expression in CDK1 (cyclin-dependent kinase 1) and cyclins B1 and B2. All these effects contribute to G2/M cell cycle arrest. Furthermore, these results coincide with the changes in molecular expressions determined by DNA-microarray analysis. Our findings indicate that IAA has an inhibitory effect on cell proliferation of MCF-7 through cell cycle arrest, giving it great potential as a future therapeutic reagent for cancers.
Keywords: Health sciences, Pharmaceutical science, Plant biology, Cancer treatment, Cancer
1. Introduction
Almost all parts of J. curcas trees have long been used as traditional folk medicine in African and Asian countries for a variety of sicknesses [1, 2]. The latex has been used as an antibacterial agent. Apigenin, vitexin, and isovitexin are contained in the leaves and work well against fever, rheumatic and muscular pains, infection, and malaria [1]. Its leaves and roots are used as antidotes for snake venom [3]. The roots also serve as treatment for sexually transmitted diseases [2]. Its seeds are used in soap oil and biodiesel production, as well as to fight infectious disease [3]. There are some reports that show anticancer activity in vitro by the applications of J. curcas phorbol esters from its kernel meal [4], and curcin from its seeds [5].
In Thailand, J. curcas seed extract is a source of oil for biodiesel energy [6, 7], but its processing involves a great amount of seed waste. It would be worthwhile to find a medical use for residual byproducts. IAA from the seeds of Phytolacca americana was reported to exhibit neurotrophic and antioxidant properties [8, 9]. However, much about its medicinal potential as a cancer treatment has not been investigated. As a goal of this study, neo-lignan IAA ((2R)-3β-(3,4-Dihydroxyphenyl)-6-[(E)-3-hydroxy-1-propenyl]-2,3-dihydro-1,4-benzodioxin-2α-methanol, Fig 1) was tested on the cancer cell lines of MCF-7 (human breast cancer cell line), MDA-MB231 (human breast cancer cell line), HuH-7 (human hepatocellular carcinoma cell line), and HeLa (human cervical cancer cell line), in search of anti-carcinogenic ability. Later, we focused on the study of molecular mechanisms of IAA treatment that cause an inhibitory effect on MCF-7. Our findings shed light on the potential of IAA from the J. curcas extract as a breast cancer treatment.
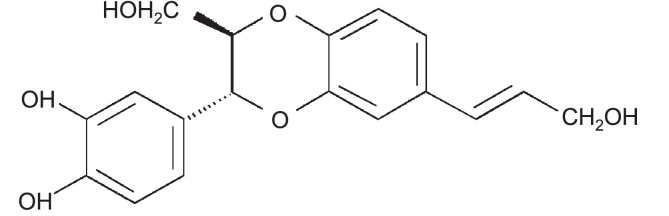
Fig. 1.
Chemical structure of isoamericanol A.
2. Materials and methods
2.1. Cell culture
At 37 °C under a humidified atmosphere of 5% CO2, all tested cells (MCF-7, MDA-MB-231, HuH-7, and HeLa) were cultured in D-MEM (Sigma) with 10% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin (GE Healthcare, Buckinghamshire, UK).
2.2. IAA preparation
IAA from the residues of expeller-pressed J. curcas seeds were successfully purified from the organic layer of the MeOH extract with a purity of over 99% by the Department of Agriculture at Kagawa University [9, 10]. The extraction methods from J. curcas seeds and IAA purification has been patented from the Japan patent office [11]. However, the experiment requires a great amount of IAA; it was mass-produced as previously described by Chen et al. [12]. To obtain enough amount of IAA, IAA was synthesized from radical coupling reaction of caffeyl alcohol with silver carbonate [12].
IAA stock of a 10 mg/ml IAA concentration was prepared in 100% pure dimethyl sulfoxide (DMSO, Sigma, St. Louis, MO, USA). It was later diluted with D-MEM to prepare 0, 25, 50, and 100 μg/ml IAA solutions. 1% DMSO was maintained for all different concentrations of tested IAA medium (0, 25, 50, and 100 μg/ml). These solutions were then filtered with a 0.45 μm filter syringe.
2.3. Cell proliferation assay
MCF-7 and HuH-7, MDA-MB-231, and HeLa were cultured in 96-well plates for up to four days. The concentrations of IAA tested were 0, 25, 50, and 100 μg/ml for cell proliferation assays. Cancer cell proliferation rate was studied using Cell Counting Kit-8 (Dojindo, Kumamoto, Japan). This kit uses WST-8, a water-soluble tetrazolium salt, as a coloring agent.
Cellular dehydrogenases reduces WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt], which yields orange colored formazan. The total number of cells that exist in each well is directly proportionate to the amount of the formazan produced by enzymes in the cell. The cell count is then translated to absorbance values. The absorbance measurements were taken at a wavelength of 450 nm with the SH-9000 Lab microplate reader (Corona, Ibaragi, Japan). Absorbance measurements were taken 0, 24, 48, and 72 h after IAA treatment.
2.4. DNA microarray analysis
MCF-7 subjected to 0 and 25 μg/ml IAA treatment for three days was used for microarray analysis. It was performed by Filgen Inc. (Nagoya, Japan) with GeneChip® Human Gene 2.0 ST Array (Affymetrix Inc., Santa Clara, CA, USA), containing 25 mer oligonucleotide probes. The fluorescence images of hybridized microarrays were obtained with GeneChip® scanner 3000 7G (Affymetrix Inc.). Scanned data were transformed into numerical values and analyzed using Expression Console™ Software (Affymetrix Inc.). The method of Robust Multichip Array (RMA) was applied to adjust the data. Microarray Data Analysis Tool Ver3.2 software (Filgen Inc.) was used to further classify the data into functional subgroups.
2.5. Cell synchronization, drug treatment, and cell cycle analysis by flow cytometry
After 24 h of cell seeding, the cells were synchronized with serum free medium for another 24 h. The following day, additives mixed in fresh 10% FCS-containing medium were applied. MCF-7 samples were harvested after three days of 0 or 25 μg/ml IAA treatments. On the third day, the samples were fixed in 70% ethanol at 4 °C for 24 h. The fixed samples were washed with PBS and incubated in 100 μg/ml propidium iodide (Wako, Yokohama, Japan) and 200 μg/ml RNase (Sigma) for 30 min at 37 °C in the dark. The flow cytometric cell analysis was performed, using a Beckman Coulter Cytomics FC500 Flow Cytometry Analyzer (Beckman Coulter, Fullerton, CA, USA). MultiCycle AV (Phoenix Flow Systems) DNA analysis software enabled determination of the phase of cell cycle arrest by comparing percentages of each cell stage between the control and treatment groups (G1, S, G2/M).
2.6. TUNEL staining
MCF-7 cells were tested for occurrence of apoptosis due to 0, 25 or 50 μg/ml IAA treatment after three days. Staurosporine (Sigma) was used as a positive control and applied 24 h prior to fixation at a concentration of 0.1 μg /ml. TUNEL staining was performed with the DeadEndTM Colorimetric TUNEL System kit (Promega, Madison, WI, USA). The cells were seeded on poly-L-lysine (Sigma) coated LAB-TEK®II (Nalge Nunc International, Rochester, NY, USA) chamber slides. The cells were fixed and TUNEL stained as per manufacturer's protocol (Promega). Percentages of apoptotic cells were quantified by manually counting cells on a well under each condition. Counting was continued to between 2,000 to 3,000 total cells.
2.7. Western blot analysis
MCF-7 cells treated with 0 or 25 μg/ml IAA for three days were lysed and centrifuged. The supernatant was used for western blot analysis. Proteins were separated on a 12.5% SDS-PAGE gel and electrotransferred onto a nitrocellulose membrane (Millipore, Billerica, MA, USA). After blocking, immunoblotting was performed using anti-BTG2 and anti-β-actin antibodies (Sigma), anti-p21 and GADD45A antibodies (Cell Signaling Technology or CST, Danvers, MA, USA), and anti-CDK1, cyclins B1 and cyclin B2 antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). Membranes were probed with a horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (CST). Proteins were detected by an enhanced chemiluminescence system (TAITEC, Saitama, Japan). The signals were analyzed with ImageJ [13].
2.8. Quantitative real-time PCR
After three days of 0 or 25 μg/ml IAA treatments, mRNA was extracted using RNeasy Protect Mini Kit (Qiagen). With the Omniscript® Reverse Transcription Kit (Qiagen), the collected RNA was reverse transcribed to cDNA. PCR was done for six target genes: BTG2, p21, GADD45A, CDK1, and cyclins B1 and B2. As per manufacturer's protocol, the Applied Biosystems® 7300 real-time PCR (Life TechnologiesTM, Carlsbad, CA, USA) was performed. Beta-actin was used for normalizing data results in each PCR reaction. The Ct values were considered to be an expression level of the gene of interest.
2.9. Statistical analysis
Data are expressed as mean ± standard deviation (SD) from at least three independent trials. The significance of difference between the control and drug-treated groups was evaluated by ANOVA and/or Student's t-test, using Microsoft Excel. Statistical significance was set at p < 0.05.
3. Results
3.1. Effect of IAA on growth of various cancer cells
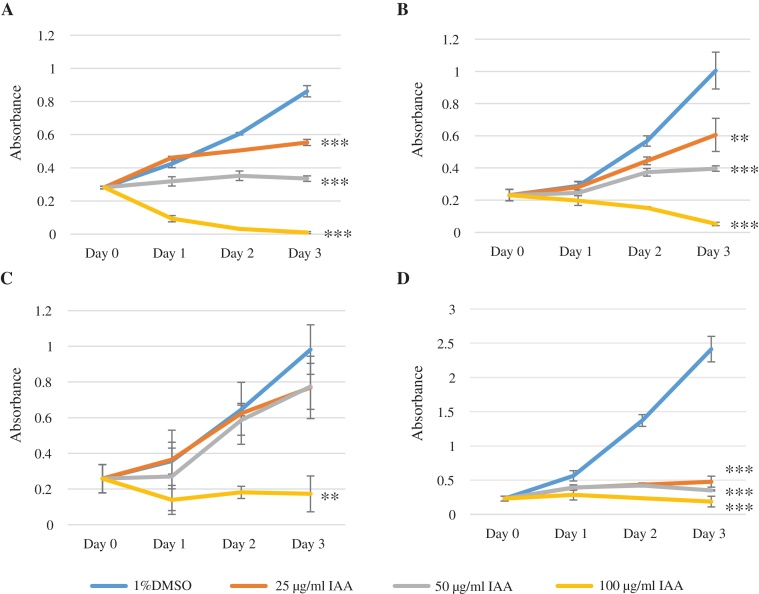
MCF-7, MDA-MB-231, HuH-7, and HeLa were tested to determine the effects of IAA. The inhibition of the growth in all tested cell lines occurred in a dose-dependent manner at 25, 50, and 100 μg/ml IAA. The IAA inhibition became apparent on Day 2, then definitive on Day 3 (Fig. 2). The differences between the control growth rate and those of the three IAA treatment groups were all statistically significant on Day 3. Because the IAA inhibitory effect was observed starting at a 25 μg/ml dose in the cell proliferation assay, we decided to focus on the comparisons between this concentration of IAA and 0 μg/ml IAA on MCF-7 for further experiments (excepting TUNEL assays, for which both 25 μg/ml and 50 μg/ml IAA concentrations were used).
Fig. 2.
Effect of IAA on a series of human cancer cell lines. MCF-7 (A), MDA-MB-231 (B), HuH-7 (C), and HeLa (D) were used to study the effects of IAA. Cell growth inhibition was observed in all tested cell lines in a dose-dependent manner. Each value was obtained by an average of 4 wells ± SD (control vs IAA; **p < 0.01, ***p < 0.001).
3.2. Changes in gene expressions
Microarray was used to determine the changes in genes that may occur in MCF-7 with IAA treatment. Using Microarray Data Analysis Tool Ver3.2, the number of genes of interest was narrowed down to 137 with a ratio (test/control) of ≤ 0.5 and 54 genes with a ratio of ≥ 2.0, a limitation was recommended by Filgen Inc. In support of the Gene-Ontology database, our selected genes were categorized into three groups (Table 1): cell cycle-related, apoptosis-related, and breast cancer-related.
Table 1.
Classification of the Candidate Genes that have changed due to 25 μg/ml IAA treatment.
| Function | Gene | Gene Description | Ratio Flag |
|---|---|---|---|
| Cell Cycle Related | GADD45A | growth arrest and DNA-damage-inducible, alpha | 3.49 |
| BTG2 | BTG family, member 2 | 2.24 | |
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 2.02 | |
| CCNB2 | cyclin B2 | 0.50 | |
| CDK1 | cyclin-dependent kinase 1 | 0.45 | |
| Apoptosis Related | FAS | TNF receptor superfamily, member 6 | 2.17 |
| BIRC5 | baculoviral IAP repeat containing 5 | 0.49 | |
| Breast Cancer Related | HMMR | hyaluronan-mediated motility receptor (RHAMM) | 0.50 |
| SGOL1 | shugoshin-like 1 (S. pombe) | 0.47 | |
| TOP2A | topoisomerase (DNA) II alpha 170kDa | 0.45 | |
| BRCA2 | breast cancer 2, early onset | 0.44 | |
| TP73 | tumor protein p73 | 0.43 | |
| KIAA1524 | KIAA1524 | 0.40 | |
| PBK | PDZ binding kinase | 0.39 | |
| KIF14 | kinesin family member 14 | 0.37 |
As described in the materials and method section, DNA microarray analysis was performed with GeneChip® Human Gene 2.0 ST Array (Affymetrix Inc.). With or without 3-day IAA treatment, any changes in mRNA expression was observed using DNA microarray analysis. Values of ratio indicates increase or decrease relative to 0 μg/ml IAA treatment.
As Table 1 shows, among cell cycle related genes, GADD45A, BTG2, and p21 expressions were enhanced by 3.49 fold, 2.24 fold, and 2.02 fold, respectively. On the contrary, cyclin B2 and CDK1 were reduced by 0.50 fold and 0.45 fold, respectively. As for a cyclin B2 related gene, cyclin B1 was reduced by 0.64 fold though it was not quite ranked within our set ratio change. FAS was enhanced by 2.17 fold, but BIRC5 was decreased by 0.49 fold for apoptosis related genes. Known breast cancer related genes, receptor for RHAMM, SGOL1, TOP2A, BRCA2, TP73, KIAA1524, PBK, and KIF14 were also reduced with IAA treatment.
3.3. Effect of IAA on the cell cycle progression
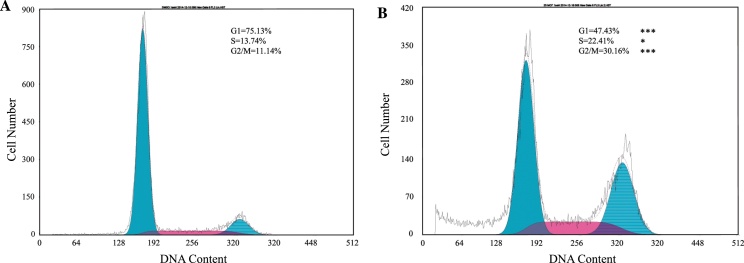
Because microarray analysis resulted in the indication of both cell cycle arrest and apoptosis induced by IAA treatment, flow cytometry was used first to test if IAA treatment interfered with MCF-7 cell growth by cell cycle arrest (Fig. 3). From the control, 75.13 ± 2.95% of the cells were at G1 phase, compared to 47.43 ± 4.03% of the cells from 25 μg/ml IAA treatment (p < 0.001). 13.74 ± 2.35% of cells were at S phase from the control, compared to 22.41 ± 3.82% from the treatment (p < 0.05). 11.14 ± 0.7% of cells were at G2/M phase from the control, compared to 30.16 ± 1.85% from the treatment (p < 0.001). Although there was an accumulation of cells at both S and G2/M stages in MCF-7, the IAA treatment induced arrest mainly at the G2/M stage.
Fig. 3.
Dynamics of cell cycle regulation. MCF-7 cells were treated for 25 μg/ml IAA application for 3 days (control, A; 25 μg/ml IAA, B). Drug treatment induced MCF-7 (1 × 106 cells) to arrest at the G2/M stage (control vs. IAA; *p < 0.05, ***p < 0.001).
3.4. Effect of IAA on induction of apoptosis
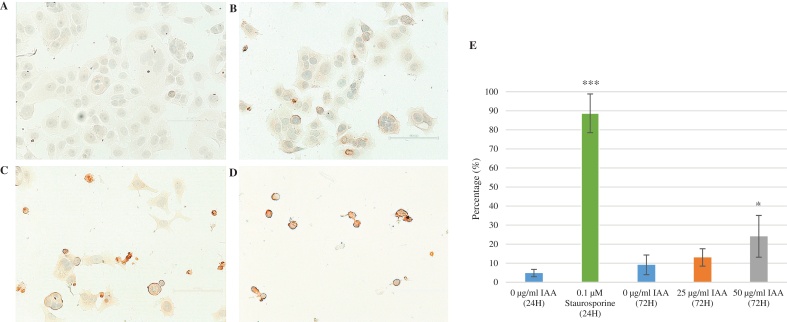
TUNEL assay was performed to test if IAA triggered apoptosis in MCF-7 cells. In comparison to the control, 25 and 50 μg/ml IAA treatment resulted in more apoptotic cells (Fig. 4). The percentages of apoptotic cells from 0, 25, and 50 μg/ml IAA treatments after 72 h were 9.16 ± 5.16%, 13.04 ± 4.56% and 24.12 ± 10.97%, respectively. A significant increase of apoptotic cells compared to the control was observed at 50 μg/ml IAA (p < 0.05), but not at 25 μg/ml IAA. Staurosporine, a well-known apoptotic inducer, was used as the positive control in the concentration of 0.1 μg/ml. A significantly higher percentage of apoptotic cells (88.69 ± 10.16%, p < 0.001) were observed even at the 24 h incubation point compared to the negative control (4.84 ± 1.92%, pictures not shown).
Fig. 4.
Apoptosis detection (Scale bar: 100 μm). Dark brown coloring indicates TUNEL-positive apoptotic cells. The cells were treated with 1% DMSO (A), and 25 (B) & 50 μg/ml (C) IAA for 3 days (control vs IAA; *p < 0.05). The cells were also treated with 0.1 μg/ml staurosporine as a positive control (D) (control vs staurosporine; ***p < 0.001). Percentages of apoptosis induced by IAA and staurosporine are shown (E).
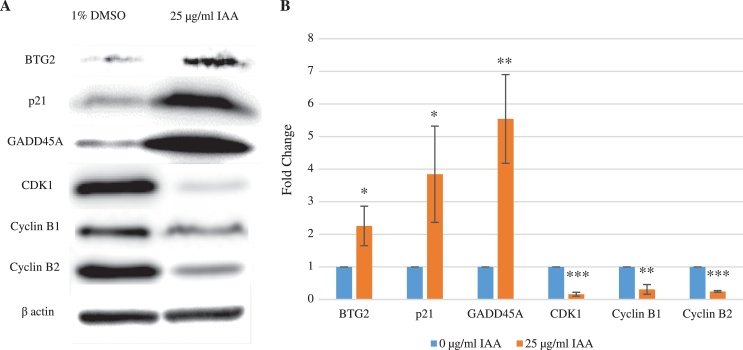
3.5. Effect of IAA on cell cycle regulatory proteins
Of the molecules with a test/control ratio of ≤ 0.5 and ≥ 2.0 from microarray results, BTG2, p21, GADD45A, CDK1, and cyclin B2 were chosen for western blot analysis as candidates for general G2/M cell cycle arrest regulatory molecules. Cyclin B1 was also included in this study because, similar to cyclin B2, it is known to be a major activator of CDK1 [14, 15]. The result of our western blot analysis agrees with previous findings [14, 15], (Fig. 5A) and the microarray analysis of those six molecules. The results were quantified and exhibited by the fold changes in protein expression (Fig. 5B). With 25 μg/ml IAA treatment, the expression of BTG2, p21, and GADD45A were 2.26 ± 0.61 fold (p < 0.05), 3.84 ± 1.48 fold (p < 0.001), and 5.54 ± 1.36 fold (p < 0.05) higher, respectively, than the control. Contrarily, the expression of CDK1 and cyclins B1and B2 were 0.16 ± 0.06 fold (p < 0.001), 0.31 ± 0.15 fold (p < 0.01) and 0.25 ± 0.02 fold (p < 0.001) lower than the control, respectively.
Fig. 5.
Changes in expressions of cell cycle regulatory proteins. BTG2, p21, and GADD45A protein expression increased due to 25 μg/ml IAA application. On the other hand, CDK1 and cyclin B2 expression decreased in comparison to 1% DMSO control (A). Protein expressions were quantified for ratio comparison (B), (control vs IAA; *p < 0.05, **p < 0.01, ***p < 0.001).
3.6. Changes in cell cycle regulators at mRNA level
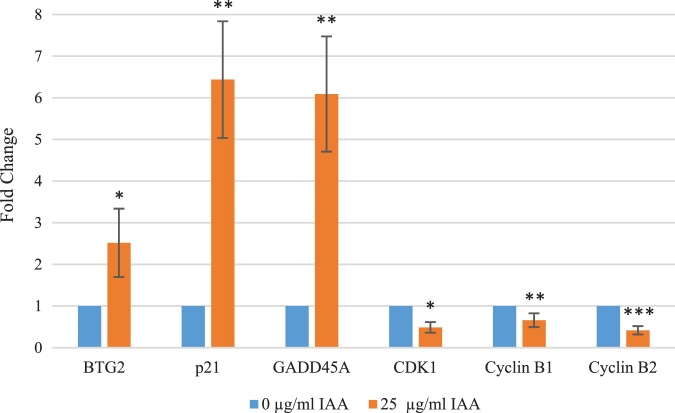
As the results showed in microarray analysis (Table 1), flow cytometry (Fig. 3), and western blot analysis (Fig. 5) on MCF-7, real-time PCR (Fig. 6) also verified both increased and decreased changes in expression for six of the cell cycle regulatory proteins. Compared to the control, BTG2, p21, and GADD45A increased by 2.52 ± 0.82 fold (p < 0.01), 6.44 ± 1.41 fold (p < 0.01), and 6.09 ± 1.38 fold (p < 0.01), respectively with IAA treatment. On the other hand, CDK1 and cyclins B1 and B2 decreased by 0.49 ± 0.13 fold (p < 0.01), 0.66 ± 0.16 fold (p < 0.05) and 0.42 ± 0.10 fold (p < 0.001), respectively.
Fig. 6.
Changes in mRNA expressions of cell cycle regulators. BTG2, p21, and GADD45A mRNA expression increased due to 25 μg/ml IAA application, whereas CDK1 and cyclin B2 expression decreased in comparison to 1% DMSO control (control vs IAA; *p < 0.05, **p < 0.01, ***p < 0.001).
4. Discussion
Many plants have been widely used in traditional medicine due to their unique properties [16]. TAXOL, a substance isolated from the bark of a Pacific yew tree over twenty years ago [17], is still in modern use as a chemotherapy treatment. Another natural product, IAA (Fig. 1) has been successfully extracted from J. curcas seeds. In this experiment, we became the first to link IAA function to possible cancer treatment. As our results show, IAA is capable of inhibiting growth of several cancer cell lines in vitro (Fig. 2). Starting at the concentration of 25 μg/ml, IAA exhibited its inhibitory effect on cancer cell growth, which was statistically significant compared to the control (p < 0.05).
We further investigated the molecular mechanisms of IAA that inhibit cell proliferation on the breast cancer cell line MCF-7. Among nearly 200 up- and down-regulated genes (ratio ≥ 2.0 or ≤ 0.5) selected for microarray analysis, we focused on three up-regulated genes; BTG2, p21 and GADD45A, and two down-regulated genes; CDK1, and Cyclin B2, all of which are considered to correlate with cell cycle regulation of the G2/M stage (Table 1). Our microarray analysis also showed decreased expression in cyclin B1, in addition to cyclin B2. Both were chosen for observation as G2/M cell cycle regulatory genes, in spite of cyclin B1's reduction value falling outside of the accepted range. The real-time PCR result of the decreasing trends for cyclin B1 and B2 matched the microarray results, confirming that both B1 and B2 cyclins were reduced in response to IAA treatment.
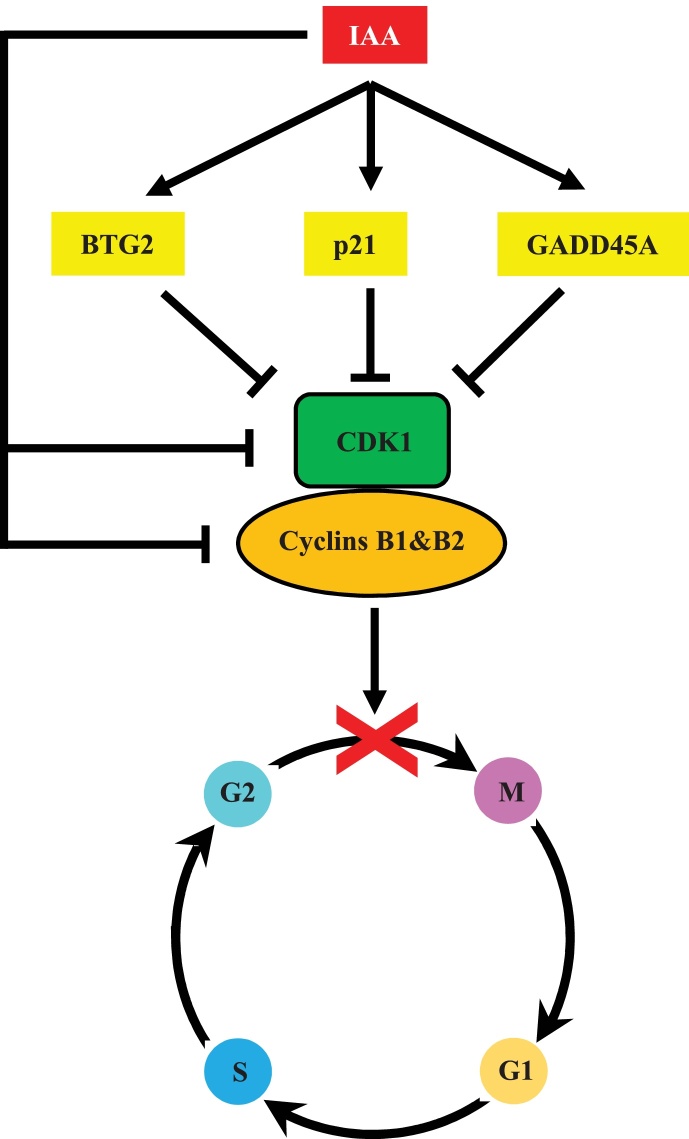
The two proteins of cyclin B1 and B2 are suggested to localize at different places [18, 19]. Cyclin B1/CDK1 complexes can move between the cytoplasm and nucleus, and regulate cell rounding, chromatin condensation, aster formation, and nuclear membrane breakdown in mitotic events [18, 19]. Cyclin B2, on the other hand, exists in the Golgi apparatus, where the complex it forms with CDK1 is essential for the mitotic reorganization of the Golgi apparatus [19, 20]. Because activation of cyclin B/CDK1 complex initiates entry into the M phase, G2/M transition usually requires a functional cyclin B/CDK1 complex [21, 22, 23]. In human breast cancer cell lines, such as MDA-MB-231 or MCF-7, BTG2 mRNA expression is generally undetectably low [24], and it is one of the tumor suppressor genes that inactivates the cyclin B1/CDK1 complex [25, 26]. GADD45A and p21 bind to CDK1, resulting in dissociation of the cyclin B/CDK1 complex and eventually induction of cell cycle arrest at G2/M [18, 27, 28]. As clearly shown in our data of cell cycle analysis (Fig. 3), western blot analysis (Fig. 5) and real-time PCR (Fig. 6), IAA treatment initiated G2/M arrest on MCF-7 by the increased expression of BTG2, p21, and GADD45A. These molecules may have caused the reduction of CDK1 activity by dissociating the cyclin B1/CDK1 and cyclin B2/CDK1 complexes as indicated in the scheme (Fig. 7). Interestingly, the IAA treatment also showed a direct reducing effect on the expression of CDK1 and cyclins B1 and B2, in addition to up-regulation of CDK1 inhibitors of BTG2, p21, and GADD45A (Fig. 5, Fig. 6). IAA may have induced G2/M cell cycle arrest by two mechanisms. The first is by the indirect CDK1/cyclin B complex inhibition through the increased expression of CDK1 inhibitors BTG2, p21, and GADD45A. The second is by the direct reduction/inhibition of CDK1 and cyclin B1 and B2. Therefore, IAA has the ability to create a double threat to halt the cell cycle at the G2/M stage (Fig. 7).
Fig. 7.
Possible IAA-induced molecular mechanism on MCF-7. IAA reduces MCF-7 cell proliferation by G2/M cell cycle arrest through stimulation of BTG2, p21, and GADD45A, along with direct reduction of CDK1 and cyclin B1& B2 expression.
Our findings suggest that cell cycle regulatory proteins may have played a major role in the inhibitory effect of IAA treatment on MCF-7. As shown in both microarray analysis and TUNEL assay however, IAA seems to exhibit additional characteristics that induce apoptosis. Although 25 μg/ml IAA treatment did not result in a statistically higher rate of apoptosis than 0 μg/ml IAA treatment in TUNEL assay, 50 μg/ml IAA (p < 0.05) treatment triggered a stronger apoptotic effect on MCF-7. As shown by the microarray results (Table 1), up-regulated FAS or down-regulated BIRC5 may be involved in apoptotic mechanisms. FAS expression was speculated to have been induced by IAA treatment, just as UV-irradiation can cause apoptosis by activation of cell surface death receptors like FAS [29]. It is also hypothesized that IAA treatment induced apoptosis by down-regulating BIRC5, the smallest of the inhibitory apoptosis proteins [30]. More in-depth molecular studies are required to ascertain if apoptosis from enhanced FAS and/or BIRC5 expression is involved in inhibition of MCF-7 cell growth.
Shown also in results of our microarray analysis, IAA treatment decreased expression in eight breast cancer-related genes (KIF, PBK, KIAA1524, BRCA2, TP73, TOP2A, SGOL, and HMMR). These genes are usually elevated in breast tumors and function in carcinogenesis and metastasis ([31, 32, 33, 34, 35, 36, 37, 38], refer to Table 1 for gene descriptions).
In conclusion, our series of studies clearly indicate that IAA has anti-carcinogenic effects on several human cancer cell lines, and cell proliferation was greatly inhibited by IAA treatment in a dose-dependent manner. Our results showed its application triggered molecular mechanisms that lead to increased expression of BTG2, p21, and GADD45A, and decreased expression of CDK1 and cyclins B1and B2, changes which all resulted in G2/M cell cycle arrest. These findings suggest that IAA has high potential for development as a new anti-cancer drug through its inhibitory effect on cancer cell proliferation.
Declarations
Author contribution statement
Ayako Katagi; Li Sui: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Kazuyo Kamitori; Akram Hossain; Chisato Noguchi; Youyi Dong: Performed the experiments.
Toshisada Suzuki; Takeshi Katayama: Contributed reagents, materials, analysis tools or data.
Fuminori Yamaguchi: Performed the experiments; Analyzed and interpreted the data.
Masaaki Tokuda: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
The authors received no funding from an external source.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Thomas R., Sah N.K., Sharma P.B. Therapeutic Biology of Jatropha curcas: A Mini Review. Curr. Pharm. Biotechnol. 2008;9:315–324. doi: 10.2174/138920108785161505. [DOI] [PubMed] [Google Scholar]
- 2.Aiyelaagbe O.O., Hamid A.A., Fattorusso E., Taglialatela-Scafati O., Schröder H.C., Müller W.E.G. Cytotoxic Activity of Crude Extracts as well as of Pure Components from Jatropha Species, Plants Used Extensively in African Traditional Medicine. Evid. Based. Complement. Alternat. Med. 2011;134:134954. doi: 10.1155/2011/134954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahidin S., Nakazibwe S., Taher M., Saxena A.K., Ichwan S.J.A. Antiproliferative activity of Curcusone B from Jatropha Curcas on human cancer cell lines. Aust. J. Basic Appl. Sci. 2011;5:47–51. [Google Scholar]
- 4.Oskoueian E., Abdullah N., Ahmad S. Phorbol esters from Jatropha meal triggered apoptosis, activated PKC-δ, caspase-3 proteins and down-regulated the proto-oncogenes in MCF-7 and HeLa cancer cell lines. Molecules. 2012;17(10):10816–10830. doi: 10.3390/molecules170910816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin J., Yan F., Tang L., Chen F. Antitumor effects of curcin from seeds of Jatropha curcas. Acta Pharmacol. Sin. 2003;24:241–246. [PubMed] [Google Scholar]
- 6.Kumar N., Sharma P.B. Jatropha curcus-A sustainable for production of biodiesel. J. Sientific Ind. Res. 2005;64:883–889. [Google Scholar]
- 7.Akbar E., Yaakob Z., Kamarudin S.K., Ismail M., Salimon J. Characteristic and Composition of Jatropha Curcas Oil Seed from Malaysia and its Potential as Biodiesel Feedstock Feedstock. Eur. J. Sci. Res. 2009;29:396–403. [Google Scholar]
- 8.Fukuyama Y., Hasegawa T., Toda M., Kodama M., Okazaki H. Structures of Americanol A and Isoamericanol A Having a Neurotrophic Property from the Seeds of Phytolacca americana. Chem. Pharm. Bull. (Tokyo). 1992;40:252–254. doi: 10.1248/cpb.40.252. [DOI] [PubMed] [Google Scholar]
- 9.Eto K., Koike Y., Suzuki T., Katayama T., Pankasemsuk T. Isolation of antioxidant catechol lignan/neolignans from Jatropha curcas, s shrub for biodiesel fuel production. Proceeding of the 57th lignin symposium. 2012:6–9. [Google Scholar]
- 10.Eto K., Matsuda Y., Hirata C., Suzuki T., Katayama T., Pankasemsuk T. Investigation of effective ingredients from plant for biodiesel fuel production. Proceeding of the 57th lignin symposium. 2011:70–73. [Google Scholar]
- 11.Suzuki T., Eto K., Katayama T, Jatropha extract, Jatropha antioxidants, and production methods for the antioxidants. JP-A-2013–064123.
- 12.Chen F., Tobimatsu Y., Havkin-Frenkel D., Dixon R.A., Ralph J. A polymer of caffeyl alcohol in plant seeds. PNAS. 2015;109:1772–1777. doi: 10.1073/pnas.1120992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider C.A. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paruthiyil S., Parmar H., Kerekatte V., Cunha G.R., Firestone G.L., Leitman D.C. Estrogen receptor beta inhibits human breast cancer cell proliferation and tumor formation by causing a G2 cell cycle arrest. Cancer Res. 2004;64:423–428. doi: 10.1158/0008-5472.can-03-2446. [DOI] [PubMed] [Google Scholar]
- 15.Paruthiyil S., Cvoro A., Tagliaferri M., Cohen I., Shtivelman E., Leitman D.C. Estrogenreceptor β causes a G2 cell cycle arrest by inhibiting CDK1 activity through the regulation of cyclin B1 GADD45A and BTG2. Breast Cancer Res. Treat. 2011;129:777–784. doi: 10.1007/s10549-010-1273-5. [DOI] [PubMed] [Google Scholar]
- 16.Ivanova D., Gerova D., Chervenkov T., Yankova T. Polyphenols and antioxidant capacity of Bulgarian medicinal plants. J. Ethnopharmacol. 2005;96:145–150. doi: 10.1016/j.jep.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 17.Nicolaou K.C., Yang Z., Liu J.J., Ueno H., Nantermet P.G., Guy R.K. Total synthesis of taxol. Nature. 1994;367:630–634. doi: 10.1038/367630a0. [DOI] [PubMed] [Google Scholar]
- 18.Liebermann D.A., Hoffman B. Gadd45 in stress signaling. J. Mol. Signal. 2008;3:15. doi: 10.1186/1750-2187-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y., Sramkoski R.M., Jacobberger J.W. The kinetics of G2 and M transitions regulated by B cyclins. PLoS One. 2013;8:e80861. doi: 10.1371/journal.pone.0080861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satyanarayana A., Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 21.King R.W., Jackson P.K., Kirschner M.W. Mitosis in Transition. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 22.Hunter T. Protein Kinases and Phosphatases: The Yin and Yang of Protein Phosphorylation and Signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 23.Yang J., Bardes E.S.G., Moore J.D., Brennan J., Powers M.A., Kornbluth S. Control of Cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawakubo H., Carey J.L., Brachtel E., Gupta V., Green J.E., Walden P.D. Expression of the NF-kappaB-responsive gene BTG2 is aberrantly regulated in breast cancer. Oncogene. 2004;23:8310–8319. doi: 10.1038/sj.onc.1208008. [DOI] [PubMed] [Google Scholar]
- 25.Ryu M.S., Lee M.S., Hong J.W., Hahn T.-R., Moon E., Lim I.K. TIS21/BTG2/PC3 is expressed through PKC-delta pathway and inhibits binding of cyclin B1-Cdc2 and its activity, independent of p53 expression. Exp. Cell Res. 2004;299:159–170. doi: 10.1016/j.yexcr.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Park T.J., Kim J.Y., Park S.H., Kim H.S., Lim I.K. Skp2 enhances polyubiquitination and degradation of TIS21/BTG2/PC3 tumor suppressor protein at the downstream of FoxM1. Exp. Cell Res. 2009;315:3152–3162. doi: 10.1016/j.yexcr.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Jin S., Antinore M.J., Lung F.D., Dong X., Zhao H., Fan F. The GADD45 inhibition of Cdc2 kinase correlates with GADD45-mediated growth suppression. J. Biol. Chem. 2000;275:16602–16608. doi: 10.1074/jbc.M000284200. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H., Jin S., Antinore M.J., Lung F.-D., Fan F., Blanck P. The central region of Gadd45 is required for its interaction with p21/WAF1. Exp. Cell Res. 2000;258:92–100. doi: 10.1006/excr.2000.4906. [DOI] [PubMed] [Google Scholar]
- 29.Rehemtulla A., Hamilton C.A., Chinnaiyan A.M., Dixit V.M. Ultraviolet Radiation-induced Apoptosis Is Mediated by Activation of CD-95 (Fas/APO-1) J. Biol. Chem. 1997;272:25783–25786. doi: 10.1074/jbc.272.41.25783. [DOI] [PubMed] [Google Scholar]
- 30.Boidot R., Vegran F., Jacob D., Chevrier S., Gangneux N., Taboureau J. The expression of BIRC5 is correlated with loss of specific chromosomal regions in breast carcinomas. Genes. Chromosomes Cancer. 2008;47:299–308. doi: 10.1002/gcc.20533. [DOI] [PubMed] [Google Scholar]
- 31.Corson T.W., Gallie B.L. KIF14 mRNA expression is a predictor of grade and outcome in breast cancer. Int. J. Cancer. 2006;119:1088–1094. doi: 10.1002/ijc.21954. [DOI] [PubMed] [Google Scholar]
- 32.Dou X., Wei J., Sun A., Shao G., Childress C., Yang W. PBK/TOPK mediates geranylgeranylation signaling for breast cancer cell proliferation. Cancer Cell Int. 2015;15:27. doi: 10.1186/s12935-015-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puustinen P., Jäättelä M. KIAA1524/CIP2A promotes cancer growth by coordinating the activities of MTORC1 and MYC. Autophagy. 2014;10:1352–1354. doi: 10.4161/auto.29076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wooster R., Neuhausen S.L., Mangion J., Quirk Y., Ford D., Collins N. Localization of a breast cancer susceptibility gene BRCA2, to chromosome 13q12-13. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 35.Marzese D.M., Hoon D.S.B., Chong K.K., Gago F.E., Orozco J.I., Tello O.M. DNA methylation index and methylation profile of invasive ductal breast tumors. J. Mol. Diagn. 2012;14:613–622. doi: 10.1016/j.jmoldx.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Capranico G., Tinelli S., Austin C., Fisher M., Zunino F. Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim. Biophys. Acta. 1992;1132(1):43–48. doi: 10.1016/0167-4781(92)90050-a. [DOI] [PubMed] [Google Scholar]
- 37.Lee M.Y., Marina M., King J.L., Saavedra H.I. Differential expression of centrosome regulators in Her2+ breast cancer cells versus non-tumorigenic MCF10A cells. Cell Div. 2014;9:3. doi: 10.1186/1747-1028-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bose N., Masellis A.M. Secretory products of breast cancer cells upregulate hyaluronan production in a human osteoblast cell line. Clin. Exp. Metastasis. 2005;22:629–642. doi: 10.1007/s10585-006-9003-4. [DOI] [PubMed] [Google Scholar]