Abstract
C2-aryl- and C2-alkyl-7-deazahypoxanthines as analogues of marine alkaloid rigidins were prepared utilizing novel synthetic methods developed for the construction of the pyrrolo[2,3-d]pyrimidine ring system. The new compounds exhibited submicromolar to nanomolar antiproliferative potencies against a panel of cell lines including in vitro models for drug resistant tumors, such as glioblastoma, melanoma and non-small-cell lung cancer. A selected representative C2-methyl-7-deazahypoxanthine was found to inhibit microtubule dynamics in cancer cells, lending evidence for tubulin targeting as a mode of action for these compounds in cancer cells. The results of the docking studies utilizing the colchicine site on β-tubulin were consistent with the observed SAR data, including an important finding that derivatization at C2 with long alkyl groups leads to the retention of activity, thus permitting the attachment of a biotin-containing linker for the subsequent proteomics assays. Because many microtubule-targeting compounds are successfully used to fight cancer in the clinic, the reported antitubulin rigidin analogues have significant potential as new anticancer agents.
Keywords: alkaloid; 7-deazapurine; drug discovery; privileged structure; pyrrolo[2,3-d]pyrimidine
Graphical abstract
Novel analogues of marine alkaloid rigidins were prepared utilizing new synthetic methods developed for the construction of the pyrrolo[2,3-d]pyrimidine ring system. The synthesized compounds exhibited submicromolar to nanomolar antiproliferative potencies against drug resistant cells, such as glioblastoma, melanoma and non-small-cell lung cancer.
Introduction
Due to their useful biological properties, marine pyrrole-derived alkaloids have been the subject of numerous studies by organic and medicinal chemistry research groups.1-9 One such investigation conducted in the authors’ laboratories is aimed at the development of synthetic chemistry and elucidation of biological properties, associated with the marine alkaloid rigidins. This group of natural products, which includes rigidins A, B, C, D (Figure 1) and E, was isolated from the tunicate Eudistoma cf. rigida found near Okinawa and New Guinea and has remained scantly explored despite initial reports of promising biological activities.10-12 The first general total synthesis of rigidins A, B, C and D, which involved only four steps from commercially available materials, was reported by the authors in 201113 and allowed for a thorough investigation of their biological properties. However, despite the earlier reports of promising antiproliferative activity against murine leukemia L1210 cells,11 the rigidins were found to have very little, if any, activity against cultured human cancer cells.14 Further synthetic investigations led to the development of approaches allowing for the modification of the 7-deazaxanthine skeleton present in the rigidins to obtain the corresponding 7-deazahypoxanthine (A), 7-deazaadenine (B) and 7-deazapurine (C) frameworks (Figure 1). These studies culminated in the discovery of potent antiproliferative activities associated with the synthesized 7-deazahypoxanthines (A) and their ability to disrupt microtubule organization in cancer cells by binding to the colchicine site of β-tubulin.14
Figure 1.
Structures of rigidins A, B, C, D, their synthetic analogues based on 7-deazahypoxanthine (A), 7-deazaadenine (B) and 7-deazapurine (C) skeletons, and proposed C2-substituted 7-deazahypoxanthines (D)
Because the only difference between the rigidins’ 7-deazaxanthine skeleton and the newly discovered 7-deazahypoxanthines A is the absence of the carbonyl group at C2 in the structure of the latter, modifications at this position seemed crucial for activity. Thus, it was further decided to prepare a series of C2-aryl- and C2-alkyl-deazahypoxanthines (D, Figure 1) and evaluate these compounds for anticancer activities. Such a study was also warranted by the results of the computer modeling experiments revealing a favorable accommodation of the lipophilic C2-side chain at the colchicine binding site, as described in the Results and Discussion section.
Results and discussion
(a) Synthesis and SAR of C2-aryl and C2-alkyl-7-deazahypoxanthines
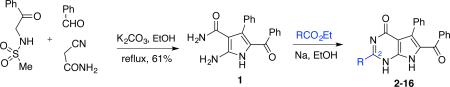
To develop synthetic access to the C2-substituted 7-deazahypoxanthines D, a previously discovered13 multicomponent reaction (MCR) of sulfonamidoacetophenones with aldehydes and cyanoacetamide was utilized to prepare pyrrole 1 (Table 1). This was followed by the pyrimidine ring closure using the reaction of 1 with a variety of aromatic and aliphatic esters catalyzed by sodium ethoxide in ethanol that was reported for a related purine system.15 The desired 7-deazahypoxanthines 2-16 were obtained in variable but acceptable yields (Table 1), given that in most cases these products precipitated after the acidification of the reaction mixtures and required no further purification.
Table 1.
Synthesis and antiproliferative activities of C2-aryl and C2-alkyl-7-deazahypoxanthines 2-16
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| # | structure | % yield | cell viabilitya GI50, μM |
# | structure | % yield | cell viabilitya GI50, μM |
||
| HeLa | MCF-7 | HeLa | MCF-7 | ||||||
| 2 |

|
NAb | >50 | >50 | 10 |

|
60 | 0.78 ± 0.05 | 0.93 ± 0.07 |
| 3 |

|
41 | >50 | >50 | 11 |

|
42 | 0.34 ± 0.01 | 0.23 ± 0.01 |
| 4 |

|
33 | >50 | >50 | 12 |

|
72 | 0.92 ± 0.11 | 1.1 ± 0.1 |
| 5 |

|
18 | 2.7 ± 0.2 | 2.5 ± 0.3 | 13 |

|
49 | 0.029 ± 0.001 | 0.035 ± 0.003 |
| 6 |

|
58 | 18.4 ± 1.8 | 23.1 ± 0.5 | 14 |

|
35 | 3.0 ± 0.2 | 3.2 ± 0.2 |
| 7 |

|
41 | >50 | >50 | 15 |

|
47 | 5.1 ± 0.2 | 4.2 ± 0.5 |
| 8 |

|
NAc | 11.1 ± 0.4 | 10.4 ± 0.8 | 16 |

|
66 | 21.5 ± 0.4 | 21.1 ± 1.1 |
| 9 |

|
21 | 9.1 ± 0.3 | 8.3 ± 0.8 | |||||
Concentration required to reduce the viability of cells by 50% after a 48 h treatment with the indicated compounds relative to a DMSO control ± SD from two independent experiments, each performed in 4 replicates, as determined by the MTT assay.
Synthesized using the method described in ref. 13.
Obtained as a side product of the reaction of 1 with CO(OEt)2.
The obtained compounds were evaluated for antiproliferative activities using the HeLa cell line as a model for human cervical adenocarcinoma and MCF-7 cells as a model for breast adenocarcinoma. The cells were treated with each compound for 48 h, and cell viability was assessed using the MTT method (Table 1). The results revealed that in comparison with the rigidin-related 7-deazaxanthine 2, containing the carbonyl group at C2, most of the 7-deazahypoxanthines showed antiproliferative activity. Although the activity was found to be only double to single digit micromolar for C2-aryl (5, 6), C2-OEt (8) and branched C2-alkyl (9), the linear C2-alkyl derivatives 10-12 showed submicromolar potency and the C2-methyl compound 13 was nanomolar. Of note, when fluorine was incorporated into the C2-Me group, as in 14, 15 and 16, the activity progressively dropped in parallel with the increasing steric size at the C2 position, i.e. CH3→CH2F→CHF2→CF3.16 The activity of the C2-CF3 derivative 16 is similar to that of C2-i-Pr compound 9, which is consistent with the comparable Taft steric parameters of the CF3 and i-Pr groups.16,17 Because the CF3 and i-Pr groups differ significantly in their electronic properties, it is thus conceivable that the steric rather than electronic properties of the C2 substituent govern the activity in this series of compounds.
(b) MCR-based synthesis and SAR of C2-methyl-7-deazahypoxanthines
Potent activity of the C2-methyl analogue 13 led to further exploration of C2-methyl compounds by varying the aldehyde-derived C7 aromatic group. To facilitate the synthesis of such compounds, a new 4-component cyclocondensation of methylsulfonamidoacetophenone, cyanoacetamide and triethyl orthoacetate with various aldehydes was developed (Table 2). In this reaction, the entire pyrrolo[2,3-d]pyrimidine skeleton is assembled in one step and its success depended on an optimized temperature regime, in which the reaction was first kept at 90 °C to allow for a clean formation of intermediate aminopyrroles (similar to 1 in Table 1), and then at 150 °C to bring the fourth component orthoacetate into the process, leading to closure of the pyrimidine portion of the molecule.
Table 2.
MCR-based synthesis and antiproliferative activities of C2-methyl-7-deazahypoxanthines 13 and 17-22

| ||||||
|---|---|---|---|---|---|---|
| # | structure | % yield | GI50
in vitro Values (μM)a |
|||
| HeLa | MCF-7 | U-87 | A549 | |||
| 13 |

|
40 | 0.029 ± 0.001 | 0.035 ± 0.003 | 0.077 ± 0.002 | 0.25 ± 0.01 |
| 17 |

|
61 | 0.27 ± 0.01 | 0.23 ± 0.00 | 0.90 ± 0.16 | 0.60 ± 0.23 |
| 18 |

|
40 | 0.29 ± 0.01 | 0.27 ± 0.03 | 0.94 ± 0.12 | 0.65 ± 0.19 |
| 19 |

|
43 | 0.29 ± 0.03 | 0.36 ± 0.02 | 1.7 ± 0.1 | 1.0 ± 0.9 |
| 20 |

|
65 | 3.1 ± 0.1 | 4.7 ± 0.8 | 9.23 ± 2.13 | 12.0 ± 1.4 |
| 21 |

|
63 | 0.49 ± 0.05 | 0.61 ± 0.01 | 2.4 ± 0.6 | 1.1 ± 0.4 |
| 22 |

|
58 | 0.29 ± 0.01 | 0.27 ± 0.01 | 0.72 ± 0.06 | 0.38 ± 0.08 |
Concentration required to reduce the viability of cells by 50% after a 48 h treatment with the indicated compounds relative to a DMSO control ± SD from two independent experiments, each performed in 4 replicates, as determined by the MTT assay.
The antiproliferative effects of compounds 13 and 17-22 were studied in a more challenging cell line panel that, in addition to the previously utilized HeLa and MCF-7, contained in vitro models for cancers known to be associated with poor prognoses, such as the U-87 human glioblastoma and A549 human non-small-cell lung (NSCLC) carcinoma. Although the newly synthesized compounds 17-22 were found to be inferior to the original C7-phenyl analogue 13, in general the submicromolar potencies were retained with the exception of the pyridine-containing variant 20 exhibiting single-digit micromolar potencies. The drop in activity of this analogue could be also explained by its polar character, impairing its cell permeability.
(c) Microtubule organization in cells
Because the original 7-deazahypoxanthines were found to have strong effects on the microtubule cytoskeleton in cells,14 it was important to confirm that these properties remained unchanged in these novel analogues. To this end, cultured HeLa cells were treated with 13 over a range of concentrations and microtubule morphology was examined (Figure 2, left). Results revealed a pronounced change in mitotic microtubule organization at concentrations between 1 and 2 μM, where 13 caused a complete collapse of the mitotic spindle (panels J and K). The interphase microtubules affected to a lesser extent at these concentrations (panels D and E) suggesting that this compound was primarily affecting dynamic microtubules. While there appeared to be little effect on stable interphase or spindle microtubules at lower concentrations, a distinct displacement of the mitotic spindle could be observed (panels H and I), suggesting a defect in astral microtubules. The central localization of the mitotic spindle is made possible by interactions between dynamic astral microtubules emanating from the spindle poles and the cell cortex, often described as Hertwig's rule.18 During mitosis, astral microtubules are the most dynamic microtubule population, and, therefore, they would be most sensitive to tubulin-targeting drugs such as 13. The quantification of spindle eccentricity with the increasing concentrations of 13 is shown in Figure 2 (right). Together, these results lend further evidence for tubulin-targeting is a likely mechanism responsible for the antiproliferative effects of the 7-deazahypoxanthines.
Figure 2.
Microtubule organization is altered with the increasing concentrations of 13. Left. HeLa cells were treated with 0.1% DMSO or 13 at different concentrations for three hours, fixed and probed for the presence of microtubules (green), F-actin (red) and DNA (blue). Scale bar, 10μm. Right. Line graph showing an increase in the eccentric spindles with the increasing concentrations of 13, 150 mitotic cells scored per condition.
(d) Computer modeling
In the authors'previous work, 7-deazahypoxanthines were proposed to bind to the colchicine site on β-tubulin based on their potent inhibitory effects on the binding of [3H]colchicine to tubulin.14 In the current study, molecular docking simulations were performed on this series of compounds to understand the contribution of the C2-subtstituent toward the binding at this site. Figure 3 shows the proposed binding mode of the 7-deazahypoxanthines in the colchicine pocket.19 Of particular interest, is the accommodation of alkyl substituents at the C2 position. A small channel in the region of Asn258 and Lys352 accommodates the colchicine methoxy group protruding from the 7-membered C-ring system. Docking results of 7-deazahypoxanthine 13 suggest that the C2 methyl group is located in a nearly identical position to the above-mentioned colchicine methoxy group (Figure 3A). Moreover, this narrow channel is able to accommodate linear alkyl substituents at C2, such as the butyl group in 11 (Figure 3B). Sterically demanding substituents at C2 are, however, too bulky for the channel, perhaps explaining the lack of activity of 3-6, 9 and 16. A superimposition of the co-crystalized colchicine and the docked 7-deazahypoxanthine 13 reveals the similar binding modes of the two compounds, with the C7-aryl substituent on the 7-deazahypoxanthine scaffold accommodated in a near identical fashion to that of the trimethoxy-aryl moeity of colchicine (Figure 3C).
Figure 3.
Docking studies (PDB ID 3UT5) of 13 (A) and 11 (B) reveal that linear C2-alkyl substituents on the 7-deazahypoxanthine scaffold can be accommodated in a small channel in the region of of Asn258 and Lys352. Co-crystallized colchicine (green) and 13 (orange) adopt similar binding poses in the colchicine pocket (C).
(e) Quantitative videomicroscopy
Poor prognoses of glioblastoma, melanoma and NSCLC patients are generally attributed to the drug resistant nature of these malignancies20-22 and therefore it was encouraging that 7-deazahypoxanthines 13 and 17-22 were active against the U87 human glioblastoma and A549 NSCLC carcinoma cell lines. Since tubulin-targeting agents are generally poorly effective against cancer cells resistant to the induction of apoptosis,23-25 compound 22 was further evaluated against U373 human glioblastoma and SKMEL human melanoma cells, reported to display apoptosis resistance.26,27 The obtained MTT-based GI50 value of 0.3 μM in both of these cell lines indicates that this compound maintains similar levels of activity in these cell types (compare with GI50 values in Table 2). Computer-assisted phase-contrast microscopy (quantitative videomicroscopy) was utilized to confirm cell death as the principal mechanism of action associated with this compound's in vitro growth inhibitory effects in these cell lines. Figure 4 shows that 22 indeed inhibited cancer cell proliferation by inducing cell death when assayed at its MTT-based GI50 values in U373 glioblastoma and SKMEL melanoma cells.
Figure 4.
Cellular imaging of 22 against U373 glioblastoma and SKMEL melanoma cells illustrating cell death at MTT colorimetric assay-related GI50 value of 0.3 μM.
(e) Synthesis of a biotinylated protein pulldown reagent
Microtubules represent an important target for a number of clinically successful drugs, but so far, there are no FDA-approved anticancer agents targeting the colchicine binding domain on β-tubulin, although many colchinoids inhibit the growth of multidrug resistant (MDR) tumor cell lines and have reduced neurotoxicity and good oral bioavailability. A number of colchinoids have been examined in human clinical trials with generally promising results, but having drawbacks that include narrow therapeutic windows and unacceptable toxicities.28 It is thus important to understand and predict possible causes of potential toxicity in the clinic by elucidating other intracellular targets of these agents during their preclinical evaluation. The importance of identifying additional targets interacting with the 7-deazahypoxanthines could also help understand their effectiveness against drug resistant cancer cell lines used in the current study, since sole tubulin targeting may not be sufficient to induce death of these cancer cells. The current study revealed that long chain alkyl substitution at C2 of the 7-deazahypoxanthine skeleton is tolerated and thus this position appeared ideal for the probe preparation to be used in proteomic pulldown assays. To this end, using the developed chemistry, pyrrole 1 was successfully converted to the C2-alkyne-containing 7-deazahypoxanthine 23, which was then coupled with a commercially available biotin-azide reagent utilizing click chemistry (Figure 5).29 Mode of action studies utilizing the prepared biotinylated probe 24 are in progress and will be reported in due course.
Figure 5.
Synthesis of the biotinylated 7-deazahypoxanthine probe 24
Conclusion
Synthetic chemistry was developed to access C2-aryl- and C2-alkyl-deazahypoxanthines as analogues of the marine alkaloid rigidins containing a carbon group instead of the carbonyl present in the natural products. The new 7-deazahypoxanthines were found to have submicromolar to nanomolar antiproliferative potencies and inhibit microtubule dynamics in cancer cells. This activity was most plausibly explained by binding of these compounds to the colchicine site on β-tubulin, a proposal consistent with the experimental findings of their effects on microtubule organization and a theoretical docking model developed for these compounds. These agents are also effective against cancer cells representing drug resistant cancers with poor prognoses raising the possibility that additional intracellular targets may be involved and the current investigation set the stage for further mode of action studies utilizing biotinylated protein pull down probes. Since many microtubule-targeting compounds have been successfully used to fight cancer in the clinic, the antitubulin agents represented by the 7-deazahypoxanthine rigidin analogues have significant potential as new anticancer agents.
Experimental Section
(a) Synthetic chemistry
General
All reagents, solvents and catalysts were purchased from commercial sources (Acros Organics and Sigma-Aldrich) and used without purification. All reactions were performed in oven-dried flasks open to the atmosphere or under nitrogen and monitored by thin layer chromatography (TLC) on TLC precoated (250 μm) silica gel 60 F254 glass-backed plates (EMD Chemicals Inc.). Visualization was accomplished with UV light. Flash column chromatography was performed on silica gel (32-63 μm, 60 Å pore size). 1H and 13C NMR spectra were recorded on a Bruker 400 spectrometer. Chemical shifts (δ) are reported in ppm relative to the TMS internal standard. Abbreviations are as follows: s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet). HRMS analyses were performed using Waters Synapt G2 LCMS. The > 95% purity of the synthesized compounds was ascertained by UPLC/MS analyses.
2-Amino-5-benzoyl-4-phenyl-1H-pyrrole-3-carboxamide (1)
To a stirred solution of N-methylsulfonamidoacetophenone13 (0.084 g, 0.4 mmol), benzaldehyde (0.055 g, 0.52 mmol) and cyanoacetamide (0.044 g, 0.52 mmol) in EtOH (3 mL) was added anhydrous granulated K2CO3 (0.028 g, 0.2 mmol). The mixture was then refluxed for 14 h. After this time the reaction mixture was cooled to room temperature and concentrated. The crude product was purified by flash chromatography on silica gel using CH2Cl2/AcOEt (2:1) as eluent to obtain 74 mg of the desired pyrrole 1 (61% yield).
2-Amino-5-benzoyl-4-phenyl-1H-pyrrole-3-carboxamide (1)
61%; 1H NMR (DMSO-d6): δ 10.89 (brs, 1H), 7.18-7.05 (m, 8H), 6.97 (t, J = 7.8 Hz, 2H), 6.70 (brs, 1H), 6.39 (s, 2H), 4.59 (brs, 1H); 13C NMR (DMSO-d6): δ 184.3, 167.6, 149.8, 139.7, 134.6, 132.6, 130.9, 129.9, 128.3, 128.2, 128.0, 127.5, 121.5, 99.4; HRMS m/z (ESI) calcd for C18H16N3O2 (M+H) 306.1243, found 306.1248.
General procedure for the synthesis of deazahypoxanthines 3-16
A selected ethyl ester (1.28 mmol) and pyrrole 1 (50 mg, 0.16 mmol) were added to the solution of EtONa in EtOH prepared by dissolving sodium metal (30 mg, 1.3 mmol) in EtOH (2 mL) . The mixture was then refluxed for 10 h overnight. After that time the reaction mixture was diluted with H2O and neutralized with 1M HCl. The formed precipitate was collected by filtration and dried under vacuum over night. Although in most cases the productdeazahypoxanthines were >95% pure, they could be further purified using column chromatography (5% MeOH in CHCl3).
Selected characterization data of compounds in Table 1
6-Benzoyl-2-phenyl-5-phenyl-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (3)
41%; 1H NMR (DMSO-d6): δ 12.21 (brs, 1H), 11.64 (brs, 1H), 8.01-7.96 (m, 2H), 7.74 (t, J = 7.3 Hz, 1H), 7.67 (t, J = 7.5 Hz, 2H), 7.37-7.09 (m, 10H); 13C NMR (DMSO-d6): δ 185.8, 167.5, 165.5, 138.4, 133.6, 133.1, 132.6, 131.3, 131.0, 129.8, 129.7, 129.0, 128.7, 128.6, 128.5, 127.9, 127.6, 123.3, 103.4; HRMS m/z (ESI) calcd for C25H20DN4O2 (M+NH4) 410.1727, found 410.1722.
6-Benzoyl-2-(2-ethoxycarbonylethyl)-5-phenyl-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (10)
60%; 1H NMR (DMSO-d6): δ 12.59 (brs, 1H), 11.91 (brs, 1H), 7.42 (d, J = 8.0 Hz, 2H), 7.32 (t, J = 8.0 Hz, 1H), 7.20-6.99 (m, 7H), 4.09 (q, J = 8.0 Hz, 2H), 2.98-2.75 (m, 4H), 1.15 (t, J = 8.0 Hz, 3H); 13C NMR (DMSO-d6): δ 188.1, 173.9, 172.4, 159.7, 159.6, 158.6, 150.1, 138.0, 133.0, 131.6, 129.5, 128.1, 127.3, 104.9, 60.5, 31.1, 30.4, 14.5; HRMS m/z (ESI) calcd for C24H21N3NaO4 (M+Na) 438.1430, found 438.1431.
6-Benzoyl-2-butyl-5-phenyl-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (11)
42%; 1H NMR (DMSO-d6): δ 12.54 (brs, 1H), 11.86 (brs, 1H), 7.43 (d, J = 6.7 Hz, 2H), 7.30 (t, J = 6.8 Hz, 1H), 7.23-6.85 (m, 7H), 2.62 (t, J = 6.9 Hz, 2H), 1.81-1.60 (m, 2H), 1.36-1.28 (m, 2H), 0.92 (t, J = 7.0 Hz, 3H); 13C NMR (DMSO-d6): δ 189.1, 160.7, 138.9, 133.9, 133.0, 132.4, 130.3, 128.9, 128.0, 105.6, 35.0, 30.4, 22.9, 14.9; HRMS m/z (ESI) calcd for C23H21N3NaO2 (M+Na) 394.1531, found 394.1528.
6-Benzoyl-2-methyl-5-phenyl-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (13)
49%; 1H NMR (DMSO-d6): δ 12.57 (brs, 1H), 11.90 (brs, 1H), 7.42 (dd, J = 6.7, 0.4 Hz, 2H), 7.31 (t, J = 7.4 Hz, 1H), 7.17-7.01 (m, 7H), 2.36 (s, 3H); 13C NMR (DMSO-d6): δ 188.1, 159.8, 156.8, 150.5, 138.1, 133.0, 132.1, 131.6, 129.5, 128.1, 127.6, 127.4, 127.2, 127.1, 104.7, 21.5; HRMS m/z (ESI) calcd for C20H15N3NaO2 (M+Na) 352.1062, found 352.1068.
6-Benzoyl-2-(difluoromethyl)-5-phenyl-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (15)
47%; 1H NMR (DMSO-d6): δ 13.12 (brs, 1H), 12.80 (brs, 1H), 7.46 (d, J = 7.0 Hz, 2H), 7.35 (t, J = 7.4 Hz, 1H), 7.24-7.05 (m, 7H); 13C NMR (DMSO-d6): δ 188.3, 159.0, 148.2, 137.6, 132.6, 132.5, 131.6, 129.6, 129.2, 128.2, 127.4, 127.0, 112.9, 110.5, 108.1, 106.9; HRMS m/z (ESI) calcd for C20H14F2N3O2 (M+H) 366.1054, found 366.1054.
6-Benzoyl-5-phenyl-2-(trifluoromethyl)-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (16)
66%; 1H NMR (DMSO-d6): δ 13.34 (brs, 1H), 13.25 (brs, 1H), 7.48 (d, J = 7.0 Hz, 2H), 7.36 (t, J = 7.4 Hz, 1H), 7.22-7.04 (m, 7H); 13C NMR (DMSO-d6): δ 188.4, 137.4, 132.8, 132.4, 131.5, 131.3, 131.1, 130.2, 129.6, 129.2, 128.9, 128.3, 128.1, 128.0, 127.5, 127.2; HRMS m/z (ESI) calcd for C20H12F3N3NaO2 (M+Na) 406.0779, found 406.0777.
General procedure for the synthesis of deazahypoxanthines 13 and 17-23
To a solution of N-(2-oxo-2-arylethyl)methanesulfonamide (0.676 mmol), the selected aldehyde (0.879 mmol) and cyanoacetamide (0.072 g, 0.879 mmol) in a mixture of EtOH (2.5 mL) and MeC(OEt)3 (2.5 mL) was added anhydrous granulated K2CO3 (0.052g, 0.372 mmol) in one portion. The mixture was purged with N2 for 5 min and then heated at 90 °C for 24 hours under the nitrogen atmosphere. The formation of the intermediate pyrrole was monitored by TLC. After that the reaction temperature was increased to 150 °C, and the reaction mixture was heated for 3-6 h. The mixture was cooled to room temperature, and the formed precipitate was collected by filtration and washed with EtOH (2 mL) and Et2O (2 mL) to give the desired 7-deazahypoxanthine 13 and 17-23. An additional amount of the product was obtained by the evaporation of the mother liquor and purification of the residue by column chromatography with MeOH/CH2Cl2=1/40 to 1/20 gradient.
Characterization data of compounds in Table 2
6-Benzoyl-5-(3-chlorophenyl)-2-methyl-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (17)
61%, 1H NMR (DMSO-d6): δ 12.67 (brs, 1H), 11.93 (brs, 1H), 7.44 (d, J = 7.2 Hz, 2H), 7.36 (t, J = 7.4 Hz, 1H), 7.17 (t, J = 7.3 Hz, 2H), 7.13-7.09 (m, 3H), 7.04 (t, J = 7.6 Hz, 1H), 2.37 (s, 3H); 13C NMR (DMSO-d6): δ 187.9, 159.7, 156.9, 150.5, 138.1, 135.2, 132.3, 132.0, 131.2, 130.2, 129.4, 129.0, 128.2, 127.9, 126.9, 125.7, 104.8, 21.5; HRMS m/z (ESI) calcd for C20H15ClN3O2 (M+H) 364.0853, found 364.0851.
6-Benzoyl-5-(3-bromophenyl)-2-methyl-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (18)
40%, 1H NMR (DMSO-d6): δ 12.67 (brs, 1H), 11.93 (brs, 1H), 7.43 (d, J = 7.0 Hz, 2H), 7.36 (t, J = 7.5 Hz, 1H), 7.30 (t, J = 1.8 Hz, 1H), 7.25-7.15 (m, 4H), 6.99 (t, J = 7.8 Hz, 1H), 2.37 (s, 3H); 13C NMR (DMSO-d6): δ 187.9, 159.7, 157.0, 150.4, 138.1, 135.4, 134.0, 132.3, 130.6, 129.8, 129.3, 129.2, 128.2, 127.9, 125.6, 120.5, 104.8, 21.6; HRMS m/z (ESI) calcd for C20H14BrN3NaO2 (M+Na) 430.0167, found 430.0168.
6-Benzoyl-5-(3-fluorophenyl)-2-methyl-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (19)
43%; 1H NMR (DMSO-d6): δ 12.66 (brs, 1H), 11.93 (brs, 1H), 7.44 (d, J = 7.0 Hz, 2H), 7.35 (t, J = 7.4 Hz, 1H), 7.17 (t, J = 7.8 Hz, 2H), 7.06-6.99 (m, 2H), 6.94 (dt, J = 7.8, 1.2 Hz, 1H), 6.91-6.85 (m, 1H), 2.37 (s, 3H); 13C NMR (DMSO-d6): δ 187.3, 162.0, 159.6, 159.1, 156.2, 149.8, 137.5, 134.8, 131.6, 128.8, 128.3, 127.5, 127.2, 125.1, 117.6, 113.3, 104.1, 20.9; HRMS m/z (ESI) calcd for C20H14FKN3O2 (M+K) 386.0707, found 386.0708.
6-Benzoyl-2-methyl-5-(pyridin-3-yl)-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (20)
65%; 1H NMR (DMSO-d6): δ 12.74 (brs, 1H), 11.97 (brs, 1H), 8.30 (dd, J = 2.2, 0.7 Hz, 1H), 8.22 (dd, J = 4.8, 1.6 Hz, 1H), 7.61-7.57 (m, 1H), 7.47-7.42 (m, 2H), 7.38-7.33 (m, 1H), 7.20-7.14 (m, 2H), 7.07 (ddd, J = 7.8, 4.8, 0.8 Hz, 1H), 2.37 (s, 3H); 13C NMR (DMSO-d6): δ 187.2, 159.4, 156.5, 150.8, 150.2, 147.2, 138.0, 137.5, 131.9, 129.1, 128.7, 127.8, 127.6, 123.2, 121.9, 104.6, 21.1; HRMS m/z (ESI) calcd for C19H14N4NaO2 (M+Na) 353.1014, found 353.1019.
6-Benzoyl-5-(5-bromopyridin-3-yl)-2-methyl-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (21)
63%; 1H NMR (DMSO-d6): δ 12.90 (s, 1H), 12.09 (s, 1H), 8.36 (d, J = 9.0 Hz, 2H), 7.81 (s, 1H), 7.52-7.11 (m, 5H), 2.40 (s, 3H); 13C NMR (DMSO-d6): 187.6, 159.9, 157.4, 150.7, 150.0, 148.4, 140.8, 138.1, 132.6, 131.2, 129.6, 128.5, 122.2, 119.1, 105.2, 21.7; HRMS m/z (ESI) calcd for C19H14BrN4O2 (M+H) 409.0300, found 409.0287.
6-Benzoyl-5-(3,5-dibromophenyl)-2-methyl-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (22)
58%; 1H NMR (DMSO-d6) δ 12.82 (s, 1H), 11.94 (s, 1H), 7.48 (s, 1H), 7.44 (d, J = 6.0 Hz, 2H), 7.37 (t, J = 7.2 Hz, 1H), 7.33 (s, 2H), 7.21 (t, J = 6.8 Hz, 2H), 2.28 (s, 3H); 13C NMR (DMSO-d6): δ 187.9, 160.0, 157.0, 151.1, 138.2, 137.2, 133.2, 132.5, 131.8, 129.2, 128.2, 124.2, 121.1, 104.8, 21.7; HRMS m/z (ESI) calcd for C20H14Br2N3O2 (M+H+) 485.9453, found for C20H14Br2N3O2 (M+H) 485.9453, found 485.9438.
6-Benzoyl-2-(pent-4-ynyl)-5-phenyl-1H-pyrrolo[2,3-d]pyrimidin-4(7H)-one (23)
72%; 1H NMR (DMSO-d6) δ 12.57 (brs, 1H), 11.91 (brs, 1H), 7.42 (d, J = 7.4 Hz, 2H), 7.30 (t, J = 7.4 Hz, 1H), 7.19-6.97 (m, 7H), 2.82 (s, 1H), 2.72 (t, J = 6.9 Hz, 2H), 2.28 (t, J = 6.8 Hz, 2H), 1.98-1.85 (m, 2H); 13C NMR (DMSO-d6): δ 187.6, 159.4, 158.8, 149.8, 137.6, 132.6, 131.7, 131.1, 129.0, 127.6, 127.3, 126.8, 126.6, 104.5, 83.8, 71.7, 32.9, 25.7, 17.3; HRMS m/z (ESI) calcd for C24H19N3NaO2 (M+Na) 404.1375, found 404.1374.
N-(2-(2-(2-(2-(4-(3-(6-benzoyl-4-oxo-5-phenyl-4,7-dihydro-1H-pyrrolo[2,3-d]pyrimidin-2-yl)propyl)-1H-1,2,3-triazol-1-yl)ethoxy)ethoxy)ethoxy)ethyl)-5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamide (24)
10%; 1H NMR (DMSO-d6) δ 12.58 (brs, 1H), 11.89 (brs, 1H), 7.87 (s, 1H), 7.79 (t, J = 5.6 Hz, 1H), 7.41 (dd, J = 8.3, 1.3 Hz, 2H), 7.31 (t, J = 7.4 Hz, 1H), 7.18-6.99 (m, 7H), 6.36 (d, J = 21.2 Hz, 1H), 5.02 (s, 1H), 4.47 (t, J = 5.2 Hz, 2H), 4.32-4.26 (m, 1H), 4.15-4.07 (m, 1H), 3.80 (t, J = 5.5 Hz, 2H), 3.55-3.41 (m, 8H), 3.19-3.15 (m, 4H), 3.11-3.06 (m, 3H), 2.82 (dd, J = 12.4, 5.1 Hz, 2H), 2.68-2.65 (m, 2H), 2.58-2.52 (m, 2H), 2.46-2.42 (m, 1H), 2.35-2.31 (m, 1H), 1.62-1.41 (m, 6H); HRMS m/z (ESI) calcd for C42H51N9NaO7S (M+Na) 848.3530, found 848.3528.
(b) Cell culture
Human cancer cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA), the European Collection of Cell Culture (ECACC, Salisbury, UK) and the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany). Human cervical adenocarcinoma HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). Human mammary carcinoma MCF-7 cells were cultured in RPMI supplemented with 10% FBS. The U87 and U373 cells were cultured in DMEM culture medium (Lonza code 12-136F, Vervier, Belgium), while the SKMEL-28 and A549 cells were cultured in RPMI culture medium (Lonza; code 12-115F) supplemented with 10% heat-inactivated FBS (Lonza, FBS South America code DE14-801F). Cell culture media were supplemented with 4 mM glutamine (Lonza code BE17-605E), 100 μg/mL gentamicin (Lonza code 17-5182), and penicillin-streptomycin (200 units/ml and 200 μg/ml) (Lonza code 17-602E). All cell lines were cultured in T25 flasks, maintained and grown at 37° C, 95% humidity, 5% CO2.
(c) Antiproliferative Properties
To evaluate antiproliferative properties of the synthesized compounds, the MTT assay was used. The cell lines were assessed by trypsinizing each cell line and seeding 4 × 103 cells per well into 96-well plates. All compounds were dissolved in DMSO at a concentration of either 100 mM or 50 mM prior to cell treatment. The cells were grown for 24 h and then treated with compounds at concentrations ranging from 0.04 to 100 μM and incubated for 48 h in 200 μL media. 20 μL of MTT reagent in serum free medium (5 mg/mL) was added to each well and incubated further for 2 h. Media was removed, and the resulting formazan crystals were re-solubilized in 200 μL of DMSO. A490 was measured using a Thermomax Molecular Device plate reader. The experiments were performed in quadruplicate and repeated at least twice for each compound per cell line. Cells treated with 0.1% DMSO were used as a control, and 1 μM phenyl arsine oxide (PAO) was used as a positive killing control.
(d) Morphological Analysis of Microtubule Organization in HeLa Cells
HeLa cells were cultured in EMEM (Lonza, Walkersville, MD) supplemented with 10% FBS (Atlanta Biologicals, Lawrenceville, GA), sodium pyruvate and sodium bicarbonate. Cells were treated for 3 h with either carrier (0.1% DMSO) or 13 solubilized in DMSO and prediluted in media. Following fixation by immersion in methanol at −20°C for 30 min, cells were subsequently rehydrated in phosphate-buffered saline (1XPBS), and blocked by incubation in 3% bovine serum albumin (dissolved in 1XPBS) for one hour at room temperature. Cells were then incubated overnight at 4 °C with mouse anti-tubulin antibody (Sigma) and rabbit anti-pericentrin antibody (Abcam, Cambridge, MA) in blocking buffer. Primary antibodies were detected using Alexafluor-conjugated secondary antibodies (Molecular Probes), while DNA was detected using Hoechst 33342 (Invitrogen). All the images were acquired using a Zeiss Axiovert 200M inverted microscope equipped with epifluorescence optics and an Apotome Structured Illumination module (Carl Zeiss, Thornwood, NY). Acquired 8-bits images were exported and figures were prepared using Adobe Photoshop CS5 software.
(e) Molecular Modeling
Multiple crystal structures of tubulin co-crystallized with ligands at the colchicine binding site were downloaded from the PDB and compared in terms of resolution and incomplete residues. The structure 3UT5 was found to be the most suitable receptor for modelling purposes. From this structure chain B was retained, along with the corresponding co-crystallized colchicine ligand. All other chains, ligands and water molecules were deleted. A minimization was performed on the receptor ligand complex using Accelrys Discovery Studio 4.0 (Smart Minimizer) and the CHARMm forcefield. Fixed atom constraints were applied to all non-hydrogen atoms, and a GBSW solvent model was employed. Docking studies were performed using the CDocker algorithm in Accelrys Disovery Studio 4.0, employing 150 starting ligand conformations and 75 structures for refinement per ligand.
(f) Quantitative videomicroscopy
The effects of 22 on the viability of human U373 glioblastoma and SKMEL melanoma cells were characterized in vitro using computer-assisted phase contrast video microscopy, as described elsewhere.30
Supplementary Material
Acknowledgment
This project was supported by the Texas State University start-up funding and National Institute of General Medical Sciences (P20GM103451). The modeling studies were made possible by the Centre for High Performance Computing (Rosebank, Cape Town), who provided the access to Accelrys Discovery Studio. SCP and WALvO acknowledge funding from the National Research Foundation (NRF) and the University of Stellenbosch, South Africa.
References
- 1.Urban S, Hickford SJH, Blunt JW, Munro MHG. Curr. Org. Chem. 2000;4:765. [Google Scholar]
- 2.Hill RA. Annu. Rep. Prog. Chem., Sect. B. 2005;1001:124. [Google Scholar]
- 3.Simmons TL, Andrianasolo E, McPhail K, Flatt P, Gerwick WH. Mol. Cancer Ther. 2005;4:333. [PubMed] [Google Scholar]
- 4.Dembitsky VM, Gloriozova TA, Poroikov VV. Mini-Rev. Med. Chem. 2005;5:319. doi: 10.2174/1389557053175362. [DOI] [PubMed] [Google Scholar]
- 5.Nakao Y, Fusetani N. J. Nat. Prod. 2007;70:679. doi: 10.1021/np060600x. [DOI] [PubMed] [Google Scholar]
- 6.Sugumaran M, Robinson WE. Mar. Drugs. 2010;8:2906. doi: 10.3390/md8122906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan H, Peng J, Hamann MT, Hu J-F. Chem. Rev. 2008;108:264. doi: 10.1021/cr078199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ploypradith M, Batsomboon P, Ruchirawat S, Ploypradith P. ChemMedChem. 2009;4:457. doi: 10.1002/cmdc.200800339. [DOI] [PubMed] [Google Scholar]
- 9.Mabuchi S, Hisamatsu T, Kawase C, Hayashi M, Sawada K, Mimura K, Takahashi K, Takahashi T, Kurachi H. T. Kimura. Clin. Cancer Res. 2011;17:4462. doi: 10.1158/1078-0432.CCR-10-2987. [DOI] [PubMed] [Google Scholar]
- 10.Kabayashi J, Cheng J, Kikuchi Y, Ishibashi M, Yamamura S, Ohizumi Y, Ohta T, Nozoe S. Tetrahedron. Lett. 1990;31:4617. [Google Scholar]
- 11.Tsuda M, Nozawa K, Shimbo K, Kobayashi J. J. Nat. Prod. 2003;66:292. doi: 10.1021/np020393a. [DOI] [PubMed] [Google Scholar]
- 12.Davis RA, Christensen LV, Richardson AD, Moreira da Rocha R, Ireland CM. Mar. Drugs. 2003;1:27. [Google Scholar]
- 13.Frolova LV, Evdokimov NM, Hayden K, Malik I, Rogelj S, Kornienko A, Magedov IV. Org. Lett. 2011;13:1118. doi: 10.1021/ol103149b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frolova LV, Magedov IV, Romero AE, Karki M, Otero I, Hayden K, Evdokimov NM, Banuls LMY, Rastogi SK, Smith WR, Lu SL, Kiss R, Shuster CB, Hamel E, Betancourt T, Rogelj S, Kornienko A. J. Med. Chem. 2013;56:6886. doi: 10.1021/jm400711t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isobe Y, Tobe M, Ogita H, Kurimoto A, Ogino T, Kawakami H, Takaku H, Sajiki H, Hirota K, Hayashi H. Bioorg. Med. Chem. 2003;11:3641. doi: 10.1016/s0968-0896(03)00369-9. [DOI] [PubMed] [Google Scholar]
- 16.Macphee JA, Panaye A, Dubois JE. Tetrahedron. 1978;34:3553. [Google Scholar]
- 17.Bott G, Field LD, Sternhell S. J. Am. Chem. Soc. 1980;102:5618. [Google Scholar]
- 18.Gillies TE, Cabernard C. Curr. Biol. 2011;21:599. doi: 10.1016/j.cub.2011.06.055. [DOI] [PubMed] [Google Scholar]
- 19.Ranaivoson FM, Gigant B, Berritt S, Joullie M, Knossow M. Acta Crystallogr., Sect. D. 2012;68:927. doi: 10.1107/S0907444912017143. [DOI] [PubMed] [Google Scholar]
- 20.Haar CP, Hebbar P, Wallace GC, IV, Das A, Vandergrift WA, III, Smith JA, Giglio P, Patel SJ, Ray SK, Banik NL. Neurochem. Res. 2012;37:1192. doi: 10.1007/s11064-011-0701-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallego MA, Ballot C, Kluza J, Hajji N, Martoriati A, Castera L, Cuevas C, Formstecher P, Joseph B, Kroemer G, Bailly C, Marchetti P. Oncogene. 2008;27:1981. doi: 10.1038/sj.onc.1210833. [DOI] [PubMed] [Google Scholar]
- 22.Soengas MS, Lowe SW. Oncogene. 2003;22:3138. doi: 10.1038/sj.onc.1206454. [DOI] [PubMed] [Google Scholar]
- 23.For an example of apoptosis resistant glioma cells, see: Mircic A, Vilimanovic U, Brajuskovic G, Bumbasirevic V. Acta Veterin. 2012;62:17.
- 24.For an example of apoptosis resistant melanoma cells, see: Merighi S, Mirandola P, Varani K, Gessi S, Capitani S, Leung E, Baraldi PG, Tabrizi MA, Borea PA. Biochem. Pharmacol. 2003;66:739. doi: 10.1016/s0006-2952(03)00400-3.
- 25.For an example of induction of a non-apoptotic cell death in colon cancer and melanoma cells, see: Biggers JW, Nguyen T, Di X, Gupton JT, Henderson SC, Emery SM, Alotaibi M, White KL, Jr., Brown R, Almenara J, Gewirtz DA. Cancer Chemother. Pharmacol. 2013;71:441. doi: 10.1007/s00280-012-2024-6.
- 26.Branle F, Lefranc F, Camby I, Jeuken J, Geurts-Moespot A, Sprenger S, Sweep F, Kiss R, Salmon I. Cancer. 2002;95:641. doi: 10.1002/cncr.10710. [DOI] [PubMed] [Google Scholar]
- 27.Mathieu V, Pirker C, Martin de Lasalle E, Vernier M, Mijatovic T, De Neve N, Gaussin JF, Dehoux M, Lefranc F, Berger W, Kiss R. J. Cell. Mol. Med. 2009;13:3960. doi: 10.1111/j.1582-4934.2009.00708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y, Chen J, Xiao M, Li W, Miller DD. Pharm. Res. 2012;29:2943. doi: 10.1007/s11095-012-0828-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Best MD. Biochemistry. 2009;48:6571. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- 30.Debeir O, Megalizzi V, Warze N, Kiss R, Decaestecker C. Exp. Cell Res. 2008;314:2985. doi: 10.1016/j.yexcr.2008.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








