Abstract
Background
Circulating tumor cell (CTC)-filtration methods capture high numbers of CTCs in non-small-cell lung cancer (NSCLC) and metastatic prostate cancer (mPCa) patients, and hold promise as a non-invasive technique for treatment selection and disease monitoring. However filters have drawbacks that make the automation of microscopy challenging. We report the semi-automated microscopy method we developed to analyze filtration-enriched CTCs from NSCLC and mPCa patients.
Methods
Spiked cell lines in normal blood and CTCs were enriched by ISET (isolation by size of epithelial tumor cells). Fluorescent staining was carried out using epithelial (pan-cytokeratins, EpCAM), mesenchymal (vimentin, N-cadherin), leukocyte (CD45) markers and DAPI. Cytomorphological staining was carried out with Mayer-Hemalun or Diff-Quik. ALK-, ROS1-, ERG-rearrangement were detected by filter-adapted-FISH (FA-FISH). Microscopy was carried out using an Ariol scanner.
Results
Two combined assays were developed. The first assay sequentially combined four-color fluorescent staining, scanning, automated selection of CD45− cells, cytomorphological staining, then scanning and analysis of CD45− cell phenotypical and cytomorphological characteristics. CD45− cell selection was based on DAPI and CD45 intensity, and a nuclear area >55 μm2. The second assay sequentially combined fluorescent staining, automated selection of CD45− cells, FISH scanning on CD45− cells, then analysis of CD45− cell FISH signals. Specific scanning parameters were developed to deal with the uneven surface of filters and CTC characteristics. Thirty z-stacks spaced 0.6 μm apart were defined as the optimal setting, scanning 82 %, 91 %, and 95 % of CTCs in ALK-, ROS1-, and ERG-rearranged patients respectively. A multi-exposure protocol consisting of three separate exposure times for green and red fluorochromes was optimized to analyze the intensity, size and thickness of FISH signals.
Conclusions
The semi-automated microscopy method reported here increases the feasibility and reliability of filtration-enriched CTC assays and can help progress towards their validation and translation to the clinic.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-016-2461-4) contains supplementary material, which is available to authorized users.
Keywords: Circulating tumor cells, Filtration enrichment, Fluorescent staining, FA-FISH, Predictive biomarkers
Background
Circulating tumor cells (CTCs) captured from blood as a liquid biopsy are currently a very active and promising area in translational cancer research [1]. CTCs are very rare cells – occurring at rates as low as one cell per 106 or 107 leukocytes – which detach from a primary tumor or metastatic site, circulate in the peripheral blood and may colonize secondary sites to form metastasis [2, 3]. To date, identification and characterization of CTCs has been hampered by their rarity, their phenotypical and genetic heterogeneity and the technical limitation of current assays [4]. As a consequence, CTC detection methods commonly rely on two steps, a first enrichment step based on either CTC phenotypical or physical properties, and a secondary step of detection to increase the sensitivity of the assay. The semi-automated CellSearch platform (Janssen Diagnostics, LLC, Raritan, USA) enriches CTC candidates through ferromagnetic beads coated with epithelial cell adhesion molecule (EpCAM) specific antibody and defines CTCs according to the presence of cytokeratins (CK8, CK18 and CK19) and the absence of the common leukocyte antigen CD45 [5]. In prospective multicenter studies in metastatic breast, prostate (mPCa) and colon cancers [6–8], CTC levels using this platform were shown to have prognostic significance, and extensive analytical validation and clinical qualification led the U.S. Food and Drug Administration (FDA) to approve CellSearch as an aid to prognosis in these tumors. In non-small-cell lung cancer (NSCLC), CTC levels were also reported to have a prognostic value, but the number of CTCs caught by the CellSearch remains low in this tumor type even in patients with metastatic advanced disease [9]. The potential reason for this is that CTCs have lost epithelial features, expressing markers of the epithelial-mesenchymal transition (EMT) which can be missed by the CellSearch [10–12]. In addition to prognostic utility, CTCs are currently extensively studied to detect predictive biomarkers and screen eligible patients for targeted therapies [13–20]. Moreover analyses performed on CTCs could be repeated at different time-points during treatment to monitor response and guide therapeutic decisions. Performing molecular analysis in CTCs analyzed by the CellSearch is relatively challenging in cancers such as NSCLC due to the low numbers of CTCs captured and the processing conditions of the system.
The limitations of the CellSearch and other antibody-based capture methods have stimulated the development of other technologies for CTC enrichment and detection [4]. An alternative approach consisting of distinguishing CTCs from normal hematopoietic cells according to size is being actively explored. Several filter devices have been described including ISET (ISET, isolation by size of epithelial tumor cells, Rarecells, Paris, France) [21–23], ScreenCell [24], the Accucyte system [25], CellSieve [26], the Portable Filter-based Microdevice [27], VyCAP [28], the Siemens prototype [29] and the Parsortix system [30]; allowing identification of CTCs according to cytomorphological, phenotypical or molecular characteristics. Ours and other groups have reported that higher numbers of CTCs are captured in cancers such as NSCLC and mPCa using a filtration system compared to CellSearch [10, 26, 31, 32]. CTCs expressing mesenchymal and epithelial markers were reported in NSCLC, mPCa and breast cancer patients using filtration enrichment [11, 12, 26, 33, 34] and CTCs enriched by ISET were reported to have a prognostic value in resected NSCLC patients [31]. By capturing CTCs independent of surface-marker expression, filtration methods may offer a significant advantage for covering the phenotypic and genetic heterogeneity of CTCs, and identification of predictive markers. We and another group reported the detection of ALK-rearrangement in CTCs ISET-enriched by filtration in NSCLC patients with an ALK-rearranged tumor [17, 18, 35] as well ROS1-rearrangement in CTCs from NSCLC patients with an ROS1-rearranged tumor [20].
While offering important advantages such as sensitivity of CTC capture and flexibility for CTC characterization following filtration, filtration systems have drawbacks. One problem is that it is difficult to design a filter membrane which sits entirely flat, regardless of the material used: virtually all filter membranes developed today are not microscopically flat. Another is that pores inevitably retain white blood cell debris and fluorescence signals which disturb microscopy analysis, thus cells placed on pores are frequently difficult to analyze. These two problems make the automation of microscopy challenging to implement. Given these difficulties the microscopy analysis of CTCs enriched on filters remains manual, time-consuming due to the high numbers of white blood cells present on filters, and highly operator-dependent. Although automation is an essential step for filtration-enriched CTC assays to progress to the clinic, there is today no published method reporting the semi-automation of microscopy and image analysis of filtration-enriched CTCs.
Here we report the semi-automated method established to analyze filtration-enriched CTCs from NSCLC and mPCa according to two combined assays i.e (i) fluorescent staining and high-resolution cytomorphology; (ii) fluorescent staining and fluorescent in situ hybridization (FISH). The first method aims to identify CTCs according to both phenotypical and cytomorphological parameters and includes the establishment of scanning parameters for selecting and creating an image gallery of CD45− cells, and characterizing CTCs. The second relies on the detection of molecular biomarkers by establishing FISH scanning parameters (z-stacking, step i.e. distance between two z-stacks, exposure time) for optimal FISH signal identification in filtration-enriched CTCs.
Methods
Patients
NSCLC and mPCa patients were recruited at the Gustave Roussy, Paris, France. Informed written consent for blood sample collection was obtained from all patients (IDRCB2008-A00585-50). The study was approved by local institutional board and ethics committees. Blood was collected into EDTA tubes.
Blood sample collection and enrichment of CTCs by ISET
CTC enrichment by the ISET filtration system (RareCells, Paris, France) was carried out according to the manufacturer’s protocol, as previously reported [10, 11]. To preserve cell integrity, the filtration pressure was optimized to -7 kPa. After processing, filters were dried, wrapped in an aluminum sheet and stored frozen in plastic bag containing a silica gel desiccant at -20 °C until use.
Fluorescent staining of filtration-enriched CTCs
ISET filters are composed of 10 spots. Each spot (corresponding to filtration of 1 mL blood) was cut out for independent analysis. Filters were thawed and individual spots were immobilized on glass slides using adhesive ribbon. A ‘snick’ was made on each spot to allow the precise relocation of cells between fluorescent staining and cytomorphological staining. After rehydratation in TBS 1X (Thermo Fisher Scientific Inc., Waltham, MA, USA), cell permeabilization was carried out by incubating filters for 7 min at room temperature in TBS 1X-Triton X-100 0.2 % (Roche, Sigma-Aldrich Co. LLC., Saint-Louis, MO, USA). After a wash with TBS 1X, saturation was carried out by incubating filters for 25 min at room temperature in TBS 1X-normal goat serum 5 % (Thermo Fisher Scientific Inc.). Epithelial markers were employed in the “green” channel including mouse anti-pancytokeratin monoclonal antibodies (clone A45-B/B3, AS Diagnostik, Hueckeswagen, Germany; clone C11, Novus Biological, Littleton, CO, USA; clone KL1, Beckman Coulter, Brea, CA, USA; clone OV-TL 12/30, Dako, Les Ulis, France) directly conjugated to Alexa Fluor (AF) 488 using the Zenon Mouse IgG Labeling Kit (Thermo Fisher Scientific Inc.) and EpCAM/CD326 AF488 (clone VU1D9, Novus Biological). An anti-vimentin (clone V9, Santa Cruz Biotechnology, Heidelberg, Germany) or an anti-N-cadherin (clone 32/N-Cadherin, BD Biosciences, Franklin Lakes, NJ, USA) conjugated in AF546 and allophycocyanin (APC)-conjugated anti-CD45 (clone HI30, BD Biosciences) were used. Antibodies incubation was carried out 25 min in a humidity dark chamber. After two washes with TBS 1X-Tween20 0.05 % (Dako) and TBS 1X, 4′,6-diamidino-2-phenylindole (DAPI) or Hoechst 33342 (Sigma-Aldrich) was added for 10 min. ISET spots were mounted between slide and coverslip using Ibidi mounting medium (Biovalley, Nanterre, France). Slides were stored at +4 °C until scanning.
Cytomorphological staining of filtration-enriched CTCs
After fluorescence scanning, the coverslip and the mounting medium were removed using a wash of PBS 1X, filters were stained with Mayer Hemalun (RAL Diagnostics, Martillac, France) at room temperature for 30 min or with Diff-Quik (Siemens Healthcare diagn., Munich, Germany) according to the manufacturer's protocol. ISET spots were mounted using Ibidi mounting medium and stored at +4 °C until scanning.
Scanning and image analysis of combined fluorescent and cytomorphological staining in filtration-enriched CTCs
Scanning and image analysis were carried out using an Ariol scanning system (Leica Biosystems Richmond Inc., Richmond, IL, USA) including a Leica DM6000 B microscope with multibay stages (MB 8). Single interference filter sets for blue (DAPI), green (FITC), red (Texas Red) and dark red (Cy5) filters were used. Calibrations were performed using the Ariol Scan application 4.0.1.5 (Leica Biosystems Richmond Inc.). After delineation of the scanning area (i.e. one entire ISET spot) at ×5 magnification, gain was set at maximum (255) to eliminate risk of fluorochrome bleaching. Exposure time was calibrated for each channel at ×20 magnification. Using only one parameter (i.e. exposure time for adjusting fluorochrome exposure) allowed to compare settings between scans done at different times or by different users. Exposure time for epithelial markers was adjusted to have a very low signal on CD45+ cells (at limit of the background noise) while the exposure time of the mesenchymal marker was set to have a saturated signal on CD45+ cells which are known to be strongly positive. Offset was calibrated as high as possible in the DAPI channel to individualize at best nucleus. Selection of CD45− from CD45+ elements was carried out in real time by the Ariol scanning system. The fluorescence scan takes ~1 h30 for one ISET spot. For cytomorphological staining scanning a pre-scan of the whole spot was performed at ×5 in the brightfield channel, the snick being used to precisely relocate cells. The cytomorphological staining scan takes ~45 min at x 20 magnification. All images were analyzed with the Ariol Review application 4.0.1.5 (Leica Biosystems Richmond Inc.).
Combination of fluorescent staining and FA-FISH of filtration-enriched CTCs
Filters were thawed and individual spots were immobilized on glass slides using adhesive ribbon. A snick was made on each spot for precise CD45− cell relocation for filter-adapted-FISH (FA-FISH) scanning. After rehydratation in TBS 1X (Dako), filters were incubated for 5 min at 98 °C in TBS 1X-EDTA (Dako) and then 30 min in TBS 1X-normal mouse serum 5 % (Thermo Fisher Scientific Inc.). Fluorescent staining was performed overnight with anti-CD45 in a dark humidity chamber. After three washes with TBS 1X-Tween20 0.05 % followed by TBS 1X, nuclear staining was performed using DAPI for 15 min. For FA-FISH, filters were then washed with TBS 1X-Tween 20 0.05 %, incubated for 30 min at room temperature in methanol:acetic acid (9:1) solution and then digested at 37 °C with a pepsin (Sigma-Aldrich) solution at 10 % in 0.01 N HCL. After washing with PBS 1X, filters were fixed at room temperature in formaldehyde solution (Sigma-Aldrich) and dehydrated in successive baths containing increasing concentrations of ethanol (VWR International, Radnor, PA, USA). FA-FISH experiments were performed using several probes according to the manufacturer’s protocol: the Vysis LSI ALK Break Apart Rearrangement Probe Kit (Abbott Molecular Inc., Des Plaines, IL, USA); the Vysis 6q22 ROS1 Break Apart FISH probe RUO Kit (Abbott Molecular Inc.); the ZytoLight SPEC RET Dual Color Break (ZytoVision GmbH, Bremerhaven, Germany); the Vysis EGFR/CEP7 FISH Probe Kit (Abbott Molecular Inc.); the Cytocell c-met amplification Probe Kit (Cytocell Ltd., Cambridge, UK); the ZytoLight SPEC FGFR1/CEN8 Dual Color Probe (ZytoVision GmbH); the FIP1L1-PDGFRA FISH DNA Probe (Dako) with the TelVysion 4q (Abbott Molecular Inc.); the Vysis AR amplification Probe Kit (Abott Molecular Inc.); the Kreatech ERG Break Apart Rearrangement Probe Kit (Kreatech Diagnostics, Amsterdam, Netherlands). After hybridization, filters were washed in stringent wash buffer 1X (Dako) and wash buffer 1X (Dako), then dehydrated again in successive ethanol solutions. Filters were finally mounted using DAPI mounting medium (Dako). All FISH probes were optimized for FA-FISH using cancer cell lines spiked in normal blood and filtered by ISET (Additional file 1: Figure S1).
Scanning and image analysis of combined fluorescent staining and FA-FISH in filtration-enriched CTCs
Scanning and image analysis of fluorescent staining were carried out as described above. For scanning FA-FISH, a pre-scan of the whole ISET spot was first performed at ×5 magnification in the DAPI channel to precisely relocate cells. Thanks to the snick made on the spot, the fluorescence and the FA-FISH scans were linked. A scan pass in the DAPI channel at ×10 magnification was developed for adjustment of focus and FISH capture was performed at ×63 magnification in the small regions containing CD45− cells. The z-stack number, step, gain, offset and exposure time (ranging from 5 to 100 ms) were calibrated at ×63 magnification. The Ariol scanning system adjusted the focus on DAPI every frame. All images were analyzed with the Ariol Review application 4.0.1.5. To determine the best number of z-stacks, filter enriched-CTCs from patients with ALK-, ROS1- and ERG-rearranged tumors were scanned six times from 5 z-stacks to 30 z-stacks. For the best step determination, filter enriched-CTC were scanned four times with steps ranging from 0.5 to 0.8 μm. No bleaching was observed.
Cell lines
The NCI-H2228, HCC78, NCI-H661, A549, H1975 and LNCaP cell lines were cultured in RPMI medium 1640 (Thermo Fisher Scientific Inc.) and TPC1, A-431, MB-MDA-134, Res259, VCaP in DMEM medium (Thermo Fisher Scientific Inc.) supplemented with 10 % fetal bovine serum and maintained in a humidified incubator in 5 % CO2 at 37 °C. Cell lines were spiked at various dilutions into blood from healthy donors filtered on ISET, as done for patient samples.
Results
Semi-automated microscopy method to identify filtration-enriched CTCs by combined phenotypical and cytomorphological analysis
CTCs are most often identified phenotypically through the detection of epithelial markers by immunofluorescent staining. Filtration-enriched CTCs may also be identified according to true cytomorphological criteria such as nucleus size (compared with the size of the filter pores (8 μm) corresponding to the internal control) and irregularity of the nuclear membrane, the presence of a visible cytoplasm and a high nuclear-to-cytoplasmic ratio [10, 31, 36]. To further characterize filtration-enriched CTCs, we established a method where CTCs were identified by combining phenotypical (immunofluorescent staining) and cytomorphological characterization, and report here the semi-automated scanning and image analysis method developed to analyze filters. Using cancer cell lines spiked in normal blood and filtered by ISET, we developed a multi-step process where filters were (i) treated by four-color fluorescent staining, (ii) scanned using an Ariol scanning system and automatically analyzed to select CD45− from CD45+ cells, (iii) treated by a cytomorphological dye, (iv) scanned and analyzed again using the Ariol scanning system to gather for cells of interest phenotypical and cytomorphological images, and cell characteristics (Fig. 1a). To identify CTCs potentially undergoing EMT, four-color fluorescent staining was carried out using epithelial (pan-cytokeratins, EpCAM), mesenchymal (vimentin or N-cadherin) markers, CD45 and nuclear staining (DAPI or Hoechst 33342).
Fig. 1.
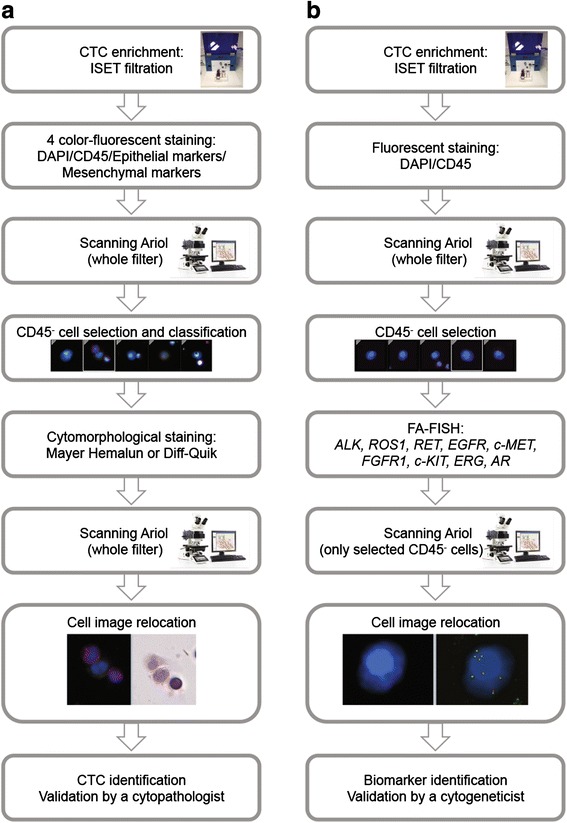
Experimental process for the two combined assays. a Schematic workflow for the identification of filtration-enriched CTCs by combined fluorescent staining and high-resolution cytomorphology. b Schematic workflow for detection of molecular biomarkers in filtration-enriched CTCs by combined fluorescent staining and fluorescent in situ hybridization (FISH)
Three scanning parameters including gain and exposure time – which operate on fluorochrome intensity – and offset – which operates on the fluorescent background of the membrane – were critical for the automated selection of CD45− cells. CD45 exposure time was systematically adjusted as high as possible to optimally discriminate CD45− and CD45+ cells (Fig. 2a). The exposure time of epithelial and mesenchymal markers was adjusted to detect low marker expression as expected in CTCs undergoing EMT (Fig. 2a). Offset calibration in the DAPI channel was critical to fully demarcate nuclei and automatically select individual CD45− from CD45+ cells. To adjust for the uneven surface of the membrane, the focus was determined before scanning using nine focus points across the filter, then readjusted every frame during scanning (~350 times). Using this scanning method, the total number of cells (mostly CD45+) counted per 1 mL ISET spot was highly variable, ranging from 6000 to 20000. The automated selection of CD45− cells was based on three criteria: (i) intensity of DAPI, (ii) intensity of CD45, (iii) a nuclear area superior to 55 μm2 (an area of 50 μm2 corresponds to a pore of 8 μm diameter). In order to select clusters, and especially mixed clusters which contain both CTCs and CD45+ cells, a nuclear area superior to 150 μm2 (equivalent of three pores) was added as a fourth criteria, and the CD45 intensity was not considered in this situation. All selected DAPI+/CD45− elements (usually ranging from 10 to 150 elements per 1 mL ISET spot) were reviewed manually and classified according to their phenotype and nucleus size. Eight categories were established including exclusively epithelial cells (E+/M−), cells in EMT (E+/M+), mesenchymal cells (E−/M+), cells with no markers and a nucleus diameter superior to 16 μm (E−/M− > 2 pores), cells with no markers and a nucleus diameter inferior to 16 μm (E−/M− < 2 pores), CTC clusters (number of CTCs ≥ 4), mixed clusters (number of CD45+ cells and CTCs ≥ 4), and microclusters (number of CD45− and/or CTC = 3). The characteristics of each selected element such as cell area and fluorescence intensities were detailed in an image gallery. An example of A549 cancer cells spiked in normal blood and treated as mentioned above is shown in Fig. 2.
Fig. 2.
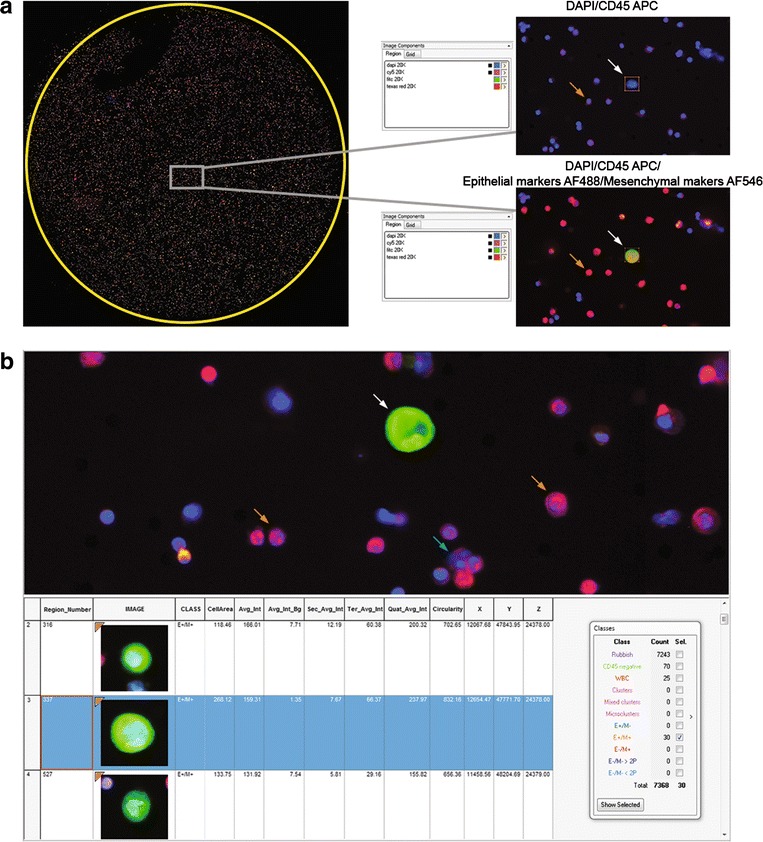
Scanning and image gallery of A459 cells spiked in normal blood and filtered by ISET. a Images of the whole ISET filter (left panel). Grey windows and right panels show DAPI/CD45 APC and DAPI/CD45 APC/Epithelial markers AF488/Mesenchymal markers AF546 channels. b Zoom in images for DAPI/CD45 APC/Epithelial markers AF488/Mesenchymal markers AF546 channels. The image gallery of A549 cells and cell characteristics are shown. Cell categories after automated selection of CD45− cells and manual review of images are shown. The white arrow indicates an A549 cell, orange arrows indicate CD45+ cell into pores, the blue arrow a CD45+ cell outside a pore. Abbreviations: Avg_Int, DAPI average intensity; Avg_Int_Bg, background average intensity, Sec_Avg_Int, CD45 average intensity; Ter_Avg_Int, AF488 average intensity; Quat_Avg_Int, AF546 average intensity
High-resolution cytomorphology was carried out by staining filters with either Mayer Hemalun or Diff-Quik, and scanning in brightfield. Cytomorphological images were precisely relocated within CD45− cell subpopulations using the Ariol scanning system and validated by an experienced cytopathologist (PV). Five criteria were used to define an epithelial CTC including i) presence of a well-defined and non-damaged nucleus inside the cytoplasm, ii) absence of CD45 expression, iii) expression of epithelial markers, iv) nuclear size superior to 1.5 pore diameter (i.e. 12 μm), v) presence of a well-defined cytoplasm. Examples of isolated CTCs from NSCLC and mPCa patients identified using these five criteria and harboring only epithelial markers or both epithelial and mesenchymal markers are shown in Fig. 3 (a and b). In the absence of epithelial marker expression, CTCs were defined as cells presenting all the following cytomorphological criteria: i) nucleus size superior to two pores (i.e. ≥ 16 μm), ii) nucleus irregularity, iii) high nuclear-to-cytoplasmic ratio. CTCs with no detectable epithelial marker expression but cytomorphological characteristics of CTCs were consistently detected in both NSCLC and mPCa patients as shown in Fig. 3 (a and b). Examples of clusters from NSCLC and/or mPCa patients are shown in Fig. 3c.
Fig. 3.
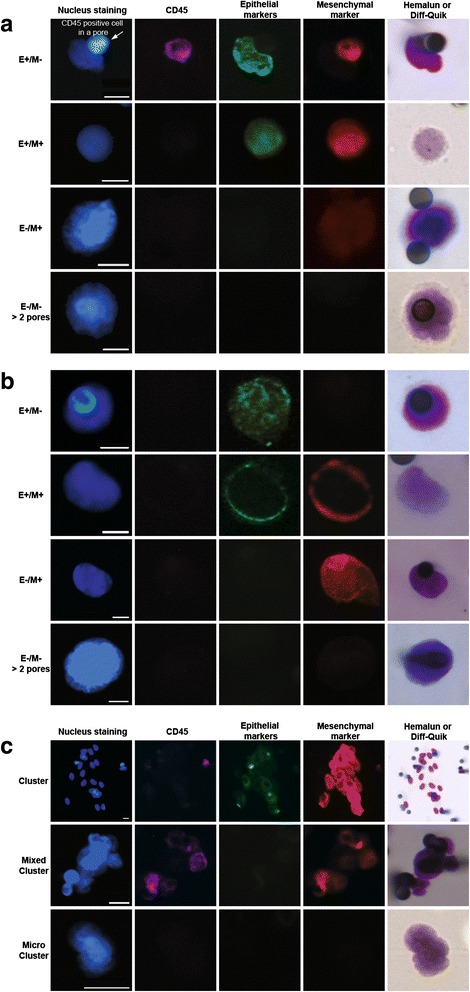
Examples of isolated CTCs (E+/M−, E+/M+, E−/M+, E−/M− > 2 pores respectively) identified by combined fluorescent staining and high-resolution cytomorphology in representative a NSCLC and b mPCa patients. c Examples of CTC clusters, mixed clusters and microclusters respectively in NSCLC and/or mPCa patients. Scale: white bars = 10 μm
Semi-automated microscopy method to identify filtration-enriched CTCs by combined phenotypical and FA-FISH analysis
We previously reported a method for FA-FISH on ISET filters, optimized to decrease non-specific fluorescent probe binding to the filter membrane and confer high cell recovery [18, 20]. Scanning software is commonly able to analyze around 100 cells per tumor sample by FISH, and it was therefore not possible to scan all cells (6000 to 20,000, as mentioned above) present on a 1 mL ISET spot. To decrease the numbers of cells to scan, we limited the FISH analysis to CD45− cells: this option provided the additional advantage of excluding CD45+ subpopulations which could occasionally harbor unspecific FISH rearrangement signals when damaged (data not shown). We established a second multi-step process where filters were (i) treated by fluorescent staining, (ii) scanned on the Ariol scanning system and analyzed automatically to locate CD45− cells as described above, (iii) treated by FA-FISH, (iv) scanned in the small regions containing the CD45− cells, (v) and analyzed for detection, interpretation and validation of FISH signals within CD45− cells (Fig. 1b).
FISH scanning in tumor specimens depends on three parameters: z-stacking (usually ranging from 7 to 11 stacks), the step i.e. the distance between z-stacks (usually 0.6 μm or 0.7 μm), and the exposure time of red and green fluorochromes. FISH scanning of filter enriched-CTCs must take into consideration two particular characteristics of CTCs and filters: first, CTCs are not cut as tissue sections and may have a large nucleus (superior to 30 μm in diameter). These characteristics add to the uneven surface of the filter and strongly influence the above three parameters. Second, CTCs are frequently located in pores which contain DAPI positive debris and fluorescent signals, rendering their analysis challenging.
We first determined the best number of z-stacks to scan filter enriched-CTCs from patients with an ALK-rearranged tumor. Six independent scans of the same filters were performed with an increasing number of z-stacks ranging from 5 to 30, at an arbitrary step of 0.6 μm. Thirty was the maximum z-stack number tolerated by the scanner. As shown in Table 1, 30 z-stacks was the optimal setting to scan CTC FISH spots (82 % detection rate) in an patient with ALK-rearranged tumor, while fewer spots were detected at lower numbers of z-stacks. Thirty z-stacks were also optimal to scan CTC FISH spots in patients with a ROS1-rearranged tumor (91 % detection rate) (Table 2). In a patient with ERG-rearranged tumor, 30 stacks also allowed a 95 % detection rate of CTC FISH spots (Table 3). Thirty z-stacks were therefore established as the optimal setting for scanning filter-enriched CTCs.
Table 1.
FISH spots detected per CTC in a patient with an ALK-rearranged tumor depending on the number of z-stacks
| CTC ID | Number of FISH spots | |||||
|---|---|---|---|---|---|---|
| 5 z-stacks | 10 z-stacks | 15 z-stacks | 20 z-stacks | 25 z-stacks | 30 z-stacks | |
| 1 | n.ia | 9 | 9 | 13 | 10 | 18 |
| 2 | 3 | 8 | 6 | 16 | 15 | 16 |
| 3 | n.ia | 3 | 2 | 8 | 7 | 12 |
| 4 | 4 | 11 | 2 | 4 | 13 | 13 |
| 5 | n.ia | 5 | 6 | 8 | 8 | 17 |
| 6 | n.ia | 12 | 4 | 4 | 2 | 16 |
| 7 | 8 | 6 | 8 | 19 | 1 | 27 |
| 8 | 2 | 4 | 4 | 8 | 10 | 15 |
| 9 | 2 | 2 | 3 | 13 | 7 | 16 |
| 10 | n.ia | 22 | 6 | 14 | 17 | 27 |
| 11 | n.ia | 4 | 8 | 8 | 10 | 16 |
| 12 | 1 | 10 | 6 | 16 | 12 | 16 |
| 13 | 1 | 4 | 2 | 16 | 2 | 3 |
| 14 | n.ia | 6 | 9 | 14 | 14 | 14 |
| 15 | 2 | 4 | n.ia | 4 | 4 | 16 |
| 16 | 1 | 10 | 8 | 2 | 7 | 16 |
| 17 | n.ia | 6 | 9 | 8 | 20 | 9 |
| 18 | 6 | 7 | 7 | 16 | 14 | 16 |
| 19 | 6 | 7 | n.ia | 10 | 12 | 18 |
| 20 | 3 | 8 | 6 | 6 | 8 | 18 |
| 21 | 1 | 3 | 9 | 6 | 8 | 8 |
| 22 | 2 | 5 | 6 | n.i.a | 14 | 8 |
| %b | 0 % | 0 % | 4 % | 23 % | 18 % | 82 % |
Abbreviations: ALK anaplastic lymphoma kinase gene; CTC circulating tumor cell; FISH fluorescence in situ hybridization; n.i. non-interpretable
aNumber of FISH spots is uncountable due to non-optimal focus in the DAPI channel
bPercentage of cases where a higher number of FISH spots were observed
The numbers in bold correspond to the highest number of spots for this CTC
Table 2.
FISH spots detected per CTC in a patient with a ROS1-rearranged tumor depending on the number of z-stacks
| CTC ID | Number of FA-FISH spots | |||||
|---|---|---|---|---|---|---|
| 5 stacks | 10 stacks | 15 stacks | 20 stacks | 25 stacks | 30 stacks | |
| 1 | 9 | n.ia | 6 | 14 | 14 | 30 |
| 2 | 4 | 10 | 10 | 10 | 10 | 10 |
| 3 | 12 | 22 | 18 | 28 | 28 | 28 |
| 4 | 3 | 2 | n.ia | 13 | 14 | 14 |
| 5 | 4 | 4 | 4 | 4 | 4 | 4 |
| 6 | 26 | 28 | 26 | 34 | 34 | 34 |
| 7 | 6 | 15 | 11 | 20 | 28 | 28 |
| 8 | 36 | 42 | 48 | 52 | 62 | 62 |
| 9 | 10 | 14 | 18 | 20 | 30 | 30 |
| 10 | 13 | 20 | 22 | 26 | 30 | 30 |
| 11 | 2 | 12 | 12 | 13 | 14 | 14 |
| 12 | 6 | 6 | 8 | 10 | 14 | 14 |
| 13 | 4 | 6 | 6 | 8 | 14 | 14 |
| 14 | 8 | 13 | 20 | 32 | 32 | 32 |
| 15 | 10 | 14 | 14 | 16 | 16 | 16 |
| 16 | 10 | 12 | 13 | 16 | 16 | 16 |
| 17 | 2 | 6 | 4 | 15 | 16 | 16 |
| 18 | 4 | 11 | 16 | 25 | 25 | 25 |
| 19 | 20 | 33 | 26 | 46 | 55 | 64 |
| 20 | 12 | 14 | 12 | 14 | 15 | 14 |
| 21 | 11 | n.ia | 20 | 24 | 28 | 28 |
| 22 | 10 | 12 | 12 | 20 | 19 | 12 |
| %b | 4 % | 9 % | 9 % | 41 % | 86 % | 91 % |
Abbreviations: CTC circulating tumor cell; FISH fluorescence in situ hybridization; n.i. non-interpretable; ROS1 c-ros oncogene 1
aNumber of FISH spots is uncountable due to non-optimal focus in the DAPI channel
bPercentage of cases where a higher number of FISH spots were observed
The numbers in bold correspond to the highest number of spots for this CTC
Table 3.
FISH spots detected per CTC in a patient with an ERG-rearranged tumor depending on the number of z-stacks
| CTC ID | Number of FA-FISH spots | |||||
|---|---|---|---|---|---|---|
| 5 z-stacks | 10 z-stacks | 15 z-stacks | 20 z-stacks | 25 z-stacks | 30 z-stacks | |
| 1 | 5 | 9 | 2 | 10 | 16 | 22 |
| 2 | 23 | 32 | 22 | 32 | 33 | 36 |
| 3 | 4 | 12 | 6 | 15 | 16 | 16 |
| 4 | 11 | 20 | 10 | 24 | 30 | 32 |
| 5 | 10 | 6 | 12 | 16 | 14 | 16 |
| 6 | 26 | 14 | 2 | 30 | 31 | 45 |
| 7 | 34 | 4 | 2 | 39 | 59 | 59 |
| 8 | 15 | 4 | 4 | 27 | 31 | 31 |
| 9 | 14 | 8 | 2 | 31 | 33 | 34 |
| 10 | 6 | 2 | 16 | 26 | 26 | 31 |
| 11 | 2 | 2 | 4 | 15 | 16 | 22 |
| 12 | 14 | 32 | 2 | 60 | 58 | 61 |
| 13 | 13 | 0 | 20 | 28 | 28 | 28 |
| 14 | 10 | 10 | 12 | 35 | 32 | 35 |
| 15 | 17 | 10 | 10 | 56 | 53 | 65 |
| 16 | 16 | 6 | 8 | 28 | 30 | 32 |
| 17 | 6 | 6 | 10 | 14 | 15 | 16 |
| 18 | 5 | 14 | 2 | 14 | 14 | 20 |
| 19 | 8 | 4 | 6 | 16 | 26 | 31 |
| 20 | 30 | 20 | 26 | 52 | 52 | 44 |
| 21 | 4 | 16 | 6 | 12 | 18 | 27 |
| 22 | 7 | n.ia | 6 | 30 | 29 | 30 |
| %b | 0 % | 0 % | 0 % | 23 % | 23 % | 95 % |
Abbreviations: CTC circulating tumor cell; ERG v-ets avian erythroblastosis virus E26 oncogene homolog; FISH fluorescence in situ hybridization; n.i. non-interpretable
aNumber of FISH spots is uncountable due to non-optimal focus in the DAPI channel
bPercentage of cases where a higher number of FISH spots were observed
The numbers in bold correspond to the highest number of spots for this CTC
We next examined the influence of the step on the number of FISH spots. Scanning of the same filters was performed with an increasing step, ranging from 0.5 μm to 0.8 μm. As shown in a patient with ALK-rearranged tumor (Table 4), a maximal number of FISH spots was detected at a step of 0.6 μm (92 % detection rate). This was confirmed in filters from patients with ROS1- and ERG-rearranged tumors (Additional file 2: Table S1 and Additional file 3: Table S2).
Table 4.
FISH spots detected per CTC in a patient with an ALK-rearranged tumor depending on the step
| CTC ID | Number of FA-FISH spots | |||
|---|---|---|---|---|
| 0.5 μm | 0.6 μm | 0.7 μm | 0.8 μm | |
| 1 | 16 | 16 | 16 | 16 |
| 2 | 16 | 16 | 16 | 16 |
| 3 | 32 | 32 | 32 | 28 |
| 4 | 32 | 32 | 28 | 27 |
| 5 | 16 | 16 | 14 | 13 |
| 6 | 32 | 32 | 32 | 32 |
| 7 | 14 | 14 | 14 | 14 |
| 8 | 13 | 16 | 12 | 16 |
| 9 | 29 | 28 | 32 | 32 |
| 10 | 16 | 16 | 16 | 16 |
| 11 | 16 | 16 | 16 | 16 |
| 12 | 28 | 26 | 31 | 30 |
| 13 | 32 | 32 | 32 | 30 |
| 14 | 32 | 32 | 32 | 32 |
| 15 | 14 | 14 | 14 | 14 |
| 16 | 27 | 28 | 18 | 27 |
| 17 | 16 | 16 | 16 | 16 |
| 18 | 14 | 14 | 14 | 12 |
| 19 | 30 | 32 | 30 | 27 |
| 20 | 16 | 16 | 16 | 14 |
| 21 | 16 | 16 | 16 | 16 |
| 22 | 32 | 32 | 31 | 22 |
| 23 | 32 | 32 | 32 | 27 |
| 24 | 32 | 32 | 32 | 32 |
| %a | 79 % | 92 % | 75 % | 58 % |
Abbreviations: ALK anaplastic lymphoma kinase gene; CTC, circulating tumor cell; FISH, fluorescence in situ hybridization
aPercentage of cases where a higher number of FISH spots were observed
The numbers in bold correspond to the highest number of spots for this CTC
The exposure time of red and green signals was observed to strongly influence the intensity, size and thickness of FISH spots. Because FISH signal intensities were highly variable from one ISET filter region to another one, we developed a multi-exposure protocol to analyze FISH signals consisting of three exposure times for both the green and red fluorochromes (Fig. 4). Twenty-seven settings of exposure time were in theory possible allowing for each candidate CTC the selection of the best setting. As shown in Fig. 4a, the highest exposure was not systematically the best setting, as thick signals can give a false idea of the distance between two spots. To identify CTCs harboring abnormal FISH patterns, signals present in DAPI+/CD45− cells were systematically validated by an experienced cytogeneticist (NA). Examples of filtration-enriched CTCs harboring ALK-, ROS1-, RET-, or ERG-gene alterations from NSCLC and mPCa are illustrated in Fig. 5.
Fig. 4.
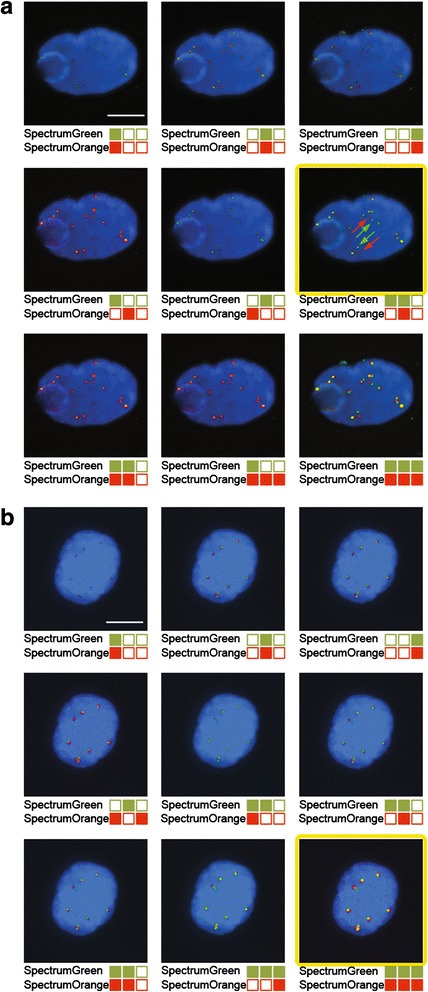
Detection of ALK-rearrangement and gain of ALK-native copies in CTCs from a patient with an ALK-rearranged tumor using the multi-exposure protocol. ALK-gene status was tested in filtration enriched-CTCs by combined fluorescent staining and fluorescent in situ hybridization (FISH). FISH signals were captured with the multi-exposure protocol and 9 out of the 27 possibilities are shown. Images of best exposure level for red and green signals are framed in yellow. a Detection ALK-rearrangement. Green and red arrows indicate the break apart signal. b Detection of gain of ALK-native copies. Scale: white bars = 10 μm
Fig. 5.
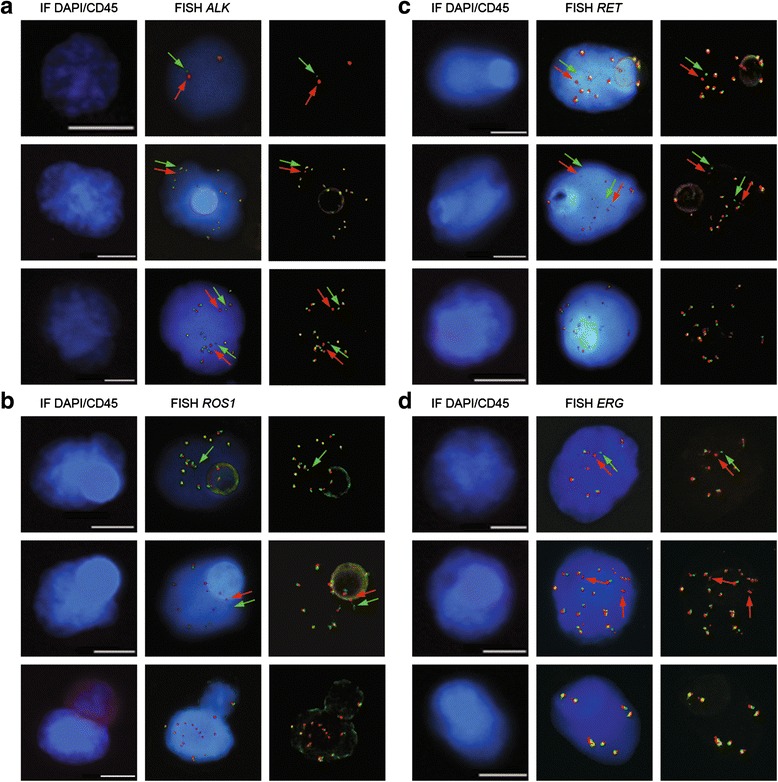
Examples of gene-rearrangement detection in filtration enriched-CTCs from NSCLC and mPCa patients by combined fluorescent staining and fluorescent in situ hybridization (FISH). a Example of ALK-rearrangement detection in NSCLC patients with an ALK-rearranged tumor. b Example of ROS1-rearrangement detection in NSCLC patients with a ROS1-rearranged tumor. c Example of RET-rearrangement detection in NSCLC patients with a RET-rearranged tumor. d Example of ERG-rearrangement detection in mPCa patients with an ERG-rearranged tumor. Gene rearrangements are shown by green and red. Scale: white bars = 10 μm
Discussion
Here we reported the development of a semi-automated microscopy method for analyzing filtration-enriched CTCs using two combined assays. The first assay allowed identification of CTCs by combining four-color fluorescence to high-resolution cytomorphological analysis. The microscopy method of this first assay included establishment of scanning parameters, definition of criteria for creating an image gallery of CD45− cells, and definition of criteria to identify CTCs among CD45− cells based on both phenotypical and true cytomorphological characteristics. The second process was developed to identify molecular biomarkers in CTCs by combining fluorescent staining and FISH. After identification and location of CD45− cells on filters as established in the first assay, the method included the establishment of parameters for scanning FA-FISH into filter regions containing CTC candidates, and the analysis of FISH using a multi-exposure protocol for optimal definition of signals. These two assays were possible due to the capacity of the Ariol scanning system to perfectly relocate images i.e. images of fluorescence and cytomorphology, and images of fluorescence and FA-FISH signals in CD45− cells.
In contrast to antibody-based immunomagnetic enrichment, high numbers of CD45+ cells are retained by most filtration systems [10, 26, 32, 37]. The automated selection of CD45− cells greatly reduces the number of cells to analyze (here approximately 100 cells instead of 6000 to 20000) thereby improving the feasibility of the assay and the reliability of the results. CTCs were identified among selected CD45− cell candidates by examining high-resolution images from both four-color fluorescence and cytomorphology. High-resolution cytomorphology is only possible when cells collected on filters are intact and their morphology well preserved during the filtration process. Although very time-consuming given the number of spots to analyze and the high number of CD45+ cells retained per spot, ISET offers the advantage of preserving cellular integrity. In both NSCLC and mPCa patients, we observed CTCs that were negative for epithelial and mesenchymal markers but were validated as true CTCs by a cytopathologist (PV). One cannot completely exclude that CTCs undergoing EMT and expressing a low level of epithelial and mesenchymal markers may be missed by our immunofluorescent assay. Furthermore as shown here, the level of both epithelial and/or mesenchymal marker expression detected in actual filtration-enriched CTCs can be substantially lower than that of spiked cell lines, a difference we also observed using manual microscopy. Although we have no evident explanation for this, the combination of four-color fluorescence to high-resolution cytomorphology contributes to a better characterization of such CTCs and reliability of the results. Therefore by combining phenotypic and cytomorphological data and high-resolution images, the approach described here enables us to better characterize CTCs as well as detect more CTCs than with phenotypic analyses alone, further highlighting the considerable phenotypic and cytomorphological heterogeneity of CTCs.
FISH analysis of tumor specimens for diagnostic testing of biomarkers such as ALK- or ROS1-rearrangement is in most cases still done manually by cytogeneticists. Tumor cells are identifiable by their morphology and the manual analysis offers the possibility to perform as many stacks as necessary for capturing all FISH spots. Manual FISH analysis of filters is laborious given the number of CD45+ cells retained per filter. It may also be a source of errors since it is well known by cytogeneticist that apoptotic CD45+ cells may harbor non-specific break apart FISH signals. Given the size of CTCs and the absence of an even filter surface, 30 z-stacks spaced 0.6 μm apart were required to capture a maximum of FISH spots and identify ALK-, ROS1- and ERG-rearrangement in CTCs from NSCLC and mPCa patients. Pre-analytical steps during ISET filtration were found critical for the quality of FA-FISH and optimal scanning, especially the possibility to filter at a very low pressure enabling preservation of cell integrity. Unlike tissue sections, standard scanning settings were unable to capture all FISH spots present in filtration-enriched CTCs. The multi-exposure protocol offering twenty-seven exposure settings was therefore essential for optimal capture and definition of FISH signals that were systematically validated by an experienced cytogeneticist (NA).
Multiple studies have highlighted the important phenotypic and genetic heterogeneity of CTCs which cannot be covered by a single CTC isolation technique. Some of these studies have also shown the capacity of filtration to enrich CTCs harboring distinct epithelial-to-mesenchymal phenotypes, capturing higher numbers of CTCs compared to antibody-based methods, mostly in NSCLC and mPCa. As a consequence of these advantages and in particular because the number of CTCs is critical for assessing molecular biomarkers, filtration based enrichment methods may be a preferred technique to progress CTCs as a non-invasive approach for selecting specific therapies. Given this clinical perspective, automation of microscopy analysis is a necessary step to improve reproducibility of analyses, reduce risks of errors, inter-operator differences, and progress towards a standardization of filtration-enriched CTC analysis. Furthermore, a second advantage of the automated analysis of filtration-enriched CTC is the possibility to combine successive assays on the same filter. Automation allows precise location of cells on filters, thus relocating cell data and images from successive experiments performed on the same filter. The combination of assays increases the amount of available information, contributing to a better characterization of CTCs and reliability of the results.
Conclusions
While offering important advantages such as sensitivity of CTC capture and flexibility for CTC characterization, filtration systems have disadvantages that make the automation of microscopy challenging to implement. Manual analysis of filtration-enriched CTCs is time-consuming and highly operator-dependent for large scale analyzes and clinical validation of CTC filtration assays. Here we report a semi-automated microscopy method established to identify filtration-enriched CTCs from NSCLC and mPCa, and detect molecular biomarkers such as ALK-, ROS1- and ERG-rearrangements in CTCs. This method may help to improve reproducibility, reduce inter-operator difference and duration of filtration-enriched CTC assays. In addition, data issued from successive experiments may be gathered, further improving and refining CTC characterization. By increasing the feasibility and reliability of filtration-enriched CTC assays, the present method may be helpful to progress towards their standardization and validation. This will contribute to advancing these assays towards the clinic using a routine clinical test.
Abbreviations
AF, alexa fluor; ALK, anaplastic lymphoma kinase; APC, allophycocyanin; AR, androgen receptor; CK, cytokeratin; c-MET, MET proto-oncogene; CTC, circulating tumor cell; Cy5, cyanine 5; DAPI, 4′,6-diamidino-2-phenylindole; DMEM, dulbecco/vogt modified eagle’s minimal essential medium; EDTA, ethylenediaminetetraacetic acid; EGFR, epidermal growth factor receptor; EMT, epithelial-mesenchymal transition; EpCAM, epithelial cell adhesion molecule; ERG, v-ets avian erythroblastosis virus E26 oncogene homolog; FA-FISH, filter-adapted-FISH; FDA, U.S. Food and Drug Administration; FGFR1, fibroblast growth factor receptor 1; FIP1L1, FIP1 like 1; FISH, fluorescence in situ hybridization; FITC, fluorescein isothiocyanate; HCL, hydrogen chloride; IgG, immunoglobulin G; ISET, isolation by size of epithelial tumor cells; MB, multibay; mPCa, metastatic prostate cancer; NSCLC, non-small-cell lung cancer; PBS, phosphate buffered saline; PDGFRA, platelet-derived growth factor receptor, alpha polypeptide; RET, RET proto-oncogene; ROS1, c-ros oncogene 1; RPMI, roswell park memorial institute medium; TBS, tris- buffered saline
Acknowledgements
We thank the patients and their families; Robert Sykes, Matthew Gorton and Karl Laczko (Leica Biosystems Richmond Inc.) for helping in the semi-automated method development.
Funding
EP is supported by the LabEx LERMIT (grant no ANR-10-LABX-0033-LERMIT) and the Fondation pour la Recherche Médicale (grant no FDT20150532072). The authors are grateful for the research support of the Fondation de France (grant no 201300038317), the Fondation ARC pour la Recherche sur le Cancer (grant no 20131200417), Innovative Medicines Initiative 11th Call CANCER ID (IMI-JU-11-2013, 115749), Institut National du Cancer (PRT-K14-032) and Agence Nationale de la Recherche (ANR-CE17-0006-01). CRL was supported by ESMO [translational research fellowship] with the aid of a grant from Hoffmann-La Roche. Any views, opinions, findings, conclusions, or recommendations expressed in this material are those solely of the author (s) and do not necessarily reflect those of ESMO or Hoffmann-La Roche. CRL is also a recipient for the grant DUERTECC/EURONCO (Diplôme Universitaire Européen de Recherche Translationelle et Clinique en Cancérologie).
Availability of data and materials
The datasets supporting the conclusions of this article are stored in Gustave Roussy lab archive directories named immunomonitorage srv-fs-01 and nas-01 for images.
Authors’ contributions
EP performed laboratory analysis, participated to Ariol development and data interpretation. MO performed laboratory analysis, participated to Ariol development and data interpretation. FB contributed to laboratory analysis. AG participated to Ariol development. NA validated FISH experiments. VF contributed to laboratory analysis. CLB contributed to fluorescence staining development. BB recruited NSCLC patients. YL recruited mPCa patients. MN was in charge of blood sample collection. MH participated to Ariol development. CRL was responsible for critical revision of the manuscript. JCS recruited NSCLC patients and contributed to the conception of the study. PV validated cytomorphological images patients and contributed to the conception of the study. FF conceived and supervised the study and the drafting of the manuscript. All authors have read and approved the final version of the manuscript.
Author’s information
We have no further relevant information about authors to include in this section.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The IDRCB2008-A00585-50 study was approved by the Agence Nationale de Sécurité du Médicament (ANSM), ethics committees (Ile de France VI, Groupe Hospitalier Pitié Salepétrière) and local institutional board. According to the protocol informed written consent for blood sample collection was obtained from all patients.
Endnotes
Not applicable.
Additional files
Examples of gene rearrangement and gain/amplification detection in filtration enriched-cell lines by filter-adapted-FISH (FA-FISH). (A) Example of gene rearrangement detection. (B) Example of gain/amplification detection. Scale: white bars = 10 μm. (TIF 8523 kb)
FISH spots detected per CTC in a patient with a ROS1-rearranged tumor depending on the step. (DOC 68 kb)
FISH spots per CTC of a patient with an ERG-rearranged tumor depending on the step. (DOC 63 kb)
Contributor Information
Emma Pailler, Email: emma.pailler@gustaveroussy.fr.
Marianne Oulhen, Email: marianne.oulhen@gustaveroussy.fr.
Fanny Billiot, Email: fanny.billiot@gustaveroussy.fr.
Alexandre Galland, Email: alexandre.galland@leica-microsystems.com.
Nathalie Auger, Email: nathalie.auger@gustaveroussy.fr.
Vincent Faugeroux, Email: vincent.faugeroux@gustaveroussy.fr.
Corinne Laplace-Builhé, Email: corinne.laplace@gustaveroussy.fr.
Benjamin Besse, Email: benjamin.besse@gustaveroussy.fr.
Yohann Loriot, Email: yohann.loriot@gustaveroussy.fr.
Maud Ngo-Camus, Email: maud.ngocamus@gustaveroussy.fr.
Merouan Hemanda, Email: merouan.hemamda@leicabiosystems.com.
Colin R. Lindsay, Email: colin.lindsay@gustaveroussy.fr
Jean-Charles Soria, Email: jean-charles.soria@gustaveroussy.fr.
Philippe Vielh, Email: philippe.vielh@lns.etat.lu.
Françoise Farace, Phone: +33-142115198, Email: francoise.farace@gustaveroussy.fr.
References
- 1.Krebs MG, Metcalf RL, Carter L, et al. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol. 2014;11:129–144. doi: 10.1038/nrclinonc.2013.253. [DOI] [PubMed] [Google Scholar]
- 2.Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer. 2004;4:448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 3.Kim MY, Oskarsson T, Acharyya S, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 5.Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 6.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 7.De Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 8.Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 9.Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29:1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 10.Farace F, Massard C, Vimond N, et al. A direct comparison of Cell Search and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer. 2011;105:847–853. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou JM, Krebs M, Ward T, et al. Circulating tumor cells as a window on metastasis biology in lung cancer. Am J Pathol. 2011;178:989–996. doi: 10.1016/j.ajpath.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lecharpentier A, Vielh P, Perez-Moreno P, et al. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer. 2011;105:1338–1341. doi: 10.1038/bjc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes DF, Walker TM, Singh B, et al. Monitoring expression of HER-2 on circulating epithelial cells in patients with advanced breast cancer. Int J Oncol. 2002;21:1111–1117. doi: 10.3892/ijo.21.5.1111. [DOI] [PubMed] [Google Scholar]
- 14.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attard G, Swennenhuis JF, Olmos D, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–2918. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 16.Leversha MA, Han J, Asgari Z, et al. Fluorescence in situ hybridization analysis of circulating tumor cells in metastatic prostate cancer. Clin Cancer Res. 2009;15:2091–2097. doi: 10.1158/1078-0432.CCR-08-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol. 2012;23:2907–2913. doi: 10.1093/annonc/mds137. [DOI] [PubMed] [Google Scholar]
- 18.Pailler E, Adam J, Barthelemy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2013;31:2273–2281. doi: 10.1200/JCO.2012.44.5932. [DOI] [PubMed] [Google Scholar]
- 19.Ross K, Pailler E, Faugeroux V, et al. The potential diagnostic power of circulating tumor cell analysis for non-small-cell lung cancer. Expert Rev Mol Diagn. 2015;15:1605–1629. doi: 10.1586/14737159.2015.1111139. [DOI] [PubMed] [Google Scholar]
- 20.Pailler E, Auger N, Lindsay CR, et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann Oncol. 2015;26:1408–1415. doi: 10.1093/annonc/mdv165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vona G, Sabile A, Louha M, et al. Isolation by size of epithelial tumor cells : a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paterlini-Brechot P, Benali NL. Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 2007;253:180–204. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 23.Young R, Pailler E, Billiot F, et al. Circulating tumor cells in lung cancer. Acta Cytol. 2012;56:655–660. doi: 10.1159/000345182. [DOI] [PubMed] [Google Scholar]
- 24.Desitter I, Guerrouahen BS, Benali-Furet N, et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011;31:427–441. [PubMed] [Google Scholar]
- 25.Campton DE, Ramirez AB, Nordberg JJ, et al. High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining. BMC Cancer. 2015;15:360. doi: 10.1186/s12885-015-1383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams DL, Stefansson S, Haudenschild C, et al. Cytometric characterization of circulating tumor cells captured by microfiltration and their correlation to the Cell Search ((R)) CTC test. Cytometry A. 2015;87:137–144. doi: 10.1002/cyto.a.22613. [DOI] [PubMed] [Google Scholar]
- 27.Lin HK, Zheng S, Williams AJ, et al. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Wit S, Van Dalum G, Lenferink AT, et al. The detection of EpCAM (+) and EpCAM (-) circulating tumor cells. Sci Rep. 2015;5:12270. doi: 10.1038/srep12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magbanua MJ, Pugia M, Lee JS, et al. A Novel Strategy for Detection and Enumeration of Circulating Rare Cell Populations in Metastatic Cancer Patients Using Automated Microfluidic Filtration and Multiplex Immunoassay. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0141166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hvichia GE, Parveen Z, Wagner C, et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int J Cancer. 2016;138:2894–2904. doi: 10.1002/ijc.30007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofman V, Bonnetaud C, Ilie MI, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res. 2011;17:827–835. doi: 10.1158/1078-0432.CCR-10-0445. [DOI] [PubMed] [Google Scholar]
- 32.Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol. 2012;7:306–315. doi: 10.1097/JTO.0b013e31823c5c16. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong AJ, Marengo MS, Oltean S, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kallergi G, Papadaki MA, Politaki E, et al. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011;13:R59. doi: 10.1186/bcr2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faugeroux V, Pailler E, Auger N, et al. Clinical Utility of Circulating Tumor Cells in ALK-Positive Non-Small-Cell Lung Cancer. Front Oncol. 2014;4:281. doi: 10.3389/fonc.2014.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofman VJ, Ilie MI, Bonnetaud C, et al. Cytopathologic detection of circulating tumor cells using the isolation by size of epithelial tumor cell method: promises and pitfalls. Am J Clin Pathol. 2011;135:146–156. doi: 10.1309/AJCP9X8OZBEIQVVI. [DOI] [PubMed] [Google Scholar]
- 37.Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the Cell Search Assay and the isolation by size of epithelial tumor cell method. Int J Cancer. 2011;129:1651–1660. doi: 10.1002/ijc.25819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are stored in Gustave Roussy lab archive directories named immunomonitorage srv-fs-01 and nas-01 for images.


