Abstract
Condensins associate with DNA and shape mitotic chromosomes. Condensins are enriched nearby highly expressed genes during mitosis, but how this binding is achieved and what features associated with transcription attract condensins remain unclear. Here, we report that condensin accumulates at or in the immediate vicinity of nucleosome‐depleted regions during fission yeast mitosis. Two transcriptional coactivators, the Gcn5 histone acetyltransferase and the RSC chromatin‐remodelling complex, bind to promoters adjoining condensin‐binding sites and locally evict nucleosomes to facilitate condensin binding and allow efficient mitotic chromosome condensation. The function of Gcn5 is closely linked to condensin positioning, since neither the localization of topoisomerase II nor that of the cohesin loader Mis4 is altered in gcn5 mutant cells. We propose that nucleosomes act as a barrier for the initial binding of condensin and that nucleosome‐depleted regions formed at highly expressed genes by transcriptional coactivators constitute access points into chromosomes where condensin binds free genomic DNA.
Keywords: condensin, Gcn5, nucleosome remodelling, RSC
Subject Categories: Cell Cycle; Chromatin, Epigenetics, Genomics & Functional Genomics
Introduction
In most eukaryotes, chromatin fibres metamorphose into compact and individualized rod‐shaped chromosomes during mitosis and meiosis. This profound reorganization, called chromosome condensation, is a strict prerequisite for the accurate segregation of chromosomes. From yeasts to human, chromosome condensation relies upon condensin complexes and topoisomerase II‐α (Topo II). It is widely accepted that Topo II ensures decatenation of sister chromatids and chromosomes (Baxter et al, 2011; Charbin et al, 2014). In contrast, how condensins reconfigure chromosome structure in a cell cycle‐regulated manner remains poorly understood.
Condensins are ring‐shaped ATPases that belong to the family of SMC (Structural Maintenance of Chromosomes) protein complexes, which also include cohesin, responsible for sister chromatid cohesion, and the Smc5/Smc6 complex, which is implicated in DNA damage repair (Thadani et al, 2012; Aragon et al, 2013; Hirano, 2016). Eukaryotic condensins are composed of the Smc2 and Smc4 ATPase subunits, called Cut14 and Cut3 in fission yeast, and three non‐SMC subunits. Smc2 and Smc4 form a V‐shaped heterodimer in which two ATPase heads face each other at the apices of two 50‐nm‐long coiled‐coil arms. A kleisin subunit (called Cnd2 in fission yeast) associates with the ATPase heads, thereby creating a tripartite ring‐like structure, and recruits two HEAT‐repeat containing subunits. Most eukaryotes possess two condensins, called condensin I and II, which are made of the same Smc2/Smc4 heterodimer but contain different sets of the three non‐SMC subunits. Budding and fission yeasts possess a single condensin complex, which is similar to condensin I by protein sequence. Condensin II is nuclear throughout the cell cycle and accumulates on chromosomes at the beginning of prophase. In contrast, condensin I is cytoplasmic during interphase and gains access to chromosomes only from prometaphase to telophase (Ono et al, 2003; Hirota et al, 2004). Fission yeast condensin shows a similar localization pattern as condensin I, with the bulk of the complexes binding chromosomes from prophase to telophase (Sutani et al, 1999). However, a fraction seems to persist on chromosomes during interphase (Aono et al, 2002; Nakazawa et al, 2015).
Studies performed in a wide range of systems have substantiated the idea that condensins modify the topology of chromosomal DNA by introducing positive supercoils (Kimura & Hirano, 1997), by topological entrapment of DNA molecules within their ring‐like structure (Cuylen et al, 2011) and/or by promoting the re‐annealing of unwound genomic DNA segments (Sutani et al, 2015). It remains unclear, however, to what extent each of these molecular activities contribute to the shaping of chromosomes. Equally unclear is how condensins associate with and manipulate chromosomal DNA in the intricate context of the chromatin fibre.
From yeasts to mammals, condensins are enriched at centromeres, telomeres and, along chromosome arms, nearby genes that are highly transcribed by either of the three RNA polymerases (D'Ambrosio et al, 2008; Schmidt et al, 2009; Dowen et al, 2013; Kim et al, 2013; Kranz et al, 2013; Nakazawa et al, 2015; Sutani et al, 2015). Several factors have been implicated in the recruitment of condensins along chromosomes. In budding yeast, the cohesin loading factor Scc2/4 has been reported to promote the full level of association of condensin at tRNA genes and at genes encoding ribosomal proteins (D'Ambrosio et al, 2008). The replication fork blocking protein Fob1, in a complex with the monopolin subunits Csm1 and Lrs4, recruits condensin at rDNA repeats (Johzuka & Horiuchi, 2009). Fission yeast monopolin associates with condensin and contributes to its localization at the rDNA repeats and also at the kinetochore, but plays no role along chromosome arms (Tada et al, 2011). In chicken DT40 cells, the chromokinesin Kif4 has been implicated in the localization of condensins (mostly condensin I) along the longitudinal axes of mitotic chromosomes (Samejima et al, 2012). In human cells, the zinc finger protein AKAP95 directly interacts with the kleisin CAP‐H and takes part in the recruitment of condensin I onto chromatin (Steen et al, 2000; Eide et al, 2002). More recently, it has been reported that condensins I and II bind the N‐terminal tail of histones H2A and H4, respectively (Liu et al, 2010; Tada et al, 2011). However, contacts with histones do not explain the pattern of condensins along chromosomes, and contrasting results have been obtained regarding the role played by histone tails, if any, in the chromosomal association of condensin I (Tada et al, 2011; Shintomi et al, 2015). Thus, the mechanisms through which condensins associate with chromatin remain unclear.
The enrichment of condensin nearby highly expressed genes suggests a crucial link between the localization of condensins and a feature associated with high transcription levels. In line with this, chemical inhibition of RNA Pol II reduces condensin association with mitotic chromosomes during early mitosis in fission yeast (Sutani et al, 2015), and transcription factors have been implicated in the loading of condensin (D'Ambrosio et al, 2008; Iwasaki et al, 2015; Nakazawa et al, 2015). Paradoxically, transcription of DNA repeats by RNA Pol I or Pol II obstructs the stable association of condensin in budding yeast (Johzuka & Horiuchi, 2007; Clemente‐Blanco et al, 2009, 2011). Fission yeast condensin appears excluded from gene bodies and accumulates towards the 3′ ends of highly expressed genes (Nakazawa et al, 2015; Sutani et al, 2015). Moreover, transcription by all three RNA Pols is usually repressed during mitosis in most eukaryotes (Gottesfeld & Forbes, 1997), when the association of condensin with chromosomes reaches its maximum. In budding yeast, the inhibition of RNA Pol I and Pol II by the Cdc14 phosphatase during anaphase is necessary for condensin binding (Clemente‐Blanco et al, 2009, 2011). Thus, the association of condensin with chromatin appears both positively and negatively linked with transcription. What remains unclear, however, is the molecular determinant(s) that defines condensin association sites and what feature(s) associated with transcription takes part in condensin binding.
Active gene promoters are associated with histone H3 acetylated at lysines 9 and 14 (H3K9ac and H3K14ac) (Pokholok et al, 2005; Roh et al, 2005; Wiren et al, 2005). Although the bulk of chromatin is deacetylated during mitosis in mammals (Kruhlak et al, 2001; Patzlaff et al, 2010), traces of H3K9ac and H3K14ac persist at some gene promoters (Wang & Higgins, 2013). Gcn5 is the histone acetyltransferase (HAT) subunit of the SAGA complex, an evolutionarily conserved modular transcription coactivator that acetylates nucleosomes, notably H3K9, H3K14 and H3K18 (Koutelou et al, 2010; Weake & Workman, 2012). Gcn5‐containing SAGA occupies the promoters and coding regions of active genes. At promoters, Gcn5 occupancy increases with transcription rate (Robert et al, 2004; Govind et al, 2007; Johnsson et al, 2009; Xue‐Franzen et al, 2013; Bonnet et al, 2014). By acetylating nucleosomes, SAGA promotes the local formation of an “open” chromatin structure where the transcription pre‐initiation complex assembles.
Using fission yeast as a model system, we show that condensin binding to chromosomes and mitotic chromosome condensation rely upon Gcn5 HAT activity. Although the majority of Gcn5 is transiently displaced from chromosomes in early mitosis and the bulk of chromatin is deacetylated, Gcn5 and acetylated H3 persist at promoters adjoining a number of highly expressed genes. There, Gcn5 and the ATP‐dependent chromatin‐remodelling complex RSC (remodels the structure of chromatin) evict nucleosomes and promote the efficient binding of condensin at these nucleosome‐depleted regions. Our results suggest that nucleosomes constitute a barrier for the localization of condensin, which is overcome by Gcn5‐mediated histone acetylation and chromatin remodelling. Besides providing unanticipated insights into the mechanism of condensin binding to chromatin, our study suggests that the presence of exposed, non‐nucleosomal DNA may be an important feature that attracts condensins to highly expressed genes in eukaryotes.
Results
Gcn5 takes part in mitotic chromosome condensation
Fission yeast cells carrying the thermosensitive cut3‐477 mutation in the Smc4 condensin subunit cease to divide at 36°C, but continue to proliferate at the semi permissive temperature of 32°C, even though condensin binding to chromosomes is reduced and mitotic chromosome condensation is partly impaired (Saka et al, 1994; Tada et al, 2011; Robellet et al, 2014). To identify the factors that collaborate with condensin, we screened for mutations synthetically lethal with cut3‐477 at 32°C (Robellet et al, 2014). We isolated gcn5‐47, a nonsense G765A mutation in the gcn5 open reading frame that is predicted to eliminate the C‐terminal bromodomain of the protein (Fig 1A and B). Deletion of the complete gcn5 open reading frame was also colethal with cut3‐477 at 32°C (Fig 1A) and with the top2‐250 allele of Topo II (Appendix Fig S1A). In sharp contrast, neither gcn5‐47 nor gcn5Δ lowered the restrictive temperature of mutations eso1‐H17 and rad21‐K1, which affect sister‐chromatid cohesion (Appendix Fig S1B). This indicates that lack of Gcn5 function does not confer a blind hypersensitivity to any perturbation in the structure of chromosomes. Thus, Gcn5 interacts positively and specifically with key chromosome condensation factors.
Figure 1. Gcn5 is required for mitotic chromosome condensation.
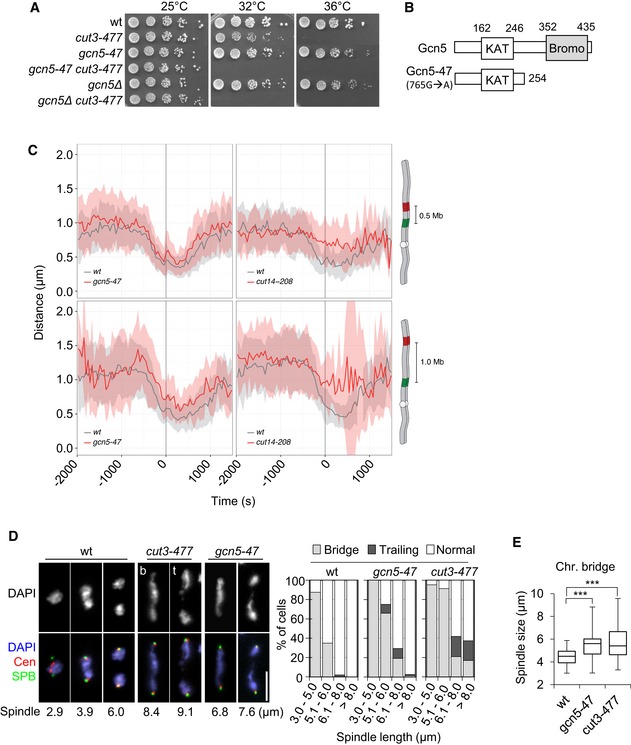
- Genetic interaction between condensin and Gcn5. Fivefold serial dilutions of fission yeast strains were spotted onto complete medium.
- Scheme of Gcn5‐47. KAT: lysine acetyltransferase. Bromo: bromodomain.
- Condensation assay. 3D distances between two fluorescently marked loci on chromosome I were measured by live cell microscopy in fission yeast cells progressing from late G2 through mitosis. Time lapse recording for n > 32 cells for one (wild type) or two (gcn5‐47 and cut14‐208) biological replicates was aligned to anaphase onset (t = 0) and average distances (± s.d.) plotted. Values are listed in Appendix Table S1.
- Chromosome segregation in gcn5‐47 or cut3‐477 mutant cells. Cells growing at 32°C were fixed and processed for immunofluorescence against Mis6‐HA and Cdc11‐GFP to reveal centromeres (Cen) and spindle pole bodies (SPB), respectively. DNA was stained with 4′,6‐diamidino‐2‐phenylindole (DAPI). Left panel: single mutant cells cut3‐477 or gcn5‐47 exhibiting chromatin bridges (b) or trailing chromatin (t). Bar: 5 μm. Right panel: frequencies of chromatin bridges and trailing chromatin as a function of spindle length (SPB‐SPB distance, n > 40 for each category).
- Spindle lengths in cells showing chromatin bridges. Boxes indicate 25th, median and 75th percentile. Whiskers are the minimum and maximum (n > 50). ***P < 0.001.
To assess whether Gcn5 plays a role in mitotic chromosome condensation, we used a quantitative condensation assay that measures the three dimensional distances between two fluorescently labelled loci located on the left arm of chromosome I as cells pass through mitosis (Petrova et al, 2013). In wild‐type cells, the distance between two loci separated by 0.5 or 1.0 Mb of DNA decreased about twofold from G2 phase until late anaphase (Fig 1C). In gcn5‐47 mutant cells, the distances between the two loci combinations remained larger throughout the mitotic time course (Fig 1C), even though condensation was not as severely affected as in the cut14‐208 condensin mutant. The distances between the fluorescently labelled loci were also slightly enlarged during interphase in gcn5‐47 cells (Fig 1C). Reduced acetylation of nucleosomes might relax chromatin fibres during interphase. Alternatively, or additionally, lack of Gcn5 might impair condensin‐mediated chromosome shaping throughout the cell cycle.
Consistent with impaired condensation, gcn5‐47 mutant cells exhibited frequent chromatin bridges or chromatin trailing in anaphase (Fig 1D) and failed to efficiently disentangle the rDNA repeats located on the arms of chromosome III (Fig EV1). These phenotypes are frequently observed as a consequence of defects in mitotic chromosome condensation (Tada et al, 2011), for example in the cut3‐477 condensin mutant (Figs 1D and EV1). However, most chromatin bridges disappeared during late anaphase B in gcn5‐47 mutant cells, unlike in the more severe cut3‐477 mutant. Plotting the number of chromatin bridges as a function of spindle length suggested that chromosome arms eventually achieved complete separation in gcn5‐47 cells, at a time when the mitotic spindle was 25% longer compared to wild‐type cells (Fig 1E). Taken together, these data indicate that Gcn5 is required for the efficient condensation of chromosome arms during mitosis. Because condensation is partly impaired in the absence of Gcn5 function, the complete separation of chromosome arms necessitates a longer mitotic spindle.
Figure EV1. Chromosomes III remain untangled during anaphase in gcn5 mutant cells.
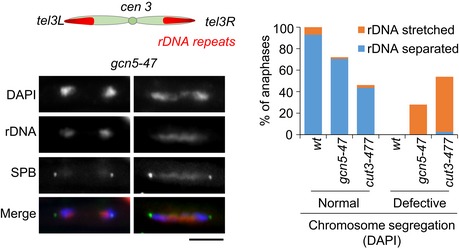
Scheme of chromosome III in which ~100 copies of 10 kb rDNA repeats, each consisting of 5.8S, 18S and 28S rDNA genes, are located adjacent to the telomeres. Fib1, which binds the rDNA repeats, was used to monitor the segregation of the arms of chromosome III in late anaphase cells. Fission yeast cells exponentially growing at 32°C and expressing Fib1‐RFP (rDNA) and Cdc11‐GFP (SPB) were fixed, stained with DAPI and examined for chromosome segregation (DAPI) and rDNA segregation (n > 70). Bar: 5 microns.
Figure EV2. The link between Gcn5 and Cut3 is direct.
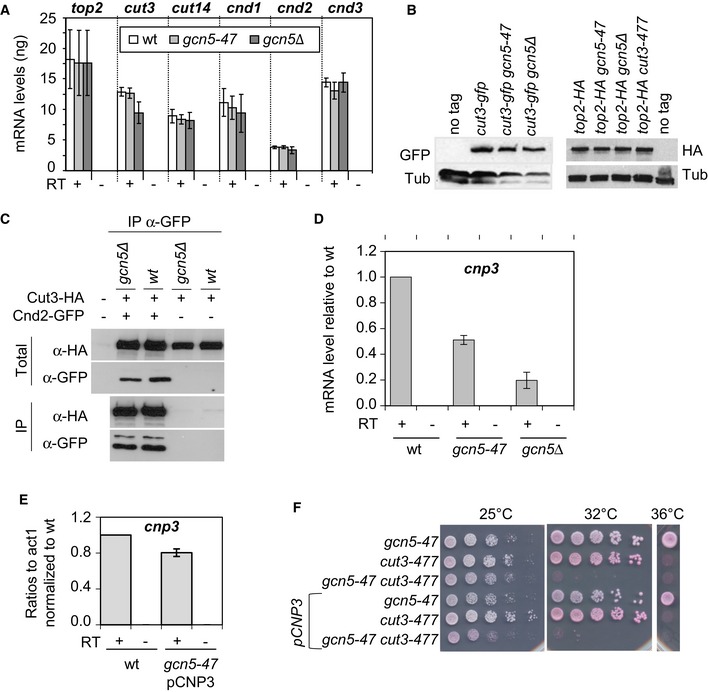
-
ASteady‐state level of condensin and Topo II mRNA in cells lacking functional Gcn5. About 500 ng of total RNA extracted from fission yeast cells exponentially growing at 32°C was reverse‐transcribed in the presence (+) or absence (−) of reverse transcriptase (RT) and cDNAs were quantified by real‐time qPCR. Indicated values correspond to the average and mean deviation from two independent experiments.
-
BSteady‐state levels of Cut3‐GFP and Top2‐HA detected by Western blotting. α‐Tubulin (Tub) was used a loading control.
-
CIntegrity of the condensin complex as judged by co‐immunoprecipitation. Cnd2‐GFP was immunoprecipitated from indicated strains arrested in mitosis (septation indexes <4%). Levels of Cnd2‐GFP and Cut3‐HA in total and immunoprecipitated fractions were assessed by Western blotting.
-
D, ESteady‐state level of cnp3 mRNA measured by RT‐qPCR. About 500 ng of total RNA was reverse‐transcribed in the presence (+) or absence (−) of RT. pCNP3 indicates that the eponym plasmid bearing the cnd3 gene was inserted into the genome. Averages and s.d. calculated from 2 biological replicates are shown.
-
FRestoring cnp3 mRNA level does not suppress the negative genetic interaction between gcn5‐47 and cut3‐477. Cells of indicated genotype were serially diluted fivefold and spotted onto complete media supplemented with phloxin B.
The role of Gcn5 in chromosome condensation relies on its acetyltransferase activity
The function of Gcn5 as a transcriptional coactivator raised the possibility that its effect on chromosome condensation might be indirect, that is that Gcn5 controls the transcription of a bona fide condensation factor. However, our data suggest that this is unlikely. The mRNA levels of Topo II and all five condensin subunits were not notably reduced in gcn5‐47 or gcn5Δ cells (Fig EV2A), nor were Cut3, Cnd2 or Topo II protein levels (Fig EV2B and C). Moreover, efficient co‐immunoprecipitation of Cut3‐HA with Cnd2‐GFP suggested that the integrity of the condensin complex was not affected in the absence of Gcn5 (Fig EV2C). We also re‐analysed available transcriptome data for wild‐type and gcn5Δ mutant cells (Helmlinger et al, 2008) by comparing the mRNA levels of 236 genes that have been reported in pombase (www.pombase.org) to be required for chromosome segregation and/or condensation, or to genetically or physically interact with condensin. Using a threshold of at least 1.5‐fold up‐ or downregulation, we found a single hit: the cnp3 gene, which encodes a centromeric protein (Fig EV2D). However, restoring the level of cnp3 mRNA by ectopic expression did not suppress the colethality between gcn5‐47 and cut3‐477 at 32°C (Fig EV2E and F), indicating that the functional interaction between Gcn5 and condensin was independent of cnp3. We conclude that Gcn5 plays a genuine role in mitotic chromosome condensation.
The Gcn5‐containing SAGA complex consists of three independent functional modules (Weake & Workman, 2012; see Fig 2A): the HAT module, which is composed of Gcn5, Ada2 and Ada3, the Spt module, which includes the Spt8 subunit implicated in the recruitment of the TATA‐binding protein to certain promoters, and the ubiquitin protease module, which contains Sgf11 and Sgf73. To test whether SAGA components other than Gcn5 genetically interact with condensin, we combined deletions of representative subunits of each of the three modules with cut3‐477 (Fig 2A). Lack of the ada2 or ada3 subunits of the HAT module was colethal with cut3‐477 at 32°C, but deletion of subunits of the two other modules had no effect. This finding strongly suggests that condensin is closely linked to the Gcn5 HAT activity. We confirmed this conclusion by testing the catalytically inactive gcn5‐E191Q mutant (Helmlinger et al, 2008) (Fig 2B).
Figure 2. The link between condensin and Gcn5 relies on its ability to acetylate nucleosomes.
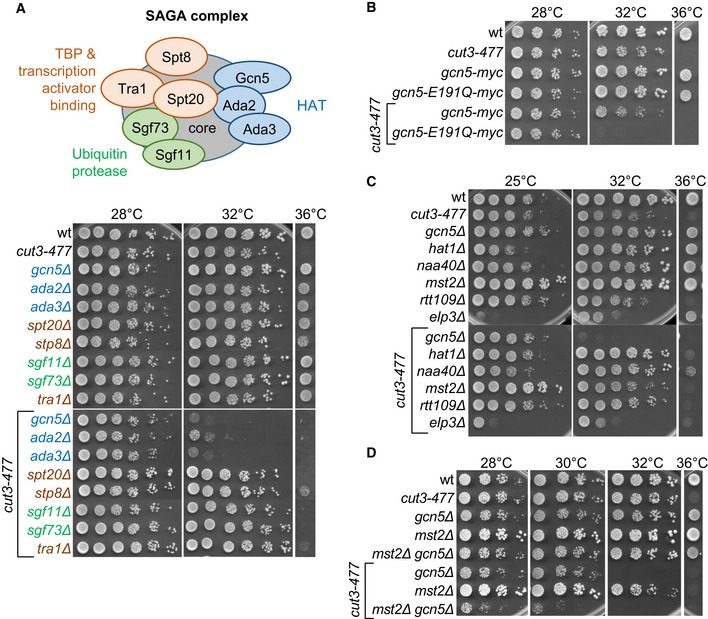
-
A–DGenetic interactions between condensin and (A) subunits of the SAGA complex, (B) catalytically inactive Gcn5, (C) multiples HATs, or (D) Gcn5 and Mst2. Fivefold serial dilutions of fission yeast strains were spotted onto complete medium.
Different HATs can have overlapping functions and Mst2 and Elp3 are partially redundant with Gcn5 (Nugent et al, 2010). Of five additional HATs—Hat1, Naa40, Rtt109, Mst2 and Elp3—tested, none showed an obvious negative genetic interaction with cut3‐477 (Fig 2C). However, simultaneous deletion of Mst2 and Gcn5 reduced viability of the cut3‐477 mutant even further than depletion of Gcn5 alone, which suggests that the two HATs might be partially redundant (Fig 2D). Thus, Gcn5 functionally interacts with condensin through its acetyltransferase activity, possibly in the context of the SAGA complex, and Mst2 is partly redundant with Gcn5 for this function.
Gcn5 and Mst2 facilitate condensin binding to mitotic chromosomes
To test whether Gcn5 and Mst2 might affect the association of condensin with chromosomes, we assessed chromosomal condensin levels using chromatin immunoprecipitation (ChIP) against the kleisin subunit Cnd2 tagged with GFP. Previous genomewide mapping experiments have shown that the condensin‐binding profile along chromosome arms in mitosis consists of low‐occupancy binding sites and hot spots of association, which correlate with highly expressed genes (D'Ambrosio et al, 2008; Schmidt et al, 2009; Nakazawa et al, 2015; Sutani et al, 2015). We quantified Cnd2‐GFP binding using qPCR at the kinetochore (cnt1), pericentric heterochromatin (dh1) and 14 loci along chromosome arms that represent 9 high‐occupancy and 5 low‐occupancy binding sites. Since highly expressed genes can be vulnerable to misleading ChIP enrichments (Teytelman et al, 2013), we first verified that the association of Cnd2‐GFP with these genes was reduced in the condensin mutant cut3‐477 (Fig EV3A–C), as we would expect for bona fide condensin‐binding sites. We then arrested wild‐type or gcn5 mutant cells in pro/metaphase by the nda3‐KM311 tubulin mutation and determined the chromosomal association of Cnd2‐GFP by ChIP. In cells lacking Gcn5, binding of Cnd2‐GFP was significantly reduced at the kinetochore domain and at all high‐occupancy condensin‐binding sites (Fig 3A). ChIP signals were even further decreased in cells lacking both Gcn5 and Mst2 (Fig 3A). The reduced ChIP signals were not due to a decrease in Cnd2‐GFP protein levels in gcn5Δ mst2Δ cells (Fig 3B). In sharp contrast, Cnd2‐GFP occupancy at pericentric heterochromatin (dh1) and at all low‐occupancy condensin‐binding sites remained unchanged.
Figure EV3. Condensin association sites in fission yeast and their transcription.
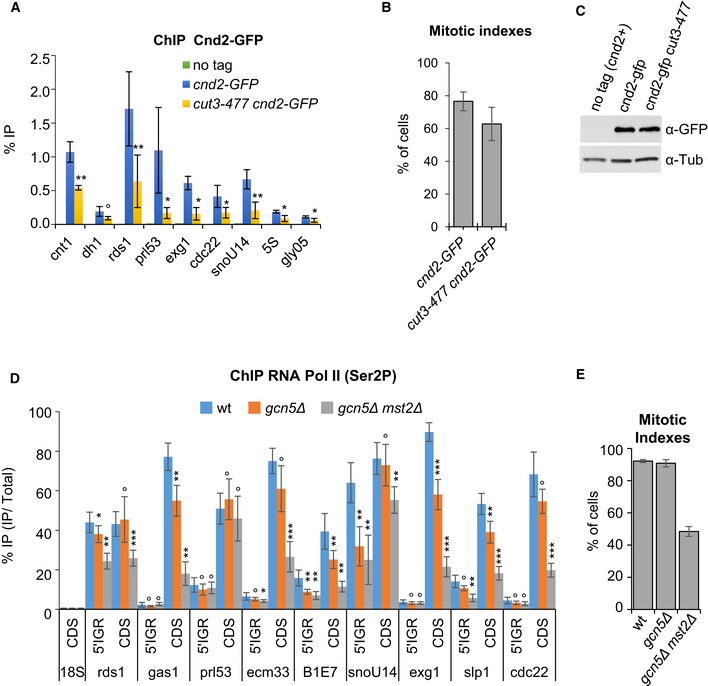
-
ACondensin binding assessed by ChIP. Fission yeast cells were arrested in pro/metaphase at 19°C by the nda3‐KM311 mutation and processed for ChIP against Cnd2‐GFP. % IP correspond to the averages, and s.d. calculated from 6 ChIPs performed on 3 biological replicates.
-
BMitotic indexes. Averages and s.d. calculated from the 3 biological replicates used in (A) are shown.
-
CSteady‐state level of Cnd2‐GFP. Exponentially growing cells were shifted at 36°C for 2.5 h to inactivate cut3‐477, and whole cell extracts assessed for Cnd2‐GFP levels by Western blotting using anti‐GFP (A11122) antibody. Tubulin (Tub) served as loading control.
-
D, ERNA Pol II occupancy assessed by ChIP in mitotically arrested cells. Cells expressing Cnd2‐GFP were arrested in mitosis and processed for ChIP against RNA Pol II phosphorylated on serine 2 (Ser2P). % IP correspond to averages, and s.d. calculated from 8 ChIPs performed on 4 biological replicates. (E) Mitotic indexes. Averages and s.d. calculated from the 4 biological replicates used in (D) are shown.
Data information: ***P < 0.001, **P < 0.01, *P < 0.05 and °P > 0.05.
Figure 3. Gcn5 and Mst2 assists condensin binding to chromatin in mitosis.
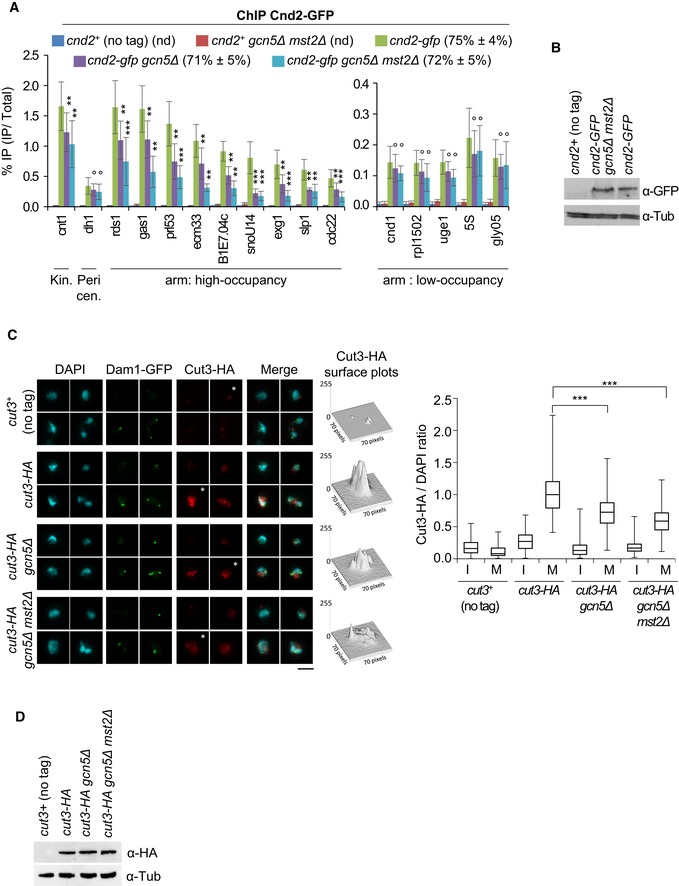
- Condensin binding assessed by ChIP against Cnd2‐GFP. Mitotic indexes are indicated in parentheses. % IP are averages and s.d. calculated from 12 ChIPs on 6 biological replicates. For repeated 5S and gly05 genes, qPCR primers were designed within adjacent, unique 5′ intergenic sequences. Note the use of different scales in the arm: high‐occupancy and arm: low‐occupancy panels. ***P < 0.001, **P < 0.01, °P > 0.05.
- The level of Cnd2‐GFP by Western blotting.
- Condensin binding assessed by chromosome spreading. Cut3‐HA immunofluorescence signals were quantified on n > 100 chromosome spreads exhibiting Dam1‐GFP at kinetochores (mitotic, M) or lacking Dam1‐GFP (interphase, I). For each strain, four representative nuclei extracted from a same image acquired with same settings are shown. Asterisks indicate nuclei corresponding to Cut3‐HA surface plots. Bar: 5 μm. Boxes indicate the 25th, median and 75th percentile and whiskers the min and max values from 3 independent experiments. ***P < 0.001.
- Cut3‐HA level by Western blotting.
To assess the total levels of chromosome‐bound condensin, we measured the amount of Cut3‐HA on mitotic chromosome spreads. To identify mitotic chromosomes, we used Dam1‐GFP, which is recruited to kinetochores specifically during mitosis (Liu et al, 2005). Cut3‐HA was markedly enriched on chromosomes positive for Dam1‐GFP in wild‐type cells (Fig 3C). The amount of Cut3‐HA bound to mitotic chromosomes was reduced by ~30% in the absence of Gcn5 and by ~40% in the absence of both Gcn5 and Mst2, even though total Cut3‐HA protein levels were not reduced (Fig 3D). This result confirms the ChIP experiment and, importantly, rules out the possibility that condensin merely relocates from its canonical high‐occupancy binding sites in the absence of Gcn5 and Mst2. Moreover, we observed no reduction in the association of Topo II or the cohesin loader Mis4 with mitotic chromosomes of gcn5Δ mst2Δ cells (Fig EV4), which reinforces the conclusion that Gcn5 is specifically linked to condensin. Together, these data indicate that Gcn5 and Mst2 specifically assist condensin binding to mitotic chromosomes at core centromeres and at its high‐occupancy sites near genes highly transcribed by RNA Pol II.
Figure EV4. Gcn5 and Mst2 are not required for the chromosomal association of Top2 and Mis4.
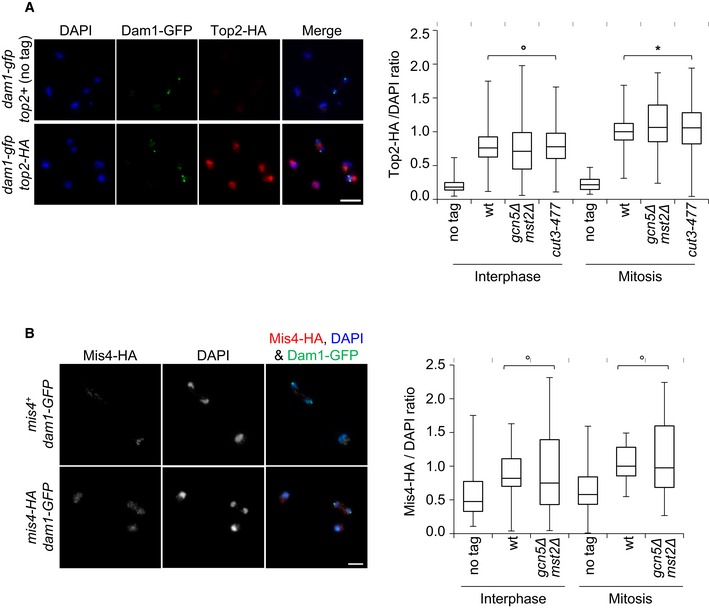
-
A, BThe chromosomal association of Topo II (A) or of the cohesin loader Mis4 (B) assessed by chromosome spreading. Fission yeast cells exponentially growing at 32°C were processed for chromosome spreading and immunofluorescence against Dam1‐GFP and Top2‐HA or Mis4‐HA. Chromatin was stained with DAPI. Dam1‐GFP, recruited at kinetochores during mitosis, was used as a mitotic marker of isolated nuclei. Top2‐HA or Mis4‐HA signals associated with interphase (Dam1‐GFP negative) or mitotic nuclei (Dam1‐GFP positive) were quantified and divided by the intensity of the DAPI signal. Ratios were normalized to the median value given by the wt control in mitosis. Boxes indicate the 25th percentile, median and 75th percentile and whiskers the min and max values calculated from n = 3 experiments. °P > 0.05 and *P < 0.05. Bars: 5 microns.
Gcn5 and acetylated H3 co‐occupy condensin‐binding sites during mitosis
To investigate how Gcn5 could act to regulate condensin binding during mitosis, we assessed transcription of condensin‐binding sites by RNA Pol II, since active transcription is believed to antagonize condensin binding (Johzuka & Horiuchi, 2007; Clemente‐Blanco et al, 2009, 2011). We observed no increase in the occupancy of transcriptionally engaged RNA Pol II throughout condensin‐binding sites during mitosis in the absence of Gcn5 or both Gcn5 and Mst2 (Fig EV3D and E), arguing against this possibility. Next, we investigated the localization of Gcn5 during mitosis. Immunofluorescence studies have shown that Gcn5 dissociates from mitotic chromosomes in vertebrate cells (Orpinell et al, 2010). Using chromosome spreading, we measured the amount of Gcn5‐myc and Cut14‐HA (condensin) on interphase and mitotic chromosomes (Fig 4A). We categorized spread nuclei as interphase, prophase or prometaphase/metaphase by judging the Cut14‐HA/DAPI ratio. We detected less Gcn5 associated with prophase chromosomes as compared to interphase chromosomes, but levels on prometaphase/metaphase chromosomes were comparable to those of interphase ones. These results imply that Gcn5 might only temporarily dissociate from chromosomes during entry into mitosis but then re‐associate at a time when condensin levels further increase. We confirmed by ChIP the presence of Gcn5 on mitotic chromosomes by comparing Gcn5‐myc levels at promoters adjoining condensin‐binding sites in asynchronous cells (80% of which are in G2) and cells arrested in pro/metaphase (Fig 4B). 5 promoters were chosen among high‐occupancy condensin‐binding sites and 4 among low‐occupancy binding sites. In asynchronous cells, we detected Gcn5 in variable amounts at all promoters. In cells arrested in mitosis, the levels of promoter‐bound Gnc5 were considerably higher at all condensin high‐occupancy sites compared to condensin low‐occupancy sites. Remarkably, Gcn5 binding decreased at all low‐occupancy sites during mitosis. In sharp contrast, Gcn5 levels remained unchanged or even increased at all promoters adjoining high‐occupancy sites (Fig 4B), where condensin binding depends on functional Gcn5 (Fig 3A). Whether Gcn5 occupancy increases or drops at promoters during mitosis is therefore linked to condensin occupancy, but unrelated to its absolute binding levels in interphase (compare Gcn5‐myc levels at the slp1 high‐occupancy binding site to the four low‐occupancy sites, Fig 4B). Despite its enrichment at promoters, we failed to detect Gcn5‐myc within adjacent gene bodies or at their 3′ ends during mitosis (Fig EV5A), which might reflect a more dynamic association of Gcn5 with transcribed regions (Bonnet et al, 2014). These data suggest that Gcn5 is specifically retained or even enriched during mitosis at promoters adjoining high‐occupancy condensin association sites, where condensin binding in return relies upon Gcn5.
Figure 4. Gcn5 and acetylated H3 persist at condensin‐binding sites in mitosis.
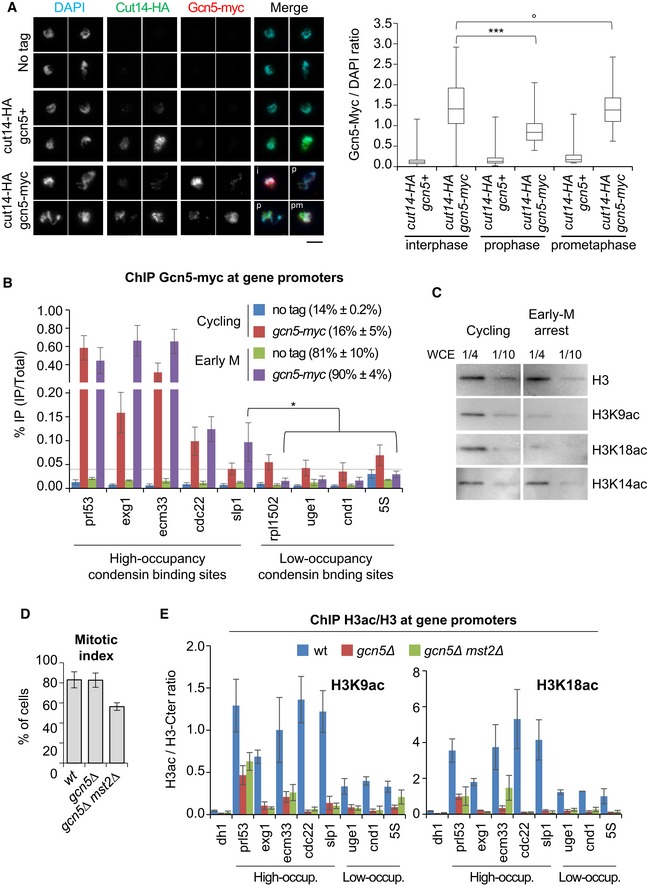
-
AGcn5 binding to chromosomes assessed by chromosome spreading. Fission yeast nda3‐KM311 cells were shifted at 19°C for 4 h to enrich the population for the early stages of mitosis and processed for chromosome spreading and immunofluorescence against Gcn5‐myc and Cut14‐HA. The Cut14/DAPI ratio served as marker for mitotic progression, with interphase (i): ratio < 0.5; prophase (p): 0.5 < ratio < 1.5; and prometaphase/metaphase (pm): ratio > 1.5. For each strain, four representative nuclei extracted from a same image acquired with same settings are shown. Bar: 5 μm. Boxes indicate the 25th, median and 75th percentile and whiskers the min and max values (n > 150 nuclei per strain).
-
BChIP against Gcn5‐myc on cycling cells or cells arrested in pro/metaphase. Mitotic indexes in parentheses were determined by scoring binucleated cells and hypercondensed nuclei. % IP are averages and s.d. calculated from 6 ChIPs on 3 biological replicates.
-
CTotal H3 and acetylated isoforms in cycling or mitotically arrested cells. Whole cell extracts (WCE) prepared from cycling cells or prometaphase cells (mitotic index 89%) were loaded on a same gel and simultaneously revealed.
-
D, EChromosomal occupancy of acetylated H3 during mitosis. Cells expressing Cnd2‐GFP were blocked in early mitosis, mitotic indexes measured (D) and processed for ChIP against total H3, H3K9ac or H3K18ac. Heterochromatin (dh1) served as negative control. H3ac/H3 average ratios and s.d. calculated from 3 ChIPs on 3 biological replicates are shown.
Figure EV5. Gcn5 localization across transcribed genes during mitosis.
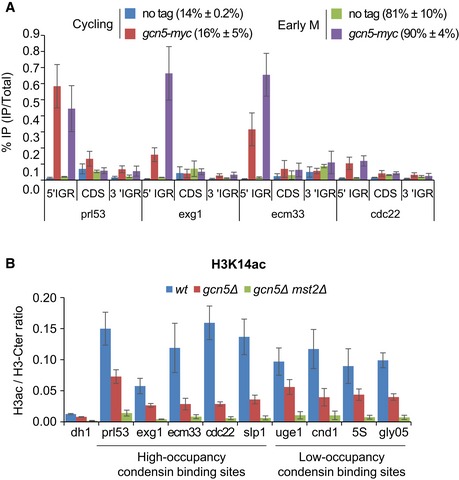
- ChIP against Gcn5‐myc on fission yeast cells cycling or arrested in prometaphase. Mitotic indexes indicated in parentheses were determined by scoring binucleated cells and hypercondensed nuclei. % IP are averages, and s.d. calculated from 6 ChIPs performed on 3 biological replicates.
- The chromosomal occupancy of H3K14ac during mitosis. Cells expressing Cnd2‐GFP were blocked in early mitosis, mitotic indexes measured (see Fig 4D) and cells processed for ChIP against total H3 (H3‐Cter) and H3K14ac. Transcriptionally silent pericentric heterochromatin (dh1) served as negative control. H3ac/H3‐Cter average ratios and standard deviations calculated from 6 ChIPs performed on 3 biological replicates are shown.
To explore the state of chromatin at these promoters, we monitored H3K9ac, H3K18ac and H3K14ac in asynchronous and mitotic cells. Reminiscent of mammalian cells, the bulk of H3K9ac and H3K18ac was reduced in fission yeast cells arrested in pro/metaphase, whilst steady‐state levels of H3K14ac remained unaltered (Fig 4C). We used ChIP to assess whether traces of H3K9ac and H3K18ac might persist at condensin‐binding sites co‐occupied by Gcn5. To control for nucleosome occupancy, we performed in parallel ChIP against total H3 using an antibody directed against the C‐terminal part of H3 (H3‐Ct) and determined the amount of acetylated H3 (H3ac) as a ratio of H3ac/H3‐Ct in cells arrested in mitosis (Fig 4D and E). We found that H3K9ac and H3K18ac levels at promoters correlated with the occupancy of Gcn5, being markedly enriched at the promoters of high‐occupancy condensin‐binding sites when compared to the heterochromatic dh1 site used as negative control (Fig 4E). H3K9ac and H3K18ac were also detected at promoters of low‐occupancy condensin‐binding sites, but acetylation ratios were considerably lower. Moreover, Gcn5 was required for the enrichment of H3K9ac and H3K18ac at all promoters tested (Fig 4E). Deletion of Mst2 did not further reduce H3K9ac and H3K18ac, indicating that these histone modifications were mostly deposited by Gcn5. H3K14ac was also present at promoters in mitosis, but, unlike for H3K9ac and H3K18ac, levels were regulated by both Gcn5 and Mst2 (Fig EV5B). Taken together, these data suggest that the bulk of Gcn5 dissociates from chromosomes during prophase, but a fraction of Gcn5 persists at, or is recruited during prometaphase to, promoters adjoining RNA Pol II‐transcribed genes that are strongly co‐occupied by condensin, where it ensures the persistence of histone acetylation.
Gcn5 plays a role in nucleosome depletion at condensin‐binding sites
Given that Gcn5 takes part in the eviction of nucleosomes at active genes (Govind et al, 2007; Xue‐Franzen et al, 2013), we reasoned that Gcn5 might assist condensin association during mitosis by controlling nucleosome occupancy. To test this idea, we arrested cells in pro/metaphase (Fig 5A) and assessed nucleosome occupancy at condensin‐binding sites by MNase digestion of mitotic chromatin (Figs 5A and EV6A) and massive parallel sequencing of the resulting mononucleosomal DNA fragments (MNase‐seq). We considered as nucleosome‐depleted regions (NDRs) any chromosomal region of at least 150 bp in length exhibiting a normalized MNase‐seq coverage depth smaller than 0.4 (Soriano et al, 2013). With those settings, we identified 6,816 ± 732 NDRs of an average size of 275 ± 6 bp (n = 3 replicates, see Dataset EV1). Given the enrichment of Gcn5 at gene promoters, we assessed nucleosome occupancy upstream of transcription start sites (TSS) of genes bound by condensin. We found that all the 47 high‐occupancy condensin‐binding sites overlapping with Pol II‐transcribed genes, identified by Sutani et al (2015), were situated in the immediate vicinity of a NDR preceding a TSS (Figs 5B, EV6B and EV7, and Dataset EV1). Moreover, 40 out of these 47 high‐occupancy condensin‐binding sites clearly overlapped with NDRs located at the 3′ end of Pol II genes (Figs 5B, EV6B and EV7, and Dataset EV1). Permutation tests confirmed that the colocalization was highly statistically significant (Fig 5C). Furthermore, by re‐assigning all the reads from condensin ChIP‐seq experiments (Sutani et al, 2015) to NDR versus non‐NDR DNA sequences, we found NDR DNA sequences specifically enriched in the fraction co‐immunoprecipitated with condensin (Fig EV6B). This suggests that the vast majority of condensin‐binding sites, and not solely the high‐occupancy ones, overlaps with an NDR and/or resides in the immediate vicinity of an NDR.
Figure 5. Condensin accumulates at nucleosome‐depleted regions dependent upon Gcn5.
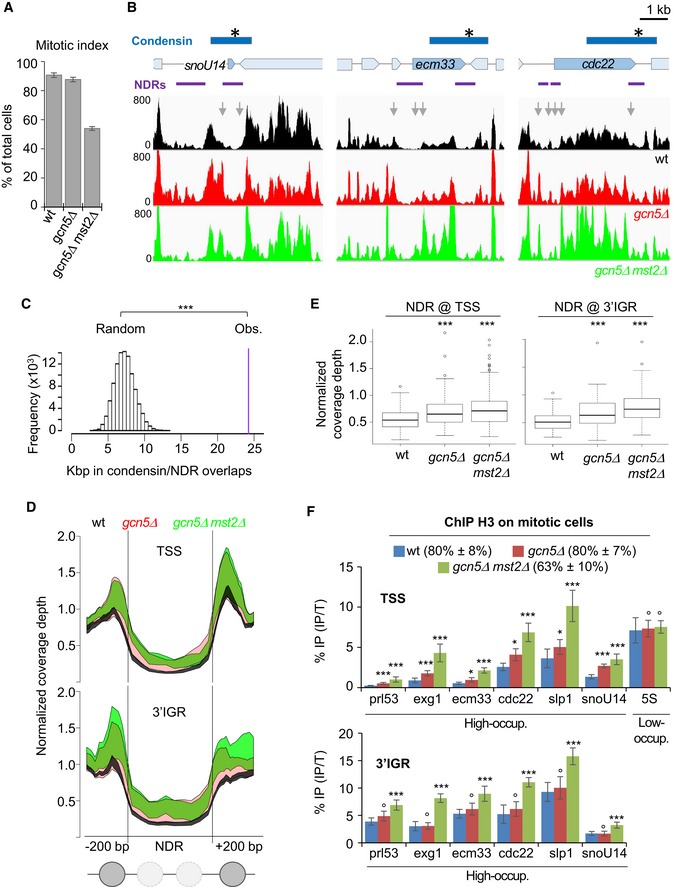
-
A, BNucleosome occupancy at condensin‐binding sites during mitosis. Fission yeast cells expressing Cnd2‐GFP were arrested in mitosis, mitotic indexes determined (A) and cells processed for MNase‐seq. (B) NDRs are shown in purple and condensin‐binding sites in blue with a star indicating the peak maximum. Nucleosome patterns shown are representative examples of 3 biological replicates.
-
CPermutation test. 100,000 sets of 47 DNA segments of the same size of condensin‐binding sites were randomly drawn from the genome and the number of bp associated with a NDR by chance compared with the experimental value (purple bar). ***P < 10−5.
-
DMetagene analysis of nucleosome occupancy within the NDR and positioning of the flanking nucleosomes.
-
ECoverage depth over NDR sequences plus upstream and downstream 200 bp. Boxes indicate the 25th, median and 75th percentile, whiskers indicate the upper and lower 1.5 IQR and circles outliers. ***P = 10−14 by Kruskal–Wallis nonparametric test.
-
FCells arrested in mitosis were processed for ChIP against histone H3. Mitotic indexes are indicated in parentheses. % IP are averages and s.d. calculated from 6 ChIPs on 3 biological replicates. ***P < 0.001, **P < 0.01, °P > 0.05. High‐occup. and Low‐occup. refer to condensin high‐occupancy and low‐occupancy binding sites, respectively.
Figure EV6. Condensin accumulates at nucleosome‐depleted regions dependent upon Gcn5.
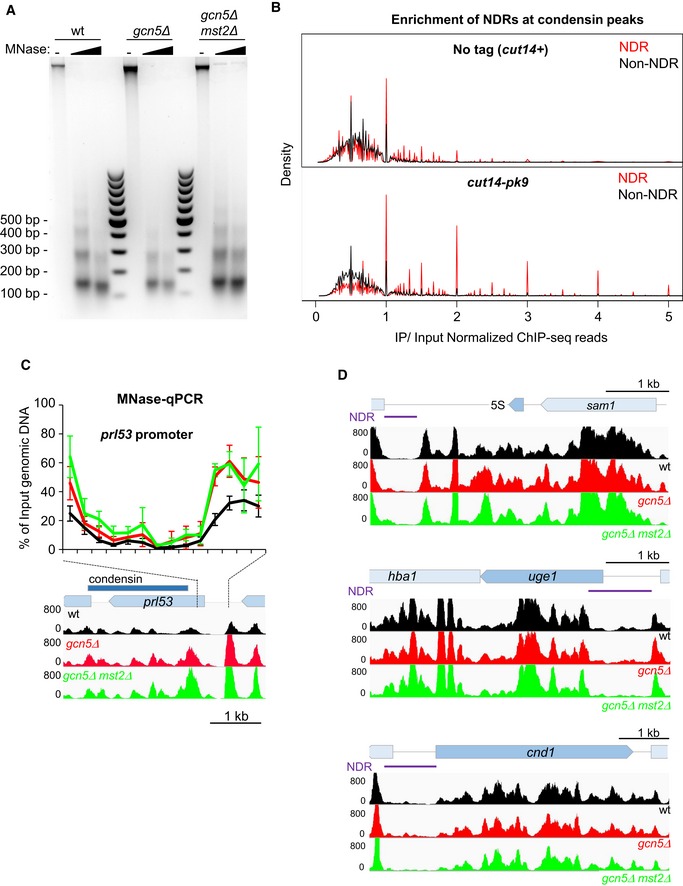
- Fission yeast cells expressing Cnd2‐GFP were arrested in early mitosis, mitotic indexes determined (see Fig 5A) and processed for MNase digestion. Mitotic chromatin was digested with increasing amounts of MNase to obtain mononucleosomes to dinucleosomes ratios of ˜80:20. Mononucleosomal DNA excised from the gel was subjected to massive parallel sequencing (MNase‐seq).
- Overlap between nucleosome‐depleted regions (NDR) or non‐NDRs identified during mitosis by MNase‐seq and condensin peaks identified by ChIP‐seq against Cut14‐pk9 (Sutani et al, 2015). NDRs (red) are enriched in the population of DNA fragments co‐immunoprecipitated with Cut14‐pk9 (IP/Input > 1) and depleted from the flow through (IP/Input < 1).
- Nucleosome scanning (MNase‐qPCR) assay for nucleosome occupancy at the prl53 promoter. Mitotic chromatin was digested by MNase as described in (A). Input (undigested) and mononucleosomal DNA was purified, and the % of input DNA, which was protected from MNase digestion, was assessed by qPCR. Averages and s.d. calculated from 3 experiments on 3 biological replicates are shown. Nucleosome patterns obtained by MNase‐seq analysis (related to Fig 5B) are indicated for comparison.
- Representative MNase‐seq nucleosome patterns at the 5S rRNA, uge1 and cnd1 genes (see Fig 5B). Lack of Gcn5 or both Gcn5 and Mst2 does not significantly modifies nucleosome occupancy at these three low‐occupancy condensin‐binding sites.
Figure EV7. MNase‐seq patterns at three high‐occupancy condensin‐binding sites in replicates.
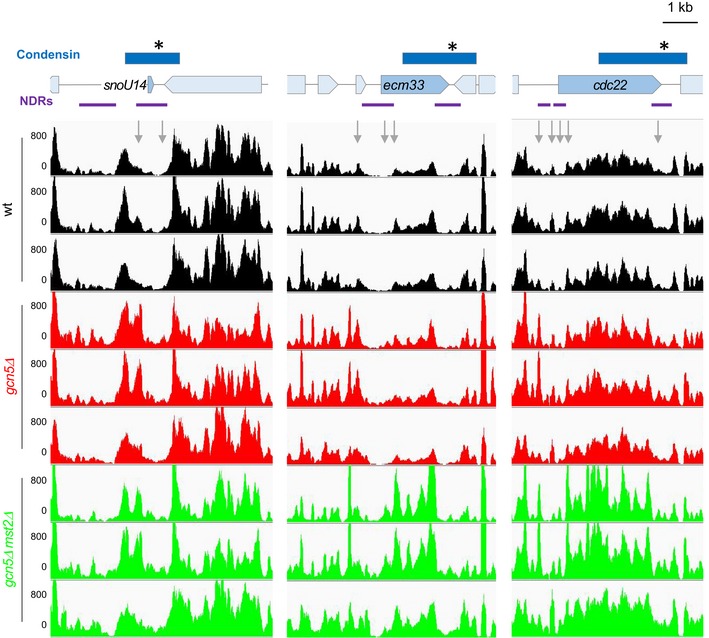
Related to Figs 5A and EV6. MNase‐seq nucleosome patterns at the snoU14, ecm33 and cdc22 genes in the three wt, gcn5Δ or gcn5Δ mst2Δ biological and technical replicates. For each gene and each replicate, results shown in Figs 5A and EV6 correspond to the first line.
The nucleosome pattern significantly changed in gcn5 mutant cells. We analysed nucleosome occupancy at promoters upstream of the 47 condensin‐binding sites and at the 40 NDRs covered by condensin at the 3′ end of genes. In both cases, we found that nucleosome occupancy increased within the core of the NDR and that the positioning of the two nucleosomes delimitating the NDR also markedly increased (Figs 5B, D and E, and EV7). Again, we observed a cumulative effect of gcn5Δ and mst2Δ mutations. We confirmed the increased nucleosome occupancy and the cumulative effect by MNase‐qPCR (Fig EV6C) and by ChIP against histone H3 (Fig 5F). Note that nucleosome occupancy appeared unchanged at the 5S rRNA, cnd1 and uge1 genes in cells lacking Gcn5 or both Gcn5 and Mst2 (Figs 5F and EV6D), where the binding of condensin remained unchanged (Fig 3A). These data indicate that Gcn5 and Mst2 collaborate during mitosis to evict nucleosomes from NDRs, both at gene promoters and at the 3′ end of genes, which constitute high‐occupancy condensin‐binding sites.
Nucleosome eviction assists condensin binding
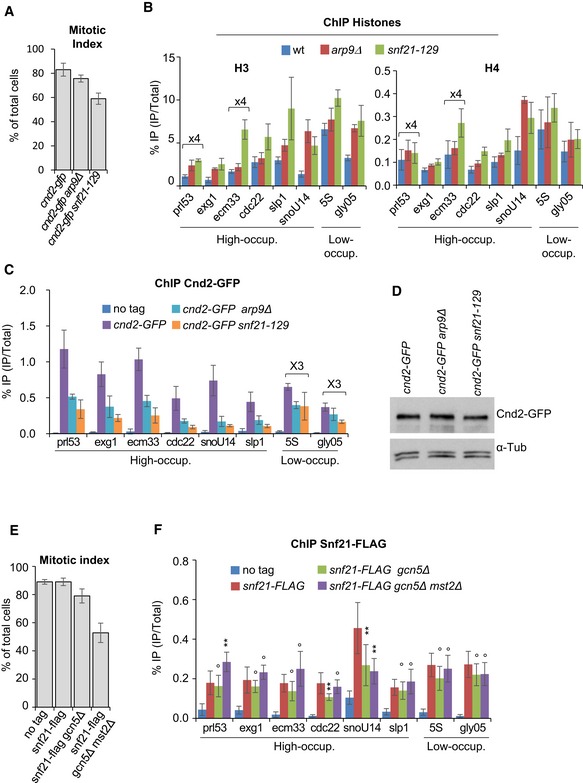
If nucleosome eviction were important for condensin binding, then increasing nucleosome occupancy by means other than Gcn5 or Mst2 inactivation should similarly reduce condensin binding. Arp9 and Snf21 are two subunits of RSC (remodels the structure of chromatin), an ATP‐dependent chromatin‐remodelling complex that evicts nucleosome from promoters (Monahan et al, 2008; Lorch et al, 2014). We had previously identified arp9‐127 and snf21‐129 loss‐of‐function mutations by screening for synthetic lethality with cut3‐477 at 32°C (Robellet et al, 2014). We therefore asked whether RSC could regulate condensin binding to chromosomes during mitosis through its function as nucleosome remodeller. We arrested wild‐type, arp9Δ and snf21‐129 cells in mitosis (Fig 6A) and processed chromatin for simultaneous ChIPs against histones H3, H4 and Cnd2‐GFP. We found that the occupancy of H3 and H4 increased at the promoters of both condensin high‐occupancy (prl53, exg1, ecm33, cdc22, slp1 and snoU14) and low‐occupancy (5S and gly05) sites in the arp9Δ and snf21‐129 mutants (Fig 6B). The occupancy of histone H3 increased also at the 3′ end of some but not all tested genes, although the effects were less dramatic (Fig EV8). This suggests that RSC evicts nucleosomes mainly at a broad range of gene promoters. Remarkably, the binding of Cnd2‐GFP in the arp9Δ and snf21‐129 mutants concomitantly dropped at all condensin‐binding sites, including 5S and gly05, although the reductions were much less dramatic at these sites compared to the other sites (Fig 6C). Note that the steady‐state level of Cnd2‐GFP remained unaffected in the arp9Δ or snf21‐129 genetic background (Fig 6D). These data strongly support the conclusion that nucleosome eviction facilitates condensin binding during mitosis.
Figure 6. Nucleosome eviction by the RSC remodelling complex facilitates condensin binding.
-
A–CNucleosome occupancy and condensin binding during mitosis upon RSC loss of function. Fission yeast cells expressing Cnd2‐GFP were arrested in early mitosis, mitotic indexes determined (A) and cells processed for ChIP against H3 and H4 (B) and Cnd2‐GFP (C). Averages and s.d. calculated from 3 ChIPs per antigen, each performed on 3 biological replicates. Values for prl53 and ecm33 were multiplied by 4 to facilitate reading.
-
DCnd2‐GFP levels by Western blotting.
-
E, FRSC localization at condensin‐binding sites during mitosis. Mitotically arrested cells were subjected to ChIP against Snf21‐FLAG. Averages and s.d. calculated from 12 ChIPs on 6 biological replicates are indicated. Values for 5S and gly05 were multiplied by 3. ***P < 0.001, **P < 0.01, °P > 0.05. High‐occup. and Low‐occup. refer to condensin high‐occupancy and low‐occupancy binding sites, respectively.
Figure EV8. The impact of RSC loss of function on histone H3 occupancy at the 3′ end of genes bound by condensin.
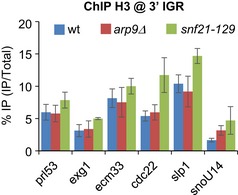
Fission yeast cells expressing Cnd2‐GFP were arrested in early mitosis, mitotic indexes determined (see Fig 6A) and cells processed for ChIP against H3. % IP correspond to averages, and s.d. calculated from 3 ChIPs performed on 3 biological replicates (see Fig 6 for additional information).
Given that the bromodomain‐containing Snf21 protein binds H3K14ac (Wang et al, 2012), we asked whether Gcn5 and Mst2 might recruit RSC at condensin‐binding sites in mitosis. Even though binding of chromatin remodellers is notoriously difficult to assay by ChIP, we detected Snf21‐flag at promoters upstream of condensin‐binding sites in cells arrested in mitosis (Fig 6E and F). The association of Snf21 at the promoter of snoU14 was reduced in cells lacking Gcn5 or both Gcn5 and Mst2, but remained unchanged at the seven other tested condensin‐binding sites. Thus, RSC is present during mitosis at gene promoters adjoining condensin‐binding sites and its localization near most, but not all, of these sites is independent of Gcn5 and Mst2.
Discussion
Condensin binding to DNA is instrumental for chromosome condensation, but how binding is achieved in the context of a chromatin environment has remained elusive. Here, we provide evidence that nucleosome eviction, promoted by the histone acetyltransferases Gcn5 and Mst2 as well as the RSC chromatin‐remodelling complex, is necessary for condensin binding to chromosomes during mitosis and for proper mitotic chromosome condensation. Thus, nucleosomes must constitute a barrier for the association of condensin. Nucleosome‐depleted regions generated at least in part by transcription cofactors, such as Gcn5, Mst2 and RSC, might therefore constitute access points into the chromosome where condensin first associates with exposed double‐stranded DNA helices.
One key finding of our study is the preferred localization of condensin at or in the immediate vicinity of NDRs (Figs 5B and EV6). This is particularly true for high‐occupancy condensin‐binding sites (see Fig 5B and C). In itself, this pattern of localization suggests that nucleosomes constitute an obstacle for the localization of condensin. In the case of transcription, the nucleosome barrier is overcome by histone acetylation and nucleosome removal (Owen‐Hughes & Gkikopoulos, 2012). We found that most Gcn5 transiently dissociates from chromatin during prophase in fission yeast, and that the bulk of H3K9ac and H3K18ac is deacetylated, reminiscent of the chromatin modifications that occurs during mitosis in mammalian cells. However, a fraction of Gcn5 remains on chromosomes during mitosis and is enriched at a number of promoters of Pol II‐transcribed genes. At these sites, Gcn5 maintains acetylation of H3K9 and H3K18 and ensures condensin binding. Thus, nucleosome acetylation by Gcn5 might play a role in regulating condensin association with chromatin in mitosis. The finding that lack of both Gcn5 and Mst2 causes a cumulative reduction of H3K14ac in addition to H3K9ac and H3K18ac, and reduces condensin occupancy further, strengthens this conclusion.
The acetylation of histone H3 is a transcription conductive modification. Gcn5 and Mst2 may therefore facilitate condensin binding by promoting transcription. Although we cannot formally rule out this possibility, we think this is unlikely, because we identified five out of nine condensin‐binding sites where the association of condensin with chromatin was reduced in the absence of Gcn5 whilst the occupancy of RNA Pol II phosphorylated on serine 2 remained unchanged (see rds1, prl53, ecm33, snoU14 and cdc22 in Figs 3A and EV3D). This suggests that transcription and condensin binding can be uncoupled.
Acetylated nucleosomes are targeted for removal by bromodomain‐containing nucleosome remodellers (Owen‐Hughes & Gkikopoulos, 2012). In line with this, nucleosome occupancy is increased at 3′ NDRs that overlap with condensin‐binding sites, and at NDR upstream of promoters, in gcn5 mutant cells, and is further increased when both Gcn5 and Mst2 are missing. Crucially, the increase in nucleosome occupancy is accompanied by a proportional reduction in condensin binding. Moreover, increasing nucleosome occupancy by directly altering RSC activity is sufficient to decrease condensin binding. Thus, nucleosome eviction most likely plays a key role in condensin binding to chromatin during mitosis. Although our results do not exclude the possibility that Gcn5 assists condensin binding at least in part by acetylating condensin and/or other non‐histone proteins, they strongly suggest that Gcn5 and Mst2 promote condensin binding in mitosis by evicting nucleosomes from condensin‐binding sites.
Like Gcn5, RSC is present at promoters adjoining condensin‐binding sites during mitosis and is necessary for condensin binding at the 3′ end of genes. However, RSC deficiency increases nucleosome occupancy strongly at gene promoters but only moderately at the 3′ end of genes (see Figs 6B and EV8). This suggests that nucleosome eviction at gene promoters plays a crucial role in the binding of condensin at the 3′ end of genes. Thus, given the enrichment of Gcn5 at gene promoters, and the physical and functional interactions between condensin and the TATA‐binding protein 1 (Iwasaki et al, 2015), it is tempting to speculate that condensin rings first associate with chromosomes at promoter NDRs and subsequently translocate towards the 3′ end of genes, as proposed for the related cohesin complex (Lengronne et al, 2004).
The central domain of centromeres (cnt1) is transcribed by RNA Pol II and is a site of high nucleosome turnover (Choi et al, 2011; Sadeghi et al, 2014). The reduced association of condensin at cnt1 in the absence of Gcn5 (Fig 3A) might therefore indicate that nucleosome eviction and/or dynamics contribute to the association of condensin, along with Monopollin (Tada et al, 2011), at centromeres.
Chromatin‐modifying activities in addition to the activities of Gcn5, Mst2 and RSC are likely to take part in condensin's binding to chromosomes. Although the vast majority of condensin‐binding sites coincides with NDRs (see Fig EV6B), Gcn5 and Mst2 seem to assist condensin binding mainly at high‐occupancy sites but play only a negligible role (if any) at low‐occupancy binding sites. RSC, in contrast, seems to play a more general role (Fig 6B and C). The fact that nucleosome residence increases at high‐occupancy condensin‐binding sites in the absence of Gcn5 and Mst2, despite the presence of RSC, implies that Gcn5 and Mst2 promote condensin binding at these sites by recruiting additional chromatin remodellers, which are at least partly redundant with RSC. The recent finding that budding yeast Gcn5 acts cooperatively, and often redundantly, with the Swi/Snf nucleosome remodelling enzyme to evict promoter nucleosomes (Qiu et al, 2016), supports our conclusion.
Note, however, that nucleosome depletion is unlikely to drive condensin binding by itself. We identified ~7,000 NDRs in mitotic chromosomes in cells arrested in pro/metaphase, but solely ~400 condensin peaks (48 high and 340 low occupancy) have been identified by ChIP‐seq at a similar cell cycle stage (Sutani et al, 2015). This suggests the existence of NDRs devoid of condensin during mitosis. The corollary, therefore, is that nucleosome eviction is necessary but not sufficient for condensin binding. Hence, additional features/activities must attract condensin. Budding yeast condensin has been shown to preferentially bind free over nucleosomal DNA in a sequence‐independent manner (Piazza et al, 2014). It has also been shown that RSC recruits the cohesin loader Scc2/Scc4 complex at NDRs of active promoters in budding yeast (Lopez‐Serra et al, 2014). The role of nucleosome eviction may therefore be to provide access to free genomic DNA for condensin and/or for DNA binding factors important for condensin association. Also condensin, cohesin and perhaps all SMC complexes may contact and subsequently entrap chromosomal DNA at NDRs.
Several lines of evidence suggest that the model of condensin loading at NDRs applies to most eukaryotes. In most species, an NDR is present around the transcription start site of active genes (Sadeh & Allis, 2011), the size of which increases with transcription rates to culminate with a disruption of chromatin for the most highly expressed genes (Lantermann et al, 2010; Soriano et al, 2013), which are occupied by condensin. Moreover, the fact that nuclease sensitivity patterns are preserved during mitosis suggests that NDRs persist despite transcriptional shut down (Gottesfeld & Forbes, 1997). Thus, the positions of condensins should, in principle, coincide with NDRs at highly expressed genes in a wide range of species. In good agreement, condensin preferentially occupies genes that are co‐occupied by RSC and Gcn5 in budding yeast (Venters et al, 2011), colocalizes with NDRs at tRNA genes (Piazza et al, 2014) and relies upon the histone chaperone Asf1 for its chromosomal association, which, together with FACT, regulates nucleosome turnover (Dewari & Bhargava, 2014). Nucleosome remodellers are components of mitotic chromosomes in vertebrates (MacCallum et al, 2002; Ohta et al, 2010) and a recent study indicates that the mobilization of embryonic nucleosomes by the histone chaperones Nap1 and FACT is necessary for the assembly of Xenopus mitotic chromosomes (Shintomi et al, 2015).
Altogether, those observations are consistent with the idea that nucleosomes constitute a barrier for the initial binding of condensins. The enrichment of condensins nearby highly expressed genes observed from yeasts to mammals may thus reflects the fact that condensin rings make their first contact with free, exposed chromosomal DNA at nucleosome‐depleted regions created by transcription‐coupled nucleosome eviction.
Materials and Methods
Media, molecular genetics and strains
Media and molecular genetics methods were as described previously (Moreno et al, 1991). Complete medium was YES + A. Gene deletions or tagging were performed using a polymerase chain reaction (PCR)‐based method (Bahler et al, 1998). All deletions were confirmed by PCR on genomic DNA. Tagging was validated by Western blotting and Sanger sequencing. Strains used in this study are listed in Appendix Table S2.
Mitotic arrest
All mitotic arrests were performed at 19°C using the cold‐sensitive nda3‐KM311 mutation. Unless otherwise stated, mitotic indexes were measured by scoring the accumulation of Cnd2‐GFP in the nucleus (Sutani et al, 1999).
Chromosome condensation assay
Cells were grown at 25°C to 0.5–1 × 107 cells/ml. Early G2 cells were purified by centrifugation through a 7–30% (w/v) lactose step gradient and pipetted onto a 35‐mm glass‐bottom dish (MatTek, No 1.5 P35G‐1.5‐10‐C) covered with 2 mg/ml BS1 lectin (Sigma, L2380), and the dish was shifted to 34°C. The glass‐bottom dish was filled with 2 ml YES + A pre‐warmed to 34°C and placed onto the sample stage of the microscope. Imaging was performed using a 100× objective (Olympus) on a DeltaVision system equipped with a Photometrics CoolSnap HQ camera (Roper Scientific) binning 2 × 2 pixels. Pixel size was about 130 × 130 nm. A GFP‐DsRed Dual dichroic mirror (art F51‐019) was used in combination with two bandpass filters to switch between excitation bands. Imaging was started 1 h after lactose gradient centrifugation, and Z‐stacks to cover a depth of 400 nm were acquired every 45 s for 6 fields of view (512 × 512 px) for 1 h. Imaging data were analysed by determining foci centroids using a custom ImageJ plugin as described (Petrova et al, 2013). Anaphase onset was defined as the frame at which the centromere‐proximal foci split. Distance time series were aligned based on anaphase onset. Average and standard deviation were calculated for each time point.
Immunofluorescence
Immunofluorescence was performed as described (Robellet et al, 2014). Images were processed and distances measured using ImageJ.
Chromosome spreading
Chromosome spreads were performed as described (Bahler et al, 1993). About 5 × 107 cells were digested with 2 mg of lysing enzymes (Sigma L‐1412), and nuclear spreading was performed in the presence of 1% formaldehyde and 1.5% lipsol. Immunofluorescence was carried out in PBS supplemented with 1% fish skin gelatin (v/v) and 0.5% BSA (w/v) using monoclonal anti‐HA 12CA5 (1/800), polyclonal rabbit anti‐GFP A11122 (1/800) or polyclonal anti‐Myc A‐14 (1/500). Images were acquired using an axioimager Z1 microscope and immunofluorescence signals quantified with ImageJ software.
Co‐immunoprecipitations
Co‐immunoprecipitations were performed as described (Vanoosthuyse et al, 2014).
Chromatin Immunoprecipitation and quantitative qPCR
ChIPs were performed as described (Vanoosthuyse et al, 2014). About 2 × 108 cells were fixed with 1% formaldehyde at 19°C for 30 min, washed with PBS and lysed using acid‐wash glass beads in a Precellys homogenizer (3 times 10″ at full speed with 30 s pauses in ice). Chromatin was sheared in 300‐ to 900‐bp fragments by sonication of whole cell extracts at 4°C using a Diagenode bioruptor (10 cycles 30 s on/30 s off), max power. Clarified chromatin was split in two equivalent fractions subjected to parallel immunoprecipitations using magnetic Dynabeads previously incubated with the appropriate antibody. Total and immunoprecipitated DNA was purified using the NucleoSpin PCR clean‐up kit (Macherey‐Nagel). DNA was analysed on a Rotor‐Gene PCR cycler using QuantiFast SYBR Green mix. Primers are listed in Appendix Table S3.
Antibodies
Antibodies used in this study are listed in Appendix Table S4.
Reverse‐transcription and quantitative (q)PCR
Total RNA was extracted from 108 cells by standard hot‐phenol method. Reverse transcription was performed on 500 ng of total RNA using Superscript II (Life Technologies) and random hexamers in the presence or absence of reverse transcriptase. cDNAs were quantified by real‐time qPCR on a Rotor‐Gene PCR cycler using QuantiFast SYBR Green mix.
MNase‐seq
MNase digestion of mitotic chromatin was performed as described (Lantermann et al, 2009) using cells arrested in early mitosis by the nda3‐KM311 mutation at 19°C and cross‐linked with 0.5% formaldehyde for 30 min. Chromatin was digested with increasing amount of MNase to reach a mononucleosome to di‐nucleosome ratio of ~80:20. Mononucleosomal DNA fragments were separated on an agarose gel, extracted from the gel and subjected to massive parallel sequencing on an Illumina NextSeq 500 Apparatus. Between 46,818,494 and 62,258,601 single‐end reads of 75 bp in length were obtained per sample and aligned to the S. pombe genome (Ensembl ASM294v2, May 2009) using Bowtie 2.2.4 with default parameters. Detection of the NDRs was performed as described (Soriano et al, 2013). Each sample was normalized by dividing signals for every base pair by the mean of coverage depth. NDRs were identified as regions of at least 150 bp in length with normalized sequence coverage inferior to 0.4 and were fused together if separated by < 15 bps.
Bioinformatic analysis of nucleosome occupancy and positioning at condensin‐binding sites
For each condensin‐binding site, adjacent NDRs of reference were manually identified and their coordinates determined by an iterative process. Manually identified NDRs were aligned with their respective counterparts calculated for each biological replicate (n = 3). When the reference NDR was overlapped by at least 70% of its length by its calculated counterpart, the two NDRs were merged, generating a new NDR of reference, which was used for the next iteration. Reference NDRs, and calculated NDRs detected in each replicate, are listed in Dataset EV1. To compare coverage depth (Fig 5E), 200‐bp upstream and 200‐bp downstream of the NDR were added to take into account the flanking nucleosomes. The sum of the normalized coverage depth at each base was calculated and divided by the total length. Resulting distributions were tested for gaussian behaviour using the Shapiro–Wilk test. The Kruskal–Wallis test was used for comparison if at least one distribution did not follow the normal law.
Metagene generation
Nucleosome‐depleted regions of reference were fractioned in 10 bins, in which the coverage depth was averaged over the bases. The 200 bp upstream and downstream of the NDR were each resumed in 20 bins following the same process as for the NDRs. The mean enrichments were calculated for each bin in each sample.
Permutation test
Positions of the 47 condensin peaks were shuffled 100,000 times using the shuffle tool of Bedtools, without any constraint, to obtain a theoretical distribution of the overlaps between condensin peaks and NDRs. The permutation test was performed on this distribution. The P‐value was calculated by dividing the number (x) of permutations giving higher numbers of bases in overlaps than the observed value increased by one (the observed value), by the total number of sets of positions (the set of the observed positions and the 100,000 permutations of the positions): (x + 1)/(100,000 + 1).
Enrichment of condensin in NDRs
Datasets of ChIP‐seq against Cut14‐pk9 (Sutani et al, 2015) n° SRR1564296, SRR1557176, SRR1559300, SRR1559301, SRR1557175 and SRR1557178 were aligned to the S. pombe genome (Ensembl ASM294v2) using bowtie 1.1.2 with the parameters indicated in the original paper. For each dataset, coverage was divided by a normalization factor, which corresponds to the ratio of the number of alignments over the size of the genome. Note that the length of the reads is unique (48 bp) for all samples. Normalized coverage of the IP sample was divided by the normalized coverage of the corresponding input (both increased by 1 to allow taking into account bases with a coverage value of 0 in the input). Thus, any enrichment gives a ratio > 1, and, reciprocally, any impoverishment gives a ratio between 0 and 1.
Statistical methods
Unless otherwise stated, we used two‐tailed Wilcoxon and Mann–Whitney test statistics when at least 6 independent values were available per sample.
Data access
The MNase‐seq data from this publication have been submitted to ArrayExpress database and assigned the identifier E‐MTAB‐4620.
Author contributions
PB involved in conceptualization; CS, CHH and PB involved in methodology; ETM, XR, LF, CS, CK, CH, PL, LNG and LM participated in investigation; SL and EC did formal analysis; DA, CHH and PB involved in supervision; PB wrote the original draft; and PB involved in funding acquisition.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Review Process File
Acknowledgements
We are very grateful to Vincent Vanoosthuyse for helpful discussions and comments on the manuscript. We thank Dom Helmlinger, Fred Winston, Jean‐Paul Javerzat, Susan Forsburg, Blerta Xhemalce and Michael Keogh for yeast strains, Keith Gull for the anti‐tubulin Tat1 antibody and the EMBL Advanced Light Microscopy Facility for help and advices. This work was supported by funding from the CNRS (ATIP grant to P.B.), the Association pour la Recherche contre le Cancer, grant SFI20111203612 (P.B.), and by a donation from Claude and Antoine Sapone to P.B. Work in Christian Haering's laboratory was supported by the German Research Foundation grant HA5853/1‐2 (C.H.H.). X.R. was supported by a postdoctoral fellowship from CNRS, L.F. and C.H.H. by PhD studentships from la Ligue Nationale Contre le Cancer and the Ministère de l'Education Nationale et de la Recherche, respectively. This publication is dedicated to the memory of Antoine Sapone.
The EMBO Journal (2016) 35: 1565–1581
References
- Aono N, Sutani T, Tomonaga T, Mochida S, Yanagida M (2002) Cnd2 has dual roles in mitotic condensation and interphase. Nature 417: 197–202 [DOI] [PubMed] [Google Scholar]
- Aragon L, Martinez‐Perez E, Merkenschlager M (2013) Condensin, cohesin and the control of chromatin states. Curr Opin Genet Dev 23: 204–211 [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR‐based gene targeting in Schizosaccharomyces pombe . Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Bahler J, Wyler T, Loidl J, Kohli J (1993) Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J Cell Biol 121: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter J, Sen N, Martinez VL, De Carandini ME, Schvartzman JB, Diffley JF, Aragon L (2011) Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science 331: 1328–1332 [DOI] [PubMed] [Google Scholar]
- Bonnet J, Wang C‐Y, Baptista T, Vincent SD, Hsiao W‐C, Stierle M, Kao C‐F, Tora L, Devys D (2014) The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev 28: 1999–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbin A, Bouchoux C, Uhlmann F (2014) Condensin aids sister chromatid decatenation by topoisomerase II. Nucleic Acids Res 42: 340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi ES, Stralfors A, Castillo AG, Durand‐Dubief M, Ekwall K, Allshire RC (2011) Identification of noncoding transcripts from within CENP‐A chromatin at fission yeast centromeres. J Biol Chem 286: 23600–23607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente‐Blanco A, Mayan‐Santos M, Schneider DA, Machin F, Jarmuz A, Tschochner H, Aragon L (2009) Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature 458: 219–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente‐Blanco A, Sen N, Mayan‐Santos M, Sacristan MP, Graham B, Jarmuz A, Giess A, Webb E, Game L, Eick D, Bueno A, Merkenschlager M, Aragon L (2011) Cdc14 phosphatase promotes segregation of telomeres through repression of RNA polymerase II transcription. Nat Cell Biol 13: 1450–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Metz J, Haering CH (2011) Condensin structures chromosomal DNA through topological links. Nat Struct Mol Biol 18: 894–901 [DOI] [PubMed] [Google Scholar]
- D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F (2008) Identification of cis‐acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22: 2215–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewari PS, Bhargava P (2014) Genome‐wide mapping of yeast histone chaperone anti‐silencing function 1 reveals its role in condensin binding with chromatin. PLoS One 9: e108652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Bilodeau S, Orlando DA, Hübner MR, Abraham BJ, Spector DL, Young RA (2013) Multiple structural maintenance of chromosome complexes at transcriptional regulatory elements. Stem Cell Rep I: 371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide T, Carlson C, Taskén KA, Hirano T, Taskén K, Collas P (2002) Distinct but overlapping domains of AKAP95 are implicated in chromosome condensation and condensin targeting. EMBO Rep 3: 426–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ (1997) Mitotic repression of the transcriptional machinery. Trends Biochem Sci 22: 197–202 [DOI] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG (2007) Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell 25: 31–42 [DOI] [PubMed] [Google Scholar]
- Helmlinger D, Marguerat S, Villen J, Gygi SP, Bahler J, Winston F (2008) The S. pombe SAGA complex controls the switch from proliferation to sexual differentiation through the opposing roles of its subunits Gcn5 and Spt8. Genes Dev 22: 3184–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T (2016) Condensin‐based chromosome organization from bacteria to vertebrates. Cell 164: 847–857 [DOI] [PubMed] [Google Scholar]
- Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM (2004) Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci 117: 6435–6445 [DOI] [PubMed] [Google Scholar]
- Iwasaki O, Tanizawa H, Kim K‐D, Yokoyama Y, Corcoran CJ, Tanaka A, Skordalakes E, Showe LC, Noma K‐I (2015) Interaction between TBP and Condensin Drives the Organization and Faithful Segregation of Mitotic Chromosomes. Mol Cell 59: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson A, Durand‐Dubief M, Xue‐Franzen Y, Ronnerblad M, Ekwall K, Wright A (2009) HAT‐HDAC interplay modulates global histone H3K14 acetylation in gene‐coding regions during stress. EMBO Rep 10: 1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K, Horiuchi T (2007) RNA polymerase I transcription obstructs condensin association with 35S rRNA coding regions and can cause contraction of long repeat in Saccharomyces cerevisiae . Genes Cells 12: 759–771 [DOI] [PubMed] [Google Scholar]
- Johzuka K, Horiuchi T (2009) The cis element and factors required for condensin recruitment to chromosomes. Mol Cell 34: 26–35 [DOI] [PubMed] [Google Scholar]
- Kim JH, Zhang T, Wong NC, Davidson N, Maksimovic J, Oshlack A, Earnshaw WC, Kalitsis P, Hudson DF (2013) Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat Commun 4: 2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hirano T (1997) ATP‐dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90: 625–634 [DOI] [PubMed] [Google Scholar]
- Koutelou E, Hirsch CL, Dent SY (2010) Multiple faces of the SAGA complex. Curr Opin Cell Biol 22: 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz AL, Jiao CY, Winterkorn LH, Albritton SE, Kramer M, Ercan S (2013) Genome‐wide analysis of condensin binding in Caenorhabditis elegans . Genome Biol 14: R112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Hendzel MJ, Fischle W, Bertos NR, Hameed S, Yang XJ, Verdin E, Bazett‐Jones DP (2001) Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J Biol Chem 276: 38307–38319 [DOI] [PubMed] [Google Scholar]
- Lantermann A, Strålfors A, Fagerström‐Billai F, Korber P, Ekwall K (2009) Genome‐wide mapping of nucleosome positions in Schizosaccharomyces pombe . Methods San Diego Calif 48: 218–225 [DOI] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Strålfors A, Yuan G‐C, Ekwall K, Korber P (2010) Schizosaccharomyces pombe genome‐wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae . Nat Struct Mol Biol 17: 251–257 [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, McLeod I, Anderson S, Yates JR 3rd, He X (2005) Molecular analysis of kinetochore architecture in fission yeast. EMBO J 24: 2919–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia‐Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG (2010) PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466: 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Serra L, Kelly G, Patel H, Stewart A, Uhlmann F (2014) The Scc2‐Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome‐free regions. Nat Genet 46: 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier‐Davis B, Kornberg RD (2014) Role of DNA sequence in chromatin remodeling and the formation of nucleosome‐free regions. Genes Dev 28: 2492–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DE, Losada A, Kobayashi R, Hirano T (2002) ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP‐aurora B. Mol Biol Cell 13: 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan BJ, Villen J, Marguerat S, Bahler J, Gygi SP, Winston F (2008) Fission yeast SWI/SNF and RSC complexes show compositional and functional differences from budding yeast. Nat Struct Mol Biol 15: 873–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe . Methods Enzym 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nakazawa N, Sajiki K, Xu X, Villar‐Briones A, Arakawa O, Yanagida M (2015) RNA pol II transcript abundance controls condensin accumulation at mitotically up‐regulated and heat‐shock‐inducible genes in fission yeast. Genes Cells 20: 481–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent RL, Johnsson A, Fleharty B, Gogol M, Xue‐Franzen Y, Seidel C, Wright AP, Forsburg SL (2010) Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation. BMC Genom 11: 59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S, Bukowski‐Wills JC, Sanchez‐Pulido L, Alves Fde L, Wood L, Chen ZA, Platani M, Fischer L, Hudson DF, Ponting CP, Fukagawa T, Earnshaw WC, Rappsilber J (2010) The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142: 810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T (2003) Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115: 109–121 [DOI] [PubMed] [Google Scholar]
- Orpinell M, Fournier M, Riss A, Nagy Z, Krebs AR, Frontini M, Tora L (2010) The ATAC acetyl transferase complex controls mitotic progression by targeting non‐histone substrates. EMBO J 29: 2381–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen‐Hughes T, Gkikopoulos T (2012) Making sense of transcribing chromatin. Curr Opin Cell Biol 24: 296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzlaff JS, Terrenoire E, Turner BM, Earnshaw WC, Paulson JR (2010) Acetylation of core histones in response to HDAC inhibitors is diminished in mitotic HeLa cells. Exp Cell Res 316: 2123–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova B, Dehler S, Kruitwagen T, Heriche JK, Miura K, Haering CH (2013) Quantitative analysis of chromosome condensation in fission yeast. Mol Cell Biol 33: 984–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza I, Rutkowska A, Ori A, Walczak M, Metz J, Pelechano V, Beck M, Haering CH (2014) Association of condensin with chromosomes depends on DNA binding by its HEAT‐repeat subunits. Nat Struct Mol Biol 21: 560–568 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA (2005) Genome‐wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Qiu H, Chereji RV, Hu C, Cole HA, Rawal Y, Clark DJ, Hinnebusch AG (2016) Genome‐wide cooperation by HAT Gcn5, remodeler SWI/SNF, and chaperone Ydj1 in promoter nucleosome eviction and transcriptional activation. Genome Res 26: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robellet X, Fauque L, Legros P, Mollereau E, Janczarski S, Parrinello H, Desvignes JP, Thevenin M, Bernard P (2014) A genetic screen for functional partners of condensin in fission yeast. G3 Bethesda 4: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA (2004) Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell 16: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K (2005) Active chromatin domains are defined by acetylation islands revealed by genome‐wide mapping. Genes Dev 19: 542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi L, Siggens L, Svensson JP, Ekwall K (2014) Centromeric histone H2B monoubiquitination promotes noncoding transcription and chromatin integrity. Nat Struct Mol Biol 21: 236–243 [DOI] [PubMed] [Google Scholar]
- Sadeh R, Allis CD (2011) Genome‐wide ‘Re’‐modeling of nucleosome positions. Cell 147: 263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M (1994) Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J 13: 4938–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima K, Samejima I, Vagnarelli P, Ogawa H, Vargiu G, Kelly DA, de Lima Alves F, Kerr A, Green LC, Hudson DF, Ohta S, Cooke CA, Farr CJ, Rappsilber J, Earnshaw WC (2012) Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIalpha. J Cell Biol 199: 755–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CK, Brookes N, Uhlmann F (2009) Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol 10: R52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K, Takahashi TS, Hirano T (2015) Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat Cell Biol 17: 1014–1023 [DOI] [PubMed] [Google Scholar]
- Soriano I, Quintales L, Antequera F (2013) Clustered regulatory elements at nucleosome‐depleted regions punctuate a constant nucleosomal landscape in Schizosaccharomyces pombe . BMC Genom 14: 813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen RL, Cubizolles F, Le Guellec K, Collas P (2000) A kinase‐anchoring protein (AKAP)95 recruits human chromosome‐associated protein (hCAP)‐D2/Eg7 for chromosome condensation in mitotic extract. J Cell Biol 149: 531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M (1999) Fission yeast condensin complex: essential roles of non‐SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev 13: 2271–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Sakata T, Nakato R, Masuda K, Ishibashi M, Yamashita D, Suzuki Y, Hirano T, Bando M, Shirahige K (2015) Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat Commun 6: 7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada K, Susumu H, Sakuno T, Watanabe Y (2011) Condensin association with histone H2A shapes mitotic chromosomes. Nature 474: 477–483 [DOI] [PubMed] [Google Scholar]
- Teytelman L, Thurtle DM, Rine J, van Oudenaarden A (2013) Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci USA 110: 18602–18607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadani R, Uhlmann F, Heeger S (2012) Condensin, chromatin crossbarring and chromosome condensation. Curr Biol 22: R1012–R1021 [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V, Legros P, van der Sar SJA, Yvert G, Toda K, Le Bihan T, Watanabe Y, Hardwick K, Bernard P (2014) CPF‐associated phosphatase activity opposes condensin‐mediated chromosome condensation. PLoS Genet 10: e1004415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck‐Walsh C, Pugh BF (2011) A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces . Mol Cell 41: 480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kallgren SP, Reddy BD, Kuntz K, Lopez‐Maury L, Thompson J, Watt S, Ma C, Hou H, Shi Y, Yates JR, Bahler J, O'Connell MJ, Jia S (2012) Histone H3 lysine 14 acetylation is required for activation of a DNA damage checkpoint in fission yeast. J Biol Chem 287: 4386–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Higgins JM (2013) Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol 23: 175–184 [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL (2012) SAGA function in tissue‐specific gene expression. Trends Cell Biol 22: 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiren M, Silverstein RA, Sinha I, Walfridsson J, Lee HM, Laurenson P, Pillus L, Robyr D, Grunstein M, Ekwall K (2005) Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J 24: 2906–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue‐Franzen Y, Henriksson J, Burglin TR, Wright AP (2013) Distinct roles of the Gcn5 histone acetyltransferase revealed during transient stress‐induced reprogramming of the genome. BMC Genom 14: 479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Review Process File


