Abstract
To assess gene signatures related to humoral response among healthy older subjects following seasonal influenza vaccination, we studied 94 healthy adults (50–74 years old) who received one documented dose of licensed trivalent influenza vaccine containing the A/California/7/2009 (H1N1)-like virus strain. Influenza-specific antibody (HAI) titer in serum samples and next-generation sequencing on PBMCs were performed using blood samples collected prior to (Day 0) and at two timepoints after (Days 3 and 28) vaccination. We identified a number of uncharacterized genes (ZNF300, NUP1333, KLK1 and others) and confirmed previous studies demonstrating specific genes/genesets that are important mediators of host immune responses and that displayed associations with antibody response to influenza A/H1N1 vaccine. These included interferon-regulatory transcription factors (IRF1/IRF2/IRF6/IRF7/IRF9), chemokine/chemokine receptors (CCR5/CCR9/CCL5), cytokine/cytokine receptors (IFNG/IL10RA/TNFRSF1A), protein kinases (MAP2K4/MAPK3), growth factor receptor (TGFBR1). The identification of gene signatures associated with antibody response represents an early stage in the science for which further research is needed. Such research may assist in the design of better vaccines to facilitate improved defenses against new influenza virus strains, as well as better understanding the genetic drivers of immune responses.
Keywords: Genetics, Immunology
1. Introduction
Influenza viruses infect, sicken, and kill humans across the world and across all age groups, but this is especially true among older adults [1]. Influenza A viruses (category C bioagents [2]) are single-stranded RNA Orthomyxoviridae viruses, characterized by specific hemagglutinin and neuraminidase transmembrane glycoproteins that generate subtype specific immune responses. Influenza A/H1N1 gained prominence during the pandemic of 1918 when a highly pathogenic novel H1N1 virus spread across the world, killing an estimated 50 million people [3]. Since its re-emergence in 1977, A/H1N1 has been included in the seasonal trivalent inactivated influenza (TIV) and in the inactivated quadrivalent influenza vaccines.
Greater than 70–90% of healthy adults seroconvert to influenza vaccine [4]. When the vaccine is well matched, TIV is 70–90% protective against laboratory-confirmed influenza infection and up to 90% protective against hospitalization in healthy adults age <65 years [5, 6]. Unfortunately, protection against infection and complications due to influenza by influenza vaccines is incomplete (with failure rates up to 50–70% in the very young, elderly, and immunocompromised individuals).
To date, few biological markers (or models) exist that explain the development of immune responses to influenza vaccine, and/or predict vaccine failure. There is evidence that host genetic factors impact the response to influenza A/H1N1 virus infections and immune responses to influenza vaccination [7, 8, 9, 10, 11]. However, a more comprehensive understanding of the cellular and molecular mechanisms of vaccine immunity is needed [12, 13, 14, 15, 16]. Several studies have applied systems biology approaches to find molecular signatures of vaccine-induced immune responses in humans [8, 9, 15, 17, 18, 19]. Utilizing gene-to-biology and biology-to-gene approaches [20], which we define herein, together with gene expression from next generation sequencing, we sought to identify biological markers (genes/genesets) that could explain humoral antibody response variations following seasonal influenza A/H1N1 vaccine in older adults.
2. Materials and methods
2.1. Participants
Recruitment of subjects described herein is similar or identical to those published by us elsewhere [21, 22]. As previously reported, healthy adults who received 2010–2011 seasonal trivalent inactivated influenza vaccine (Fluarix), containing the A/California/7/2009 (H1N1)-like, A/Perth/16/2009 (H3N2)-like, and B/Brisbane/60/2008-like viral strains, were enrolled in the study [22, 23]. Specifically, between August 2010 and October 2010, we enrolled 106 healthy adults (ages 50 to 74 years), recruitment was designed to obtain a uniform distribution across the age range. All participants underwent detailed review of their vaccination history and were in good health during the length of this study. Study participants were excluded from enrollment if they showed symptoms consistent with influenza prior to or throughout the study. Blood samples were collected prior to (Day 0, the baseline level of immune status) and after vaccination (Days 3, the innate immune response; and 28, the peak of serum antibody response). The Mayo Clinic Institutional Review Board granted approval for the study. Written, informed consent from all subjects was obtained at the time of enrollment.
2.2. HAI assay
Our description of the hemagglutination inhibition (HAI) assay is similar to those we published elsewhere [22, 23]. Serum samples from each subject on Days 0, 3 and 28 visits were used for antibody titer determination. HAI assay was performed with the influenza A/California/07/2009 (H1N1)-like virus strain, and developed with 0.6% solution of turkey red blood cells (RBC) [24]. The HAI titer was defined as the highest dilution of serum that inhibits RBC hemagglutination. Seroconversion to the influenza virus vaccine, as described elsewhere [25], was defined by either a four-fold increase in the antibody titers between the pre-vaccination and the serum samples at Day 28, or an increase of antibody titers from <10 to ≥40 for pre-vaccination and the Day 28 serum samples.
2.3. Next generation sequencing
The mRNA next generation sequencing methods are similar or identical to those published for our previous transcriptomics studies [26, 27]. In brief, total RNA was extracted from each sample of cryopreserved mixed PBMCs using RNeasy Plus mini Kit (Qiagen) and RNAprotect reagent (Qiagen; Valencia, CA). RNA quantity and quality were assessed by Nanodrop (Thermo Fisher Scientific, Wilmington, DE) and Agilent 2010 Bioanalyzer (Agilent; Palo Alto, CA), respectively. Full-length cDNA libraries were created in the Mayo Clinic’s Advanced Genomics Technology Center, Gene Sequencing Facility using the mRNA-Seq 8 Sample Prep Kit (Illumina; San Diego, CA) according to the manufacturer’s protocols. Poly-A RNA was isolated using magnetic purification with olido-dT coated beads, fragmented, reverse transcribed into cDNA, and combined with Illumina adaptor sequences. Library validation and quantification was carried out using DNA 1000 Nano Chip kits on an Agilent 2100 Bioanalyzer (Agilent). cDNA libraries (5–7pM) were loaded onto individual flow cell lanes and single-end read sequencing was performed using the Illumin HiSeq 2000 (Illumina) with Illumina’s Single Read Cluster Generation kit (v2) and 50 Cycle Illumina Sequncing Kit (v3). The sequencing reads were aligned to the human genome build 37.1 using TopHat (1.3.3) and Bowtie (0.12.7). HTSeq (0.5.3p3) was used to perform gene counting while BEDTools (2.7.1) was used to count the reads mapping to individual exons [28, 29, 30].
2.4. Statistical methods
Total number of counts per gene were obtained from the mRNA expression and used in all analyses. Quality control was assessed pre- and post-normalization graphically with minus- vs-average and box-and-whisker plots. The GC content and gene length adjustments were also evaluated graphically. Normalization of the gene counts was done with Conditional Quantile Normalization, which accounts for differences in library size and also adjusts for GC content and gene length [31]. These normalized values were used for subsequent analyses.
Our gene-to-biology analyses utilize per-gene analyses to understand the relationship of gene expression with vaccine response. Due to the overall goal of developing multivariable models of dichotomized vaccine response, the logistic modeling framework was utilized. Specifically, per-gene logistic regression models were fit with HAI response relative to non-response as the dependent variable, and the normalized gene count as the independent variable on the log2 scale. These models were fit for each timepoint (Days 0, 3 and 28) and for the differences in log2 normalized gene counts between timepoints (e.g., Day 3-Day 0, Day 28-Day 0, and Day 28-Day 3). Evaluation of these timepoints allows us to address baseline, innate and adaptive humoral gene expression associated with response, as well as potentially identifying changes in gene expression as they relate to HAI response (or non-response). Results are presented as odds ratios (OR) for the 75th percentile relative to the 25th percentile of the gene expression value. Genes that are described as upregulated are those genes that have a higher expression level on average in the subjects who responded relative to those who did not respond to vaccine (as measured by a four-fold increase in the HAI titers from Day 0 to 28). Likewise, genes that are downregulated are those genes that have lower expression levels in subjects who responded relative to those who did not respond.
Our biology-to-gene analyses utilize a priori known biological information in the form of genesets in order to maximize power and minimize false discovery. Specifically, we utilized defined genesets to group genes into putative functional groups for analyses. Genesets utilized for the biology-to-gene analyses are the genesets (MSigDB) with significant changes over time (Day 0, Day 3, Day 28) [32]. Negative binomial generalized estimating equation models were used to assess changes in gene expression over time, utilizing moderated over-dispersion parameter estimates from edgeR [33, 34, 35, 36]. These p-values were then combined for all genes within a geneset utilizing the gamma method of combining p-values, a self-contained geneset testing method shown to have greater power than Fisher’s method of combining p-values [20, 37]. Redundancy analysis [38] was used to reduce the number of genes considered within each geneset model; genes were excluded from further considerations if the R2 for the gene was greater than 0.75 when modeled as a function of the other genes from the geneset. The remaining genes from each of these genesets were then included in elastic net penalized logistic regression models (α = 0.5 and 0.8) of HAI response relative to non-response as the dependent variable [39]. Models were fit using the “glmnet” function in R [39], and the optimal model selected according to the minimum misclassification error based on 10-fold cross validation. Results are presented with misclassification error rates and coefficients from the models.
2.5. Gene regulatory interactions
We consulted databases of known regulatory interactions to further understand the biologic relationships linking the genesets that are individually informative for HAI response. The result is a regulatory network within which differential expression patterns inform HAI response. To link genesets together and generate this network, we combined NCI-Nature pathways [40], the Transcription Factor Encyclopedia [41], and a directed protein interaction network [42], where edge direction indicates passage of a signal (e.g., protein interaction leading to phosphorylation). This focuses the analysis on regulatory interactions that should be robust to subject-specific details.
Using this network, we computed a geneset interaction metric, quantifying the extent to which two genesets can be directly regulated by each other. For genesets and that contain and genes, let the neighbors of a gene, those adjacent to the gene in a pathway, be . Further, let the neighbors of set that are also members of be denoted and U the unique set union. Finally, let the unique overlap between the two sets then be: . Then the interconnectivity is formally defined as: .
3. Results
3.1. Subject demographics
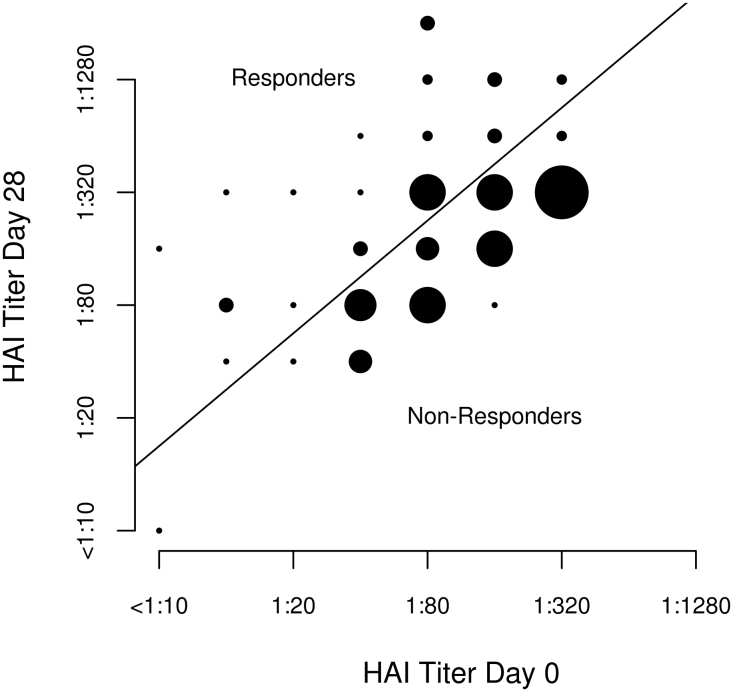
During the HAI testing, 106 subjects were assayed for HAI titer, and 12 subjects were excluded: 11 individuals based on influenza virus vaccine serology (i.e., due to a ceiling titer of 1:640 for which no subject augmented their titer from Day 0 to Day 28) [43]; and one subject based on cDNA library preparation failure. Thus, 94 of 106 participants were used in the study. Thirty-six of the 94 vaccine recipients (38.3%) had at least a four-fold increase in HAI titer to influenza A/H1N1. Demographic data and other characteristics of these subjects are shown in Table 1. Baseline (Day 0) influenza-specific HAI titer (median titer of 1:80; IQR 1:40; 1:160) demonstrated the presence of pre-existing HAI antibodies. Median HAI titer for the entire cohort increased by Day 28 (median titer of 1:320; IQR 1:80; 1:320, p < 0.001). Comparing the median HAI titer values between responders (n = 36) and non-responders (n = 58), responders had a lower baseline titer compared to non-responders (p = 0.003) (Table 1). Pre-existing immunity, as measured by HAI titers at baseline, had a strong positive correlation with Day 28 titers for subjects classified as responders (r = 0.758, p = 8.62 × 10−8) and for those classified as non-responders (r = 0.900, p = 7.72 × 10−22). The change in HAI titer from baseline to Day 28 is illustrated in Fig. 1.
Table 1.
Demographic and immunological variables of the study subjects.
| Variable | Non-responders (N = 58) |
Respondersa (N = 36) |
Overall (N = 94) |
|---|---|---|---|
| Age, years, median (IQRb) | 59.7 (55.9; 69.0) | 59.7 (55.0; 63.8) | 59.7 (55.4; 67.0) |
| Gender (n, %) |
|||
| Female | 37 (63.8) | 20 (55.6) | 57 (60.6) |
| Male | 21 (36.2) | 16 (44.4) | 37 (39.4) |
| Race (n, %) |
|||
| Caucasians | 57 (98.3) | 36 (100.0) | 93 (98.9) |
| Others | 1 (1.7) | 0 (0.0) | 1 (1.1) |
| HAI titer, median (IQRb) |
|||
| Day 0 | 1:160 (1:80, 1:160) | 1:80 (1:40, 1:80) | 1:80 (1:40, 1:160) |
| Day 3 | 1:160 (1:80, 1:160) | 1:80 (1:40, 1:160) | 1:80 (1:50, 1:160) |
| Day 28 | 1:160 (1:80, 1:320) | 1:320 (1:280, 1:1280) | 1:320 (1:80, 1:320) |
At least four-fold increase in the antibody titers between the pre-vaccination and the Day 28 sample. bIQR, interquartile range.
Fig. 1.
Comparison of baseline HAI titer values to Day 28 HAI titer values. Scatterplot of the HAI titer at Day 28 versus the HAI titer at baseline. The points on the graph are proportional to the number of individuals with the corresponding titers. The line through the plot is the divide between the responders and the non-responders and is based on a four-fold change from baseline to Day 28. Subjects above the line have a 4-fold or greater change and are responders, while the subjects below the line are classified as non-responders.
3.2. Gene expression associated with HAI response (gene-to-biology approach)
Overall transcriptomic expression (14,917 genes with a median count ≥ 32 reads at least one timepoint were used for all analyses) related to HAI response (defined as a positive four-fold change in HAI titer between Day 0 and Day 28) was analyzed at Days 0 (n = 688 genes had p < 0.05), 3 (n = 163 genes, p < 0.05) and 28 (n = 232 genes, p < 0.05). The false discovery rate (FDR) values for these genes were >0.9; we report herein a brief summary of these findings. A comparison between responders and non-responders failed to identify differentially expressed genes at Days 0, 3, and 28 when multiple-testing correlation was applied to the direct comparison analysis (FDR <0.05). At Day 0, 49 unique genes had a p-value <0.01. At Days 3 and 28, eight and seventeen genes, respectively, had a p-value <0.01 (Supplemental Table 1). The top genes with a p-value of <0.002 encode for proteins that play a central role in numerous cellular processes (such as ARHGEF10, MLST8, SPATA24, and C17orf97), which are likely to be important in immune response.
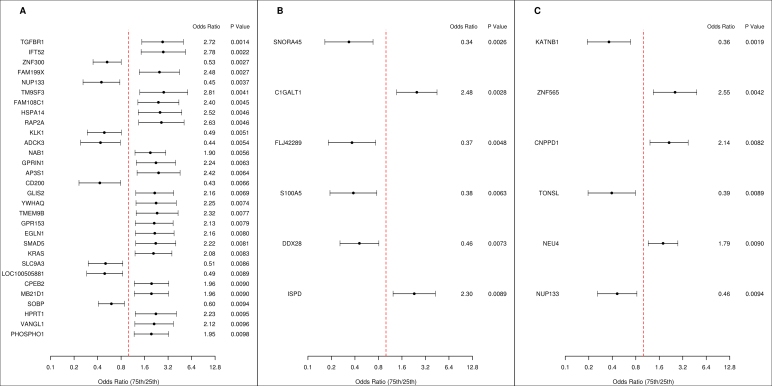
Changes in gene expression at Day 28 relative to baseline identified 328 genes (p < 0.05) significantly associated with HAI response. The top 30 genes with significant (p < 0.01) transcriptional changes encode for numerous proteins that comprise signal transduction pathways and transcriptional regulators (TGFBR1, ZNF300, GLIS2), heat shock proteins and activators of dendritic and T cells (HSPA14), transmembrane and nucleoporin complex glycoproteins (TM9SF3, NUP133, CD200), members of RAS oncogene and kallikrein families (RAP2A, KLK1), along with a number of proteins participating in a various physiological functions (GPRIN1, GPR153, EGLN1) (Fig. 2A). The majority of these differentially expressed genes were upregulated. In addition, changes in gene expression related to HAI response from Day 28 minus Day 3 and from Day 3 minus Day 0 identified 118 (p < 0.05) and 188 (p < 0.05) genes, respectively. Further, Fig. 2B and C show the results for the genes differentially expressed from Day 28 minus Day 3, p < 0.01; and the six genes differentially expressed from Day 3 minus Day 0, p < 0.01, respectively. These genes included calcium binding protein A5 (S100A5), putative RNA helicase (DDX28), zinc finger protein 565 (ZNF565), a regulator of NF-kappa-B-mediated transcription (TONSL), nucleoporin protein (NUP133), and other genes with unknown function.
Fig. 2.
Univariate HAI response. Forest plots displaying the odds ratios and confidence intervals for the genes with p < 0.01 from univariate logistic regression models with positive HAI response as the dependent variable. Odds ratios represent the change in odds of a positive HAI response as a result of moving from the 25th percentile to the 75th percentile of the gene expression. The vertical dashed line corresponds to an odds ratio of 1.0. In panel A, the change in gene expression between days 0 and 28 (Day 28 minus Day 0 delta) is the independent variable. Similarly, panel B shows results for (Day 28 minus Day 3) delta models, and panel C shows results for (Day 3 minus Day 0) delta models.
3.3. Geneset signatures associated with HAI response (biology-to-gene approach)
Our goal was to build multivariable regression models that explain variation in HAI response. When all genes were used to build multivariable models of HAI response, no genes entered the model, regardless of whether the Day 0, Day 28, or Day 28–Day 0 delta were used. We reasoned that pathways having statistically significant changes over time would most likely contain those genes changing in response to vaccine. Thus, genesets that were previously identified as having significant changes over time in our mRNA expression data (p < 0.001, FDR < 0.05) were used in models to predict HAI response (unpublished data). There were a total of 339 genesets for which models were generated. Thirteen genesets exhibited the ability to explain the odds of HAI response with models containing genes achieving a cross-validated error rate <35%. Many of the genes encompassing these 13 genesets have various and/or unidentified functions (Table 2); however, several genesets were related to immune functions (TGFBR1, CCR5, CCR9, ADAR, IRFs, CCL5, BAX, MAP2K4, MAPK3, NSMAF), including production of antibodies (IFNG, IL10RA, TNFRSF1A) (Fig. 3 and Fig. 4). For example, interferon regulatory factor 1 (IRF) and IRF7 genes function as transcriptional activators of type I IFN genes (α and β) and were found to be significantly upregulated in relation to the HAI response, and IRF2, IRF6 and IRF9 genes were found to be downregulated (Fig. 4). Interestingly, when the models were used to classify the glass ceiling subjects (i.e., due to a ceiling titer of 1:640), nine of the 11 subjects were classified as non-responders.
Table 2.
Top 13 genesets with minimum cross validated error rate <0.35 in penalized regression models of HAI response.
| Geneset Name [32] | Delta | α | Genes | Standardized Coefficients | Minimum CV Error |
|---|---|---|---|---|---|
|
NOL7: Genes down-regulated in SiHa cells by stable expression of NOL7 (HASINA_NOL7_TARGETS_DN [62]) |
Day 28 vs Day 0 | 0.8 | ANGPT1 | -0.023 | 0.29 |
| TGFBR1 | 0.628 | ||||
|
Purine: Genes involved in Purine salvage (REACTOME_PURINE_SALVAGE) |
Day 28 vs Day 0 | 0.5 | ADA | 0.008 | 0.32 |
| ADAL | -0.08 | ||||
| ADK | -0.363 | ||||
| AMPD3 | -0.08 | ||||
| HPRT1 | 0.633 | ||||
| PNP | 0.15 | ||||
|
Ceramide: Ceramide Signaling Pathway (BIOCARTA_CERAMIDE_PATHWAY) |
Day 28 vs Day 0 | 0.8 | BAX | 0.197 | 0.33 |
| MAP2K4 | -0.538 | ||||
| MAPK3 | 0.006 | ||||
| NSMAF | 0.043 | ||||
| TNFRSF1A | 0.443 | ||||
|
Kurozumi: Inflammatory cytokines and their receptors modulated in brain tumors (KUROZUMI_RESPONSE_TO_ONCOCYTIC_VIRUS_AND_CYCLIC_RGD [63]) |
Day 28 vs Day 0 | 0.8 | CCR5 | 0.065 | 0.33 |
| CCR9 | -0.198 | ||||
| IFNG | 0.317 | ||||
| IL10RA | 0.063 | ||||
|
SHH: Sonic Hedgehog Pathway (BIOCARTA_SHH_PATHWAY) |
Day 28 vs Day 0 | 0.8 | DYRK1A | 1.108 | 0.34 |
| DYRK1B | -0.098 | ||||
| GLI1 | 0.102 | ||||
| GLI3 | 0.099 | ||||
| GSK3B | 0.684 | ||||
| PRKACB | -0.393 | ||||
| PRKAR1A | 0.973 | ||||
| PRKAR2B | -0.12 | ||||
| PTCH1 | 0.41 | ||||
| SMO | 0.378 | ||||
| SUFU | -0.238 | ||||
|
CM: Genes annotated by the GO term GO:0048475. (COATED_MEMBRANE) |
Day 28 vs Day 0 | 0.8 | AFTPH | 0.035 | 0.34 |
| AP3S1 | 0.445 | ||||
|
MC: Genes annotated by the GO term GO:0030117 (MEMBRANE_COAT) |
Day 28 vs Day 0 | 0.8 | AFTPH | 0.035 | 0.34 |
| AP3S1 | 0.445 | ||||
|
ERG: Genes down-regulated in prostate cancer samples bearing the fusion of TMPRSS2 with ERG (SETLUR_PROSTATE_CANCER_TMPRSS2_ERG_FUSION_DN [64]) |
Day 28 vs Day 0 | 0.8 | ITGAD | 0.28 | 0.34 |
| KLHL21 | 0.254 | ||||
| MPPED2 | -0.102 | ||||
| RAB27A | 0.088 | ||||
|
AMI: Acute Myocardial Infarction (BIOCARTA_AMI_PATHWAY) |
Day 28 vs Day 3 | 0.5 | COL4A3 | 0.124 | 0.34 |
| F2R | 0.444 | ||||
| PLAT | -0.044 | ||||
| PROC | -0.634 | ||||
| PROS1 | 0.07 | ||||
| TFPI | 0.478 | ||||
|
EIF: Eukaryotic protein translation (BIOCARTA_EIF_PATHWAY) |
Day 28 vs Day 3 | 0.8 | EEF2 | 0.168 | 0.34 |
| EEF2 K | 0.351 | ||||
| EIF1AX | -0.557 | ||||
| EIF2S1 | -0.104 | ||||
| EIF2S2 | 0.027 | ||||
| EIF2S3 | -0.303 | ||||
| EIF3A | -0.57 | ||||
| EIF4A1 | -0.191 | ||||
| EIF4A2 | 0.056 | ||||
| EIF4E | 0.485 | ||||
| EIF4G3 | 0.35 | ||||
|
EPA: Extrinsic Prothrombin Activation Pathway (BIOCARTA_EXTRINSIC_PATHWAY) |
Day 28 vs Day 3 | 0.5 | F2R | 0.417 | 0.34 |
| F3 | 0.24 | ||||
| F5 | 0.006 | ||||
| PROC | -0.673 | ||||
| PROS1 | 0.095 | ||||
| TFPI | 0.413 | ||||
|
STING: Primary innate immune response genes induced in 293T cells by overexpression of STING (TMEM173) (ISHIKAWA_STING_SIGNALING [65]) |
Day 28 vs Day 3 | 0.8 | ADAR | 0.791 | 0.34 |
| CCL5 | 0.519 | ||||
| IRF1 | 0.552 | ||||
| IRF2 | -0.609 | ||||
| IRF6 | -0.328 | ||||
| IRF7 | 0.365 | ||||
| IRF9 | -0.498 | ||||
|
GOLGI: Genes involved in Transport to the Golgi and subsequent modification (REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION) |
Day 28 vs Day 3 | 0.8 | B4GALT1 | 0.25 | 0.34 |
| B4GALT2 | 0.439 | ||||
| B4GALT4 | -0.019 | ||||
| MAN1C1 | -0.079 | ||||
| MCFD2 | -0.282 | ||||
| MGAT2 | -0.699 | ||||
| MGAT3 | 0.465 | ||||
| MGAT4A | -0.698 | ||||
| MGAT4B | 0.111 | ||||
| SAR1B | 0.087 | ||||
| SEC13 | -0.949 | ||||
| SEC23A | 0.319 | ||||
| SEC24B | -0.504 | ||||
| ST8SIA6 | 0.286 |
Results from the top 13 genesets with minimum cross validation error rate <0.35 in the elastic net penalized logistic regression models of HAI response. The geneset name provides the abbreviation that is used for simplicity in the text, a brief description and geneset name from the MSigDB [32] and the actual gene. Delta is the time period that geneset was found to significantly change over time; α is the elastic net penalty for the model; symbol is the gene symbol of genes that remained in each model; coefficients are from the model at the minimum misclassification error rate and the minimum CV error rate is the observed error rate after 10-fold cross validation.
Fig. 3.
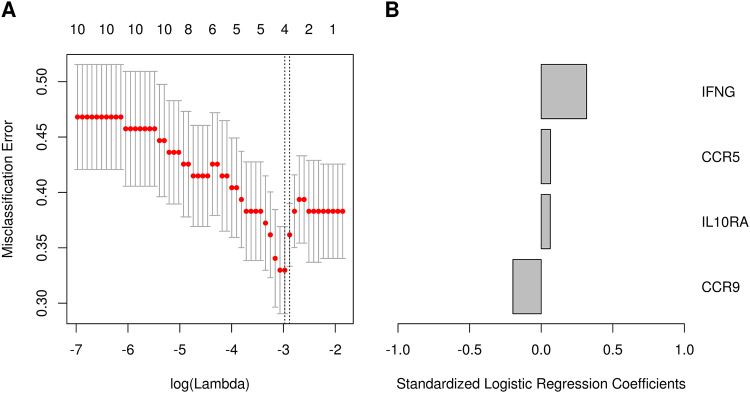
Multivariable correlates of HAI response: Geneset (Day 28 vs. Day 0). Results from the elastic net logistic regression model for association of the genes in the “KUROZUMI” geneset with HAI response. (A) The cross validated misclassification error rate (y-axis) as a function of the tuning parameter (bottom x-axis) that governs the number of variables entered into the model (top x-axis). The misclassification error rate indicates the portion of patients incorrectly classified. The error bars indicate one standard error of the misclassification error rate. The vertical dashed lines indicate either the model with the minimum cross validated error rate, or the model with cross validated error rate within one standard error of the minimum rate. (B) The logistic regression coefficients for the genes selected in the model with the minimum misclassification error. IFNG, IL10RA and CCR5 genes are positively associated with influenza HAI response, whereas CCR9 gene is negatively associated with influenza HAI response.
Fig. 4.
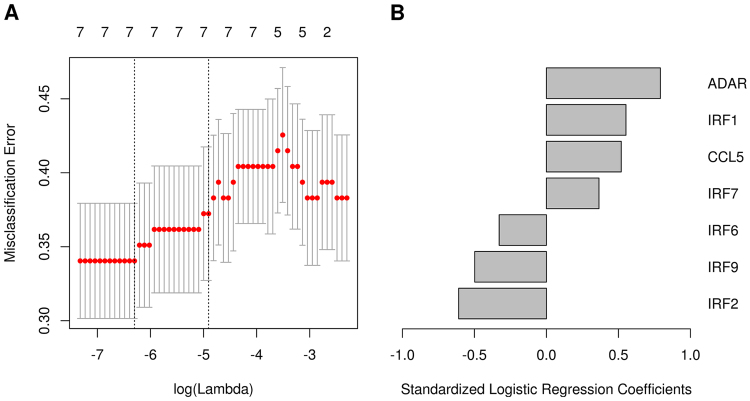
Multivariable correlates of HAI response: Geneset (Day 28 vs. Day 3). Results from the elastic net logistic regression model for association of the genes in the “STING” geneset with HAI response. (A) The cross validated misclassification error rate (y-axis) as a function of the tuning parameter (bottom x-axis) that governs the number of variables entered into the model (top x-axis). The misclassification error rate indicates the portion of patients incorrectly classified. The error bars indicate one standard error of the misclassification error rate. The vertical dashed lines indicate either the model with the minimum cross validated error rate, or the model with cross validated error rate within one standard error of the minimum rate. (B) The logistic regression coefficients for the genes selected from the model that were selected from the model at the minimum misclassification error. ADAR, IRF1, IRF7, and CCL5 genes are positively associated with influenza HAI response, whereas IRF6, IRF2, and IRF9 genes are negatively associated with influenza HAI response.
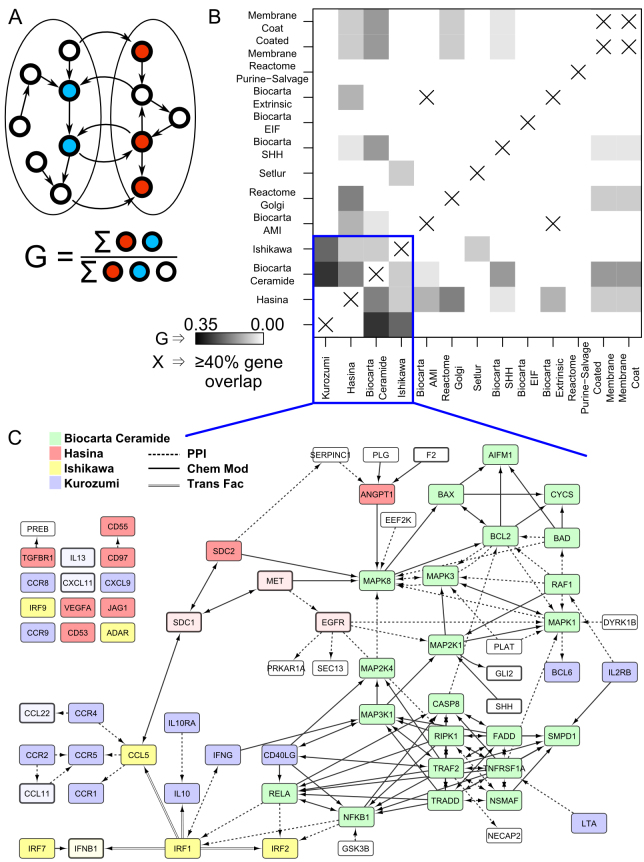
3.4. Interactions between genesets
In order to better understand the biological processes, we then identified interrelationships in these 13 genesets (Day 28 minus Day 0 and Day 28 minus Day 3). We found four genesets (NOL7, CERAMIDE, KUROZUMI, and STING) that capture the bulk of identified genes and their interactions (Fig. 5). The interconnectivity (see Methods) within these genesets is unusually high. Randomly generating 100,000 genesets of the same size and computing their interconnectivity, we generated a distribution with mean 0.24 and standard deviation 0.06, while the observed interconnectivity is 0.90, revealing the observed network density to be highly non-random. These genesets include members of the protein kinase receptors, MAPK signal cascade, regulators of NFκB, cytokines/cytokine receptors, chemokines/chemokine receptors, and interferon regulatory transcription factors. NFκB interacts with IRF1 and IRF2, which regulate expression of CCL5 (C-C motif ligand 5). CCL5 links the system to multiple C-C motif chemokine receptors, which in turn interact with more C-C motif ligands. This integrated paradigm places these diverse signaling processes into a common regulatory model.
Fig. 5.
Interactions between genesets. A) Geneset interconnectivity, the fraction of genes between two genesets that are regulatory partners, is illustrated. B) Geneset interconnectivity is shown for the 13 genesets identified with the lowest cross-validation error rate. While some gene sets are disjoint (share no genes), many are not. Some share enough genes that we simply place an ‘X’ in the plot to indicate that the sets are too similar for the metric to be meaningful. C) All genes in the 13 genesets (Table 2) are shown in their network context. The extent of interconnectivity, quantified in panel B, is evident. The majority of genes can be unambiguously colored by their inclusion in only 4 genesets. Edge type indicates the type of interaction. For clarity, at most one edge is shown between any two genes, with transcription factor regulation (Trans Fac) taking the highest precedence, followed by chemical modification (including phosphorylation), and other protein-protein interactions. Genes that are in a geneset, but not detected by mRNA-seq in our samples are colored a lighter shade with darker border.
4. Discussion
This study focuses on an examination of the genetics influencing variability in humoral immunity, with the goal of identifying gene signatures and their influence on the humoral immune response to influenza vaccine. Our primary focus was to use two balancing analytical approaches to assess transcriptomic profiling of influenza vaccine-induced humoral (HAI) response in older adults [20]. Both gene-to-biology and biology-to-gene approaches were used to assess gene expression and to characterize genesets in relation to influenza vaccine-induced humoral antibody response. The gene-to-biology approach is an inductive, evidence-based approach that utilizes individual per-gene variables. In contrast, the biology-to-gene approach is an empirical approach that relies on known biological knowledge to create genesets recognized to be involved in the immune response processes [20]. Unlike our gene-to-biology approach, the latter analyses begin at the variable set level and drill down to the per-variable level.
Our gene-to-biology per-gene analyses identified differentially expressed novel and known genes related to influenza-specific HAI response. Since the FDR values for these findings are large, we discuss them only briefly here. The majority of transcriptomic changes related to positive HAI response were observed at Day 28 relative to baseline (the peak of adaptive humoral immune response mediated by memory B cells). Twenty-two of the top 30 (73.3%) genes with significant (p < 0.01) transcriptional changes were found to be upregulated (Fig. 2A). The top two genes differentially expressed in our study from Day 28 minus Day 0 are TGFBR1 (OR 2.72; p < 0.002) and the X chromosome-linked IFT52 genes (OR 2.78; p < 0.003). Notably, in our geneset (NOL7), the TGFBR1 gene also demonstrated a positive association with HAI response (Table 2). TGFBR1 encodes a serine/threonine protein kinase protein that transmits TGF-beta signal from the cell surface into the cell, and is known to affect cell growth and division [44]. Likewise, we found several interesting genes related to HAI response, such as: ZNF300 (zinc finger protein 300); NUP133 (nucleoporin 133 kDa); KLK1 (kallikrein 1); CD200 (CD200 membrane glycoprotein); and SLC9A3 (solute carrier family 9, subfamily A) (all downregulated with the exception of HSPA14, GPRIN1, GPR153). Conversely, the lowest number of significant gene expression changes associated with influenza HAI response was observed at Day 28 relative to Day 3 (top six genes) and at Day 3 relative to Day 0 (top six genes). These included the cellular growth and division (helicase DDX28), cell cycle progression and differentiation (S100A5), as well as regulation of NF-kappa-B-mediated transcription (TONSL) genes and also genes with unknown function. The specific role of these differentially expressed genes in influenza-induced adaptive immunity is unclear. Hence, a gene expression replication study and a closer examination of transcriptional activation induced by influenza vaccine are needed.
As a variety of genes and gene pathways are involved in influenza virus-induced immune responses, we also used biology-to-gene analysis to identify genesets associated with potential for explaining the variation in influenza-induced humoral immunity in multivariable models. We discuss these in greater depth here, as the FDR values and cross-validation methods indicate these findings are more likely reproducible than the gene-to-biology findings. This analysis is based on use of gene sets, which has been shown to be both more powerful for detecting associations and less susceptible to false discoveries [32, 45]. This analysis identified 13 genesets (from existing public databases and the literature) with components (genes) related both positively and negatively to the HAI response. Most of the specific genes encompassing these genesets correspond to biological processes, including immune regulation, inflammation, signal transduction, cell cycle and proliferation, and biosynthesis (Table 2). We observed significant transcriptomic changes in genesets at both timepoints: Day 28 vs. Day 0 and Day 28 vs. Day 3. Among eight genesets that were identified at Day 28 vs. Day 0, are the five-gene (BAX, MAP2K4, MAPK3, NSMAF, and TNFRSF1A) signatures (CERAMIDE) that contain genes that are involved in processes such as P53-mediated apoptosis (BAX), proliferation, differentiation, and transcription regulation (MAP2K4, MAPK3), as well as the nuclear factor-kappa B signal transduction and tumor necrosis factor receptor superfamily (TNFRSF)-induced cellular responses such as inflammation (NSMAF, TNFRSF1A). Proteins encoded by the MAPKs (mitogen-activated protein kinases) family and TNF genes have been shown to play an important role in directing innate and adaptive (cytokine) immune responses and thus may influence influenza-specific antibody response [46, 47]. Notably, one of the important genes in the “predictive signatures” of neutralizing antibody responses to seasonal influenza vaccine and yellow fever vaccine (YF-17D) in humans was the TNFRSF17 gene, which encodes BCMA, a receptor for the B cell growth factor (BLyS-BAFF) [48, 49] and plays a significant part in B cell differentiation and B cell homeostasis [19, 50]. In fact, the B cell growth factor TNFRSF17 gene predicted the antibody response to YF-17D vaccine with up to 100% accuracy [50].
Another interesting result is the identification of the gene signature (Day 28 vs. Day 0) (KUROZUMI) related to variation in HAI titers. All genes comprising this specific geneset (CCR5, CCR9, IFNG, and IL10RA) have an important known role in immune function. Studies have also established that CCR5 expression is necessary for influenza-specific CD8+ T cell response and for the clinical outcome of respiratory influenza A virus infection [51, 52, 53]. A study of the trivalent 2004–2005 influenza vaccine found that an age-related decrease in antibody response is inversely correlated with high IL-10 secretion (p < 0.0001) [54]. The finding of these gene signatures, with a possible relation to humoral antibody response following influenza A/H1N1 vaccine, leads to the high possibility that these gene signatures may be involved in influenza virus-induced immune activation and antibody response.
Our data comparing Day 28 vs. Day 3 identified multiple differentially expressed genesets involved in immunity, cell migration and tissue remodeling, mRNA processing, coagulation pathway, protein transport and glycolipid biosynthesis. An important geneset signature (STING) was found in association with HAI response (ADAR, CCL5, IRF1, IRF2, IRF6, IRF7, and IRF9). Specifically, molecules involved in proliferation and activation of natural killer (NK) cells, such as chemotactic cytokine CCL5 (RANTES), as well as transcriptional factors that control type I interferons (IRF1, IRF2, IRF6, IRF7, IRF9), were significantly induced. Consistent with this finding, a systems biology study of yellow fever vaccine (YF-17D) in humans also found that expression of specific IRF genes is correlated with innate and adaptive (antibody) response.
As systems biology approaches have shown promise in other studies, we linked prioritized genesets together using known regulatory interactions to form a local vaccine response network. Interestingly, of the 13 genesets highlighted, only four are required to cover most of the genes involved. These four genesets (i.e., NOL7, CERAMIDE, KUROZUMI, and STING) form a regulatory core with which members of the remaining gene sets interact. The high degree of interconnectivity between these genesets points to their related function, supporting the biology-to-gene paradigm. The integration of these diverse signaling processes into a common regulatory model is a first step in identifying the dominant features involved in determining the strength of response to influenza vaccination.
Our findings are in agreement with a report demonstrating that the 2008–2009 seasonal influenza vaccination upregulated gene expression of interferon-inducible genes (IRF9, IFIT1, MX1) in the peripheral blood 24 hours after vaccination [55]. Nakaya et al. utilized a systems biology strategy to examine immune responses to vaccination against TIV and live attenuated influenza vaccine (LAIV) [19]. This study also identified the increased expression of interferon-associated genes after vaccination with LAIV, such as IRF3 and IRF7 encoding proteins involved in interferon signaling pathways [19]. In addition to IRF7 and IRF9, in our study we also found that IRF1 is positively associated with HAI response, whereas IRF2 and IRF6 are negatively associated with influenza-specific HAI. Consistent with these observations, Querec et al. examined early gene signatures that predict immune responses following the attenuated live YF-17D vaccination [50]. Their results indicated the verification of expression of several genes, including transcription factors IRF7 and IRF9 induced by YF-17D vaccine [50]. The biological understanding of these findings to vaccine-induced immune responses must be further examined.
Li et al. also applied systems biology approaches to examine gene signatures of antibody responses to five human vaccines [18]. They observed the IRF2 and DDX58-like signaling genes were strongly correlated with carbohydrate-specific antibody titers to the meningococcal quadrivalent polysaccharide (MPSV4) and meningococcal polysaccharide-protein conjugate (MCV4) vaccines [18]. In our geneset (STING), the IRF2 gene demonstrated a negative association with HAI response (Fig. 4). IRF2 is known to inhibit IRF1-facilitated transcriptional activation of IFN-α and IFN-β and other genes that use IRF1 for transcription activation [56]. This suggests vaccines against influenza and the meningococcus may use common genes, such as the IRF2, and different innate and adaptive immune pathways to produce antibody responses [11, 18].
In our study, pre-existing immunity and the relationship of pre-existing HAI antibodies with postvaccination antibody titers had an effect on the resulting humoral response following influenza vaccine. It has been shown that the history of an individual’s influenza vaccination likely influences his/her current response to vaccine [57]. Our data are consistent with other studies demonstrating that high baseline antibody titer inversely correlated with the postvaccination response to influenza A/H1N1 vaccine [15, 57, 58].
Our gene expression results are based on PBMCs, which consist of multiple cell types. We and others have observed small fold changes in mixed PBMCs [26, 27]. Geneset approaches are known to have higher power to detect changes in situations where there are many changes of small magnitude, such that they are not readily detected by individual per-gene analyses.
The strengths of our study include the use of next generation sequencing, which allows for whole transcriptome profiling, and the application of comprehensive statistics and bioinformatics algorithms. Our study focuses on older individuals (50–74 years of age), as signs of immunosenescence (increased susceptibility to infection, vaccine failure) are frequently observed in this age group [59, 60, 61]. Our analytical systems biology (gene-to-biology and biology-to-gene) approach allows us to study transcriptional associations with HAI titers that are highly likely (based on the literature and biologic plausibility) to explain variations in influenza A/H1N1-induced humoral immune responses. We performed transcriptome profiling using a heterogeneous cell population (PBMCs) without sequential isolation of neutrophils, monocytes and T cell subsets. As the immune response to influenza A/H1N1 is a complex interaction of different cells and mediators and is not controlled by a single cell type, we assessed and found genesets associated with influenza HAI response in a heterogeneous cell population. Our gene expression results are based on PBMCs, which consist of multiple cell types. We and others have observed small fold changes in mixed PBMCs [26, 27]. Geneset approaches are known to have higher power to detect changes in situations where there are many changes of small magnitude, such that they are not readily detected by individual per-gene analyses. Exploratory studies of transcriptional profiling of isolated cell types from peripheral blood before and after influenza vaccine are in progress.
A major strength of this analysis approach is that we utilized a priori information from existing genesets that group genes based on biological rationale. Our approach of filtering based on gene expression changes over time, in essence a sophisticated variance filter we believe most likely to retain genes that are altered consistently in response to vaccine, helps to control false discoveries since it was agnostic to the outcome variable. In addition, the elastic net penalized regression performs internal cross validation and shrinkage, which, in turn, should provide more reproducible results, help reduce false discovery, and provide a realistic measure of model performance. This resampling strategy for internal model validation has several advantages over splitting the data into a discovery cohort and replication cohort. Data splitting results in a costly reduction in sample size, may provide different results if split differently, and does not validate the final model fit to the full dataset. Cross validation avoids these disadvantages, and therefore is a more efficient use of the data, and the generalizability of our findings should be evaluated in an external cohort [38].
Despite these strategies that we employed to minimize false discoveries, due to a large number of tests in our gene analysis, there is always the risk of false discoveries when looking for potentially novel genes that impact immune response, and the generalizability of our findings should be evaluated in an external cohort. The examination of gene expression in a control group of unvaccinated subjects would be beneficial to this study. There might be a concern that antibody titers to the stalk of the influenza hemagglutinin and other components of the TIV (influenza A/H3N2 and B viruses) were not studied, and that possible sex-related differences in both influenza HAI response and expression of genes (located on the X and Y chromosomes) were not addressed in our study. We restricted our study to influenza A/H1N1 vaccine since new influenza A/H1N1 strains represent a potentially devastating worldwide public health threat, and because influenza A/California/7/2009/H1N1 virus is the A/H1N1 component of the 2010–2011 trivalent vaccine when subjects were enrolled in this study.
In summary, we have demonstrated that gene-to-biology and biology-to-gene approaches can be applied to elucidate host genetic influences on the antibody response to influenza vaccine. We identified a number of uncharacterized—and confirmed previously reported—specific genes and genesets that displayed associations with HAI response to seasonal influenza A/H1N1 vaccine. The ability to detect gene signatures related to HAI response may assist with the design of better vaccines and adjuvants to facilitate improved defenses against new strains of influenza as emerging infectious agents. Future studies will attempt to unravel the biological mechanisms underlying the involvement of a given gene/geneset, as well as the function of the protein produced by that gene and its role in generating immune responses.
Declarations
Author contribution statement
Inna G. Ovsyannikova, Richard B. Kennedy: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Ann L. Oberg: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Michael T. Zimmermann: Wrote the paper.
Iana H. Haralambieva: Conceived and designed the experiments; Performed the experiments.
Krista M. Goergen: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Diane E. Grill: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Gregory A. Poland: Conceived and designed the experiments; Wrote the paper.
Funding statement
This work was supported by the NIH (U01AI089859). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interest statement
The authors declare the following conflict of interests: Gregory A. Poland, Inna G. Ovsyannikova; Dr. Poland is the chair of a Safety Evaluation Committee for novel investigational vaccine trials being conducted by Merck Research Laboratories; Dr. Poland offers consultative advice on vaccine development to Merck & Co. Inc., CSL Biotherapies, Avianax, Dynavax, Novartis Vaccines and Therapeutics, Emergent Biosolutions, Adjuvance, Microdermis, Seqirus, NewLink, Protein Sciences, GSK Vaccines, and Sanofi Pasteur. Drs. Poland and Ovsyannikova hold two patents related to vaccinia and measles peptide research.
These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. This research has been reviewed by the Mayo Clinic Conflict of Interest Review Board and was conducted in compliance with Mayo Clinic Conflict of Interest policies.
Additional information
Data associated with this study has been deposited at synapse.org under the http://dx.doi.org/10.7303/syn3219180. Additional data associated with this cohort is available through NIH's ImmPort repository at https://immport.niaid.nih.gov/immportWeb/clinical/study/displayStudyDetails.do?itemList=SDY67.
Acknowledgments
We thank the Mayo Clinic Vaccine Research Group and the subjects who participated in our studies. We thank Randy A. Albrecht and Adolfo García-Sastre of the Mount Sinai School of Medicine (New York, NY) for performing the hemagglutination inhibition assay. We thank Caroline L. Vitse for her editorial assistance.
Appendix A. Supplementary data
References
- 1.Writing Committee of the Second World Health Organization Consultation on Clinical Aspects of Human Infection with Avian I Update on Avian Influenza A (H5N1) Virus Infection in Humans. N. Engl. J. Med. 2008;358(3):261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 2.Fraser C., Donnelly C.A., Cauchemez S., Hanage W.P., Van Kerkhove M.D., Hollingsworth T.D. Pandemic Potential of a Strain of Influenza A (H1N1): Early Findings. Science. 2009;324(5934):1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taubenberger J.K., Morens D.M. 1918 Influenza: the mother of all pandemics. Emerg. Infect. Dis. 2006;12(1):15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hannoun C., Megas F., Piercy J. Immunogenicity and protective efficacy of influenza vaccination. Virus Res. 2004;103(1-2):133–138. doi: 10.1016/j.virusres.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 5.Fiore A.E., Shay D.K., Broder K., Iskander J.K., Uyeki T.M., Mootrey G. Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2008. MMWR Recomm. Rep. 2008;57(RR-7):1–60. [PubMed] [Google Scholar]
- 6.Villari P., Manzoli L., Boccia A. Methodological quality of studies and patient age as major sources of variation in efficacy estimates of influenza vaccination in healthy adults: a meta-analysis. Vaccine. 2004;22(25-26):3475–3486. doi: 10.1016/j.vaccine.2004.01.068. [DOI] [PubMed] [Google Scholar]
- 7.Srivastava B., Blazejewska P., Hessmann M., Bruder D., Geffers R., Mauel S. Host genetic background strongly influences the response to influenza a virus infections. PLoS ONE. 2009;4(3):e4857. doi: 10.1371/journal.pone.0004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furman D., Hejblum B.P., Simon N., Jojic V., Dekker C.L., Thiebaut R. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc. Natl. Acad. Sci. USA. 2014 Jan 14;111(2):869–874. doi: 10.1073/pnas.1321060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furman D., Jojic V., Kidd B., Shen-Orr S., Price J., Jarrell J. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol. Syst. Biol. 2013;9:659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan Y., Tamayo P., Nakaya H., Pulendran B., Mesirov J.P., Haining W.N. Gene signatures related to B-cell proliferation predict influenza vaccine-induced antibody response. Eur. J. Immunol. 2014 Oct 17;44(1):285–295. doi: 10.1002/eji.201343657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Obermoser G., Presnell S., Domico K., Xu H., Wang Y., Anguiano E. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013 Apr 18;38(4):831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pulendran B., Li S., Nakaya H.I. Systems vaccinology. Immunity. 2010;33(4):516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat. Rev. Immunol. 2009;9(10):741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 14.Nakaya H.I., Li S., Pulendran B. Systems vaccinology: learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012;4(2):193–205. doi: 10.1002/wsbm.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsang J.S., Schwartzberg P.L., Kotliarov Y., Biancotto A., Xie Z., Germain R.N. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014 Apr 10;157(2):499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsang J.S. Utilizing population variation, vaccination, and systems biology to study human immunology. Trends Immunol. 2015 Aug;36(8):479–493. doi: 10.1016/j.it.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009;10(1):116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S., Rouphael N., Duraisingham S., Romero-Steiner S., Presnell S., Davis C. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol. 2014 Feb;15(2):195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakaya H.I., Wrammert J., Lee E.K., Racioppi L., Marie-Kunze S., Haining W.N. Systems biology of seasonal influenza vaccination in humans. Nat. Immunol. 2011;12(8):786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oberg A.L., Kennedy R.B., Li P., Ovsyannikova I.G., Poland G.A. Systems biology approaches to new vaccine development. Curr. Opin. Immunol. 2011;23(3):436–443. doi: 10.1016/j.coi.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ovsyannikova I.G., White S.J., Albrecht R.A., Garcia-Sastre A., Poland G.A. Turkey versus guinea pig red blood cells: hemagglutination differences alter hemagglutination inhibition responses against influenza A/H1N1. Viral Immunol. 2014 May;27(4):174–178. doi: 10.1089/vim.2013.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ovsyannikova I.G., White S.J., Larrabee B.R., Grill D.E., Jacobson R.M., Poland G.A. Leptin and leptin-related gene polymorphisms, obesity, and influenza A/H1N1 vaccine-induced immune responses in older individuals. Vaccine. 2014 Feb 7;32(7):881–887. doi: 10.1016/j.vaccine.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salk H.M., Haralambieva I.H., Ovsyannikova I.G., Goergen K.M., Poland G.A. Granzyme B ELISPOT assay to measure influenza-specific cellular immunity. J. Immunol. Methods. 2013 Sep 18;398-399:44–50. doi: 10.1016/j.jim.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster R., Cox N., Stohr K. WHO Manual on Animal Influenza Diagnosis and Surveillance. 2002 [Google Scholar]
- 25.Brady R.C., Treanor J.J., Atmar R.L., Keitel W.A., Edelman R., Chen W.H. Safety and immunogenicity of a subvirion inactivated influenza A/H5N1 vaccine with or without aluminum hydroxide among healthy elderly adults. Vaccine. 2009;27(37):5091–5095. doi: 10.1016/j.vaccine.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy R.B., Oberg A.L., Ovsyannikova I.G., Haralambieva I.H., Grill D.E., Poland G.A. Transcriptomic profiles of high and low antibody responders to smallpox vaccine. Genes Immun. 2013;14(5):277–285. doi: 10.1038/gene.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haralambieva I.H., Oberg A.L., Ovsyannikova I.G., Kennedy R.B., Grill D.E., Middha S. Genome-wide characterization of transcriptional patterns in high and low antibody responders to rubella vaccination. PLos ONE. 2013;8(5):e62149. doi: 10.1371/journal.pone.0062149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinlan A.R., Hall I.M. BEDTools a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010 Mar 15;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C., Pachter L., Salzberg S.L. TopHat discovering splice junctions with RNA-Seq. Bioinformatics. 2009 May 1;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen K.D., Irizarry R.A., Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13(2):204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCullagh P., Nelder J.A. Chapman and Hall; London: 1983. Generalized Linear Models. [Google Scholar]
- 34.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010 Nov 11;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardin J.W., Hilbe J.M. Chapman and Hall/ CRC; London: 2003. Generalized Estimating Equations. [Google Scholar]
- 37.Fridley B.L., Jenkins G.D., Grill D.E., Kennedy R.B., Poland G.A., Oberg A.L. Soft truncation thresholding for gene set analysis of RNA-seq data: application to a vaccine study. Sci. Rep. 2013;3:2898. doi: 10.1038/srep02898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrell F.E. Springer; New York: 2001. Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis. [Google Scholar]
- 39.Friedman J., Hastie T., Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 40.Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009 Jan;37(Database issue):D674–679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yusuf D., Butland S.L., Swanson M.I., Bolotin E., Ticoll A., Cheung W.A. The transcription factor encyclopedia. Genome Biol. 2012;13(3):R24. doi: 10.1186/gb-2012-13-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vinayagam A., Stelzl U., Foulle R., Plassmann S., Zenkner M., Timm J. A directed protein interaction network for investigating intracellular signal transduction. Sci. Signal. 2011 Sep 6;4(189):rs8. doi: 10.1126/scisignal.2001699. [DOI] [PubMed] [Google Scholar]
- 43.Jacobson R.M., Grill D.E., Oberg A.L., Tosh P.K., Ovsyannikova I.G., Poland G.A. Profiles of influenza A/H1N1 vaccine response using hemagglutination-inhibition titers. Hum. Vaccin. Immunother. 2015;11(4):961–969. doi: 10.1080/21645515.2015.1011990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Massague J. Receptors for the TGF-beta family. Cell. 1992 Jun 26;69(7):1067–1070. doi: 10.1016/0092-8674(92)90627-o. [DOI] [PubMed] [Google Scholar]
- 45.Efron B., Tibshirani R. On testing the significance of sets of genes. The Annals of Applied Statistics. 2007;1(1):107–129. [Google Scholar]
- 46.Karin M. Inflammation-activated protein kinases as targets for drug development. Proc Am Thorac Soc. 2005;2(4):386–390. doi: 10.1513/pats.200504-034SR. discussion 394-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sareneva T., Matikainen S., Kurimoto M., Julkunen I., Influenza A. Virus-induced IFN-alpha/beta and IL-18 synergistically enhance IFN-gamma gene expression in human T cells. J. Immunol. 1998;160(12):6032–6038. [PubMed] [Google Scholar]
- 48.Avery D.T., Kalled S.L., Ellyard J.I., Ambrose C., Bixler S.A., Thien M. BAFF selectively enhances the survival of plasmablasts generated from human memory B cells. J. Clin. Invest. 2003 Jul;112(2):286–297. doi: 10.1172/JCI18025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodland R.T., Schmidt M.R., Thompson C.B. BLyS and B cell homeostasis. Semin. Immunol. 2006;18(5):318–326. doi: 10.1016/j.smim.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009 Jan;10(1):116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohlmeier J.E., Miller S.C., Smith J., Lu B., Gerard C., Cookenham T. The chemokine receptor CCR5 plays a key role in the early memory CD8+ T cell response to respiratory virus infections. Immunity. 2008;29(1):101–113. doi: 10.1016/j.immuni.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cerwenka A., Morgan T.M., Harmsen A.G., Dutton R.W. Migration kinetics and final destination of type 1 and type 2CD8 effector cells predict protection against pulmonary virus infection. J. Exp. Med. 1999;189(2):423–434. doi: 10.1084/jem.189.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dawson T.C., Beck M.A., Kuziel W.A., Henderson F., Maeda N. Contrasting effects of CCR5 and CCR2 deficiency in the pulmonary inflammatory response to influenza A virus. Am. J. Pathol. 2000;156(6):1951–1959. doi: 10.1016/S0002-9440(10)65068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corsini E., Vismara L., Lucchi L., Viviani B., Govoni S., Galli C.L. High interleukin-10 production is associated with low antibody response to influenza vaccination in the elderly. J. Leukoc. Biol. 2006;80(2):376–382. doi: 10.1189/jlb.0306190. [DOI] [PubMed] [Google Scholar]
- 55.Bucasas K.L., Franco L.M., Shaw C.A., Bray M.S., Wells J.M., Nino D. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J. Infect. Dis. 2011;203(7):921–929. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chae M., Kim K., Park S.M., Jang I.S., Seo T., Kim D.M. IRF-2 regulates NF-kappaB activity by modulating the subcellular localization of NF-kappaB. Biochem. Biophys. Res. Commun. 2008 Jun 6;370(3):519–524. doi: 10.1016/j.bbrc.2008.03.136. [DOI] [PubMed] [Google Scholar]
- 57.Reber A.J., Kim J.H., Biber R., Talbot H.K., Coleman L.A., Chirkova T. Preexisting Immunity, More Than Aging Influences Influenza Vaccine Responses. Open Forum Infect. Dis. 2015 Apr;2(2):ofv052. doi: 10.1093/ofid/ofv052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller M.S., Palese P. Peering into the crystal ball: influenza pandemics and vaccine efficacy. Cell. 2014 Apr 10;157(2):294–299. doi: 10.1016/j.cell.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 59.Goronzy J.J., Fulbright J.W., Crowson C.S., Poland G.A., O'Fallon W.M., Weyand C.M. Value of immunological markers in predicting responsiveness to influenza vaccination in elderly individuals. J. Virol. 2001;75(24):12182–12187. doi: 10.1128/JVI.75.24.12182-12187.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernstein E., Kaye D., Abrutyn E., Gross P., Dorfman M., Murasko D.M. Immune response to influenza vaccination in a large healthy elderly population. Vaccine. 1999;17:82–94. doi: 10.1016/s0264-410x(98)00117-0. [DOI] [PubMed] [Google Scholar]
- 61.Lambert N.D., Ovsyannikova I.G., Pankratz V.S., Jacobson R.M., Poland G.A. Understanding the immune response to seasonal influenza vaccination in older adults: a systems biology approach. Expert Rev. Vaccines. 2012;11(8):985–994. doi: 10.1586/erv.12.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hasina R., Pontier A.L., Fekete M.J., Martin L.E., Qi X.M., Brigaudeau C. NOL7 is a nucleolar candidate tumor suppressor gene in cervical cancer that modulates the angiogenic phenotype. Oncogene. 2006 Jan 26;25(4):588–598. doi: 10.1038/sj.onc.1209070. [DOI] [PubMed] [Google Scholar]
- 63.Kurozumi K., Hardcastle J., Thakur R., Yang M., Christoforidis G., Fulci G. Effect of tumor microenvironment modulation on the efficacy of oncolytic virus therapy. J. Natl. Cancer Inst. 2007 Dec 5;99(23):1768–1781. doi: 10.1093/jnci/djm229. [DOI] [PubMed] [Google Scholar]
- 64.Setlur S.R., Mertz K.D., Hoshida Y., Demichelis F., Lupien M., Perner S. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J. Natl. Cancer Inst. 2008 Jun 4;100(11):815–825. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008 Oct 2;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.