Abstract
In this issue of Blood, Assouline et al report the clinical activity of the oral histone deacetylase inhibitor, panobinostat, in 40 patients with relapsed or refractory diffuse large B-cell lymphoma (DLBCL) not preselected on the basis of molecular subtype as well as transformed lymphomas.1 The authors found a modest overall response rate (ORR) of 28% in the entire cohort with no benefit from the addition of rituximab. Importantly, a subset of responders achieved relatively durable remissions, including 6 responders (55%) who had yet to progress.
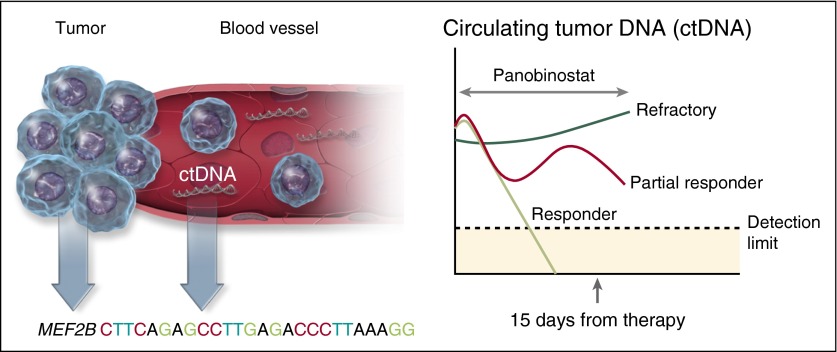
Molecular predictors of response to panobinostat in DLBCL. The presence of molecular biomarkers before therapy and after only 15 days of treatment predicted response in patients with relapsed DLBCL, and transformed lymphomas treated with panobinostat. The presence of a mutation in the histone-modifying enzyme MEF2B prior to therapy, increased the likelihood of response by almost fourfold, whereas levels of ctDNA in the plasma at day 15 was a strong predictor of response. Virtually all patients who responded to panobinostat (light green line) saw an immediate reduction in ctDNA levels by day 15, whereas patients refractory to therapy (dark green line) had an increase in ctDNA. Other patients only partially responded with ctDNA (red line), but did not have a clinical response. Professional illustration by Somersault18:24.
The most interesting findings by Assouline et al, however, relate to the comprehensive set of molecular predictive biomarkers used in this study (see figure). The authors analyzed pretreatment specimens with a combination of whole exome sequencing and targeted sequencing for known lymphoma hot spots to identify individual somatic mutations that correlated with response. They found that 6 patients (15%) harbored pretreatment MEF2B mutations and 4 of these patients (66%) achieved a clinical response to panobinostat (likelihood ratio, 3.67). In contrast, mutations of TP53, FAS, STAT6, and CD79B in pretreatment biopsies did not predict response. On-treatment tissue biopsies (N = 14) were also performed in patients at day 15. They observed a slight increase in the acetylation of the target histone H3, but this was not associated with clinical response.
A second novel molecular biomarker evaluated in this study was circulating tumor DNA (ctDNA) measured by droplet digital polymerase chain reaction in the plasma at baseline and repeated 15 days after initiation of therapy. The fraction of cell-free DNA in the plasma was identified as tumor-derived based on the presence of variant allele fractions for somatic mutations present in the baseline tumor. Almost every patient (96%) had at least one plasma sample with measurable levels of ctDNA. The relative level of ctDNA in the plasma at day 15 compared with baseline was highly predictive of response. No patients whose ctDNA increased achieved a clinical response, making it an excellent early negative predictor of responsiveness. Conversely, all patients who had a response had either a significant reduction in ctDNA by day 15 or a trend toward declining levels.
We have entered the era of molecular subtyping of DLBCL and evaluating the role of precision medicine through integration of patient-specific molecular characteristics and treatment selection.2 The benefits of this strategy over traditional and empirical approaches, however, have yet to be determined. Stronger emphasis on technologies such as next-generation sequencing (NGS) to define the mutational landscape of tumors, as well as molecular monitoring for ctDNA are important components of precision medicine. Numerous targeted therapies are being developed for DLBCL with the common goal of overcoming known mechanisms of cellular resistance.3 Precision medicine initiatives attempt to correlate individual somatic mutations within tumors that predict response or confer resistance to these targeted agents.
The first evidence for precision medicine in DLBCL comes from a multicenter study of 80 patients with relapsed DLBCL treated with single-agent ibrutinib.4 The ORR to ibrutinib was only 25%, but responses were significantly more common in patients with an activated B-cell subtype (14/38, 37%) compared with patients with a germinal center B-cell subtype (1/20, 5%), as defined by gene expression profiling. Further, individual somatic mutations were predictive of response. As predicted from preclinical models, patients with CARD11 mutations did not respond to ibrutinib (0/3, 0%). Patients with an underlying MYD88 mutation also did not respond unless they also harbored a mutation in CD79B (4/5, 80%). This proof-of-principle trial led to a multinational phase 3 trial, testing the effect of ibrutinib added to standard frontline therapy for DLBCL.
The trial by Assouline et al identified MEF2B mutations as a molecular predictor of response to panobinostat. With the caveat that definitive conclusions cannot be drawn from small numbers, the observation supports a more rigorous assessment in an expanded cohort enriched with patients who harbor MEF2B mutations. Although only a minority of patients with relapsed DLBCL and transformed lymphomas may be eligible, such studies embody the essence of precision medicine.
Assouline et al also analyzed ctDNA as an early predictive biomarker of response. Recent data in DLBCL demonstrate that NGS-based assays for ctDNA can directly measure tumor response kinetics during combination chemotherapy and predict treatment outcomes.5 In addition to measuring the dynamics of clearance, ctDNA can also be analyzed for tumor-specific mutations as a form of “liquid biopsy.” Emerging assays for ctDNA can identify panels of genetic aberrations circulating at low allele frequencies in DLBCL, and may identify mutations not found in the tumor biopsy.6-8 As a surrogate for the composite tumor genome, ctDNA as a liquid biopsy integrates all the genetic lesions from a tumor and can provide more comprehensive information than a single-tissue biopsy.9
It is notable that ctDNA was identified as an early molecular biomarker for response to panobinostat. The clearance of ctDNA after a few cycles of combination chemotherapy is predictive for response in DLBCL, but the observation of similar patterns with targeted therapy paves the way to test new agents using ctDNA as a surrogate translational end point. Two nonresponding patients who had a decrease in ctDNA at day 15 highlight another potential application of ctDNA. Could a liquid biopsy that incorporates a more extensive mutational panel or serial testing have discovered a treatment emergent resistant subclone?
As we pivot from conventional “one-size-fits-all” paradigms to treatment approaches that consider individual molecular variability, precision molecular monitoring becomes an important translational component. Certainly, both NGS and ctDNA monitoring will continue to evolve, expand, and improve. Although the clinical implications are constrained by the need for technical standardization and clinical validation, regulatory bodies recognize the importance of these approaches.10 Indeed, Assouline et al deserve commendation for conducting a clinical trial that incorporates informative translational studies, and hence provides a far more nuanced result than response rates alone.
Footnotes
Conflict-of-interest disclosure: The author declares no competing financial interests.
REFERENCES
- 1.Assouline SE, Nielsen TH, Yu S, et al. Phase 2 study of panobinostat with or without rituximab in relapsed diffuse large B-cell lymphoma. Blood. 2016;128(2):185–194. doi: 10.1182/blood-2016-02-699520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roschewski M, Staudt LM, Wilson WH. Diffuse large B-cell lymphoma-treatment approaches in the molecular era. Nat Rev Clin Oncol. 2014;11(1):12–23. doi: 10.1038/nrclinonc.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta-Shah N, Younes A. Novel targeted therapies in diffuse large B-cell lymphoma. Semin Hematol. 2015;52(2):126–137. doi: 10.1053/j.seminhematol.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Wilson WH, Young RM, Schmitz R, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roschewski M, Dunleavy K, Pittaluga S, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol. 2015;16(5):541–549. doi: 10.1016/S1470-2045(15)70106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurtz DM, Scherer F, Newman AM, et al. Dynamic noninvasive genomic monitoring for outcome prediction in diffuse large B-cell lymphoma [abstract]. Blood. 2015;126(23) Abstract 130. [Google Scholar]
- 7.Scherer F, Kurtz DM, Newman AM, et al. Noninvasive genotyping and assessment of treatment response in diffuse large B cell lymphoma [abstract]. Blood. 2015;126(23) Abstract 114. [Google Scholar]
- 8.Rasi S, Monti S, Zanni M, et al. Liquid biopsy as a tool for monitoring the genotype of diffuse large B-cell lymphoma [abstract]. Blood. 2015;126(23) Abstract 127. [Google Scholar]
- 9.Roschewski M, Staudt LM, Wilson WH. Dynamic monitoring of circulating tumor DNA in non-Hodgkin lymphoma. Blood. doi: 10.1182/blood-2016-03-635219. 2016;127(25):3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blumenthal GM, Mansfield E, Pazdur R. Next-generation sequencing in oncology in the era of precision medicine. JAMA Oncol. 2016;2(1):13–14. doi: 10.1001/jamaoncol.2015.4503. [DOI] [PubMed] [Google Scholar]