Abstract
Iron homeostasis is disrupted in multiple myeloma, a difficult-to-cure plasma cell malignancy with lytic bone lesions. Here, we systematically analyzed iron gene expression signature and demonstrated that mRNA expression of iron exporter ferroportin (FPN1) is significantly downregulated in myeloma cells and correlates negatively with clinic outcome. Restoring expression of FPN1 reduces intracellular liable iron pool, inhibits STAT3-MCL-1 signaling, and suppresses myeloma cells growth. Furthermore, we demonstrated that mRNA of FPN1 is also downregulated at the initial stages of osteoclast differentiation and suppresses myeloma cell–induced osteoclast differentiation through regulating iron regulator TFRC, NF-κB, and JNK pathways. Altogether, we demonstrated that downregulation of FPN1 plays critical roles in promoting myeloma cell growth and bone resorption in multiple myeloma.
Introduction
Iron is an essential element for almost every type of cell; it is involved in various biologic processes, such as haem synthesis, mitochondria respiratory, DNA synthesis, and cell cycle (1). Intra-cellular iron homeostasis is strictly regulated in normal cells at the level of uptake, storage, export, and even the microenvironment (2). Dysregulation of iron homeostasis results in change of cell fate and promotes diseases such as cancer (3, 4). Many types of cancer have alterations in iron metabolism, resulting in increased intracellular iron levels that facilitate malignant cell growth and disease progression. For instance, iron enhances colorectal tumorigenesis in the presence of APC mutations by augmenting WNT signaling and downregulating E-cadherin (5). Reduced iron export is another way that cancer cells use to increase concentration of iron besides increased iron uptake and decreased iron storage. Unlike multiple ways of iron uptake by different proteins such as TFRC (6) and lipocalin 2 (7), ferroportin (FPN1) is the only known exporter of iron in vertebrates (8). FPN1 encodes a multiple-transmembrane domain protein for the transfer of cellular iron to plasma, which regulates the exit of iron from enterocytes into the blood circulation (9). It is expressed in many iron-exporting cells, including placental syncytiotrophoblasts, duodenal enterocytes, hepatocytes, and reticuloendothelial macrophages (10). Mutations, resulting in loss-of-function of FPN1, cause hyperferritinemia and iron overload in macrophage or hepatocytes (11). Hepcidin, the physiologic ligand of FPN1 (12), inhibits iron export by inducing internalization and proteasome degradation of FPN1 protein (13). Although regulation of iron by FPN1 has been well studied in iron homeostasis, the role of FPN1 in the context of cancer is just beginning to emerge. FPN1 is significantly downregulated in breast cancer cells compared with their normal counterparts. Consistent with the low levels of FPN1 expression, the breast cancer cells showed a markedly higher labile iron pool than the nonmalignant breast epithelial (14). Importantly, low expression of FPN1 was linked to poor prognosis in primary breast cancer samples using gene expression profiles (14).
Multiple myeloma is a difficult-to-cure plasma cell malignancy, the growth, survival, and drug resistance of which are intimately tied to interactions with stromal cells, including osteoclasts, osteoblasts, and extracellular matrix proteins in the bone microenvironment (15). The cross-talk between the multiple myeloma cells and different cells of the bone marrow microenvironment regulates survival and proliferation of multiple myeloma cells and results in other pathologic conditions such as bone destruction by osteoclasts (16). It is very common that patients with multiple myeloma present with anemia due to systemic alteration in iron metabolism (17). The limited iron availability, resulting in impaired erythropoiesis, is probably the main reason for myeloma-associated anemia (18). Elevated IL6 and bone morphogenetic proteins (BMP) in the marrow of multiple myeloma patients are the primary cytokines that induce high expression of hepcidin (19, 20). The myeloma cells also show increased requirement of iron due to a relatively high rate of proliferation and metabolism when compared with normal plasma cells (NPC; ref. 21). In addition to suppressing osteoblastogenesis by secreting the WNT signaling inhibitor DKK1 (16), myeloma cells also promote osteoclastogenesis by eliciting stroma cells to produce RANKL, which is the major inducer of osteoclast differentiation from its macrophage precursors (22). A recent study shows that activation of osteoclasts demands high iron uptake to facilitate mitochondrial biogenesis (23).
Here, we explore the expression of FPN1 in different progression stages of multiple myeloma and correlate FPN1 expression with patient outcome. We provide evidence that downregulation of FPN1 plays an important role in accelerating malignant cell growth and osteoclast differentiation. Our data indicate that therapeutics with ability of elevating expression of FPN1 might hold promising key of improving the outcome in multiple myeloma.
Materials and Methods
Cell culture, drug treatment, and cell growth
Human myeloma cell lines (ARP1, OCI-MY5, and their derivative cell lines ARP1-FPN1, OCI-MY5-FPN1, OCI-FPN1-STAT3c, and OCI-MY5-MCL-1, and their relative controls) and murine myeloma cell lines (5TGM1 and 5TGM1 with inducible expression of FPN1) were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA), supplemented with 10% heat-inactivated FBS (Invitrogen), penicillin (100 IU/mL), and streptomycin (100mg/mL) in a humidified incubator at 37°C and 5% CO2/95% air.
The murine macrophage cells RAW264.7 were cultured in DMEM medium (Invitrogen) with the same supplements. Primary bone marrow mononuclear cells collected from wild-type C57B mice were cultured in the macrophage medium (Cell Biologics) with M-CSF (PeproTech) for 3 days with medium change every day. The attached cells were considered as bone marrow macrophage (BMM) for the following osteoclast differentiation experiments induced by myeloma conditioned medium (5%) or RANKL (50 ng/mL plus 10 ng/mL M-CSF; PeproTech). For experiments, human myeloma cells were incubated with FeCl3, deferoxamine, and FAC (Sigma-Aldrich). Cell growth was counted by the PrestoBlue assay (Invitrogen) and Trypan blue exclusion assay as reported (24).
Clonogenic assay
Clonogenic growth was performed as previous reported (25). Briefly, 10,000 myeloma cells were plated in 0.5 mL 0.33% agar/RPMI 1640 (Invitrogen) supplemented with 10% FBS. Cells were incubated (37°C, 5% CO2) and fed with the same medium twice a week. One colony was defined if more than 40 cells were observed. Plates were imaged and colonies were enumerated using Image J software.
Plasmids and virus production
Human FPN1 coding sequence was purchased from OriGene and subcloned into lentiviral vector pWPI using PacI and XhoI sites. An EGFP-N1 vector with mouse FPN1 was provided by Dr. Ivana De Domenico. The mouse FPN1 coding sequence was inserted to Age I and Mlu l sites of doxycycline-inducible expression derivate from pTRIPZ from Dr. Dana Levasseur’s lab (University of Iowa, Iowa City, IA). The constitutively active forms of STAT3 and MCL-1 expression vectors were purchased from Addgene. Lentivirus was produced in HEK293T cells using vsv-g and psPAX2 helper vectors (Addgene).
Mouse models
All mouse experiments were performed under protocols approved by the Institutional Animal Use and Care Committee of the University of Iowa. Human myeloma cells (1.5 x 106 cells in 100 mL PBS) were injected subcutaneously into the abdomen of NOD-Rag/null gamma mice. Tumor burdens were monitored by tumor volumes. In KaLwRij mouse model, 5TGM1–FPN1 (1 x 106 cells in 100 mL PBS) were injected through tail vein. After 7 days, doxycycline (2 mg/mL, Sigma-Aldrich) was added to the drinking water (contains 5% sucrose) and dextran-iron (0.5 mg/kg, Sigma-Aldrich) was injected intraperitoneally twice a week. Mice were bled every week to harvest sera for detecting IgG2b by the ELISA assay according to the manufacturer’s instructions (Bethyl Laboratories).
Western blotting
Cell lysates were equally loaded onto 4% to 15% SDS-PAGE (Bio-Rad), electrophoresed, and transferred to enhanced chemiluminescence–nitrocellulose membranes (Bio-Rad). The membranes were stained with 0.2% Ponceau S red to ensure equal protein loading. After blocking with 5% nonfat milk in TBS, the membranes were incubated with antibodies against FPN1 (Abcam), STAT3, p-STAT 3, MCL-1, ERK1/2, p-RRK, JNK, p-JNK, p38, p-p38, NfkB-pNFkB, AKT, p-AKT, BCL2, BCL-XL, BAX, BAK, BIM, NOXA, BID, and PUMA (Cell Signaling Technology) overnight at 4°C, followed by horseradish peroxidase–linked secondary antibody (Cell Signaling Technology) for 1 hour at room temperature. Detection was developed by Immobilon Western ECL substrate (Millipore), according to the manufacturer’s instructions. β-actin (Cell Signaling Technology) was used as an internal control.
Quantitative real-time PCR
For quantitative analysis of gene expression, total RNA was isolated by an RNeasy kit (Qiagen). Complementary DNA was synthesized using Iscript reverse transcription kit according to the manufacturer’s instructions (Bio-Rad). Real-time quantitative PCRs for mouse FPN1, IRF8, TFRC, PGC1b, NFATc1, CTSK, tartrate-resistant acid phosphatase (TRAP), and β-actin were performed with SYBR Green Super Mixture Reagents (Bio-Rad) on the CFX connect real-time system (Bio-Rad). The specific primers used for detecting genes are listed in Supplementary Table S2. PCR was initiated with 95°C for 3 minutes to hot-start the DNA polymerase and denature the template, and then 40 cycles consisted of denaturing at 95°C for 30 seconds, annealing and extension at 60°C for 30 seconds. Data were analyzed as previously reported (26).
Labile iron pool assay
The cellular liable iron pool (LIP) was measured with fluorescent metallosensor calcein as described (14). The SIH (salicylaldehyde isonicotinoyl hydrazone) used in the assay was synthesized by Dr. Zhendong Jin lab (University of Iowa, Iowa City, IA).
Flow cytometry
Cell apoptosis were measured by Annexin V/PI kit (eBioscience) according to the manufacturer’s instructions. To detect surface expression of TFRC, isolated BMMs were firstly blocked with anti-CD16/32 (eBioscience) and then stained with PE-conjugated TFRC (eBioscience) in PBS containing 2% FBS on ice for 20 minutes. Then cells were washed with cold PBS, resuspended, and detected by flow cytometry (University of Iowa Flow Cytometry Core, Iowa City, IA).
Prussian blue and TRAP staining
Prussian blue and TRAP staining were performed according to the manufacturer’s instructions (Sigma-Aldrich).
Microarray data sets
BMMs were harvested from both Fpn1+/+ wild-type and Fpn1+/− mice. The Affymetrix Mouse Genome 430 2.0 Arrays were applied to identify fpn1-related genes in BMMs in duplicate as reported previously (27, 28). We also used 3 publicly available datasets for human multiple myeloma microarrays to compare NPCs and multiple myeloma cells (GSE2658, GSE9782, GSE19784). The supervised cluster analysis was performed in statistical software R (Version2.6.2).
Statistical analyses
The unpaired t test was used to evaluate the difference between two different groups. In correlation of FPN1 and MCL-1 expression with disease progression, event-free survival (EFS) and overall survival (OS) were measured using the Kaplan–Meier method, and the log-rank test was used for group comparison as previously reported (29). A P value of less than 0.05 was considered statistically significant.
Results
Low expression of FPN1 is linked to poor survival in multiple myeloma
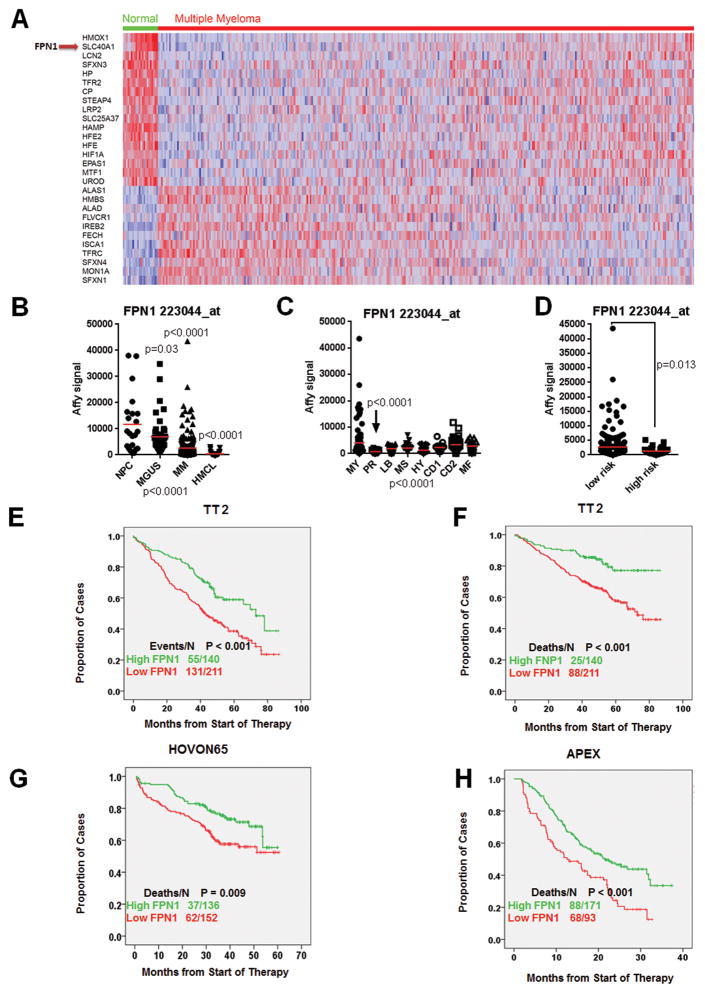
To evaluate iron regulation in multiple myeloma cells, we reanalyzed our previous gene expression profiling data containing 22 NPC samples and 351 newly diagnosed multiple myelomas from the Total Therapy 2 (TT2) cohort using the Affymetrix U133Plus 2 platform (30). Of the 61 signature genes related to iron metabolism (131 probe sets; ref. 31), we identified 29 genes significantly dysregulated by comparison of NPC versus multiple myeloma samples (Supplementary Table S1; P < 0.005; ratio 2′ 1.5-fold). A supervised hierarchical cluster in Fig. 1A showed clearly that a subset of multiple myeloma patients have 18 genes downregulated and 11 genes upregulated in multiple myeloma. These include iron-importing genes, IREB2 and TFRC, and iron-exporting gene, FPN1. We then correlated the expression of these 29 genes with patient outcome in the TT2 cohort, FPN1, the only vertebrate iron exporter, was found to be the top gene whose downregulation in multiple myeloma was associated with an inferior outcome (Supplementary Table S1; OS: P = 0.0032). Because FPN1 had a dysregulated expression and was associated with poor survival, we explored its functional role in multiple myeloma. We also evaluated the expression of FPN1 in sequential multiple myeloma samples from the same patient, in the different genetic subgroups and in the different risk-related subtypes (27, 28). FPN1 expression was significantly lower in PCs derived from multiple myeloma patients compared with PCs derived from patients with monoclonal gammopathy of undetermined significance (MGUS) and healthy donors (P < 0.0001, Fig. 1B). We also found that expression of FPN1 had the lowest expression in the proliferation subgroup (PR), which is the subgroup with the poorest prognosis (P < 0.0001; Fig. 1C), and was significantly lower in the high-risk group compared with the low-risk group based on our 70-gene model (P = 0.013; Fig. 1D). We performed survival analyses in three different datasets. Consistent with the low FPN1 expression in the aggressive multiple myeloma subgroups, decreased FPN1 in the 351 TT2 cohort showed that about 60% of such cases showed short EFS (Fig. 1E; P < 0.001) and also inferior OS (P < 0.001; Fig. 1F). The correlation of low FPN1 with inferior patient outcome was further validated in two other independent cohorts, including 270 newly diagnosed multiple myeloma enrolled in the HOVON-65 clinical trial (Fig. 1G) and 264 relapsed myeloma patients enrolled in the APEX phase III clinical trial (Fig. 1H; ref. 32).
Figure 1. Identification of FPN1 as a poor prognostic marker in multiple myeloma.
A, supervised cluster analysis of iron signature genes in NPCs and multiple myeloma cells. B, scatter plots depict the Affymetrix signal of FPN1 in NPCs, MGUS, newly diagnosed multiple myeloma (MM; TT2 cohort), and multiple myeloma cell lines (MMCL). One-way ANOVA was performed and identified the P < 0.0001 among these four groups. The P value presented in the figure wasobtainedbycomparisonbetween NPC and indicated group, respectively. C, the expression of FPN1 among 8 multiple myeloma subgroups is highly variable (P < 0.0001 by one-way ANOVA). Indicated P value was obtained among 8 subgroups by one- way ANOVA. D, expression of FPN1 between low- and high-risk subgroups in multiple myeloma. The difference was compared by the Student t test between these two groups. E–H, survival analyses were performed based on FPN1 expression in different cohorts. The EFS (E) and OS (F) were performed in the TT2 cohort, and OS was also analyzed in the HOVON65 (G) and APEX (H) clinical trials.
Restoring expression of FPN1 suppresses multiple myeloma cell growth both in vitro and in vivo
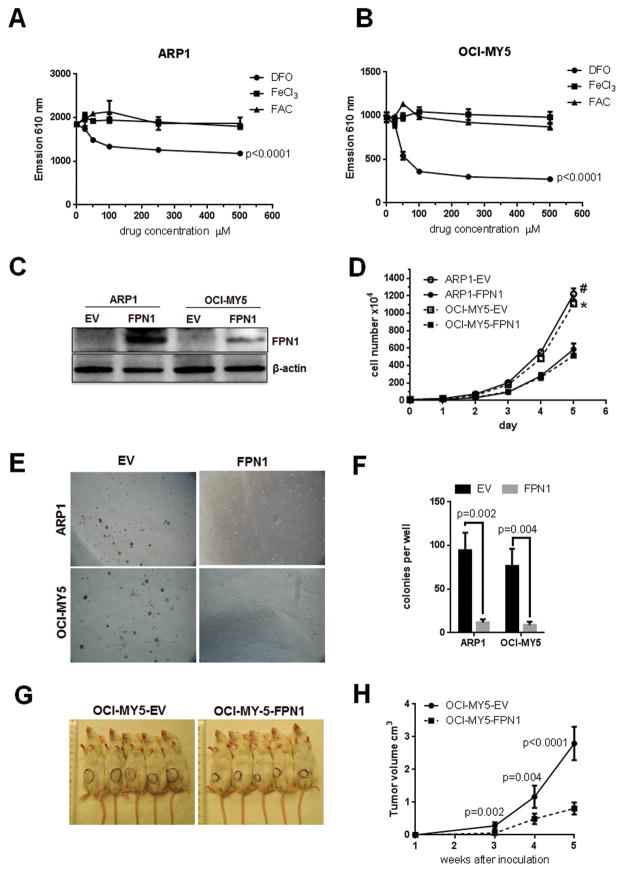
The altered iron gene expression signature strongly suggests that multiple myeloma cells have abnormal iron metabolism and harbor intracellular iron to fuel the growth of the malignant cells. To test the requirement of iron for the survival of multiple myeloma cells, two multiple myeloma cell lines ARP1 and OCI-MY5 were treated with iron supplement FAC or FeCl3, or the iron chelator deferoxamine. Supplementation of iron in the range of 0 to 500 mmol/L had minimal effect on cell growth (Fig. 2A and B), possibly because multiple myeloma cells were already fully saturated with intracellular iron. However, multiple myeloma cell growth was significantly inhibited by deferoxamine treatment starting at 50 mmol/L, which is a commonly-used concentration in vitro (Fig. 2A and B and Supplementary Fig. S1A), indicating intracellular iron is essential for multiple myeloma cell growth.
Figure 2. Ferroportin plays a role of tumor suppression in multiple myeloma cell growth, both in vitro and in vivo.
A and B, multiple myeloma cells are sensitive to iron chelator. ARP1 (A) and OCI-MY5 cells were treated with deferoxamine (DFO), FeCl3, and FAC at indicated concentration for 48 hours. Cell proliferation was measured by the Prestoblue assay, and data were collected by measuring emission at 610 nm. P value indicated significant difference between treatment and nontreatment. C, confirmation of FPN1 expression in FPN1 overexpressing multiple myeloma cells by Western blot. FPN1 cDNA was transduced into multiple myeloma cell lines using lentiviral delivery; the expression of FPN1 was examined by Western blot. β-actin served as internal control. D, overexpression of FPN1 inhibits multiple myeloma cell growth. The cell growth between FPN1 and EV cells from ARP1 and OCI-MY5 cell lines was counted by the Trypan blue assay from day 0 to day 5. E and F, overexpression FPN1 inhibits colony formation. ARP1 and OCI-MY5 multiple myeloma cells overexpressed FPN1, and their controls were plated in methylcellulose to evaluate their clonogenic potential assay for 7 days. The difference between FPN1 and EV cells was compared by the Student t test. G and H, FPN1 inhibits multiple myeloma cell growth in vivo. Indicated cells were subcutaneously injected into NOD-SCID mice (n = 5) for 5 weeks. Xenograft tumor is circled (G). H, tumor volume was measured from weeks 3, 4, and 5 and graphed; P value indicated a significant difference between EV and FPN1 overexpression xenograft tumors at indicated time points.
We then overexpressed FPN1 in two multiple myeloma cell lines ARP1 and OCI-MY5, which expressed FPN1 at nondetectable level by Western blotting, to evaluate the effect of FPN1 on multiple myeloma cell growth using lentiviral delivery (Fig. 2C). Overexpression of FPN1 significantly inhibited cell proliferation compared with cells transduced with empty vector (EV) in both cell lines at day 5 (Fig. 2D). In a soft-agar clonogenic assay that assesses the expansion of more primitive multiple myeloma cells, overexpression of FPN1 greatly inhibited the clonogenic potential of multiple myeloma cells in ARP1 and OCI-MY5 cells (Fig. 2E and F). To further elucidate the tumor-suppressing role of FPN1, the OCI-MY5 EV/FPN1 cells were subcutaneously injected into the flank immunocompromised NOD-SCID mice. Consistent with the in vitro study, FPN1 inhibited tumor growth in vivo at 5 weeks compared with its EV control (0.81 ± 0.59 cm3 vs. 2.80 ± 0.18 cm3, P < 0.0001). These data indicate that FPN1 is a tumor suppressor both in vitro and in vivo.
FPN1 regulates intracellular iron in vitro and in vivo in multiple myeloma cells
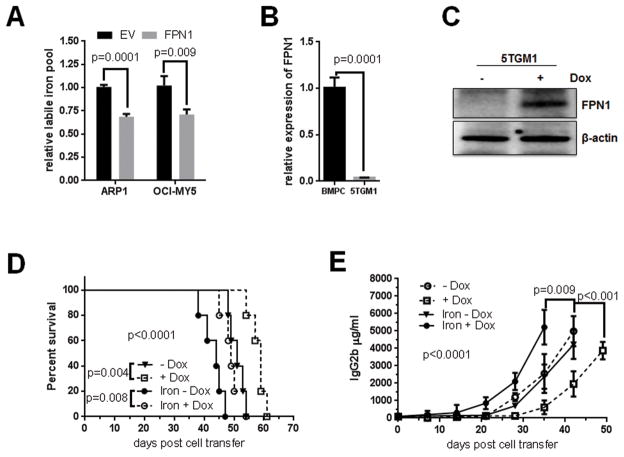
To test whether FPN1 regulates iron exportation in multiple myeloma cells, the LIP was measured with fluorescent metallosensor calcein. ARP1 and OCI-MY5 cells overexpressing FPN1 had lower LIP compared with their EV counterparts (Fig. 3A). We further employed 5TGM1-KaLwRij model to test the role of FPN1 on multiple myeloma progression in vivo. Real-time PCR confirmed that 5TGM1 myeloma cells had much lower expression of FPN1 than normal bone marrow plasma cells in KaLwRij mice (Fig. 3B). The coding region of FPN1 cDNA in a doxycycline-inducible lentiviral construct was stably transduced into the 5TGM1 cells with lentivirus, in which the expression of FPN1 could be induced upon addition of doxycycline (Fig. 3C). One week after transduced 5TGM1 cell injection, mice were administrated doxycycline and dextran-iron to increase systemic iron content in the mouse body as previously reported. In the absence of dextran-iron, overexpression of FPN1 (activated by administration of doxycycline) significantly delayed tumor progression evidenced by decreased tumor burden measured by mouse serum IgG2b level (Fig. 3D; P = 0.008) and prolonged survival (Fig. 3E; P < 0.001) compared with noninduced (no doxycycline) group. Addition of iron accelerated tumor progression of the 5TGM1 mice, resulting in a shorter survival and higher tumor burden than those without iron in drinking water (Fig. 3E; P = 0.009); the effect of iron administration on multiple myeloma progression could be blocked by activation of FPN1 expression (Fig. 3D and E).
Figure 3. FPN1 regulates multiple myeloma cell intracellular iron in vitro and in vivo.
A, high FPN1 decreases intracellular LIP. The intracellular LIP was measured in indicated cells using fluorescent metallosensor calcein. B, FPN1 expression is significantly lower in mouse multiple myeloma cells than in normal mouse plasma cells. Expression of FPN1 mRNA in normal bone marrow plasma cells from wild-type KaLwRij mice and 5TGM1 cells was measured by qRT-PCR and compared by the Student t test. C, confirmation of FPN1 expression in an inducible mouse multiple myeloma cell line. Inducible expression of FPN1 in 5TGM1 cells was detected by Western blot. D and E, high FPN1 antagonizes iron-induced cell growth in vivo. 5TGM1-FPN1 KaLwRij mice were administrated with or without doxycycline and dextran-iron as indicated 1 week after cell injection. Kaplan–Meier showed the survival curves, and P value was analyzed by the log-rank test. D, tumor burden was measured by the ELISA assay, and the significance (P < 0.0001) was determined by one-way ANOVA.
STAT3-MCL-1 mediates FPN1 effect on myeloma cell growth and survival
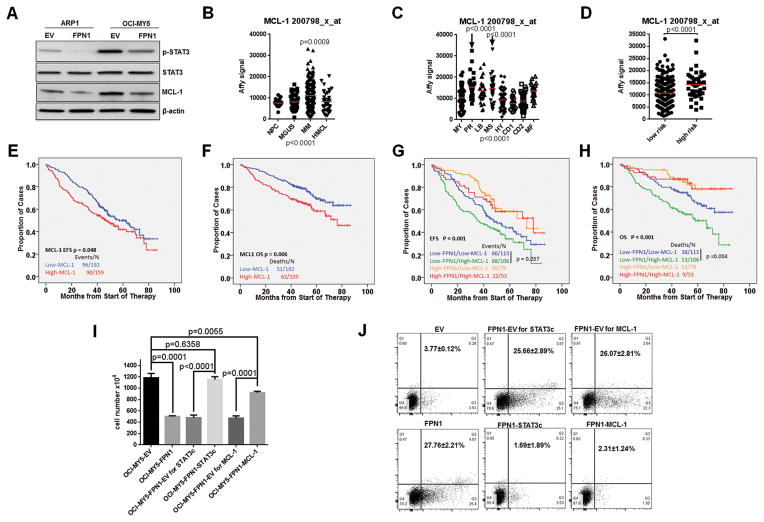
As shown in Fig. 4A and Supplementary Fig. S1B, overexpression of FPN1 predominantly suppressed tyrosine phosphorylation of STAT3 and decreased expression of MCL-1, while it had only a minimal effect on other survival factors such as BAK and BID. Fpn1+/− mice harbor a single copy of H32R mutation in Fpn1 gene, which functions as dominate-negative form (Supplementary Fig. S1C; ref. 33). Gene expression data from Fpn1+/− mice showed that MCL-1 expression was upregulated in bone marrow plasma cells and spleen B cells when compared with wild-type mice (Supplementary Fig. S1D). The expression pattern of MCL-1 was negatively correlated with that of FPN1 in NPC and myeloma patient samples. MCL-1 was increased in multiple myeloma cells comparing with NPCs (Fig. 4B), and the highest expression levels were observed in the proliferation group (Fig. 4C) and in the high-risk group designated by the 70-gene model (Fig. 4D). Consistent with the high MCL-1 expression in aggressive multiple myeloma subgroups, high expression of MCL-1 was linked to an inferior outcome, significant for both EFS and OS in newly diagnosed multiple myeloma patients (Fig. 4E, EFS P = 0.048; Fig. 4F, OS P = 0.006). Furthermore, by combining FPN1 and MCL-1, we identified that multiple myeloma patients with low expression of FPN1 and high expression of MCL-1 had the worst outcome (Fig. 4G, EFS P < 0.001; Fig. 4H, OS P < 0.001).
Figure 4. FPN1 downregulates STAT3-MCL-1 signaling in myeloma cells.
A, phosphor-STAT3 (Tyr705) and MCL-1 expression were detected by Western blot in indicated cell lines. B, expression of MCL-1 was compared among NPC, MGUS, multiple myeloma (MM), and MMCL by GEP (P < 0.0001 by one-way ANOVA). C, expression of MCL-1 was compared in myeloma subgroups. Note: the PR and MS are the most aggressive subtypes (P < 0.0001 by one-way ANOVA). D, expression of MCL-1 was compared between the low-risk and high-risk subgroups (P < 0.0001 by t test). E–H, survival analyses were performed on the TT2 cohort based on MCL-1 and FPN1 expression. Kaplan–Meier showed EFS (E) and OS (F) curves according to MCL-1 expression, and EFS (G) and OS (H) were based on combination of MCL-1 and FPN1 expression. The log-rank test was used to analyze the P values. I, determination of STAT3-MCL-1 function in FPN1-induced cell growth inhibition. Cell number of indicated cells was counted by Trypan blue on day 5 after plating. J, flow cytometry showed cell apoptosis in FPN1 signaling. Indicated cells were washed and serum starved overnight; apoptosis was measured by the Annexin V/PI assay as described in Material and Methods. Annexin V+ cells were counted as apoptotic cells.
The above data suggest that STAT3 and MCL-1 are likely involved in FPN1 signaling in multiple myeloma. To test whether STAT3 or MCL-1 can rescue cell growth inhibition induced by high FPN1, a constitutively active form of STAT3 (STAT3c) and MCL-1 was transduced into OCI-MY5-FPN1 cells (Supplementary Fig. S1E and S1F). As shown in Fig. 4I, overexpression of STAT3c almost complete reversed cell growth inhibition in the presence of ectopic expression of FPN1. Overexpression of MCL-1 only par tially recovered cell growth, indicating that STAT3 also activates other pathways than just MCL-1. As MCL-1 is the major factor involved in the survival of bone marrow plasma cells, we tested whether the STAT3-MCL-1 pathway would have an effect on the cell survival in the presence of high FPN1 levels. As shown in Fig. 4J, overexpression of FPN1 sensitized multiple myeloma cell to apoptotic stimuli, which was abrogated by overexpression of STAT3c or MCL-1.
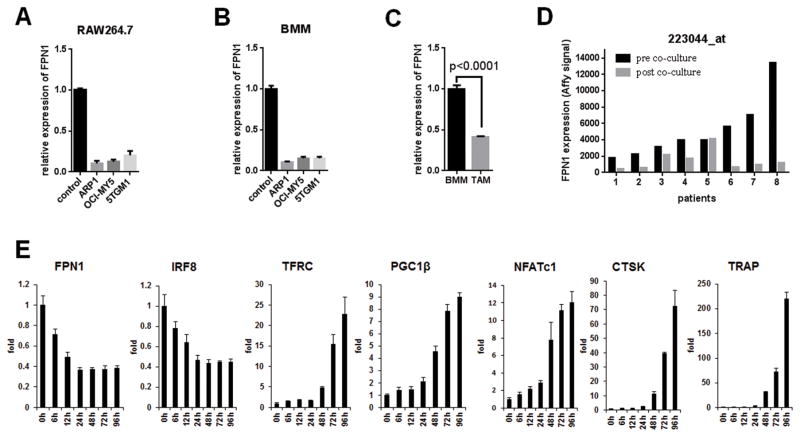
FPN1 is downregulated in the initial phases of myeloma cell– induced osteoclast differentiation
Osteolytic bone disease is a frequent complication of multiple myeloma affecting up to 80% of patients (34). It is widely known that FPN1 is highly expressed in normal macrophages, which are the precursors of osteoclast in bone marrow, therefore, we hypothesized that FPN1 had a major role in multiple myeloma cell–induced osteoclast differentiation. The murine macrophage cells RAW264.7 and primary BMMs from wild-type C57BL6 mice were cultured in the conditioned media collected from ARP1, OCI-MY5, and 5TGM1 multiple myeloma cells to induce osteoclast differentiation (Supplementary Fig. S2A and S2B). FPN1 expression was dramatically downregulated by qRT-PCR in RAW264.7 and primary BMMs cultured with conditioned media (Fig. 5A and B). To validate this observation using in vivo model(s), BMMs were isolated from both wild-type KaLwRij mice and KaLwRij mice bearing 5TGM1 multiple myeloma cells. FPN1 showed significantly lower expression in tumor-associated macrophages than in macrophages derived from wild-type mice (Fig. 5C). To clarify whether the downregulation of FPN1 in osteoclast precursors was the cause or the consequence of osteoclastogenesis, expression of FPN1 mRNA was monitored by real-time PCR in a time course of osteoclast differentiation induced by M-CSF plus RANKL (Supplementary Fig. S2C). The decreased expression of FPN1 was observed as early as 6 hours after addition of RANKL and M-CSF (Fig. 5D), which was very similar to the previously reported osteoclastogenesis inhibitory factor IRF8 (Fig. 5D; ref. 35). Previous reports show that iron uptake coordinates with mitochondrial biogenesis orchestrated by PGC-1b to induce osteoclastogenesis (23). Here, we observed that significant upregulation of TFRC and PGC-1b occurred at 48 hours after induction (Fig. 5D), indicating the downregulation of FPN1 preceded the changes of most critical driving factors (TFRC, PGC-1b, and NFATc1; refs. 23, 36, 37) or markers (CTSK and TRAP) of osteoclastogenesis (38), which is a critical event of osteoclast differentiation (Fig. 5D).
Figure 5. FPN1 is downregulated at initial phase of myeloma cell–induced osteoclast differentiation.
A and B, to determine if multiple myeloma cells regulate FPN1 expression in osteoclast precursors, RAW264.7 cells were cultured in the conditioned media from indicated myeloma cells for 72 hours; FPN1 mRNA was detected by real-time PCR (A). Primary bone marrow monocytes from wild-type KaLwRij mice were differentiated to macrophage by M-CSF and then cultured in the conditioned media from indicated myeloma cells for 72 hours; FPN1 mRNA was detected by real-time PCR (B). C, to compare the FPN1 expression between normal macrophages and myeloma-associated macrophages, primary BMMs (F4/8+ CD11b+) were isolated from wild-type KaLwRij mice and KaLwRij mice bearing 5TGM1-induced multiple myeloma; FPN1 mRNA was detected by real-time PCR. D, qRT-PCR was used to examine expression of genes related to osteoclastogenesis. Primary bone marrow monocytes were differentiated to macrophage by M-CSF and then stimulated by RANKL for indicated time. Expression of indicated genes was detected by real-time PCR.
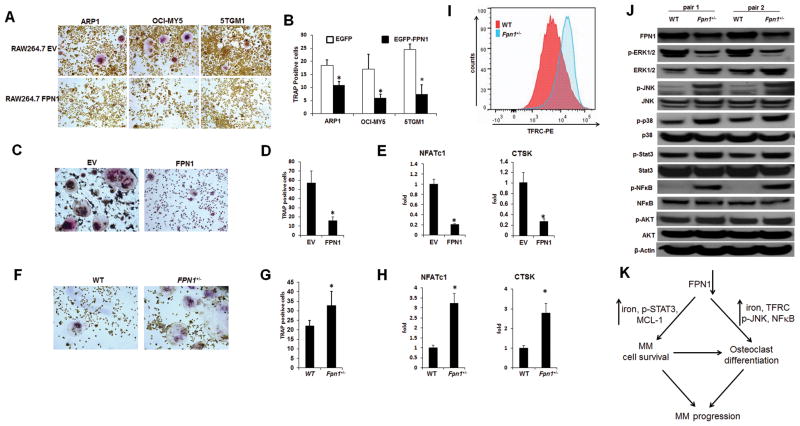
High FPN1 suppresses myeloma cell–induced osteoclast differentiation
The early downregulation of FPN1 in osteoclast precursors by multiple myeloma cells or RANKL/M-CSF–induced osteoclast differentiation suggests that high FPN1 in osteoclast precursors may block osteoclastogenesis. The RAW264.7 cells were transiently transduced with Fpn1-cDNA, and the expression of FPN1 was confirmed by qRT-PCR (Supplementary Fig. S3A). These cells were then cultured in the presence of conditioned media collected from multiple myeloma cell culture for 3 days. TRAP staining showed that overexpression of FPN1 significantly decreased osteoclast differentiation compared with control RAW264.7 cells transfected with EV (Fig. 6A and B). We further repeated this study on the primary osteoclast precursors by transducing FPN1 into mouse BMMs (Supplementary Fig. S3B). As shown in Fig. 6C and D, TRAP-positive osteoclasts were much less frequent in Fpn1-high BMMs than in the control BMMs. The real-time PCR verified that high FPN1 in BMMs downregulated the expression of NFATc1 and CTSK, reliable makers for osteoclastogenesis (Fig. 6E). In contrast, the BMMs derived from Fpn1+/− mice increased osteoclast formation (Fig. 6F and G) and had higher expression of NFATc1 and CTSK after RANKL/M-CSF induction compared with those derived from Fpn1+/+ mice (Fig. 6H).
Figure 6. High FPN1 suppresses myeloma cell–induced osteoclast differentiation.
A and B, RAW264.7 cells transfected with EV or FPN1 were cultured in the conditioned media collected from indicated myeloma cells for 72 hours. Osteoclasts were detected by TRAP staining (A) and quantified (B). C–E, primary bone marrow monocytes from wild-type KaLwRij mice were transduced with EV or FPN1 and induced by M-CSF plus RANKL for 7 days. Osteoclasts were detected by TRAP staining and quantified (D); the expression of genes related to osteoclastogenesis was examined by real-time PCR (E). F–H, primary bone marrow monocytes were isolated from wild-type Fpn1+/+ and Fpn1+/− mice and induced by M-CSF plus RANKL for 4 days. Osteoclasts were detected by TRAP staining (F) and quantified (G); the expression of genes related to osteoclastogenesis was examined by real-time PCR (H). I, BMMs were isolated and stained with TFRC-PE antibody, and surface expression of TFRC was examined by flow cytometry. J, indicated proteins were detected by Western blot in BMMs from Fpn1+/+ and Fpn1+/− mice. K, an illustration of role of decreased expression FPN1 in multiple myeloma (MM) progression. Briefly, downregulation of FPN1 in multiple myeloma cells upregulates STAT3-MCL-1 signaling, which promotes the survival of multiple myeloma cells; decreased expression of FPN1 in BMMs increases expression of TFRC, NF-kB, and p-JNK, resulting in osteoclast differentiation. The prolonged multiple myeloma cell survival and enhanced osteoclast differentiation accelerate multiple myeloma disease progression.
To probe the mechanism of FPN1 in osteoclast differentiation, gene array analysis was performed in BMMs from both Fpn1+/+ and Fpn1+/− mice. Of the iron gene signature, only TFRC was upregulated gene in Fpn1+/− BMMs (Fig. 6I and Supplementary Fig. S3C). Because TFRC mediates iron uptake, the main consequence of low FPN1 and high TFRC in BMMs and osteoclast is increasing intracellular iron. The 5TGM1 cells in KaLwRij mouse model were used to confirm this observation. PBS or dextran-iron was administrated to KaLwRij mice 1 week after injection of 5TGM1 cells. Prussian blue staining indicated an abundance of positive macrophages in mice administrated dextran-iron, but not in those control mice (Supplementary Fig. S3D). TRAP-positive osteoclasts were significantly increased in the dextran-iron–treated mice compared with controls (Supplementary Fig. S4D), demonstrating that iron overload increases osteoclastogenesis, resulting in bone loss. We screened several typical pathways involved in osteoclast differentiation by Western blot, Fpn1+/− BMMs showed significantly increased phosphorylation of NF-κB and JNK compared with Fpn1+/+ BMMs (Fig. 6J). Thus, downregulation of FPN1 promotes osteoclast differentiation probably by activating NF-κB and JNK, the major two pathways related to increased osteoclastogenesis (39). The model of our working hypothesis is summarized in Fig. 6K.
Discussion
Iron is mainly absorbed in the duodenum by duodenal cytochrome b (DCYTB) and exits enterocytes by the iron efflux pump FPN1. The high demand of iron by red blood cells (RBC) to synthesize hemoglobin makes the bone marrow the main site of iron utilization. Macrophages recycle iron by catabolizing RBCs and export iron to blood stream through FPN1. Excessive iron is primarily stored in the liver and regulated by systemic iron levels (40). Many of these iron metabolic steps are altered in cancer, resulting in iron-related clinic symptoms such as anemia (1). In addition, the cancer cell itself also has dramatic changes in iron metabolism. The storage of intracellular iron into ferritin is often disrupted by oncogenes such as MYC or RAS, leading to increased availability of iron, which promotes DNA synthesis and cell proliferation (41, 42). Decreased iron export is relatively less well studied in the context of cancer cells.
In the present study, we systemically investigated, for the first time, the expression of iron gene signature using gene expression profiling in multiple myeloma. We observed that iron metabolism in multiple myeloma is strongly dysregulated by applying an iron signature to a clinical data set from a previously studied cohort of patients (30). When compared with NPCs, multiple myeloma cells are prone to upregulate signals that maintain a high intracellular iron level. A marked reduction of the only known iron exporter FPN1 was observed in multiple myeloma cells compared with NPCs. The lowest FPN1 levels were observed in the most aggressive subgroups of myeloma, which is consistent with previous reports that FPN1 is a strong and independent predictor of prognosis in breast cancer (14).
Previous studies show that high hepcidin is responsible for myeloma-associated anemia and is correlated with multiple myeloma progression (43). Unlike breast cancer cells, hepcidin expression is significantly decreased in multiple myeloma cells compared with NPCs (Supplementary Table S1), suggesting multiple myeloma is unlikely to have an autocrine way to regulate iron by depletion of FPN1 protein through hepcidin. However, the abundance of IL6 and elevated expression of BMPs may induce high production of hepcidin in liver in multiple myeloma patients (19, 20). Our data show that the decrease of FPN1 in multiple myeloma cells happens at the transcription level. Therefore, we think that hepcidin has only a minor effect on the regulation of FPN1 in multiple myeloma cells. Elevated hepcidin may have more predominant role on other cells like in hepatocytes, duodenal enterocytes, RBCs, and macrophages, which regulate systematic iron metabolism.
We found that FPN1 downregulates STAT3-MCL-1 signaling, one of the critical pathways of plasma cell growth and survival(44, 45). Although JAK-STAT3 signaling is not required for hepcidin-induced proteasome degradation of FPN1 (46), overexpression of FPN1 indeed suppresses STAT3 signaling. One possible mechanism is that downregulation of FPN1 promotes accumulation of intracellular LIP and ROS, which might, in turn, activate STAT3-MCL-1 signaling (47). FPN1-regulated LIP and STAT3-MCL-1 signaling are likely related rather than independent events. The detailed mechanism of how FPN1 regulates STAT3-MCL-1 signaling needs further study.
Accumulation of multiple myeloma cells in bone marrow induces bone resorption, which is a hallmark of multiple myeloma. Enhanced osteoclastogenesis and suppressed osteoblastogenesis induced by multiple myeloma cells account for the lytic bone lesions. The macrophages, osteoclast precursors in the bone marrow, are the main cells to recycle iron, a process regulated by FPN1. Osteoclast differentiation requires high rates of biogenesis through respiratory mechanisms in mitochondria and demands iron (23). The importance of downregulation of FPN1 in multiple myeloma cells led us to uncover the role of FPN1 in multiple myeloma cell–induced bone resorption. Compared with other key regulators in osteoclast differentiation such as PGC1b, TFRC, and NFATc, downregulation of FPN1 is induced earlier (around 6 hours) after incubation with RANKL plus M-CSF. The pattern of downregulation is similar to that of IRF8, a critical inhibitory factor of osteoclast differentiation (35). The experiments of ectopic expression of FPN1 or loss of function mutation of Fpn1 demonstrate that FPN1 has a predominant role in suppressing osteoclast differentiation through regulation of iron deposit, JNK, and NF-κB signaling. Thus, downregulation of FPN1 in BMMs induced by multiple myeloma cells plays an important role in bone resorption in multiple myeloma. The fact that we did not observe consistent result of bone resorption in Fpn1+/− mice is possibly due to some activity of the wild-type allele of Fpn1 gene. It will be very interesting to test the physiologic role of FPN1 in bone homeostasis using conditional knockout mice model in future.
In summary, downregulation of FPN1 both in multiple myeloma cells and macrophage-osteoclast cells favors the expansion of the malignant clone and osteoclastogenesis, which both have a negative effect on the outcome of multiple myeloma. Exploring agents capable of upregulating FPN1 expression could be a novel direction in multiple myeloma treatment, which may result in a strategy that targets both the myeloma tumor cells and their microenvironment.
Supplementary Material
Acknowledgments
The authors thank Dr. James P. Kushner (University of Utah) for critical discussion of the project and Dr. Jason Weirather (University of Iowa) for helping analysis of GEP data. The data presented in this article were obtained at the Flow Cytometry Facility, which is a Carver College of Medicine/Holden Comprehensive Cancer Center core research facility at the University of Iowa. The Facility is funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, and Iowa City Veteran’s Administration Medical Center.
Grant Support
This work was supported by NIH grants R01CA115399 (G. Tricot), R01CA152105 (F. Zhan), R21CA143887 (F. Zhan), the MMRF Senior (F. Zhan, 2008 and 2010), the Leukemia and Lymphoma Society TRP (F. Zhan, 2010 and 2011), and institutional start-up funds from the Department of Internal Medicine, Carver College of Medicine, University of Iowa (F. Zhan and G. Tricot), and the National Natural Science Foundation of China, China (no. 81228016). The project described was supported by Award Number P30CA086862 from the NCI.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI or the NIH.
Authors’ Contributions
Conception and design: Z. Gu, H. Wang, Z. Jin, J. Shi, G. Tricot, F. Zhan
Development of methodology: Z. Gu, Y. Yang, Z. Jin
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Z. Gu, H. Wang, I. De Domenico, F. Zhan
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Z. Gu, H. Wang, G. Tricot, F. Zhan
Writing, review, and/or revision of the manuscript: Z. Gu, Z. Jin, G. Tricot, Zhan
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Z. Gu, J. Xia, Z. Jin, F. Zhan
Study supervision: G. Tricot, F. Zhan
Other (performed experiments and analyzed results): H. Xu
References
- 1.Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer. 2013;13:342–55. doi: 10.1038/nrc3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews NC. Forging a field: the golden age of iron biology. Blood. 2008;112:219–30. doi: 10.1182/blood-2007-12-077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell JA. Effects of precipitated silica and of iron oxide on the incidence of primary lung tumours in mice. Br Med J. 1940;2:275–80. doi: 10.1136/bmj.2.4156.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richmond HG. Induction of sarcoma in the rat by iron-dextran complex. Br Med J. 1959;1:947–9. doi: 10.1136/bmj.1.5127.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–98. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 6.Waheed A, Grubb JH, Zhou XY, Tomatsu S, Fleming RE, Costaldi ME, et al. Regulation of transferrin-mediated iron uptake by HFE, the protein defective in hereditary hemochromatosis. Proc Natl Acad Sci U S A. 2002;99:3117–22. doi: 10.1073/pnas.042701499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–21. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 8.Donovan A, Lima CA, Pinkus JL, Pinkus GS, Zon LI, Robine S, et al. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1:191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Rice AE, Mendez MJ, Hokanson CA, Rees DC, Bjorkman PJ. Investigation of the biophysical and cell biological properties of ferroportin, a multipass integral membrane protein iron exporter. J Mol Biol. 2009;386:717–32. doi: 10.1016/j.jmb.2008.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deicher R, Horl WH. New insights into the regulation of iron homeostasis. Eur J Clin Invest. 2006;36:301–9. doi: 10.1111/j.1365-2362.2006.01633.x. [DOI] [PubMed] [Google Scholar]
- 11.Hetet G, Devaux I, Soufir N, Grandchamp B, Beaumont C. Molecular analyses of patients with hyperferritinemia and normal serum iron values reveal both L ferritin IRE and 3 new ferroportin (slc11A3) mutations. Blood. 2003;102:1904–10. doi: 10.1182/blood-2003-02-0439. [DOI] [PubMed] [Google Scholar]
- 12.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–3. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 13.Qiao B, Sugianto P, Fung E, Del-Castillo-Rueda A, Moran-Jimenez MJ, Ganz T, et al. Hepcidin-induced endocytosis of ferroportin is dependent on ferroportin ubiquitination. Cell Metab. 2012;15:918–24. doi: 10.1016/j.cmet.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinnix ZK, Miller LD, Wang W, D’Agostino R, Jr, Kute T, Willingham MC, et al. Ferroportin and iron regulation in breast cancer progression and prognosis. Sci Transl Med. 2010;2:43ra56. doi: 10.1126/scisignal.3001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–60. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 16.Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N Engl J Med. 2003;349:2483–94. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- 17.VanderWall K, Daniels-Wells TR, Penichet M, Lichtenstein A. Iron in multiple myeloma. Crit Rev Oncog. 2013;18:449–61. doi: 10.1615/critrevoncog.2013007934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett CL, Silver SM, Djulbegovic B, Samaras AT, Blau CA, Gleason KJ, et al. Venous thromboembolism and mortality associated with recombinant erythropoietin and darbepoetin administration for the treatment of cancer-associated anemia. JAMA. 2008;299:914–24. doi: 10.1001/jama.299.8.914. [DOI] [PubMed] [Google Scholar]
- 19.Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood. 2006;108:3204–9. doi: 10.1182/blood-2006-06-027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–9. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 21.Eberhard Y, McDermott SP, Wang X, Gronda M, Venugopal A, Wood TE, et al. Chelation of intracellular iron with the antifungal agent ciclopirox olamine induces cell death in leukemia and myeloma cells. Blood. 2009;114:3064–73. doi: 10.1182/blood-2009-03-209965. [DOI] [PubMed] [Google Scholar]
- 22.Schramek D, Leibbrandt A, Sigl V, Kenner L, Pospisilik JA, Lee HJ, et al. Osteoclast differentiation factor RANKL controls development of progestin-driven mammary cancer. Nature. 2010;468:98–102. doi: 10.1038/nature09387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii KA, Fumoto T, Iwai K, Takeshita S, Ito M, Shimohata N, et al. Coordination of PGC-1beta and iron uptake in mitochondrial biogenesis and osteoclast activation. Nat Med. 2009;15:259–66. doi: 10.1038/nm.1910. [DOI] [PubMed] [Google Scholar]
- 24.Gu ZM, Wu YL, Zhou MY, Liu CX, Xu HZ, Yan H, et al. Pharicin B stabilizes retinoic acid receptor-alpha and presents synergistic differentiation induction with ATRA in myeloid leukemic cells. Blood. 2010;116:5289–97. doi: 10.1182/blood-2010-02-267963. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Zhou W, Xia J, Gu Z, Wendlandt E, Zhan X, et al. NEK2 mediates ALDH1A1 induced drug-resistance in multiple myeloma. Oncotarget. 2014;5:11986–97. doi: 10.18632/oncotarget.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Shi J, Tolomelli G, Xu H, Xia J, Wang H, et al. RARalpha2 expression confers myeloma stem cell features. Blood. 2013;122:1437–47. doi: 10.1182/blood-2013-02-482919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–84. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 28.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou W, Yang Y, Xia J, Wang H, Salama ME, Xiong W, et al. NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell. 2013;23:48–62. doi: 10.1016/j.ccr.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhan F, Barlogie B, Arzoumanian V, Huang Y, Williams DR, Hollmig K, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109:1692–700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller LD, Coffman LG, Chou JW, Black MA, Bergh J, D’Agostino R, Jr, et al. An iron regulatory gene signature predicts outcome in breast cancer. Cancer Res. 2011;71:6728–37. doi: 10.1158/0008-5472.CAN-11-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulligan G, Mitsiades C, Bryant B, Zhan F, Chng WJ, Roels S, et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109:3177–88. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- 33.Zohn IE, De Domenico I, Pollock A, Ward DM, Goodman JF, Liang X, et al. The flatiron mutation in mouse ferroportin acts as a dominant negative to cause ferroportin disease. Blood. 2007;109:4174–80. doi: 10.1182/blood-2007-01-066068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristinsson SY, Minter AR, Korde N, Tan E, Landgren O. Bone disease in multiple myeloma and precursor disease: novel diagnostic approaches and implications on clinical management. Expert Rev Mol Diagn. 2011;11:593–603. doi: 10.1586/erm.11.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao B, Takami M, Yamada A, Wang X, Koga T, Hu X, et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat Med. 2009;15:1066–71. doi: 10.1038/nm.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei W, Wang X, Yang M, Smith LC, Dechow PC, Sonoda J, et al. PGC1beta mediates PPARgamma activation of osteoclastogenesis and rosiglitazone-induced bone loss. Cell Metab. 2010;11:503–16. doi: 10.1016/j.cmet.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takayanagi H. The role of NFAT in osteoclast formation. Ann N Y Acad Sci. 2007;1116:227–37. doi: 10.1196/annals.1402.071. [DOI] [PubMed] [Google Scholar]
- 38.Wan Y, Chong LW, Evans RM. PPAR-gamma regulates osteoclastogenesis in mice. Nat Med. 2007;13:1496–503. doi: 10.1038/nm1672. [DOI] [PubMed] [Google Scholar]
- 39.Vaira S, Alhawagri M, Anwisye I, Kitaura H, Faccio R, Novack DV. RelA/p65 promotes osteoclast differentiation by blocking a RANKL-induced apoptotic JNK pathway in mice. J Clin Invest. 2008;118:2088–97. doi: 10.1172/JCI33392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nemeth E, Ganz T. Regulation of iron metabolism by hepcidin. Annu Rev Nutr. 2006;26:323–42. doi: 10.1146/annurev.nutr.26.061505.111303. [DOI] [PubMed] [Google Scholar]
- 41.Radulescu S, Brookes MJ, Salgueiro P, Ridgway RA, McGhee E, Anderson K, et al. Luminal iron levels govern intestinal tumorigenesis after Apc loss in vivo. Cell Rep. 2012;2:270–82. doi: 10.1016/j.celrep.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Kakhlon O, Gruenbaum Y, Cabantchik ZI. Ferritin expression modulates cell cycle dynamics and cell responsiveness to H-ras-induced growth via expansion of the labile iron pool. Biochem J. 2002;363:431–6. doi: 10.1042/0264-6021:3630431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma S, Nemeth E, Chen YH, Goodnough J, Huston A, Roodman GD, et al. Involvement of hepcidin in the anemia of multiple myeloma. Clin Cancer Res. 2008;14:3262–7. doi: 10.1158/1078-0432.CCR-07-4153. [DOI] [PubMed] [Google Scholar]
- 44.Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–15. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 45.Peperzak V, Vikstrom I, Walker J, Glaser SP, LePage M, Coquery CM, et al. Mcl-1 is essential for the survival of plasma cells. Nat Immunol. 2013;14:290–7. doi: 10.1038/ni.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross SL, Tran L, Winters A, Lee KJ, Plewa C, Foltz I, et al. Molecular mechanism of hepcidin-mediated ferroportin internalization requires ferroportin lysines, not tyrosines or JAK-STAT. Cell Metab. 2012;15:905–17. doi: 10.1016/j.cmet.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 47.Yoon S, Woo SU, Kang JH, Kim K, Kwon MH, Park S, et al. STAT3 transcriptional factor activated by reactive oxygen species induces IL6 in starvation-induced autophagy of cancer cells. Autophagy. 2010;6:1125–38. doi: 10.4161/auto.6.8.13547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.