Summary
Combining the results of a large scale proteomic analysis of human transcription factor interaction network with knowledge databases, we identified FOXR2 as one of the top-ranked candidate proto-oncogenes. Here, we show that FOXR2 forms a stable complex with MYC and MAX and subsequently regulates cell proliferation by promoting MYC's transcriptional activities. We demonstrated that FOXR2 is highly expressed in several breast, lung, and liver cancer cell lines and related patient tumor samples, while reduction of FOXR2 expression in a xenograft model inhibits tumor growth. These results indicate that FOXR2 acts with MYC to promote cancer cell proliferation, which is a potential tumor-specific target for therapeutic intervention against MYC-driven cancers.
Keywords: MYC, protein-protein interaction, forkhead box, FOXR2
Graphical abstract
Introduction
MYC is one of the best-known oncogenes; it is commonly amplified or translocated in cancers (Dang, 2012). MYC is the major downstream target of many important pathways related to cancer cell growth and proliferation, which include the MAPK/ERK, WNT, TGFβ, and SHH pathways. In mammalian cells, MYC serves as a transcription factor (TF) and binds to thousands of promoters (Lin et al., 2012; Nie et al., 2012; Zeller et al., 2006). Under some circumstances, such as in lymphocytes and embryonic stem cells, it promotes virtually all gene expression (Nie et al., 2012). However, MYC differentially activates or represses specific sets of direct target genes in MYC-transformed tumor cells (Walz et al., 2014), probably via its associations with different binding partners in cancer cells. Constitutive activation of MYC, caused by MYC amplification and mutation, results in uncontrolled proliferation and ultimately the development of cancer (Beroukhim et al., 2010; Collins and Groudine, 1982; Dalla-Favera et al., 1982; Taub et al., 1982).
MYC is involved in many cellular processes, including cell metabolism, DNA replication, cell adhesion, differentiation, and metastasis, mainly via its function as a transcription factor. Paradoxically, MYC also activates apoptosis (Askew et al., 1991; Evan et al., 1992). It can either induce or sensitize cells to apoptosis through transcriptionally regulating a series of genes involved in apoptosis (Dang, 1999). Tumors with MYC overexpression often have the activation of RAS or AKT pathway, or mutations that disable the apoptotic program, and therefore allow these tumors to proliferate (Hoffman and Liebermann, 2008; Prendergast, 1999).
Genetic analyses have revealed that MYC overexpression, which is commonly caused by genomic amplification, is present in many types of human cancer (Beroukhim et al., 2010), such as lymphoma (Dalla-Favera et al., 1982; Taub et al., 1982), breast cancer (Bieche et al., 1999), lung cancer (Little et al., 1983), and liver cancer (Schlaeger et al., 2008). Transgenic mouse models with MYC overexpression in various tissues develop tumors rapidly, underscoring the potency of MYC as an oncogene (Dang, 2012). Intriguingly, cancer cells with MYC overexpression are often addicted to MYC expression. Depletion or inhibition of MYC in mouse models triggers rapid tumor regression (Boxer et al., 2004; D'Cruz et al., 2001; Soucek et al., 2013), strongly suggesting that MYC is a therapeutic target for cancer treatment. As a transcription factor, MYC is rigorously regulated via its association with other proteins on chromatin (Blackwood and Eisenman, 1991). MYC interacts and functions with many other transcription factors, such as MAX (Amati et al., 1993). Most MYC-binding proteins contain the bHLH/LZ (basic helix-loop-helix leucine zipper) domain (Ewing et al., 2007). Through interactions with these proteins or protein complexes, MYC differentially regulates the transcriptional activities of its target genes.
Our previous proteomic analysis of transcription factors revealed the distinct protein-protein interaction networks on and off chromatin (Li et al., 2015b), where we uncovered a previously unknown interaction between MYC and FOXR2. As a member of fork-head (FOX) transcription factor family, FOXR2 was first identified in 2004 and it locates on human chromosome Xp11.21 (Katoh, 2004b). FOXR2 shares 57.7% identity with FOXR1 (Katoh, 2004a). Similar to other FOX family members, it contains a highly conserved forkhead domain at its C-terminus (Katoh, 2004b). Recent large-scale transposon mutagenesis screenings indicate that Foxr2 is a potential tumor driver gene in malignant peripheral nerve sheath tumors (Rahrmann et al., 2013) and medulloblastoma (Koso et al., 2014). However, the roles of FOXR2 in human cancer development and the underlying molecular mechanisms remain largely unknown. In this study, we demonstrated that FOXR2 is highly expressed in several human cancer cell lines and related patient cancer tissues including breast, lung, and liver. FOXR2 promotes tumor growth through the activation of MYC transcriptional activity. Together, these data not only demonstrated that FOXR2 acts with MYC to promote cancer cell proliferation, but also propose FOXR2 as a potential therapeutic target for the MYC-driven cancers.
RESULTS
FOXR2 forms a stable complex with MYC and MAX
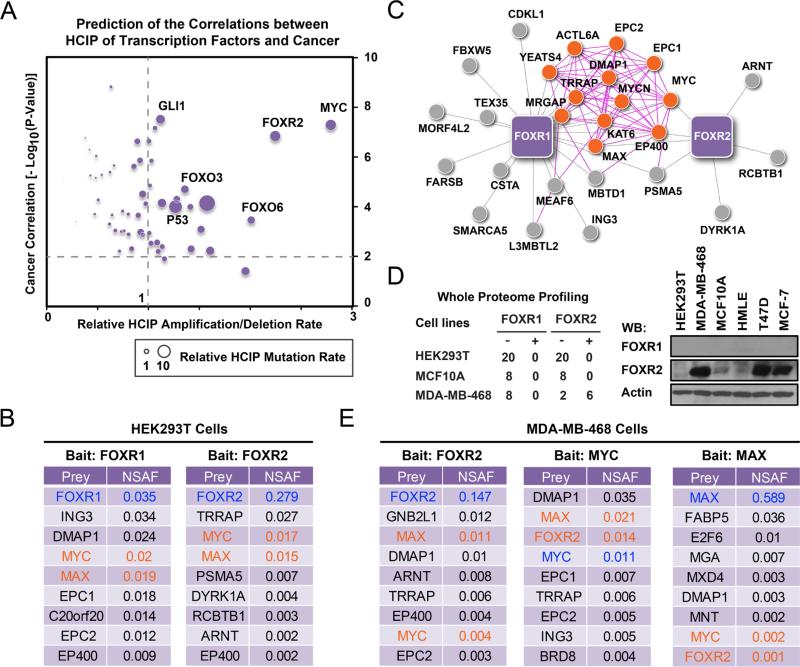
The proteins encoded by the genes mutated in inherited genetic disorders are likely to interact with proteins known to cause similar disorders, which suggest the existence of disease related protein-protein interaction subnetworks (Gandhi et al., 2006). Based on this theory, we decided to further analyze the correlations between cancer and the transcription factor interactomes we recently established (Li et al., 2015b). Since most of tumorigenic functions of transcriptional factors rely on their transcriptional activities, we analyzed the cancer correlation for each bait-associated high-confidence candidate interacting protein (HCIP) datasets in chromatin fractions (Supplementary Table S1). We also performed expression alteration and mutation profiling of these HCIP datasets in multiple TCGA databases (Cerami et al., 2012; Gao et al., 2013). Combining these datasets, we were able to characterize these transcription factors and predict their involvement in tumorigenesis (Figure 1A and Supplementary Table S1). As expected, MYC was classified as the strongest oncogene through its transcriptional activities, and the expression of many MYC-associated proteins is highly altered in TCGA database; other well-studied cancer-driver genes and tumor suppressors, such as TP53, GLI1, and FOXO3, also passed the threshold (Figure 1A). Surprisingly and excitingly, FOXR2 was identified as one of the strongest candidates involved in tumorigenesis, just next to MYC in this unbiased study (Figure 1A). The Gene Ontology annotation indicated that FOXR2 HCIPs identified from chromatin fraction are highly enriched in cancer processes (p = 1.48 × 10−7), and their gene alteration rate is much higher than the average rate of all the HCIP sets in this study (Figure 1A).
Figure 1. FOXR2 forms a stable complex with MYC/MAX.
(A) Prediction of transcription factors' involvement in tumorigenesis is based on their HCIPs identified by TAP-MS analysis. The cancer correlations were generated by searching the HCIP datasets of each transcription factors in the knowledge base to estimate the significance of these correlations. Transcription factor interactomes were searched for their alteration (numbers and rates) in multiple TCGA databases using their HCIP sets. X axis indicates the relative average expression alteration of indicated TF HCIP dataset in multiple TCGA databases. Y axis indicates cancer correlation for each transcription factor, estimated on the basis of their HCIPs identified in chromatin fractions. The size of each dot indicates the relative average mutation rate of TF HCIP dataset in multiple TCGA databases. (B) Top HCIPs of FOXR1 and FOXR2 in HEK293T cells are listed together with their NSAF values. Baits are highlighted in blue; MYC and MAX are highlighted in orange. (C) Schematic representation of FOXR1 and FOXR2 interactomes with top-ranked interacting proteins. The interaction networks were visualized using unweighted force-directed distributions. Purple lines indicate interactions defined by the literature. Grey lines indicate newly identified interactions on the basis of the results of our proteomic study. Orange dots indicate MYC-MAX complex members reported in the literature. (D) FOXR2 expression in HEK293T, MCF10A and MDA-MB-468 cells were evaluated using whole proteome profiling and WB analysis with antibodies against endogenous FOXR2. (E) TAP-MS was performed with MDA-MB-468 cells that stably expressed SFB-tagged FOXR2, MYC, or MAX. Top-ranked interacting proteins are listed together with their NSAF values. Baits are highlighted in blue; prey FOXR2, MYC, and MAX are highlighted in orange. (B and E) All the data listed here are statistical significant.
We evaluated the raw interaction data and identified MYC and MAX, as well as many MYC- or MAX-associated proteins (such as TRRAP and p300), on the HCIP lists of FOXR2 and FOXR1 in chromatin fractions, (Figures 1B, 1C). To evaluate the relative strength of the binding, we calculated the normalized spectral abundance factor (NSAF), in which spectral counts of protein identified by mass spectrometry (MS) were normalized for their theoretical numbers of tryptic peptides and represented a percentage of the total protein recovered, as described previously (Malovannaya et al., 2010; Paoletti et al., 2006; Sardiu et al., 2008; Zybailov et al., 2005). All the interactions listed were statistical significant based on the Bayesian approach described previously. MYC and MAX were identified among the top hits in FOXR1 and FOXR2 interactomes (Figures 1B). Surprisingly, although MYC and MAX were shown to be among the top interacting proteins in FOXR2 tandem affinity purification followed by mass spectrometry (TAP-MS), we did not identify FOXR2 in our initial TAP-MS analysis of MYC or MAX performed in HEK293T cells. We reasoned that this may due to the low or restricted expression of FOXR2 in HEK293T cells, which are a derivative of HEK293 cells that originally generated by transformation with the Adenovirus 5 genome of a likely neuroendocrine progenitor cell contaminant in embryonic kidney prep. Thus, we performed whole proteome profiling of HEK293T, MCF10A and MDA-MB-468 cells. FOXR2 was not detected in any of the proteome profiling experiments using HEK293T (0/20) or MCF10A (0/8) cells, but was detected in 6 out of 8 proteome profiling experiments using MDA-MB-468 cells (Figure 1D). Western analysis using FOXR2 antibody further confirmed that FOXR2 expresses in MDA-MB-468 and several other breast cancer cell lines, but not in HEK293T cells (Figure 1D). Although FOXR1 shares significant sequence similarity with FOXR2 (Katoh, 2004a, b), FOXR1 expression was not detected in any of the cell lines we have tested by MS or Western analysis (Figure 1D). Thus, we focused on FOXR2 in our subsequent studies. We performed TAP-MS analysis in the chromatin fraction of MDA-MB-468 cells stably expressing S, FLAG, Streptavidin triple-tagged- (SFB-) MYC, MAX or FOXR2. To extract chromatin-bound protein complexes while minimizing the interactions mediated by DNA, we treated the insoluble pellets from the crude lysis step with TurboNuclease, which hydrolyses both single-and double-stranded DNA or RNA to oligonucleotides of 1–4 bases in length, to release chromatin-bound proteins (i.e. the chromatin fraction). This step disrupts any protein-protein interactions that may be mediated by DNA. We detected very little histones, HMG proteins and other common chromatin components in our TAP-MS results, suggesting that we were able to eliminate most, if not all, of the non-specific interactions mediated by DNA. As predicted, all the TAP-MS studies identified all three proteins, as well as many known MYC/MAX interacting proteins (Figure 1E), indicating that FOXR2 forms a stable protein complex with MYC and MAX.
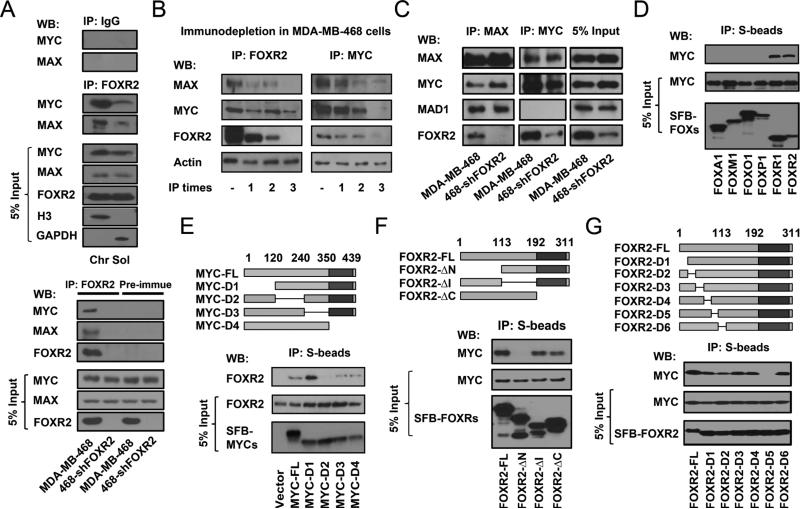
We confirmed the endogenous MYC, MAX, and FOXR2 complex in MDA-MB-468 cells using IP-Western blotting experiments. FOXR2 pulled down endogenous MAX and MYC, mainly in the chromatin fractions, while IgG or pre-immune serum failed to do so (Figure 2A). To further understand the details of this complex formation, we performed immunodepletion experiments and confirmed that these three proteins form a stable complex, since depleting one of them reduced the levels of the other two in the cell lysates (Figure 2B). The FOXR2-MYC/MAX binding may be independent of MAX-MAD1 association, since depletion of FOXR2 did not significantly change the interaction between MAX and MAD1 (Figure 2C). We confirmed the specificity of this interaction by demonstrating that MYC only binds to FOXR1 and FOXR2, but not to other FOX transcription factors (Figure 2D). We mapped the FOXR2-binding region to the mid region (residues 120-240) of MYC (Figure 2E). We also generated several deletion mutants of FOXR2 and showed that the N-terminus of FOXR2 is required for its binding to MYC (Figure 2F). Further analysis narrowed the domain required for MYC binding to a 20-peptide region of FOXR2 (Figure 2G).
Figure 2. FOXR2 associates with MYC/MAX on chromatin through its N-terminal region.
(A) Upper panel: Co-IP of endogenous MAX or MYC with FOXR2 was performed with IgG or anti-FOXR2 antibody using soluble and chromatin fractions prepared from MDA-MB-468 cells. 5% of the corresponding cell lysate used in the IP was included as input control. Immunoblotting was conducted using the indicated antibodies. Lower panel: Co-IP of endogenous MAX or MYC with FOXR2 was performed with anti-FOXR2 antibody or pre-immune serum from the same mouse, using chromatin fractions prepared from MDA-MB-468 and MDA-MB-468/sh-FOXR2 cells. (B) Immunodepletion of FOXR2 and MYC performed using extracts prepared from MDA-MB-468 cells. MDA-MB-468 cell lysates were immunoprecipitated with FOXR2 or MYC antibodies three times. FOXR2, MYC, MAX, and β-actin levels were measured after each round. (C) Cell lysates of MDA-MB-468 and MDA-MB-468/sh-FOXR2 cells were immunoprecipitated with MYC or MAX antibodies and immunoblotted with the indicated antibodies. (D) HEK293T cells were transfected with constructs encoding the indicated FOX proteins. Co-IP experiments were performed using S-protein beads and blotted with antibodies recognizing the Flag-epitope tag or endogenous MYC. 5% of the corresponding cell lysate used in the IP was included as input control. Only FOXR1 and FOXR2 were able to pull down endogenous MYC. (E) Schematic diagram of MYC mutants is shown. The bHLH domain is depicted as dark grey. The mutants were: D1: (Δ1-120); D2: (Δ120-240); D3: (Δ240-350); D4: (Δ350-439). SFB-tagged wild-type and mutants of MYC were subjected to co-precipitation experiments with endogenous FOXR2 in MDA-MB-468 cells. 5% of the corresponding cell lysate used in the IP was included as input control. (F, G) Schematic diagram of FOXR2 mutants is shown. The FOX domain appears as dark grey. The mutants were: ΔN: (Δ1-113); ΔI: (Δ113-192); ΔC: (Δ192-311); D1: (Δ1-20); D2: (Δ20-40); D3: (Δ40-60); D4: (Δ60-80); D5: (Δ80-100); D6: (Δ100-113). SFB-tagged wild-type and mutants of FOXR2 were subjected to co-precipitation experiments with endogenous MYC in HEK293T cells. 5% of the corresponding cell lysate used in the IP was included as input control.
FOXR2 regulates MYC transactivation activity and promotes cancer cell proliferation
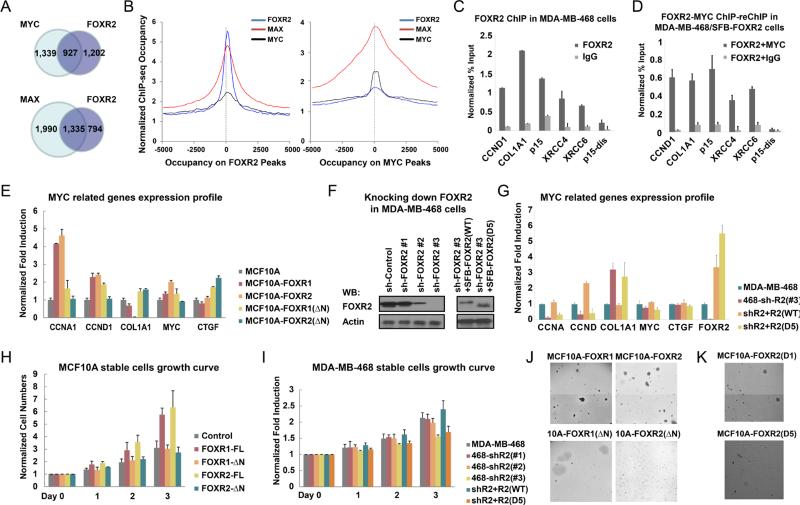
Because several transcriptional co-activators were also co-purified with FOXR2 and the MYC/MAX complex, we hypothesized that FOXR2 promotes MYC/MAX transactivation activity. Indeed, using ChIP-sequencing data on FOXR2, MYC and MAX, we observed that 43.5% and 62.7% of FOXR2 target genes overlapped with MYC and MAX target genes (p < 10−6), respectively (Figure 3A), while 64.6% and 93.8% of FOXR2 target genes overlapped with MYC and MAX target genes when using relatively loose cut-offs for MYC and MAX (p < 0.0001), respectively (Supplementary Figures S1A-B). Moreover, similar to MYC, FOXR2 binding regions mostly localized in the central promoter regions as demonstrated by a global analysis of relative peak positions, and highly overlapped with MYC and MAX peaks on gene promoters (Figure 3B). These results suggest that FOXR2 may modulate a fraction of MYC/MAX target genes. We further confirmed this by ChIP and ChIP-reChIP assays, performed with antibodies recognizing endogenous FOXR2 and showed that FOXR2 binds to the promoter regions of several MYC target genes, including CCNA1, CCND1, XRCC4, XRCC6, p15 and COL1A1 in MDA-MB-468 cells (Figures 3C-D and Supplementary Figures S1C-D). These data indicated that FOXR2 and MYC shared the similar localization on the promoter regions for a group of downstream genes.
Figure 3. FOXR2 promotes MYC transcriptional activities and cell proliferation.
(A) Chromatin IP-sequencing (ChIP-seq) assay was performed in MDA-MB-468 cells using endogenous antibodies against FOXR2, MYC or MAX. Overlap between FOXR2 and MAX or MYC target genes were evaluated. (B) The global analyses of relative peak positions of FOXR2 and MYC or MAX were shown. (C) A chromatin IP (ChIP) assay was performed in MDA-MB-468 cells using FOXR2 antibody or control IgG. The recovery of MYC downstream gene promoter regions was examined by real-time PCR. All the promoters except the control p15-distal promoter have significantly enriched in the FOXR2 (P < 0.001) but not IgG immunoprecipitants. (D) A ChIP-reChIP assay was performed in MDA-MB-468 cells overexpressing SFB-tagged FOXR2 using streptavidin-beads, eluted with biotin and re-IPed with MYC-antibody or control IgG. The recovery of MYC downstream gene promoter regions was examined by real-time PCR. All the promoters except the control p15-distal promoter have significantly enriched in the MYC (P < 0.001) but not IgG immunoprecipitants. (E) MYC target gene expression profiles were evaluated by RT PCR in MCF10A, MCF10A-FOXR1, -R2, -R1 (ΔN), and -R2 (ΔN) cells. mRNA levels were determined by real-time RT-PCR and normalized with GAPDH. CCNA, CCND1 (P < 0.01) and COL1A1 (P < 0.05) expression levels have been significantly changed in MCF10A-FOXR1/R2 cells comparing with MCF10A cells. (F) Knocking down FOXR2 in MDA-MB-468 cells and reconstitution with wild-type or D5 mutant of FOXR2 were confirmed by immunoblotting, as indicated. (G) MYC target gene expression profiles were evaluated by RT PCR in MDA-MB-468, MDA-MB-468-shFOXR2, MDA-MB-468-shFOXR2+SFB-FOXR2 and MDA-MB-468-shFOXR2+SFB-FOXR2(D5) cells. mRNA levels were determined by real-time RT-PCR and normalized with GAPDH. CCNA, CCND1 and COL1A1 expression levels have been significantly changed in MDA-MB-468-shFOXR2 and MDA-MB-468-shFOXR2+SFB-FOXR2(D5) cells (P < 0.01), comparing with MDAMB-468 cells. (H) Growth curve of MCF10A derivative cells used in (C). MCF10A-FOXR1/R2 cells have significantly more cell numbers after 3 days (P < 0.01), comparing with MCF10A cells. Data are averages (± SD) of three independent experiments. (I) Growth curves of MDA-MB-468 parental and derivative cells used in (E) are shown. MDA-MB-468-shFOXR2 and MDA-MB-468-shFOXR2+SFB-FOXR2(D5) cells have significantly less cell numbers after 3 days (P < 0.01), comparing with MDA-MB-468 cells. (J, K) Soft agar colony formation of MCF10A cells that stably expressed indicated proteins was assessed and is presented. Colony numbers: MCF10A-FOXR1: 12.4 ± 2.2; MCF10A-FOXR1(ΔN): 1.4 ± 1.3; MCF10A-FOXR2: 14.2 ± 2.4; MCF10A-FOXR2(ΔN): 0.8 ± 0.8; MCF10A-FOXR2(D1): 9.7 ± 1.8; MCF10A-FOXR2(D5): 2.8 ± 1.9. The P < 0.001 between MCF10A-FOXR1 and MCF10A-FOXR1(ΔN); MCF10A-FOXR2 and MCF10A-FOXR2(ΔN); MCF10A-FOXR2(D1) and MCF10AFOXR1(D5).
To further validate the functions of FOXR2 in promoting MYC target genes and cell proliferation, we first used MCF10A cells, which have no detectable expression of FOXR1 or FOXR2 (Figure 1D). Expression of FOXR1 or FOXR2 in MCF10A cells led to elevated expression of the MYC target genes CCNA1 and CCND1 (P < 0.01) and reduced the expression of the MYC-downregulated gene COL1A1 (P < 0.05), whereas the mRNA levels of MYC and unrelated control CTGF were largely unaffected (Figure 3E). As controls, overexpression of FOXR1-ΔN mutant or FOXR2-ΔN mutant, the two mutants that could not bind to MYC, did not result in any obvious changes in MYC target genes (Figure 3E). In addition, we downregulated FOXR2 expression in MDA-MB-468 cells (Figure 3F) and found reduced expression of the MYC target genes CCNA1, CCND1, p15, XRCC4, XRCC6, and elevated the expression of the MYC-downregulated gene COL1A1 (P < 0.01), whereas the mRNA levels of MYC and unrelated control CTGF were largely unaffected (Figure 3G and Supplementary Figure 1E). We also infected MDA-MB-468/shFOXR2 cells with lentivirus to stably express wild-type FOXR2 or the MYC-binding defective mutant FOXR2-D5 to restore the regulation of FOXR2 on MYC target genes. Wild-type FOXR2 successfully rescued the regulation of CCNA1, CCND1, p15, XRCC4, XRCC6, and COL1A1 (P < 0.01), while FOXR2-D5 failed to do so (Figure 3G). These results suggested that FOXR2 is required for the transcription of MYC downstream target genes.
Since MYC is a well-known master regulator of cell growth and proliferation, we determined whether FOXR2 would contribute to cell proliferation through regulating MYC transcriptional activities. Indeed, we found that FOXR1 or FOXR2 expression strongly promoted MCF10A cell growth (P < 0.01), whereas the expression of MYC-binding defective mutants FOXR1-ΔN or FOXR2-ΔN failed to do so (Figure 3H). Conversely, FOXR2 knockdown in MDA-MB-468 cells led to diminished cell proliferation. MDA-MB-468-shFOXR2 and MDA-MB-468-shFOXR2+SFB-FOXR2(D5) cells have significantly less cell numbers after 3 days (P < 0.01), comparing with MDA-MB-468 cells (Figure 3I). Moreover, when wild-type FOXR2 or the MYC-binding defective mutant FOXR2-D5 was expressed in MDA-MB-468/sh-FOXR2 cells, only wild-type FOXR2 could rescue the growth defects caused by FOXR2 downregulation (Figure 3I). A soft agar colony formation assay also confirmed that FOXR1 and FOXR2 have oncogenic potential that is dependent on their ability to bind to MYC (Figures 3J-K). Wild-type MCF10A cells did not form colony on soft agar, which agrees with our previous results (Wang et al., 2012). However, MCF10A cells overexpressing FOXR1, FOXR2, or FOXR2(D1), which still binds to MYC, formed significantly more colonies on soft agar (12.4 ± 2.2, 14.2 ± 2.4 and 9.7 ± 1.8, respectively), while MCF10A cells overexpressing MYC binding defective mutants FOXR1(ΔN), FOXR2(ΔN), or FOXR2(D5) formed significantly reduced numbers of colonies (1.4 ± 1.3, 0.8 ± 0.8, 2.8 ± 1.9, respectively) (Figures 3J-K). These data suggested that FOXR1/FOXR2 promoted oncogenic transformation through their association with MYC.
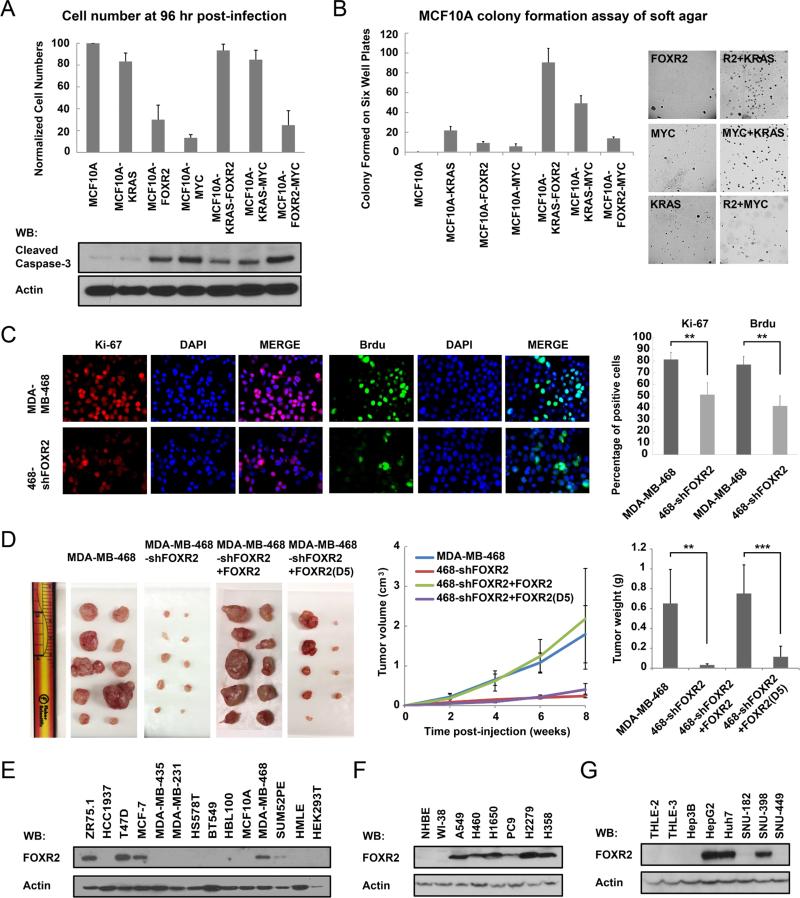
Taking advantage of the knowledge of the collaboration between MYC and RAS in promoting oncogenic transformation, we overexpressed FOXR2, MYC, RAS, or any two together in MCF10A cells, to evaluate whether FOXR2 and MYC function in the same oncogenic pathway. Overexpression of FOXR2, similar to that of MYC, led to ~70% reduction in cell number following the initial infection (96 hrs post-infection), which was likely due to MYC-induced apoptosis. An interesting observation was that cells overexpressing FOXR2 plus RAS, but not FOXR2 plus MYC, rescued this cell death phenotype (Figure 4A). Western analysis using cleaved Caspase-3 antibody as a marker for apoptosis confirmed elevated apoptosis in MCF10A cells infected by retroviruses overexpressing FOXR2 or MYC, while MCF10A cells expressing RAS with FOXR2 or MYC partially blocked this induced apoptosis (Figure 4A). Moreover, MCF10A cells expressing FOXR2 with RAS formed significantly more colonies on soft agar than did MCF10A cells expressing FOXR2, MYC, or both (Figure 4B). We knocked down MYC using shRNA in MCF10A cells and then infected these cells with retroviruses overexpressing FOXR2. The FOXR2-induced apoptosis was reduced in MCF10A-shMYC cells (Supplementary Figure S2A). Moreover, overexpression of tagged FOXR2 in MCF10A-shMYC cells failed to induce colony formation on soft agar (Supplementary Figure S2B). These results indicate that FOXR2-induced apoptosis and transformation are MYC dependent. These findings support the idea that FOXR2 and MYC function in the same oncogenic pathway.
Figure 4. FOXR2 facilitates MYC's activities and promotes tumor proliferation both in vitro and in vivo.
(A) MCF10A cells were infected in six-well plates with retroviruses encoding RAS, MYC, FOXR2, or any two together. Cells were counted after 96 hours of infection. Normalized cell numbers are presented. Data are averages (± SD) of three independent experiments. Cell lysates of each cell lines were immunoblotted with the indicated antibodies. (B) Soft agar colony formation of MCF10A cells that stably expressed the indicated proteins was assessed and is presented. (C) Ki-67 and BrdU staining of MDA-MB-468 and MDA-MB-468/sh-FOXR2 cells was performed as indicated. The percentages of positive cells are summarized on the right. **P < 0.01. (D) Xenograft tumor growth studies. 5 × 106 MDA-MB-468 and MDA-MB-468/sh-FOXR2 cells were resuspended in 100 μL of Matrigel diluted with PBS at 1:1 ratio and injected subcutaneously into left and right flanks of 10 anesthetized 6- to 8-week-old female BALB/c nude mice respectively. 5 × 106 MDA-MB-468/sh-FOXR2+SFB-FOXR2 and MDAMB-468/sh-FOXR2+SFB-FOXR2(D5) cells were resuspended in 100 μL of Matrigel diluted with PBS at 1:1 ratio and injected subcutaneously into left and right flanks of 10 anesthetized 6- to 8-week-old female BALB/c nude mice respectively. Starting from the day 0, the tumor weight and size were measured bi-weekly. Mice were euthanized after 8 weeks of injection. The tumors were excised, photographed, and weighed. *** P < 0.001. (E) Immunoblotting of FOXR2 in normal and breast cancer cell lines, with HEK293T cells as the negative control. (F) Immunoblotting of FOXR2 in normal and lung cancer cell lines. (F) Immunoblotting of FOXR2 in normal and liver cancer cell lines.
FOXR2 downregulation also led to reduced cell proliferation in MDA-MB-468 cells, as measured by Ki-67 staining (81% vs 51%; P < 0.01) and BrdU incorporation (77% vs 42%; P < 0.01) (Figure 4C). Moreover, both tumor size and tumor weight were significantly reduced in mice injected with MDA-MB-468/sh-FOXR2 cells compared with those in mice injected with control MDA-MB-468 cells (32.2 ± 11.9 mg vs 651.7 ± 342.0 mg; P < 0.01) (Figure 4D). Reconstitution with SFB-tagged wild-type FOXR2, but not MYC binding defective mutant FOXR2(D5), fully rescued tumor growth (751.1 ± 287.9 mg vs 115.4 ± 105.5 mg; P < 0.001) (Figure 4D). Together, these data suggest that FOXR2 acts with MYC and promotes cancer cell proliferation both in vitro and in vivo.
FOXR2 was overexpressed in cancer cell lines and patient tumor samples
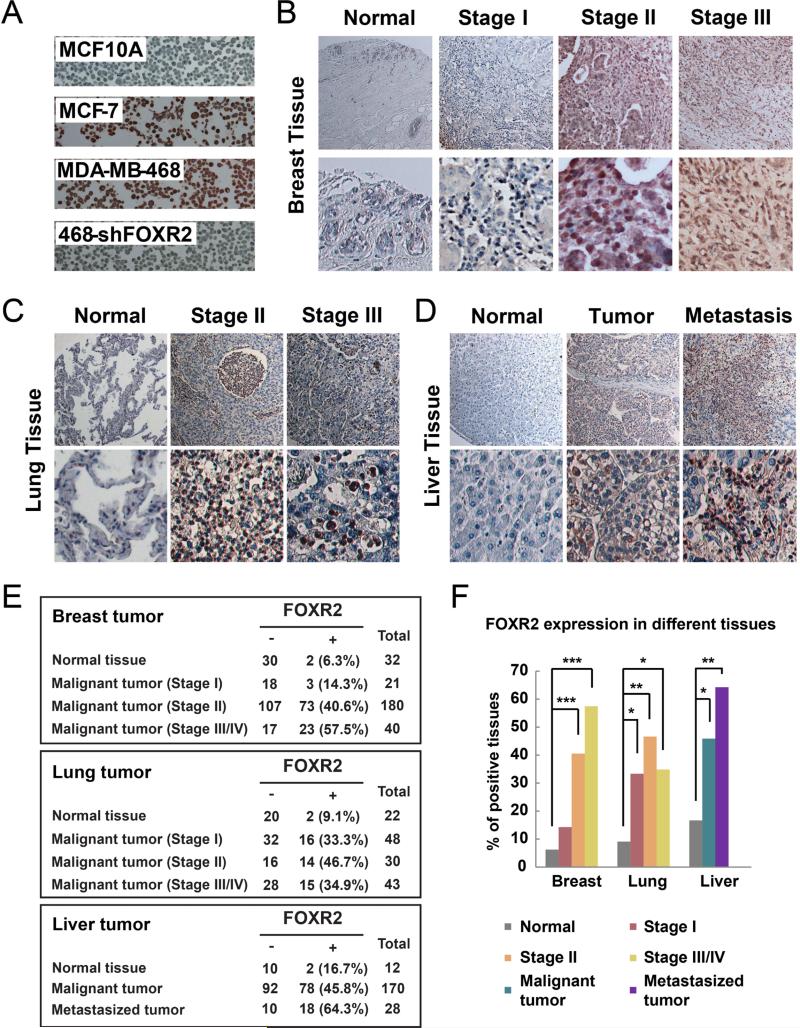
We tested 13 breast cell lines, as well as HEK293T cells as the negative control. Indeed, we found that FOXR2 was not expressed in the 3 normal human breast epithelial cell lines we tested (MCF10A, HMLE, and HBL100) but was detectable in 5 of the 11 (45%) breast cancer cell lines we assayed (ZR75.1, T47D, MCF-7, MDA-MB-468, and SUM52PE) (Figure 4E and Supplementary Figure S2C). Moreover, FOXR2 was not expressed in the 2 normal human lung cell lines we tested (Human Bronchial epithelial cells NHBE and WI-38, which are lung-derived fibroblasts) but was detectable in all the 6 lung cancer cell lines we assayed (A549, H460, H1650, PC9, H2279 and H358) (Figure 4F). Similarly, FOXR2 was not expressed in the 2 normal human liver cell lines we tested (THLE-2 and THLE-3) but was detectable in 3 of the 6 (50%) liver cancer cell lines we assayed (HepG2, Huh7 and SNU-398) (Figure 4G). MG132 treatment could stabilize FOXR2 in HBL100 cells but not MCF10A cells (Supplementary Figure S2D), indicating that FOXR2 may be regulated, at least in some cases, via a ubiquitination- and proteasome-dependent pathway. These data suggest that FOXR2 is overexpressed in human cancer cell lines. Given the potential role of FOXR2 in promoting cell proliferation, as demonstrated above, we further analyzed FOXR2 expression by immunohistochemical analysis in human primary tumor samples. We first confirmed the specificity of our FOXR2 antibody in paraffin-embedded cell lines (Figure 5A). FOXR2 antibody only stained MCF-7 and MDA-MB-468 cells but not MCF10A cells, which is consistent with Western blotting results using these cell lines (Figures 1D, 4E, 5A). Knocking down FOXR2 in MDA-MB-468 cells greatly reduced the staining (Figure 5A), indicating that the cross-reaction of this FOXR2 antibody is quite limited. Thus, we used this antibody to examine the expression of FOXR2 in 273 breast, 145 lung, and 210 liver tumor or normal tissue samples (Figures 5B-D). FOXR2 was positively stained in only 6.3% of normal breast tissue samples (2 of 32), but 14.3% of stage I (3 of 21), 40.6% of stage II (73 of 180), and 57.5% of stage III/IV (23 of 40) malignant breast tumor samples (Figures 5B, 5E, 5F). It was stained positively in only 9.1% of normal lung tissue samples (2 of 22), but in 33.3% stage I (16 of 48), 46.7% stage II (14 of 30), and 34.9% of stage III/IV (15 of 43) malignant lung tumor samples (Figures 5C, 5E, 5F). It was stained positively in 16.7% of normal liver tissue samples (2 of 12), but 45.8% in malignant liver tumor samples (Figures 5D, 5E, 5F). Therefore, we conclude that FOXR2 is frequently overexpressed in breast, lung, and liver cancers and may contribute to the proliferation of these cancers.
Figure 5. FOXR2 is overexpressed in cancers.
(A) Immunohistochemical staining of breast cell lines was performed using FOXR2 antibody to validate the specificity of our anti-FOXR2 antibody. (B-E) FOXR2 staining of normal and tumor tissue microarrays of breast (B), lung (C), and liver (D) samples. The data were summarized in (E), with case numbers and percentages. (F) Bar graph summary of data presented in (B-E). *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
In this study, we revealed that FOXR2 is overexpressed in breast, lung and liver cancers and plays a role in MYC-dependent gene transcription and cell proliferation. The potential role of FOXR2 as an oncogene was independently reported (Rahrmann et al., 2013), in which the authors identified Foxr2 as a proto-oncogene in mice and showed that it is overexpressed in human malignant peripheral nerve sheath tumors and required for tumor proliferation. Therefore, it is likely that FOXR2 overexpression contributes to human tumor growth.
We found that a subset of cancer cell lines and tumor samples showed FOXR2 upregulation (Figures 4E-G and 5). Together with the observations that FOXR2 binds to MYC (Figures 1 and 2) and activates MYC-dependent gene transcription (Figure 3), we reason that targeting FOXR2 may be a viable option for treating these cancers. Furthermore, we speculate that agents that disrupt FOXR2 or FOXR2-MYC interaction in adult tissues may have mild or limited side effects compared with agents that directly target MYC, since FOXR2 is only expressed in cancer cell lines and tumor samples (Figures 4E-G and 5), and therefore hold great therapeutic potential.
FOXR1 shares significant sequence homology with FOXR2 (Katoh, 2004a, b) and also interacts with MYC/MAX (Figures 1B, 1C and 2D). Overexpression of FOXR1 leads to similar effects as that of FOXR2 (Figures 3E, 3H and 3J). However, unlike FOXR2, FOXR1 protein was not detectable in any breast, liver, or lung cell lines we tested by MS or Western analysis (Figure 1D). These data suggest that FOXR2 is likely a protooncogene in these cancers, while FOXR1 expression may be more restricted to early stages of development as previously reported (Schuff et al., 2006).
Experimental Procedures
Western blotting, immunoprecipitation, immunostaining and immunohistochemistry assays
Whole-cell lysates were prepared by lysing cells with NETN buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40) on ice for 30 min and then boiling them in 2× Laemmli buffer (Wang et al., 2015). To extract chromatin-bound protein complexes while minimizing the interactions mediated by DNA, we treated the insoluble pellets from the crude lysis step with TurboNuclease, which hydrolyses both single- and double-stranded DNA or RNA to oligonucleotides of 1–4 bases in length, to release chromatin-bound proteins (i.e. the chromatin fraction). This step disrupts any protein-protein interactions that may be mediated by DNA. We used the same protocol to prepare lysates for our co-IP experiments in chromatin fractions. Lysates were subjected to SDS-PAGE, followed by immunoblotting with antibodies against various proteins, including MYC, FOXR2, GFP, GAPDH, MAD1 (Santa Cruz Biotechnology), cleaved Caspase-3, MYC, MAX (Cell Signaling), FOXR2 (Abcam), β-actin, FLAG (Sigma), and histone H3 (Upstate). Rabbit polyclonal antibody against human FOXR2 (residues 1–311) was generated by immunizing rabbits (Cocalico Biologicals) with GST-FOXR2 fusion protein and affinity purified before being used for Western blotting, ChIP-seq and IHC analyses.
For immunoprecipitation and co-immunoprecipitation assays, a total of 1 × 107 cells were lysed with NETN buffer on ice for 30 min, and fractionized as described above. The lysates were then incubated with 20 μl conjugated beads (for SFB-tagged pull-down) for 2 hr at 4°C; or incubated with antibodies against endogenous proteins for 1 hour at 4°C, followed by adding 20 μl of protein A/G agarose and incubated for 2 hr at 4°C. Beads were washed three times with NETN buffer and boiled in 2× Laemmli buffer.
For immunostaining assays, cells cultured on coverslips were washed with PBS, fixed with 3% paraformaldehyde for 20 min, and permeabilized with 0.5% (v/v) Triton X-100 solution for 5 min. Coverslips were washed with PBS and immunostained with primary antibodies in 5% goat serum for 60 min. Cells were then washed and incubated with rhodamine-or FITC-conjugated secondary antibodies for 60 min, and nuclei were stained with 1 g/ml 4',6-diamidino-2-phenylindole (DAPI). Slides were mounted and visualized using a Nikon ECLIPSE E800 fluorescence microscope with a Nikon Plan Fluor 40× oil objective lens (numerical aperture 1.30) at ambient temperature. Cells were photographed using a SPOT camera (Diagnostic Instruments).
For the IHC assays, 5 × 106 MCF10A, MCF-7, MDA-MB-468 and MDA-MB-468/sh-FOXR2 cells were trypsinized, PBS washed, paraffin-embedded and sliced to make slides. All of the human breast, liver and lung cancer tissue microarrays were purchased from US Biomax. The slides were deparaffinized and rehydrated and blocked for 30 min in 1% hydrogen peroxide and methanol to remove endogenous peroxidase activity. After antigen retrieval, slides were blocked in 5% goat serum in PBS and incubated overnight with anti-FOXR2 antibodies diluted (1:200) in blocking buffer. After being washed with PBS, slides were incubated in secondary antibodies diluted (1:200) in blocking buffer for 3 hr, developed with 3,3'-diaminobenzidine (DAKO), and re-stained with hematoxylin for 30 sec. Slides were mounted and visualized using the same microscope and lens.
Tandem affinity purification, mass spectrometry and data analysis
Tandem affinity purification and mass spectrometry analysis were carried out as previously described (Elias and Gygi, 2007; Li et al., 2015a; Li et al., 2015b; Shevchenko et al., 1996). To generate HCIP lists, we applied a modified SAINT algorithm, which uses a collection of TAP-MS results that follow the same protocol as the control group (Li et al., 2015b; Wang et al., 2014; Xu et al., 2014). We kept the same SAINT Score > 0.80 interactions as on the HCIP list. To calculate the normalized spectral abundance factor (NSAF), protein results from MS sequencing were converted to NCBI gene identifiers. Spectral counts of these gene products were then normalized for their theoretical numbers of tryptic peptides and represented a percentage of the total protein recovered, as described previously (Malovannaya et al., 2010; Paoletti et al., 2006; Sardiu et al., 2008; Zybailov et al., 2005). The probability of interactions within and between complexes is computed solely based upon NSAFs using Bayesian approach. Only the statistical significant results were presented. We searched the proteins identified in FOXR1 and FOXR2 TAP-MS in the protein-protein interaction knowledge databases to identify the interactions reported in the literature. The databases we used were BioGrid (http://thebiogrid.org) (Stark et al., 2006), STRING (http://string-db.org) (von Mering et al., 2003), BIND (http://bind.ca) (Bader et al., 2003), DIP (http://dip.doembi.ucla.edu) (Xenarios et al., 2000), and HPRD (http://www.hprd.org) (Prasad et al., 2009). The interaction networks were visualized by Cytoscape software (Smoot et al., 2011) using unweighted force-directed distributions. The protein points were placed based on the number of shared interacting proteins. The distances between different points have no correlation with the binding affinities between proteins.
ChIP, ChIP-reChIP, ChIP sequencing and genomic data analysis
For the ChIP assay, 1 × 106 MDA-MB-468 cells (1 × 107 cells for ChIP sequencing) were cross-linked with 1% formaldehyde in growth medium for 10 min. Cross-linking was terminated by the addition of glycine to a final concentration of 125 mM and incubation at 37°C for 10 min. Cells were rinsed with cold PBS, harvested in 200 μl of SDS lysis buffer supplemented with protease inhibitor mixture (Roche Applied Science) and 1 mM phenylmethylsulfonylfluoride (PMSF) per 1 × 106 cells, and sonicated to shear the DNA to yield fragments of 500 bp or less. Samples were then diluted with 9 volumes of ChIP dilution buffer before being precleared for 1 hour with 40 μl of a mixture of protein A/G agarose and salmon sperm DNA. Approximately 5 μg of FOXR2, MYC or MAX antibody or mouse normal immunoglobulin (IgG) were added to the precleared supernatant and incubated for 1 hour at 4°C. Forty μl of protein A/G agarose were added, and the mixture was incubated overnight at 4°C. Washes were sequentially performed with low-salt buffer, high-salt buffer, LiCl buffer, and twice with TE buffer. Immunoprecipitates were eluted in 500 μl of elution buffer (1% SDS, 0.1 M NaHCO3), followed by the addition of NaCl (20 μl). Crosslinking was reversed by overnight incubation of samples at 65°C and treatment with protease K (Sigma) for 2 hours at 45°C. The recovered DNA was extracted with phenol-chloroform and precipitated with ethanol. Quantification was performed by real-time PCR with the My IQ real-time PCR detection system and an IQ SYBR Green Supermix (Bio-Rad). Control IgG and input DNA values were used to normalize values from ChIP recovery. For the ChIP-reChIP assay, 1 × 107 MDA-MB-468/SFB-FOXR2 cells were processed followed the same ChIP protocol, but used 50 μl streptavidin beads for the initial pull-down. The beads were washed with NETN buffer three time and immunoprecipitates were eluted twice in 150 μl NETN buffer containing 2 mg/ml biotin. The elutes were then diluted with 9 volumes of ChIP dilution buffer before adding 10 μg of MYC antibody or mouse normal immunoglobulin (IgG) for the secondary immunoprecipitation.
ChIP sequencing was performed in HEK293T cells with two biological replicates. Samples were sequenced using Illumina HiSeq, and raw reads were mapped to human reference genome (hg19). Peaks were selected using MACS in Galaxy and annotated with PAVIS (Huang et al., 2013; Langmead et al., 2009; Zhang et al., 2008). Datasets were analyzed in Galaxy and mapped to genes using PAVIS, with −5000 to +1000 TSS windows.
In vivo xenograft study
All animal experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the M.D. Anderson Cancer Center. 5 × 106 MDA-MB-468 and MDA-MB-468/sh-FOXR2 cells were resuspended in 100 μL of Matrigel diluted with PBS at 1:1 ratio and injected subcutaneously into left and right flanks of 10 anesthetized 6- to 8-week-old female BALB/c nude mice respectively. Similarly, 5 × 106 MDA-MB-468/sh-FOXR2+SFB-FOXR2 and MDA-MB-468/sh-FOXR2+SFBFOXR2(D5) cells were resuspended in 100 μL of Matrigel diluted with PBS at 1:1 ratio and injected subcutaneously into left and right flanks of 10 anesthetized 6- to 8-week-old female BALB/c nude mice respectively. Starting from the day 0, the tumor weight and size were measured bi-weekly. Mice were euthanized after 8 weeks of injection. The tumors were excised, photographed, and weighed.
Soft agar colony formation assay
1 × 103 cells were added to 1.5 mL of growth medium with 0.2% agar and layered onto 2 mL of 0.5% agar bed in 6-well plates. Medium was replenished every 3 days for 30 days. Resulting colonies were fixed and stained with Crystal Violet. The numbers of colonies were counted with a GelDoc with Quantity One software (Bio-Rad).
Accession Numbers
ChIP-sequencing data are available in the Gene Expression Omnibus (GEO) database (Barrett et al., 2013; Edgar et al., 2002) under accession number GSE81381.
Statistical analysis
All experiments were repeated at least three times. Data were analyzed by Pearson χ2 analysis (for IHC) and the student's test (for all other experiments). A P-value of <0.05 was considered statistically significant.
Supplementary Material
Highlights.
-
1)
FOXR2 associates with the MYC-MAX complex;
-
2)
FOXR2 promotes cell proliferation by regulating MYC transcriptional activities;
-
3)
FOXR2 is overexpressed in multiple cancers and patient tumor samples.
ACKNOWLEDGMENTS
We thank the Junjie Chen, Jun Qin and Steven Lin laboratory for their advice and technical assistance, especially Dr. Nan Li and Dr. Yawei Qiao, and Ms. Ann Sutton for editing the manuscript. X.L. is a recipient of a Computational Cancer Biology Training Program Fellowship, supported by the Cancer Prevention and Research Institute of Texas. This work was supported in part with a startup fund from The University of Texas MD Anderson Cancer Center, Era of Hope Scholar Research Award W81XWH-09-1-0409 from the U.S. Department of Defense (J.C.), and National Institutes of Health grant R01 HG007538 (W.L.). The ChIP-sequencing and RNA-sequencing analysis was supported by a Solexa allowance through the Center for Cancer Epigenetics, MD Anderson. MD Anderson is supported by the National Institutes of Health Core Grant CA016672.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
X.L., W.W., W.L., J.Q., and J.C. designed the experiments. X.L., W.W., M.G., and K.A. performed experiments. X.L., Y.X., and M.T. analyzed data. X.L., W.W. and J.C. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
eTOC Blurb
Li et al. identify FOXR2 as a proto-oncogene through proteomics approaches. They find that FOXR2 regulates cell proliferation by interacting with MYC and MAX, and promoting MYC's transcriptional activities. FOXR2 is highly expressed in several breast, lung, and liver cancer cell lines and patient tumor samples, suggesting that overexpression of FOXR2 may contribute to tumorigenesis.
References
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- Askew DS, Ashmun RA, Simmons BC, Cleveland JL. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieche I, Laurendeau I, Tozlu S, Olivi M, Vidaud D, Lidereau R, Vidaud M. Quantitation of MYC gene expression in sporadic breast tumors with a real-time reverse transcription-PCR assay. Cancer Res. 1999;59:2759–2765. [PubMed] [Google Scholar]
- Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Boxer RB, Jang JW, Sintasath L, Chodosh LA. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6:577–586. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S, Groudine M. Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature. 1982;298:679–681. doi: 10.1038/298679a0. [DOI] [PubMed] [Google Scholar]
- D'Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, Cox JD, Ha SI, Belka GK, Golant A, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- Huang W, Loganantharaj R, Schroeder B, Fargo D, Li L. PAVIS: a tool for Peak Annotation and Visualization. Bioinformatics. 2013;29:3097–3099. doi: 10.1093/bioinformatics/btt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Identification and characterization of human FOXN5 and rat Foxn5 genes in silico. Int J Oncol. 2004a;24:1339–1344. [PubMed] [Google Scholar]
- Katoh M. Identification and characterization of human FOXN6, mouse Foxn6, and rat Foxn6 genes in silico. Int J Oncol. 2004b;25:219–223. [PubMed] [Google Scholar]
- Koso H, Tsuhako A, Lyons E, Ward JM, Rust AG, Adams DJ, Jenkins NA, Copeland NG, Watanabe S. Identification of FoxR2 as an oncogene in medulloblastoma. Cancer Res. 2014;74:2351–2361. doi: 10.1158/0008-5472.CAN-13-1523. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang W, Chen J. From pathways to networks: connecting dots by establishing protein-protein interaction networks in signaling pathways using affinity purification and mass spectrometry. Proteomics. 2015a;15:188–202. doi: 10.1002/pmic.201400147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang W, Wang J, Malovannaya A, Xi Y, Li W, Guerra R, Hawke DH, Qin J, Chen J. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol Syst Biol. 2015b;11:775. doi: 10.15252/msb.20145504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- Malovannaya A, Li Y, Bulynko Y, Jung SY, Wang Y, Lanz RB, O'Malley BW, Qin J. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci U S A. 2010;107:2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TS, Kandasamy K, Pandey A. Human Protein Reference Database and Human Proteinpedia as discovery tools for systems biology. Methods Mol Biol. 2009;577:67–79. doi: 10.1007/978-1-60761-232-2_6. [DOI] [PubMed] [Google Scholar]
- Prendergast GC. Mechanisms of apoptosis by c-Myc. Oncogene. 1999;18:2967–2987. doi: 10.1038/sj.onc.1202727. [DOI] [PubMed] [Google Scholar]
- Rahrmann EP, Watson AL, Keng VW, Choi K, Moriarity BS, Beckmann DA, Wolf NK, Sarver A, Collins MH, Moertel CL, et al. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat Genet. 2013;45:756–766. doi: 10.1038/ng.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeger C, Longerich T, Schiller C, Bewerunge P, Mehrabi A, Toedt G, Kleeff J, Ehemann V, Eils R, Lichter P, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47:511–520. doi: 10.1002/hep.22033. [DOI] [PubMed] [Google Scholar]
- Schuff M, Rossner A, Donow C, Knochel W. Temporal and spatial expression patterns of FoxN genes in Xenopus laevis embryos. Int J Dev Biol. 2006;50:429–434. doi: 10.1387/ijdb.052126ms. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB, Evan GI. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci U S A. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Huang J, Wang X, Yuan J, Li X, Feng L, Park JI, Chen J. PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 2012;26:1959–1971. doi: 10.1101/gad.192955.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li X, Huang J, Feng L, Dolinta KG, Chen J. Defining the protein-protein interaction network of the human hippo pathway. Mol Cell Proteomics. 2014;13:119–131. doi: 10.1074/mcp.M113.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li X, Lee M, Jun S, Aziz KE, Feng L, Tran MK, Li N, McCrea PD, Park JI, et al. FOXKs Promote Wnt/beta-Catenin Signaling by Translocating DVL into the Nucleus. Dev Cell. 2015;32:707–718. doi: 10.1016/j.devcel.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. DIP: the database of interacting proteins. Nucleic Acids Res. 2000;28:289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Li X, Gong Z, Wang W, Li Y, Nair BC, Piao H, Yang K, Wu G, Chen J. Proteomic analysis of the human cyclin-dependent kinase family reveals a novel CDK5 complex involved in cell growth and migration. Mol Cell Proteomics. 2014;13:2986–3000. doi: 10.1074/mcp.M113.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.