Abstract
Background
Autophagy is the degrading process of protein and organelles mediated by lysosomes. This process is involved in purging senescent organelles and subversive proteins while maintaining the stability of the intracellular environment. This phenomenon is highly conservative, existing in nearly every species, and is involved in cell growth, proliferation and tumorigenesis.
Summary
In recent decades, with the discovery of autophagy-related genes and proteins in conjunction with the improvement in detection methods, the study of autophagy is constantly achieving new breakthroughs. It has been discovered that multiple regulatory mechanisms, including Atg protein and its conjugation system, mammalian target of rapamycin upstream and downstream pathways, complex of B-cell lymphoma-2 and Beclin-1c, cellular stress and dual regulation of p53 protein, jointly mediate the process of autophagy. Aberrant autophagy can cause impairment of resident kidney cells and development of various renal diseases.
Key Message
In this paper, we summarize recent discoveries regarding the development and regulatory mechanisms of autophagy. We also highlight the role of autophagy in the pathogenesis of some kidney diseases, such as diabetic nephropathy, obstructive nephropathy, IgA nephropathy, nephropathic cystinosis, aristolochic acid nephropathy, autoimmune kidney diseases and chronic cyclosporin A-induced nephrotoxicity. These findings provide new insights into the mechanisms of renal diseases and are useful for designing novel therapeutic approaches for the treatment of chronic kidney disease.
Key Words: Autophagy, Autophagy-related genes or proteins, Chronic kidney diseases, Mammalian target of rapamycin, Regulatory mechanisms
Introduction
Autophagy is an intracellular degrading system that is highly dependent on lysosomes and has existed universally in eukaryotic cells. It is an adaptive metabolism that can eliminate impaired and senescent organelles and biological macromolecules. It is an important and highly conservative regulatory mechanism to maintain intracellular stability. The autophagy phenomenon was first observed by Ashford and Portein in 1962 in liver cells through electron microscopes, but following that discovery, no breakthroughs on autophagy research were made until Tsukada and colleagues discovered the autophagy-related gene (Atg) in 1993 in Saccharomycetes [1,2]. Recent studies have found that autophagy is associated with kidney aging and the occurrence of some kidney diseases including acute kidney injury, drug-induced renal impairment, hereditary renal diseases and diabetic nephropathy. Accordingly, autophagy could possibly be considered as a new target for treating kidney diseases in the future. In this review article, we briefly introduce the key molecules and signaling pathways associated with autophagy development and highlight the role of autophagy in various chronic kidney disease processes.
Overview of Autophagy
Classification of Autophagy
Based on the difference between channels that deliver substances to lysosomes, autophagy is categorized into macroautophagy, microautophagy and chaperone-mediated autophagy (CMA) [3]. Macroautophagy is a catabolic process in which portions of the cytoplasm are sequestered with double-membraned vesicles, termed autophagosomes, and then delivered to lysosomes for bulk degradation. Microautophagy is a process in which lysosome membranes invaginate themselves directly to enclose and degrade the substrates. CMA occurs after fusing with chaperones, where the protein inside the cytoplasm is transferred to lysosomes and degraded by enzymes inside the lysosomes. This process is highly selective. In CMA, proteins containing KFERQ motif are identified by heat shock cognate protein of 70 kDa (HSC70), thus forming the HSC70/substrate protein complex which interacts with the lysosomal membrane and is mediated by lysosome-associated membrane protein 2A (LAMP-2A) and is later engulfed and degraded by the lysosome [4]. Macroautophagy is the most common of these three types, and its degree of usage in diseases is the highest, so the autophagy discussed in this article is mainly macroautophagy (hereafter referred to simply as autophagy). In terms of different control approaches concerning cell quality, autophagy could be further classified into selective autophagy and nonselective autophagy. Even with adequate nutrition in the liver, around 1-1.5% of cellular protein degrades and metabolizes each hour. As the quality control tool of the cytoplasm, autophagy plays a fundamental part in the homeostasis of anaphase cells such as neurocytes, hepatocytes and the like. This quality control is partly accomplished by nonselective autophagy. However, selective autophagy is also able to degrade particular proteins, organelles and foreign bacteria [5,6] and can also be induced by cellular stress, including selective degradation mediated by p62 and selective degradation related to ubiquitinated cargos [7,8,9].
Pathological and Physiological Significance of Autophagy
The organism accomplishes organelle regeneration through the elimination of subversive proteins, peroxidase, mitochondrion and other unnecessary or impaired cells and organelles through autophagy, thus maintaining a stable intracellular environment. Meanwhile, free amino acids and aliphatic acids generated in the process of protein degradation are reused by cells under the stimulus of hunger and certain factors, providing necessary elements for metabolism. Furthermore, the gene-oriented researches of tissue specificity have shown that autophagy plays a role in the differentiation of many specific cells, such as adipocytes, erythrocytes and T cells, and participates in the surface-active substance composition of alveolar type II cells. In addition, autophagy is closely related to such diseases as tumorigenesis, pathogen infection, neurodegeneration and myocardial ischemia reperfusion injury.
Formation Process of Autophagy
The formation process of autophagy falls into four stages [10]. (1) Formation of a separation film: stimulated by various autophagy-inducing factors to develop a cup-shaped isolated double-membrane structure, i.e. phagophore, in the surroundings of organelles or proteins to be degraded in the cytoplasm. Many factors inside and outside cells can induce autophagy. The most common are nutrient deficiency, unfolded and incorrectly folded proteins, impaired or senescent organelles, growth factor deficiency and hypoxia; also, contact with cytotoxic substances, such as cisplatin, cyclosporine and cadmium, can stimulate the activity of cell autophagy [11]. (2) Formation of autophagosomes: the separation film gradually stretches and increases the degree of bend and devours protein, organelles and the like, forming a closed spherical structure, i.e. autophagosome, surrounded by a double membrane. The half-life period of an autophagosome is around 8 min, which indicates that autophagy is a fast and effective reaction of cells to changing environments. (3) Formation of autophagic lysosomes: under the guidance of ESCRT and monomeric GTPase (Rabs), autophagosomes transfer what they have engulfed to lysosomes and then fuse with them into autophagic lysosomes in the function of SNARE and Rabs [12]. (4) Degradation of autophagosome contents: after the fusion of autophagosomes and lysosomes, the intima and contents of the autophagosome are degraded by various proteolytic enzymes in lysosomes and later released into the metabolism and recirculation system. The proteins involved in this process include Atg1, Atg13, Atg15, Atg 22, etc. [13,14].
Autophagy Assays
Today, there are mainly the following three methods for detection of autophagy. (1) Morphological detection with electron microscopes, which serves as a golden standard of autophagy detection: the autophagy formation process and the ultimate nondegradable residues inside the autophagosome can be seen in the impaired organelles [15]. (2) Detection of iconic proteins in the autophagic membrane, including two submethods, namely chemical detection of the immune system and Western imprint detection: the iconic proteins in the autophagic membrane include microtubule-associated protein 1 light chain 3, Beclin-1, Atg7, Atg12 and autophagy-adjusted protein and the like [16]. (3) The monodansylcadaverine (MDS) staining method is a unique way to detect autophagy, mainly through the fusion of Atg8 in the autophagic membrane surface and MDS. After fluorescence gets stained, perinuclear positive coloration can be seen. Most autophagy models, autophagy-related diseases and the autophagy-related chronic kidney diseases which will be introduced in the following paragraphs utilize these three detection methods to conduct quantitative and qualitative experimental analyses. Emphasis is put on the chemical detection of the immune system and Western imprint detection. Furthermore, the establishment of a mouse model with autophagy-related genes knocked out is also convenient for studying autophagy. For example, in transgenic mice expressing green fluorescent protein (GFP)-LC3, experiments can be conducted to assess the role of autophagy in various diseases through examining the level of LC3 [3].
Regulatory Mechanism of Autophagy
Regulation of Atg
So far, scientists have discovered over 30 kinds of Atgs and most of their orthologs in high eukaryotes. By function, Atg proteins can be divided into five groups, namely Atg1 kinase complex [Atg1/Unc-51-like kinase (ULK) 1/2], Atg9, class III phosphoinositide 3-kinase complex (PI3KC3), Atg12 conjugation system and Atg8 conjugation system. Atg1/ULK 1/2 complexes in mammals (including Atg13, FIP200 and Atg101) incorporate other Atg proteins into the preautophagosomal structure and facilitate the formation of autophagosomes through specific substrate phosphorylation [17]. The complexes formed by a subset of Atg proteins (Atg6/Beclin-1, Atg14/Atg14L) and PI3KC3 and UV radiation resistance-associated gene (UVRAG) can generate phosphatidylinositol 3-phosphate (PI3P). Atg18, Atg20, Atg21 and Atg24 are positioned with autophagic vacuoles by binding with PI3P and PI (3 and 5) so as to take part in the formation of autophagosomes [2,18]. Furthermore, the Atg12 conjugation system and the Atg8/LC3 conjugation system can coregulate autophagosome spreading. In the Atg12 conjugation system, Atg12 is activated by Atg7, forming covalent binding with its target protein Atg5 by catalyzing Atg10 and then binding with Atg16 to form the Atg12-Atg5-Atg16 complex. This complex forms a polymer through isotype oligomerization and is positioned in autophagic vacuoles. On the other hand, in the Atg8/LC3 conjugation system, Atg8 protein C-terminal arginine (R) is hydrolyzed by Atg4 and activated through binding with Atg7; the activated Atg8 is mediated by Atg3 and combines with phosphatidylethanolamine (PE) to form an Atg8-PE/LC3-II complex [19]. The formation of the Atg12-Atg5-Atg16 complex and the Atg8-PE/LC3-II complex is critical for the process of autophagy, as the mutation of any complex or other components participating in this process will result in an autophagy defect; hence, knocking out these genes is a common method to inhibit autophagy in experimental research.
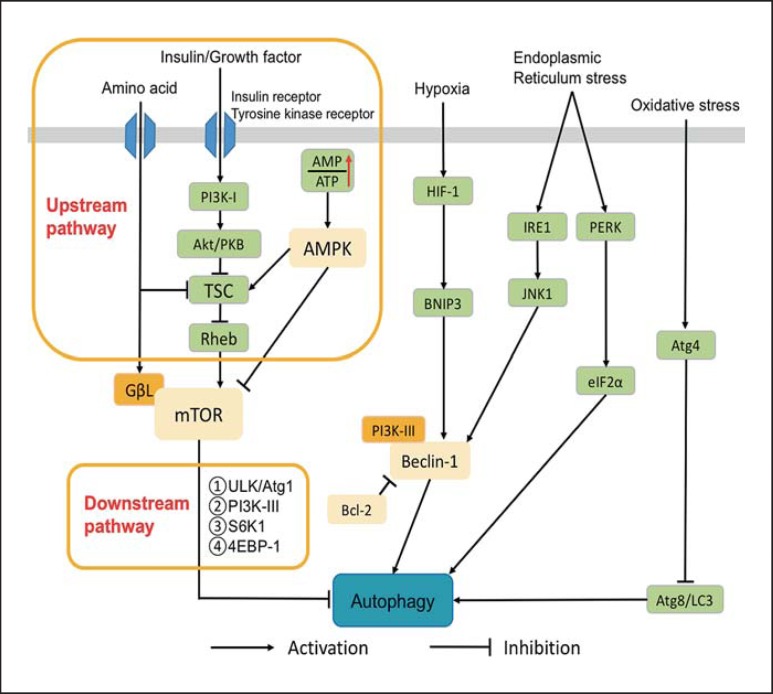
Autophagic Signaling Pathways (fig. 1)
Fig. 1.
Regulatory mechanisms of autophagy. PI3K-I = Phosphatidylinositol 3-kinase I; Akt/PKB = protein kinase B; TSC = tuberous sclerosis complex; Rheb = Ras homologue enriched in brain; GΒL = G protein β-like; AMPK = AMP-activated protein kinase; HIF-1 = hypoxia-inducible factor-1; BNIP3 = Bcl-2/E1B 19 kDa-interacting protein 3; JNK1 = c-Jun n-terminal kinase 1; IRE1 = inositol-requiring enzyme 1; PERK = PKR-like endoplasmic reticulum kinase; eIF2α = eukaryotic initiation factor 2α; Bcl-2 = B-cell lymphoma-2; S6K1 = subunit 6 kinase 1; 4EBP1 = eIF4E-binding protein 1; ULK = Unc-51-like kinase.
Mammalian Target of Rapamycin Signaling Pathway
Downstream Signaling Components of Mammalian Target of Rapamycin. Mammalian target of rapamycin (mTOR) acts as an autophagy inhibitor. When there are sufficient nutrients in cells (such as rich amino acids and growth factors), mTOR is activated to block autophagy pathways; but when there is nutrient deficiency, cells are in a starvation condition and mTOR's activity is inhibited; thus, its inhibiting effect on autophagy pathways is suppressed. After activation, mTOR can inhibit the formation of autophagy-related genes or protein complexes including ULK/autophagy gene (Atg1) complex and PI3KC3, both being important complexes in the formation of autophagy isolation membranes. Furthermore, activated mTOR can regulate the two downstream factors, ribosome protein subunit 6 kinase 1 (S6K1) and eIF4E-binding protein 1 (4EBP-1), to inhibit autophagy through affecting the transcription and translation of related proteins [20,21,22].
Upstream Signaling Components of mTOR. (1) Phosphatidylinositol 3-kinase I (PI3K-I)/protein kinase B (Akt/PKB): PI3K-I is activated by insulin or growth factors binding with transmembrane insulin receptors or tyrosine kinase receptors that phosphorylate PIP into PI3P. PI3P then binds with Akt/PKB and its activated molecule PDK1 in order to inhibit downstream tuberous sclerosis complex 1 and 2 (TSC1/2) proteins, thus activating mTOR to exert its inhibiting effect on autophagy [23,24,25]. (2) AMP-activated protein kinase (AMPK): AMPK regulates the energy condition of cells through monitoring the AMP/ATP ratio [26]. When glucose is deficient, the content of ATP concentration will decrease, thus activating the AMPK signaling pathway in cells [27]. Similarly, at the condition of low glucose, the expression of autophagy will increase. The activated AMPK inhibits mTOR activity by directly phosphorylating mTOR or activating TSC (TSC1/2 heterodimer), thus enhancing autophagy. (3) Amino acids [28]: amino acids are the major end-products of autophagy pathways. Some amino acids including alanine, leucine, glutamate and phenylalanine are also important regulatory factors of autophagy. By negative regulation of autophagy levels, amino acids can maintain a stable intracellular environment.
B-Cell Lymphoma-2/Yeast Atg6 Homolog (Beclin-1) Pathway
In mammals, Beclin-1 is Atg6's homolog, and the most important positive regulatory factor in the process of autophagy. It usually plays a role in regulating the positioning of other Atg proteins in the autophagy precursor structure. For example, assembling a Beclin-1/PI3K-III complex promotes the formation of autophagy. B-cell lymphoma-2 (Bcl-2) is an antiapoptotic protein that can bind to the BH3 domain of Beclin-1 to inhibit the interaction between Beclin-1 and PI3K-III, thereby affecting the formation of autophagy [29]. In starvation conditions, the decreased interaction between Beclin-1 and PI3K-III results in increased autophagy levels, whereas in nonstarvation conditions, Bcl-2 reduces autophagy levels by binding with Beclin-1 [30].
Cellular Stress Pathway
Hypoxia. In hypoxia, autophagy levels increase in mitochondria in order to decrease reactive oxygen species (ROS) and maintain cell integrity. Hypoxia-inducible factor 1 (HIF-1) in hypoxia can activate the target gene BNIP3 (Bcl-2 family protein), which competes with Beclin-1 for binding with Bcl-2, releasing Beclin-1 to activate autophagy [31,32,33].
Oxidative Stress. ROS are a common autophagy inducer in cell stress, facilitating Atg8/LC3 lipidation to activate autophagy through inhibiting the proteinase activity of Atg4.
Endoplasmic Reticulum Stress. PKR-like endoplasmic reticulum kinase (PERK) and inositol-requiring enzyme 1 (IRE1) are the main substances for mediating the formation of autophagy in mammalian endoplasmic reticulum stress [34,35]. Each, respectively, participating in the pathway of IRE1-XBP1/IRE1-JNK and the pathway of PERK/eukaryotic translation initiation factor 2α (eIF2α) to induce the formation of autophagy.
Dual Regulation of p53
p53 is the sensor protein for all sorts of intracellular pressure, controlling the process of autophagy and regulating cell apoptosis by its own activation or deactivation. By different intracellular positioning, p53 plays diverse roles in autophagy regulation: p53 facilitates autophagy when positioned in the cell nuclei because it can transactivate the two subunits of AMPK as well as TSC2. Meanwhile, p14ARF that exists in the cell nuclei can easily bind with ubiquitin hydrolase MDM2 of p53 to effectively prevent p53 from the degradation caused by MDM2, thus preserving the role of p53 in the nucleus [36]; when positioned in the cytoplasm, p53 inhibits autophagy in three ways [37]: activating autophagy inhibitor mTOR, inhibiting the effect of AMPK and exerting a direct effect. Based on the specific characteristics of p53's positioning, regulatory roles of p53 in autophagy can be controlled. For example, blocking the cell nucleus positioning signals of p53 can result in more p53 distributed in the cytoplasm, thereby resulting in autophagy inhibition. Conversely, if the nuclear export signals of p53 are destroyed, more p53 will be accumulated in the nucleus, losing its effect on autophagy inhibition.
Autophagy in Chronic Kidney Diseases
Diabetic Nephropathy
Diabetic nephropathy is one of the most common chronic microvascular complications of diabetes mellitus [38]. In roughly 35-40% of patients suffering from diabetes, the disease is further complicated by diabetic nephropathy [39]. Recent researches have demonstrated that the occurrence and development of type 2 diabetes and its complications are closely associated with the change of nutrient-sensitive pathways. The major related nutrient-sensitive signal proteins include mTOR [40], AMPK [41] and oxidized coenzyme-dependent histone deacetylase (Sirt1) [42]. As nutritional conditions change (high glucose environment), nutrient signal pathways inflict damage on the autophagy functions of cells so that the cells are unable to react normally to all sorts of extracellular stress, and organelle functions are further disordered, resulting in diabetic nephropathy. (1) mTOR is a target molecule of rapamycin and can combine with other proteins to form two types of complexes with different functions (mTORC1 and mTORC2) in mammals, with mTORC1 sensitive to the immunosuppressive agent rapamycin. In models of type 1 and 2 diabetic nephropathy, inhibiting mTORC1 signaling pathways via rapamycin plays a protective role for the kidney [43,44]. Recent studies have reported that mTORC1 plays an important role in maintaining sertoli cell homeostasis. However, it has been discovered in models of type 1 and 2 diabetic nephropathy that excessive activation of mTORC1 is one of the causes of glomerulus damage [45]. (2) The AMPK signaling pathway is a protective regulatory mechanism. Its activity is inhibited in diabetic nephropathy, resulting in reduced renal autophagy function. Both resveratrol and metformin can be used to activate AMPK. Reports have demonstrated that resveratrol can alleviate renal damage in early-stage diabetic nephropathy by activating AMPK [41]. In the glomeruli and renal tubules in animal models of type 1 and 2 diabetic nephropathy, AMPK activity is inhibited. However, activating AMPK via human intervention can reduce glomerulus and renal tubule damage [46]. (3) Sirt1 plays an important role in regulating the aging of mammals and the pathogenesis of aging-related metabolic diseases like type 2 diabetes. Researchers have discovered that Sirt1 expression decreases in animal models of diabetic nephropathy [47,48]. Diabetes causes significant glomerular cell apoptosis, while Sirt1 resists mesangial cell apoptosis incurred by diabetes through inhibiting oxidative stress and transforming growth factor β (TGF-β), to protect the kidneys [42]. Autophagy might become a new diabetic nephropathy pathogenesis-based target for prevention and treatment. Protecting the kidneys by reversing the above-mentioned change of nutrient signal pathways through human intervention demonstrates great therapeutic promise.
Obstructive Nephropathy
Nephron number decrease, renal tubular atrophy and renal interstitial fibrosis can be observed in obstructive nephropathy, but the pathogenesis remains unclear. Existing research has pointed out that autophagy plays a part in both renal tubular epithelial cell (TEC) apoptosis and renal tubular epithelial-mesenchymal transition [49,50,51]. Li et al. [52], in models of obstructive nephropathy in mice induced by unilateral ureteral obstruction, discovered that within 7-14 days after obstruction, the levels of autophagosome, LC3 and Beclin-1 increased significantly, the damage of proximal tubules deepened and the cell death percentage increased from 2.9 to 7.6%. Therefore, it is speculated that autophagic cell death is closely related to urinary tract obstruction. On this basis, Kim et al. [53], in further research on models of obstructed kidneys in rats, discovered that applying 3-methyladenine to rats at the postoperative stage of ureteroureterostomy to inhibit autophagy will result in increased renal tubular cell apoptosis and severer renal interstitial fibrosis, while inducing cell proliferation in the contralateral kidney. This indicates that in ureteroureterostomy models, autophagy plays a protective role in the obstructed kidney and has regulating effects on compensatory cell proliferation in the contralateral kidney [53].
In terms of mechanism regulation, autophagy induction in obstructed kidneys is related to the mTOR signaling pathway. In earlier-stage obstruction, due to regulating mechanism response in the organisms, mTOR is inhibited and cell autophagy activity is enhanced in order to remove abnormal protein molecules, peroxide, etc. caused by obstruction. Thus, the disease is characterized by increasing expression of LC3, which incurs protective response to stressful environments. However, as obstruction continues to exist and cell autophagy capacity is limited, mTOR is activated and autophagy activity is decreased at the advanced stage of obstruction. Nonetheless, the factors with damaging capabilities on cells continue to exist and increase with time, resulting in the gradual development of the disease. This is evident in the increase in expression of fibrosis factor TGF-β1 and the decrease in the expression of LC3, further indicating progressive renal damage. Hence, it is believed that autophagy activated by urinary tract obstruction plays a protective role at the early stage, but when stimulated by continuous urinary obstruction, the protective function of autophagy is altered and it begins to accelerate renal TEC apoptosis [53,54]. Based on this mechanism, it has been found that rapamycin, an mTOR signaling pathway blocker, is capable of inhibiting excessive expression and biological activity of mTOR, TGF-β1, etc., as well as improving LC3 expression. Therefore, it can partly alleviate the inhibition of cell autophagy borne at the advanced stage of obstruction and enhance autophagy activity, reducing renal interstitial fibrosis and inflammatory cell infiltration. Rapamycin has also been shown to have clinical efficacy in delaying the progress of chronic renal diseases. However, its specific mechanism of action still requires further research.
IgA Nephropathy
IgA nephropathy (IgAN) is a chronic progressive disease, and about 25-50% of patients are at risk for the development of end-stage renal disease within 25 years following their confirmed diagnosis. Further, IgAN demonstrates diversified clinical and pathological manifestations as well as differences in prognosis, with a 10-year survival rate of 63-96% according to foreign reports and 85% in China [55,56]. Therefore, how to evaluate IgAN prognosis is of particular importance in clinical treatment. Sato et al. [57] discovered that the autophagy type of sertoli cells is closely connected with IgAN prognosis. There are two types of autophagy in sertoli cells: type I autophagy and type II autophagy. Autophagosomes (diameter about 1 μm) of type I autophagy contain a large number of dense ribosomes and a small quantity of lipid vacuoles with incomplete membranes; therefore, they are unable to form functional autophagosomes. However, autophagosomes (diameter about 3-8 μm) of type II autophagy contain a small number of dense ribosomes and a large quantity of lipid vacuoles; therefore, they are able to complete the process of autophagy and effectively remove damaged protein and liposomes in the cells. The authors also compared 16 cases of IgAN patients and ensured the autophagy types in these cases remained unchanged through repeated (two or three samples per patient) renal biopsy. The results implied that, in the pathological categories of type I autophagy in IgAN patients, moderate glomerular mesangial proliferation is mainly displayed and is associated with mild to moderate focal segmental glomerulosclerosis and renal tubular atrophy. These findings demonstrate severer degrees of pathological damage than in IgAN patients with type II autophagy. This implies that IgAN patients dominated by type I autophagy tend to have severer renal damage, more recurrence and faster progression, which leads to a poorer prognosis [58]. This research finding provides evidence for the efficacy of delayed autophagy activity as an indicator for the early diagnosis and prognosis of IgAN.
Autoimmune Renal Disease
Among autoimmune renal diseases, such as glomerular basement membrane renal disease, anti-neutrophil cytoplasmic antibody (ANCA)-associated renal damage of vasculitis as well as lupus nephritis, autophagy may provide the sources of immune activators and autologous antigens for organs. Anders and Schlondorff [59] thought that the damage-associated molecular pattern molecules (DAMPs) and pathogen-associated molecular pattern molecules (PAMPs) generated by renal TECs or dendritic cells and macrophages were mainly degraded through autophagy. In autophagic lysosomes, DAMPs and PAMPs can be recognized by the Toll-like receptors (TLRs) in the cell surface and their own immune receptors; this causes the nuclear transcription factor NF-κB and Jun/Fos to be activated by a series of cascade reactions of proteins, hence leading to the effective activation of immune reactions. If DAMPs and PAMPs in autophagic lysosomes serve as intracorporal concealed antigens and become the autologous antigens through TLR recognition and main histocompatible compounds (MHC-I or MHC-II), then their presence will result in the absence of immune tolerance and autoimmune renal diseases.
Lupus nephritis is the most common chronic kidney disease and is associated with severe complications in systemic lupus erythematosus patients. Among all the known autophagy-related genes, apolipoprotein L1 (APOL1)[60] and myotubularin-related phosphatase 3 (MTMR3)[61] have been verified as being closely related to lupus nephritis. APOL1 is a BH3-only protein; its overexpression can lead to the death of autophagocytes [62]. MTMR3 is a kind of PI3P, regulating the level of PI3P in cells with PI kinases, that mediates the formation of autophagosomes and the occurrence of autophagy [63,64]. In the northern Han Chinese population, Zhou et al. [61] have found that the gene mutation of MTMR3 (rs9983A) is closely related to the morbidity of lupus nephritis. To explore the relationship between lupus nephritis and the mTOR inhibitor rapamycin, a Greek experimental team conducted a study on mice. They divided 32 female NZBW/F1 mice into four groups, namely a healthy control group, an untreated group, a preventive group and a therapeutic group. The results showed that the total and phosphorylated forms of Akt and mTOR level in the kidney cortex of lupus nephritis mice had a remarkably higher upregulation than those of healthy control mice. Furthermore, they found that the intervention with rapamycin in the preventive and therapeutic groups could prolong the life span of mice and decrease the levels of urinary protein, serum creatinine and anti-ds DNA titer [65]. This indicates the existence of an abnormally activated PI3K/Akt/mTOR pathway in mice suffering from lupus nephritis, suggesting that autophagy is involved in the pathological process of lupus nephritis.
Nephropathic Cystinosis
Nephropathic cystinosis falls into the category of lysosomal storage diseases and is a common autosomal and hereditary disease. The main disease characteristics are abnormal function of the kidney tubules and progressive development of renal insufficiency, the etiology of which at present is still unknown. Recently, researchers have discovered that mitophagy has an upregulated expression in cystinosis fibroblasts and proximal tubular cells [66,67]. They also verified that there is the phenomenon of autophagy dysfunction in cystinosis. Sansanwal and Sarwal [68] detected four cases of biopsy specimen in patients with nephropathic cystinosis syndrome and confirmed a significant increase in LC3 and p62 expression levels. Meanwhile, Sansanwal et al. [66], in studying nephropathic cystinosis syndrome, discovered an increase in autophagic vacuoles of proximal renal TECs and a growing expression of autophagic labeled proteins of LC3 and Beclin-1. They also found that the apoptosis rate of proximal renal TECs of cystinosis can be reduced by 3-MA-hindering autophagy, implying that autophagy may serve as the molecular mechanism for nephropathic cystinosis [66].
Aristolochic Acid Nephropathy
Aristolochic acid (AA), commonly seen in herbal medicine, has the effect of relieving cough and asthma, antibiosis and inflammation and reducing pressure. However, it also possesses the ability to induce renal toxicity that can cause AA nephropathy which results in the apoptotic damage of renal TECs. Zeng et al. [69], in studying the renal toxicity of AA, discovered that an increased expression of autophagic labeled proteins of LC3-II and Beclin-1 can be detected after incubating renal TECs for 3-6 h with the main ingredient of AA, namely AAI. The apoptotic rate of TECs induced by AAI after application of Wortmannin and 3-MA (autophagy hinderers) as well as silent Atg7 significantly increased when compared to the control group. Results showed that autophagy plays a protective role in TECs injured by AAI, that is, autophagy can improve the damage of AA nephropathy.
Chronic Cyclosporin A Renal Toxicity
Chronic cyclosporin A (CsA) renal toxicity is caused by a pathological change featuring belt-shaped fibrosis of renal tubules that can cause the apoptosis of renal TECs as a result of oxidative stress. Meanwhile, CsA can activate autophagy by inducing reticulum stress of renal tubules in order to reduce renal toxicity. Pallet et al. [70], in applying CsA to human renal tubular cells, discovered that it could induce autophagy by promoting LC3-II expression and the formation of autophagosomes. They also found that after siRNA of Beclin-1-hindering autophagy, the activity of TECs treated by CsA decreased. This implies that in the treating process with CsA, if the autophagy is inhibited, the death of renal TECs will increase, suggesting that autophagy plays a protective role in tubular damages induced by CsA.
Concluding Remarks
Autophagy is an important process involved in the growth, development, physiology and pathology of cells. It is typically considered as a cell self-protection mechanism. However, as the research on autophagy continues to progress, more and more discoveries reveal that autophagy is a ‘double-edged sword’ - exerting both therapeutic and/or detrimental effects on cells and tissues. These two-way effects may be explained by differences in tissue, time period, type of autophagy, activation method and stress severity. Similar to other diseases, autophagy is also implicated in the pathological process of chronic kidney disease. Although our knowledge on the molecular mechanisms of autophagy in relation to chronic kidney disease has been greatly improved in recent years, there is still a lot of missing information necessary for a full establishment of clinical settings of using autophagy as a target for patients with kidney disease. Thus, we must continue to uncover the pathogenesis and regulatory mechanisms of autophages in various underlying renal diseases in order to ensure autophagy as a novel target for the treatment of chronic kidney diseases with diverse underlying causes.
Conflict of Interest Statement
No authors have any conflicts of interest, financial or otherwise.
Acknowledgements
This study was supported by the National Nature Science Foundation of China (grants 81270778 and 81470920 to S.Z.; 81200492 and 81470991 to N.L.) and Key Discipline Construction Project of Pudong Health Bureau of Shanghai (PWZx2014-06 to S.Z.).
References
- 1.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 2.Nakatogawa H, Suzuki K, Kamada Y, Yoshinori O. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, Wang K, Chu C. After the banquet: mitochondrial biogenesis, mitophagy and cell survival. Autophagy. 2013;9:1–14. doi: 10.4161/auto.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki K. Selective autophagy in budding yeast. Cell Death Differ. 2012;20:43–48. doi: 10.1038/cdd.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichimura Y, Komatsu M. Selective degradation of p62 by autophagy. Semin Immunopathol. 2010;32:431–436. doi: 10.1007/s00281-010-0220-1. [DOI] [PubMed] [Google Scholar]
- 8.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riley BE, Kaiser SE, Shaler TA, et al. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J Cell Biol. 2010;91:537–552. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue K, Kuwana H, Shimamure Y, et al. Cisplatin-induced macroautophagy occurs prior to apoptosis in proximal tubules in vivo. Clin Exp Nephrol. 2009;14:112–122. doi: 10.1007/s10157-009-0254-7. [DOI] [PubMed] [Google Scholar]
- 12.Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956–965. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- 13.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abeliovich H, Zhang C, Dunn WA, et al. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinet W, De Meyer GR, Audries L, et al. In situ detection of starvation induced autophagy. J Histochem Cytochem. 2006;54:85–96. doi: 10.1369/jhc.5A6743.2005. [DOI] [PubMed] [Google Scholar]
- 16.Kim JS, Nitta T, Mohuczy D, et al. Impaired autophagy: a mechanism of mitochondrial dysfunction in anoxic rat hepatocytes. Hepatology. 2008;47:1725–1736. doi: 10.1002/hep.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiuri MC, Criollo A, Tasdemir E, et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L). Autophagy. 2007;3:374–376. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 20.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganley IG, Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoki K, Li Y, Xu T, Guan KL. Rheb GTpase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signaling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 26.Yun CL, Zierath JR. AMP-activated protein kinase signaling in metabolic regulation. J Clin Invest. 2006;116:1776–1783. doi: 10.1172/JCI29044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plomp PJ, Gordon PB, Meijer AJ, Hoyvik H, Seglen PO. Energy dependence of different steps in the autophagic-lysosomal pathway. J Biol Chem. 1989;264:6699–6704. [PubMed] [Google Scholar]
- 28.Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinha S, Levine B. The autophagy effector Beclin-1: a novel BH3-only protein. Oncogene. 2008;27((suppl 1)):137–148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majmundar J, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol. 2010;22:177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Bellot G, Garcia-Medina R, Gounon P, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouroku Y, Fujita E, Tanida I, et al. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 35.Ogata M, Hino S, Saito A, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Z, Zhang H, Levine AJ, et al. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine B, Abrams J. p53: the Janus of autophagy. Nat Cell Biol. 2008;10:637–639. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molitch ME, Defronzo RA, Franz MJ, et al. Nephropathy in diabetes. Diabetes Care. 2004;27((suppl 1)):S79–S83. doi: 10.2337/diacare.27.2007.s79. [DOI] [PubMed] [Google Scholar]
- 39.Raptis AE, Viberti G. Pathogenesis of diabetic nephropathy. Exp Clin Endocrinol Diabetes. 2001;109((suppl 2)):S424–S437. doi: 10.1055/s-2001-18600. [DOI] [PubMed] [Google Scholar]
- 40.Inoki K. Role of TSC-mTOR pathway in diabetic nephropathy. Diabetes Res Clin Pract. 2008;82((suppl 1)):S59–S62. doi: 10.1016/j.diabres.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 41.Ding DF, You N, Wu XM, et al. Resveratrol attenuates renal hypertrophy in early-stage diabetes by activating AMPK. Am J Nephrol. 2010;31:367–374. doi: 10.1159/000300388. [DOI] [PubMed] [Google Scholar]
- 42.Kume S, Uzu T, Kashiwagi A, et al. Sirt1, a calorie restriction mimetic, in a new therapeutic approach for type 2 diabetes mellitus and diabetic vascular complications. Endocr Metab Immune Disord Drug Targets. 2010;10:16–24. doi: 10.2174/187153010790827957. [DOI] [PubMed] [Google Scholar]
- 43.Sakaguchi M, Isono M, Isshiki K, et al. Inhibition of mTOR signaling with rapamycin attenuates renal hypertrophy in the early diabetic mice. Biochem Biophys Res Commun. 2006;340:296–301. doi: 10.1016/j.bbrc.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y, Wang JJ, Qin L, et al. Rapamycin prevents early steps of the development of diabetic nephropathy in rats. Am J Nephrol. 2007;27:495–502. doi: 10.1159/000106782. [DOI] [PubMed] [Google Scholar]
- 45.Inoki K, Mori H, Wang JY, et al. mTORC1 activation in podocytes is a critical step in the development of diabetic nephropathy in mice. J Clin Invest. 2011;121:2181–2196. doi: 10.1172/JCI44771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CC, Chang CY, Wu YT, et al. Resveratrol retards progression of diabetic nephropathy through modulations of oxidative stress, proinflammatory cytokines, and AMP-activated protein kinase. J Biomed Sci. 2011;18:47. doi: 10.1186/1423-0127-18-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tikoo K, Tripathi DN, Kabra DG, et al. Intermittent fasting prevents the progression of type 1 diabetic nephropathy in rats and changes the expression of Sir2 and p53. FEBS Lett. 2007;581:1071–1078. doi: 10.1016/j.febslet.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Tikoo K, Singh K, Kabra D, et al. Change in histone H3 phosphorylation, MAP kinase p38, SIR 2 and p53 expression by resveratrol in preventing streptozotocin induced type 1 diabetic nephropathy. Free Radic Res. 2008;42:397–404. doi: 10.1080/10715760801998646. [DOI] [PubMed] [Google Scholar]
- 49.Chevalier RL. Pathogenesis of renal injury in obstructive uropathy. Curr Opin Pediatr. 2006;18:153–160. doi: 10.1097/01.mop.0000193287.56528.a4. [DOI] [PubMed] [Google Scholar]
- 50.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112:1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y. Epithelial to mesenchymal transition in renal fibrogenesis: pathologic significance, molecular mechanism, and therapeutic intervention. J Am Soc Nephrol. 2004;15:1–12. doi: 10.1097/01.asn.0000106015.29070.e7. [DOI] [PubMed] [Google Scholar]
- 52.Li L, Zepeda-Orozco D, Black R, et al. Autophagy is a component of epithelial cell fate in obstructive uropathy. Am J Pathol. 2010;176:1767–1778. doi: 10.2353/ajpath.2010.090345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim WY, Nam SA, Song HC, et al. The role of autophagy in unilateral ureteral obstruction rat model. Nephrology (Carlton) 2012;17:148–159. doi: 10.1111/j.1440-1797.2011.01541.x. [DOI] [PubMed] [Google Scholar]
- 54.Xu Y, Ruan S, Wu X, et al. Autophagy and apoptosis in tubular cells following unilateral ureteral obstruction are associated with mitochondrial oxidative stress. Int J Mol Med. 2013;31:628–636. doi: 10.3892/ijmm.2013.1232. [DOI] [PubMed] [Google Scholar]
- 55.Geddes CC, Rauta V, Gronhagen-Riska C, et al. A tricontinental view of IgA nephropathy. Nephrol Dial Transplant. 2003;18:1541–1548. doi: 10.1093/ndt/gfg207. [DOI] [PubMed] [Google Scholar]
- 56.Le W, Liang S, Hu Y, et al. Long-term renal survival and related risk factors in patients with IgA nephropathy: results from a cohort of 1,155 cases in a Chinese adult population. Nephrol Dial Transplant. 2012;27:1479–1485. doi: 10.1093/ndt/gfr527. [DOI] [PubMed] [Google Scholar]
- 57.Sato S, Yanagihara T, Ghazizadeh M, et al. Correlation of autophagy type in podocytes with histopathological diagnosis of IgA nephropathy. Pathobiology. 2009;76:221–226. doi: 10.1159/000228897. [DOI] [PubMed] [Google Scholar]
- 58.Sato S, Kitamura H, Adachi A, et al. Two types of autophagy in the podocytes in renal biopsy specimens: ultrastructural study. J Submicrosc Cytol Pathol. 2006;38:167–174. [PubMed] [Google Scholar]
- 59.Anders HJ, Schlondorff DO. Innate immune receptors and autophagy: implications for autoimmune kidney injury. Kidney Int. 2010;78:29–37. doi: 10.1038/ki.2010.111. [DOI] [PubMed] [Google Scholar]
- 60.Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, Garner NE, Birmingham DJ, Hebert LA, Hicks PJ, Segal MS, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol. 2014;66:390–396. doi: 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou XJ, Nath SK, Qi YY, Cheng FJ, Yang HZ, Zhang Y, Yang W, Ma JY, Zhao MH, Shen N, et al. Identification of MTMR3 as a novel susceptibility gene for lupus nephritis in northern Han Chinese by shared-gene analysis with IgA nephropathy. Arthritis Rheumatol. 2014;66:2842–2848. doi: 10.1002/art.38749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wan G, Zhaorigetu S, Liu Z, Kaini R, Jiang Z, Hu CA. Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J Biol Chem. 2008;283:21540–21549. doi: 10.1074/jbc.M800214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vergne I, Deretic V. The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett. 2010;584:1313–1318. doi: 10.1016/j.febslet.2010.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 2010;11:468–478. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- 65.Stylianou K, Petrakis I, Mavroeidi V, Stratakis S, Vardaki E, Perakis K, Stratigis S, Passam A, Papadogiorgaki E, Giannakakis K, et al. The PI3K/Akt/mTOR pathway is activated in murine lupus nephritis and downregulated by rapamycin. Nephrol Dial Transplant. 2011;26:498–508. doi: 10.1093/ndt/gfq496. [DOI] [PubMed] [Google Scholar]
- 66.Sansanwal P, Yen B, Gahl WA, Ma Y, Ying L, Wong LJ, Sarwal MM. Mitochondrial autophagy promotes cellular injury in nephropathic cystinosis. J Am Soc Nephrol. 2010;21:272–283. doi: 10.1681/ASN.2009040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sansanwal P, Sarwal MM. Abnormal mitochondrial autophagy in nephropathic cystinosis. Autophagy. 2010;6:971–973. doi: 10.4161/auto.6.7.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sansanwal P, Sarwal MM. p62/SQSTM1 prominently accumulates in renal proximal tubules in nephropathic cystinosis. Pediatr Nephrol. 2012;27:2137–2144. doi: 10.1007/s00467-012-2227-4. [DOI] [PubMed] [Google Scholar]
- 69.Zeng Y, Yang X, Wang J, et al. Aristolochic acid I induced autophagy extenuates cell apoptosis via ERK1/2 pathway in renal tubular epithelial cells. PLoS One. 2012;7:e30312. doi: 10.1371/journal.pone.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pallet N, Bouvier N, Legendre C, et al. Autophagy protects renal tubular cells against cyclosporine toxicity. Autophagy. 2008;4:783–791. doi: 10.4161/auto.6477. [DOI] [PubMed] [Google Scholar]